|
1
|
International Agency for Research on
CancerGlobal cancer statistics. http://globocan.iarc.fr/Pages/fact_sheets_population.aspx
|
|
2
|
De Las Rivas J and Fontanillo C:
Protein-protein interactions essentials: Key concepts to building
and analyzing interactome networks. PLoS Comput Biol.
6:e10008072010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wiggans AJ, Cass GK, Bryant A, Lawrie TA
and Morrison J: Poly(ADP-ribose) polymerase (PARP) inhibitors for
the treatment of ovarian cancer. Cochrane Database Syst Rev:
CD007929. 2015. View Article : Google Scholar
|
|
6
|
Olopade OI and Wei M: FANCF methylation
contributes to chemoselectivity in ovarian cancer. Cancer Cell.
3:417–420. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malati T: Tumour markers: An overview.
Indian J Clin Biochem. 22:17–31. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kurman RJ and Shih Ie M: Pathogenesis of
ovarian cancer: Lessons from morphology and molecular biology and
their clinical implications. Int J Gynecol Pathol. 27:151–160.
2008.PubMed/NCBI
|
|
9
|
Lee E and Moon A: Identification of
biomarkers for breast cancer using databases. J Cancer Prev.
21:235–242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin CM and Feng W: Microarray and
synchronization of neuronal differentiation with pathway changes in
the Kyoto Encyclopedia of Genes and Genomes (KEGG) databank in
nerve growth factor-treated PC12 cells. Curr Neurovasc Res.
9:222–229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:(Database Issue). D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Connectivity Map. https://portals.broadinstitute.org/cmap
|
|
15
|
Kaplan EL and Meier P: Nonparametric
estimation from incomplete observations. J Am Statist Assoc.
53:457–481. 1958. View Article : Google Scholar
|
|
16
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data from 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
The Human Protein Atlas. https://www.proteinatlas.org/about
|
|
18
|
Protein Atlas version 18. https://www.proteinatlas.org/about/releases
|
|
19
|
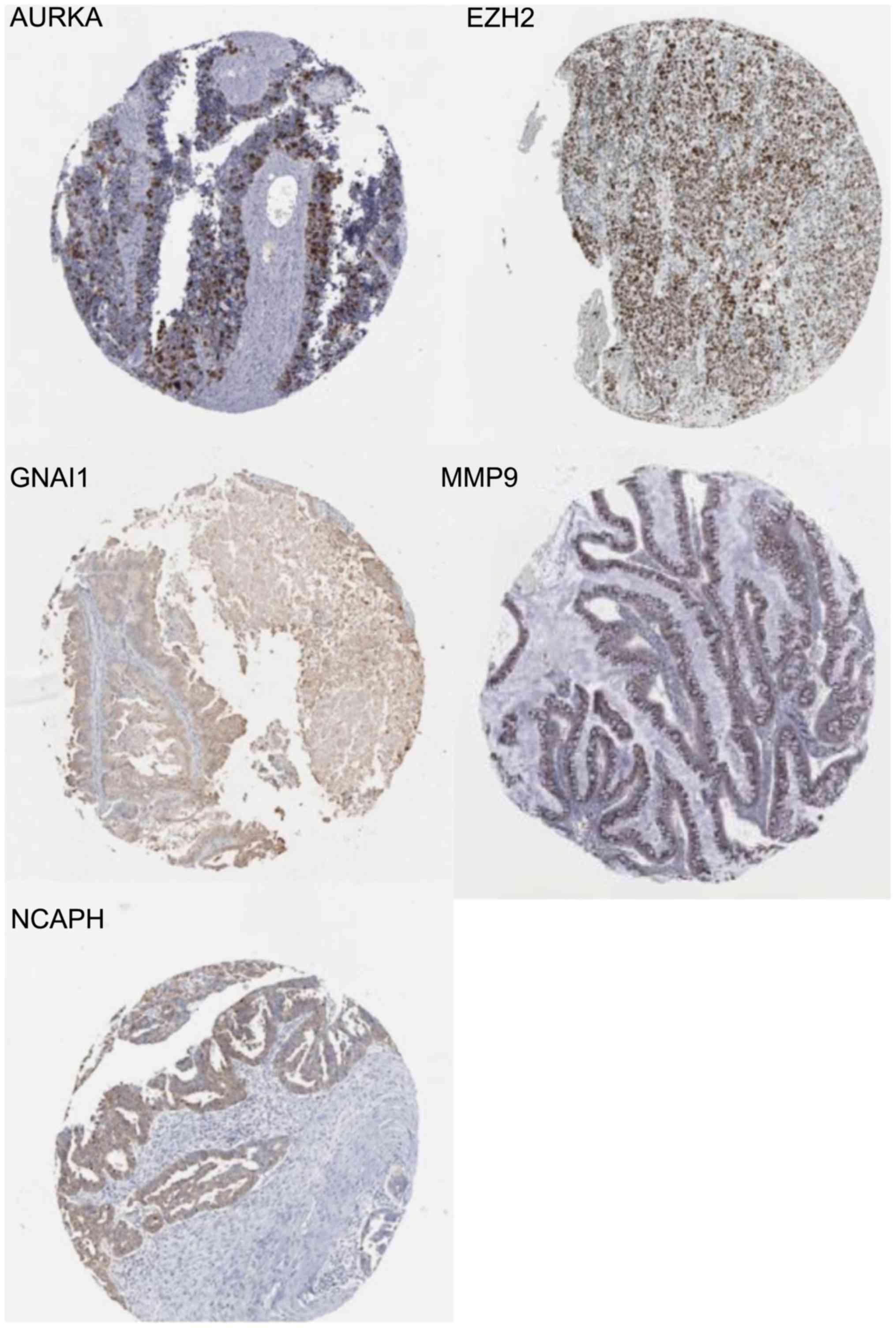
GNAI1/pathology/tissue/ovarian+cancer.
https://www.proteinatlas.org/ENSG00000127955-GNAI1/pathology/tissue/ovarian+cancer
|
|
20
|
NCAPH/pathology/tissue/ovarian+cancer.
https://www.proteinatlas.org/ENSG00000121152-NCAPH/pathology/tissue/ovarian+cancer
|
|
21
|
MMP9/pathology/tissue/ovarian+cancer.
https://www.proteinatlas.org/ENSG00000100985-MMP9/pathology/tissue/ovarian+cancer
|
|
22
|
AURKA/pathology/tissue/ovarian+cancer.
https://www.proteinatlas.org/ENSG00000087586-AURKA/pathology/tissue/ovarian+cancer
|
|
23
|
EZH2/pathology/tissue/ovarian+cancer.
https://www.proteinatlas.org/ENSG00000106462-EZH2/pathology/tissue/ovarian+cancer
|
|
24
|
Ochoa CE, Mirabolfathinejad SG, Ruiz VA,
Evans SE, Gagea M, Evans CM, Dickey BF and Moghaddam SJ:
Interleukin 6, but not T helper 2 cytokines, promotes lung
carcinogenesis. Cancer Prev Res. 4:51–64. 2011. View Article : Google Scholar
|
|
25
|
Lesina M, Kurkowski MU, Ludes K, Rose-John
S, Treiber M, Klöppel G, Yoshimura A, Reindl W, Sipos B, Akira S,
et al: Stat3/Socs3 activation by IL-6 transsignaling promotes
progression of pancreatic intraepithelial neoplasia and development
of pancreatic cancer. Cancer Cell. 19:456–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tan X, Carretero J, Chen Z, Zhang J, Wang
Y, Chen J, Li X, Ye H, Tang C, Cheng X, et al: Loss of p53
attenuates the contribution of IL-6 deletion on suppressed
tumor progression and extended survival in Kras-driven
murine lung cancer. PLoS One. 8:e808852013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yanagawa H, Sone S, Takahashi Y, Haku T,
Yano S, Shinohara T and Ogura T: Serum levels of interleukin 6 in
patients with lung cancer. Br J Cancer. 71:1095–1098. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yeh HH, Lai WW, Chen HH, Liu HS and Su WC:
Autocrine IL-6-induced Stat3 activation contributes to the
pathogenesis of lung adenocarcinoma and malignant pleural effusion.
Oncogene. 25:4300–4309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao SP, Mark KG, Leslie K, Pao W, Motoi N,
Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B and Bromberg
JF: Mutations in the EGFR kinase domain mediate STAT3 activation
via IL-6 production in human lung adenocarcinomas. J Clin Invest.
117:3846–3856. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Haura EB, Livingston S and Coppola D:
Autocrine interleukin-6/interleukin-6 receptor stimulation in
non-small-cell lung cancer. Clin Lung Cancer. 7:273–275. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brooks GD, McLeod L, Alhayyani S, Miller
A, Russell PA, Ferlin W, Rose-John S, Ruwanpura S and Jenkins BJ:
IL6 trans-signaling promotes KRAS-driven lung carcinogenesis.
Cancer Res. 76:866–876. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
GeneSupport Center of NCBI. https://www.ncbi.nlm.nih.gov/gene/
|
|
33
|
Neuwald AF and Hirano T: HEAT repeats
associated with condensins, cohesins, and other complexes involved
in chromosome-related functions. Genome Res. 10:1445–1452. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Wu Y, Guo J, Fei X, Yu L and Ma S:
Adipocyte-derived exosomes promote lung cancer metastasis by
increasing MMP9 activity via transferring MMP3 to lung cancer
cells. Oncotarget. 8:81880–81891. 2017.PubMed/NCBI
|
|
35
|
Chen Q, Yin D, Zhang Y, Yu L, Li XD, Zhou
ZJ, Zhou SL, Gao DM, Hu J, Jin C, et al: MicroRNA-29a induces loss
of 5-hydroxymethylcytosine and promotes metastasis of
hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis.
Cell Death Dis. 8:e29062017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie H, Li L, Zhu G, Dang Q, Ma Z, He D,
Chang L, Song W, Chang HC, Krolewski JJ, et al: Correction:
Infiltrated pre-adipocytes increase prostate cancer metastasis via
modulation of the miR-301a/androgen receptor (AR)/TGF-β1/Smad/MMP9
signals. Oncotarget. 7:83829–83830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Limoge M, Safina A, Beattie A, Kapus L,
Truskinovsky AM and Bakin AV: Tumor-fibroblast interactions
stimulate tumor vascularization by enhancing cytokine-driven
production of MMP9 by tumor cells. Oncotarget. 8:35592–35608. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reiner AT, Tan S, Agreiter C, Auer K,
Bachmayr-Heyda A, Aust S, Pecha N, Mandorfer M, Pils D, Brisson AR,
et al: EV-associated MMP9 in high-grade serous ovarian cancer is
preferentially localized to Annexin V-Binding EVs. Dis Markers.
2017:96531942017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bischoff JR, Anderson L, Zhu Y, Mossie K,
Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, et
al: A homologue of Drosophila aurora kinase is oncogenic and
amplified in human colorectal cancers. EMBO J. 17:3052–3065. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW,
Sahin A, Brinkley BR and Sen S: Tumour amplified kinase STK15/BTAK
induces centrosome amplification, aneuploidy and transformation.
Nat Genet. 20:189–193. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tanner MM, Grenman S, Koul A, Johannsson
O, Meltzer P, Pejovic T, Borg A and Isola JJ: Frequent
amplification of chromosomal region 20q12-q13 in ovarian cancer.
Clin Cancer Res. 6:1833–1839. 2000.PubMed/NCBI
|
|
42
|
Bao Z, Lu L, Liu X, Guo B, Zhai Y, Li Y,
Wang Y, Xie B, Ren Q, Cao P, et al: Association between the
functional polymorphism Ile31Phe in the AURKA gene and
susceptibility of hepatocellular carcinoma in chronic hepatitis B
virus carriers. Oncotarget. 8:54904–54912. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang L, Arras J, Katsha A, Hamdan S,
Belkhiri A, Ecsedy J and El-Rifai W: Cisplatin-resistant cancer
cells are sensitive to Aurora kinase A inhibition by alisertib. Mol
Oncol. 11:981–995. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Do TV, Hirst J, Hyter S, Roby KF and
Godwin AK: Aurora A kinase regulates non-homologous end-joining and
poly(ADP-ribose) polymerase function in ovarian carcinoma cells.
Oncotarget. 8:50376–50392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang G, Chang B, Yang F, Guo X, Cai KQ,
Xiao XS, Wang H, Sen S, Hung MC, Mills GB, et al: Aurora kinase A
promotes ovarian tumorigenesis through dysregulation of the cell
cycle and suppression of BRCA2. Clin Cancer Res. 16:3171–3181.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Wang Z, Qi Z, Yin S, Zhang N, Liu
Y, Liu M, Meng J, Zang R, Zhang Z and Yang G: The negative
interplay between Aurora A/B and BRCA1/2 controls cancer cell
growth and tumorigenesis via distinct regulation of cell cycle
progression, cytokinesis, and tetraploidy. Mol Cancer. 13:942014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brodie KM and Henderson BR:
Characterization of BRCA1 protein targeting, dynamics, and function
at the centrosome: A role for the nuclear export signal, CRM1, and
Aurora A kinase. J Biol Chem. 287:7701–7716. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dann RB, DeLoia JA, Timms KM, Zorn KK,
Potter J, Flake DD II, Lanchbury JS and Krivak TC: BRCA1/2
mutations and expression: Response to platinum chemotherapy in
patients with advanced stage epithelial ovarian cancer. Gynecol
Oncol. 125:677–682. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Muggia F and Safra T: ‘BRCAness’ and its
implications for platinum action in gynecologic cancer. Anticancer
Res. 34:551–556. 2014.PubMed/NCBI
|
|
50
|
Rao ZY, Cai MY, Yang GF, He LR, Mai SJ,
Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, et al: EZH2 supports
ovarian carcinoma cell invasion and/or metastasis via regulation of
TGF-beta1 and is a predictor of outcome in ovarian carcinoma
patients. Carcinogenesis. 31:1576–1583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao J, Zhu Y, Nilsson M and Sundfeldt K:
TGF-β isoforms induce EMT independent migration of ovarian cancer
cells. Cancer Cell Int. 14:722014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Villanueva MT: Anticancer drugs: All roads
lead to EZH2 inhibition. Nat Rev Drug Discov. 16:2392017.
View Article : Google Scholar : PubMed/NCBI
|