Introduction
Human breast epithelial cells are frequently
characterized based on the expression of cell surface markers,
including CD44 and CD24 (1–8). CD44 is expressed in basal-like and
CD24 in luminal-like cell lines. CD44 is a transmembrane
glycoprotein that normally regulates cell-cell adhesion and
cell-matrix interactions, as well as cell migration. This
glycoprotein binds mainly to hyaluronic acid, collagen,
fibronectin, laminin and chondroitin-sulfate, which are all
important epithelial and mesenchymal components (6–11).
Either independently or in collaboration with other cell surface
receptors, CD44 can promote uncontrolled growth, evasion of
apoptosis, angiogenesis, cell motility and invasion, which are
hallmarks of cancer progression (10,12,13).
The association of CD44 with breast cancer progression has been
evaluated in vivo using mouse models. One of the earliest
indications of the role of CD44 in metastasis derived from
pancreatic cancer. The transfection of CD44 variants into a
non-metastatic rat pancreatic carcinoma cell line conferred
metastatic potential in these cells when injected into syngeneic
rats (14).
CD24 is a small, heavily-glycosylated protein core
that consists of 27 amino acids attached to cell membranes. It has
been shown to be expressed at lower levels in progenitor cells
compared with differentiated cells (15). CD24 is a cell surface protein that,
depending on the cell or tissue type, presents highly variable
glycosylation (4). CD24 is
overexpressed during cancer progression, supporting its usefulness
as a marker for diagnosis and prognosis in breast, ovarian,
prostate and pancreatic cancers (15–19).
Previous studies have indicated that CD24 may promote cell
proliferation (20) and invasion of
cancer cells (21). In breast
cancer cells, CD24 increased cell proliferation, motility and
invasiveness (22).
Curcumin (diferuloylmethane) is a natural yellow
pigment derived from the rhizome of the herb Curcuma longa.
It has been demonstrated that curcumin has a variety of
pharmacological effects, including potent antiproliferative,
anti-inflammatory, antioxidant and anti-carcinogenic activities,
conferring chemopreventive potential (9–11,23).
Recent evidence indicated that curcumin had a diverse range of
molecular targets, supporting the concept that it acted upon
numerous biochemical and molecular cascades. Curcumin can induce
apoptosis-like changes in some cells, whereas it can inhibit
proliferation in others (23,24).
Since research has demonstrated the importance of developing novel
preventive strategies that can target cancer cells, the aim of the
present study was to investigate the effect of curcumin on breast
epithelial cell lines with regard to the expression of the cell
surface markers CD44 and CD24, to assess its potential capacity for
breast cancer prevention and to determine whether curcumin can
increase the ratio of CD44+/CD24+ cells and
decrease that of CD44+/CD24− cells.
Materials and methods
Breast cancer cell lines
An in vitro experimental breast cancer model,
termed the Alpha model, was used in these studies; it was derived
from MCF-10F (ATCC®; CRL-10318™), an immortalized human
epithelial cell type (25), which
was exposed to low doses of high linear energy transfer (LET)
α-particle radiation (150 keV/µm) and subsequently, to
17β-estradiol (25). The cell lines
used in this model were as follows: i) MCF-10F, a normal cell line
used as a control; ii) Alpha5, a pre-malignant and tumorigenic cell
line; and ii) Tumor2, derived from Alpha5 after the injection of
that cell line into nude mice. These cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM)/F-12 (1:1) supplemented
with antibiotics [100 U/ml penicillin, 100 µg/ml streptomycin, 2.5
µg/ml amphotericin B (all from Life Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA)], 10 µg/ml 5% equine serum
(Biofluids, Rockville, MD, USA), 0.5 µg/ml hydrocortisone
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 0.02 µg/ml
epidermal growth factor (Collaborative Research, Bedford, MA, USA).
Two other breast carcinoma cell lines, MDA-MB-231
(ATCC®; HTB26™) and MCF-7 (ATCC®; HTB22™),
were cultured in RPMI-1640 and minimum essential medium (MEM) (Life
Technologies; Thermo Fisher Scientific, Inc.), respectively. All
the cell lines were maintained at 37°C in a humidified atmosphere
containing 5% CO2. Curcumin at 30 µM was applied for 48
h in all the experimental designs.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol (Invitrogen,
Carlsbad, CA, USA) from untreated and treated breast cell lines,
following the manufacturer's instructions. A UV spectrophotometer
was used to determine RNA purity and concentration (Thermo Fisher
Scientific, Inc., Rochester, NY, USA). A High-Capacity cDNA Reverse
Transcription kit (Applied Biosystems, Carlsbad, CA, USA) and 10
units of RNase inhibitor (Applied Biosystems) were used to
reverse-transcribe the RNA into cDNA, according to the
manufacturer's protocol. For qPCR, we used 2 µl cDNA with
SYBR-Green PCR Master Mix (Agilent Technologies, Inc., La Jolla,
CA, USA) and primers for target gene (CD44 and CD24; Table I) at a concentration of 5 µM. A
CFX96 Real-Time PCR Detection system (Bio-Rad Laboratories,
Hercules, CA, USA) was used to perform the reactions under the
following conditions: 95°C for 10 min; and 40 cycles of a 2-step
program consisting of 95°C for 10 sec and 61°C for 45 sec, when
fluorescence-reading occurred. PCR products were monitored through
dissociation curve analysis (measurement of fluorescence during
stepped-increases in temperature of 2°C/min from 61 to 95°C).
Bio-Rad CFX Manager 2.1 software was used to obtain the threshold
cycle (Ct) and a reference housekeeping gene (β-actin) was used to
normalize the average gene expression.
 | Table I.Primers for genes selected to develop
cDNA probes. |
Table I.
Primers for genes selected to develop
cDNA probes.
| Gene name | Product length
(bp) | Primer
sequence |
|---|
| CD44 | 116 | F:
CGGACACCATGGACAAGTTT |
|
|
| R:
CACGTGGAATACACCTGCAA |
| CD24 | 16 | F:
AACTAATGCCACCACCAAGG |
|
|
| R:
GACGTTTCTTGGCCTGAGTC |
Determination of the protein
expression of cell surface markers CD44 and CD24 in cells by
immunoperoxidase staining
Cells were placed on a glass chamber slide (Nunc
Inc., Naperville, IL, USA) at a density of 1×104
cells/ml of medium and allowed to grow until 70% confluent.
Subsequently, they were fixed with buffered paraformaldehyde,
incubated with 1% H2O2 in methanol for 20 min
to block endogenous peroxidase and washed with buffer solution, and
then covered with normal horse serum for 30 min. The cells were
then incubated with mouse monoclonal antibodies anti-CD44 (DF1485)
(cat. no. sc-7297) and anti-CD24 (BA-1) (cat. no. sc-65257; both
were obtained from Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) at 1:500 dilution overnight at 4°C. Subsequently they were
incubated for 45 min with diluted 1:500 biotinylated secondary
antibody solution (cat. no. PK-6102 mouse IgG; Vector Laboratories,
Burlingame, CA, USA) and Vectastain Elite ABC reagent (Vector
Laboratories). The localization of bound antibodies was visualized
with DAB (Peroxidase substrate kit; Vector Laboratories) for 5 min
and counterstaining with Mayer's hematoxylin solution (MHS128-4L;
Sigma-Aldrich; Merck KGaA) for 30 sec was performed.
Flow cytometric analysis of cell
surface markers CD24 and CD44 in different cell lines
Breast cells were washed with Dulbecco's
phosphate-buffered saline (DPBS/modified; HyClone, Logan, UT, USA)
and then harvested with Trypsin, 2.2 mM of
ethylenediaminetetraacetic acid (EDTA; Corning, Manassas, VA, USA).
Detached cells were re-suspended in phosphate-buffered saline
supplemented with 0.5% fetal bovine serum (1×106 cells/50 µl).
Fluorochrome-conjugated antibodies (1:1) against human CD44
(FITC-conjugated; cat. no. 555478) and CD24 (PE-conjugated; cat.
no. 555428) were obtained from BD Biosciences (San Diego, CA, USA).
Antibodies were added to the cell suspension, as recommended by the
manufacturer, and incubated at 4°C in the dark for 30–40 min. The
labeled cells were analyzed in a Cytomics FC500 flow cytometer
(Beckman Coulter, Inc., Fullerton, CA, USA), and the data were
analyzed using CXP software (Beckmann Coulter, Inc.).
Determination of protein expression of
cell surface markers CD24 and CD44 in breast tissue samples by
immunoperoxidase staining
Breast tissue samples were obtained from the
archives of the School of Medicine, Saint-Luc Hospital, IMAG Unit
(IREC), of University of Louvain (Brussels, Belgium), entrusted by
Professor P. Maldague and Professor A. Trouet. The study was
approved by the Ethics Committee of the same Institution and
conducted in accordance with Institutional Guidelines. Tissue
samples from primary surgery were used in the present study, and
none of the patients received neo-adjuvant therapy prior to the
surgery. Hematoxylin/eosin-stained sections from specimens were
evaluated to confirm the diagnosis and to select the most
representative area for each sample. Lesions were diagnosed and
classified according the World Health Organization guidelines
(26). Samples were obtained from
33 patients (all patients had provided written informed consent
prior to obtaining the samples), including 3 normal tissues, 8
dysplastic tissues, 9 ductal carcinomas and 13 reduction
mammoplasty tissues.
Immunocytochemical studies were
performed using a streptavidin-biotin immunoperoxidase method
Tissue sections (5-µm-thick), obtained after
formalin fixation and paraffin-embedding, were deparaffinized in
xylene for 1 h and rehydrated with TBS. Each section was treated
with citrate buffer for 10 min (pH 6.0) for antigen retrieval and
with hydrogen peroxide for quenching of the endogenous peroxidase
activity, then washed with blocking reagent for 20 min. The
aforementioned monoclonal antibodies were applied to all samples,
and the tissues were then placed in a moist chamber overnight at
4°C. A biotinylated secondary antibody was applied, then the
tissues were washed and streptavidin-peroxidase was added. The
localization of bound antibodies was visualized with DAB for 7 min,
and counterstaining with Mayer's hematoxylin solution for 2 min was
performed. Table III describes
the analysis of CD44 and CD24 surface markers in breast biopsy
specimens.
 | Table III.Analysis of CD44 and CD24 surface
markers in breast biopsy specimens. |
Table III.
Analysis of CD44 and CD24 surface
markers in breast biopsy specimens.
| No. | Breast
biopsies | CD44 | CD24 |
|---|
| 1 | Normal tissue | − | − |
| 2 | Normal tissue | − | − |
| 3 | Normal tissue | − | − |
| 4 | Dysplasia | + | + |
| 5 | Dysplasia | + | + |
| 6 | Dysplasia | + | + |
| 7 | Dysplasia | + | + |
| 9 | Dysplasia | + | + |
| 10 | Dysplasia | + | + |
| 11 | Dysplasia | + | + |
| 12 | Dysplasia | + | + |
| 13 | Ductal
carcinoma | + | − |
| 14 | Ductal
carcinoma | + | + |
| 15 | Ductal
carcinoma | + | − |
| 16 | Ductal
carcinoma | − | + |
| 17 | Ductal
carcinoma | + | + |
| 18 | Ductal
carcinoma | − | + |
| 19 | Ductal
carcinoma | − | + |
| 20 | Ductal
carcinoma | − | + |
| 21 | Ductal
carcinoma | − | + |
Statistical analysis
Numerical data are expressed as the mean ± standard
error of the mean (SEM). Comparisons between treated groups and
controls were carried out by ANOVA and Dunnett's test. P<0.05,
P<0.01, P<0.001 were considered to indicate statistically
significant differences. All the experiments were performed at
least three times.
Results
The potential effect of curcumin on the gene and
protein expression of CD24 and CD44 was examined in an established
breast cancer model as explained above (25). MCF-10F, the normal breast cell line,
did not demonstrate any of the features that characterize malignant
cells, such as anchorage-independent growth in soft agar, invasion
or tumor growth in nude mice, whereas the Alpha5 cell line formed
colonies in soft agar, had invasive capability and formed mammary
gland tumors in immunosuppressed mice after injection. Tumors
derived from such animals gave rise to the Tumor2 cells (25).
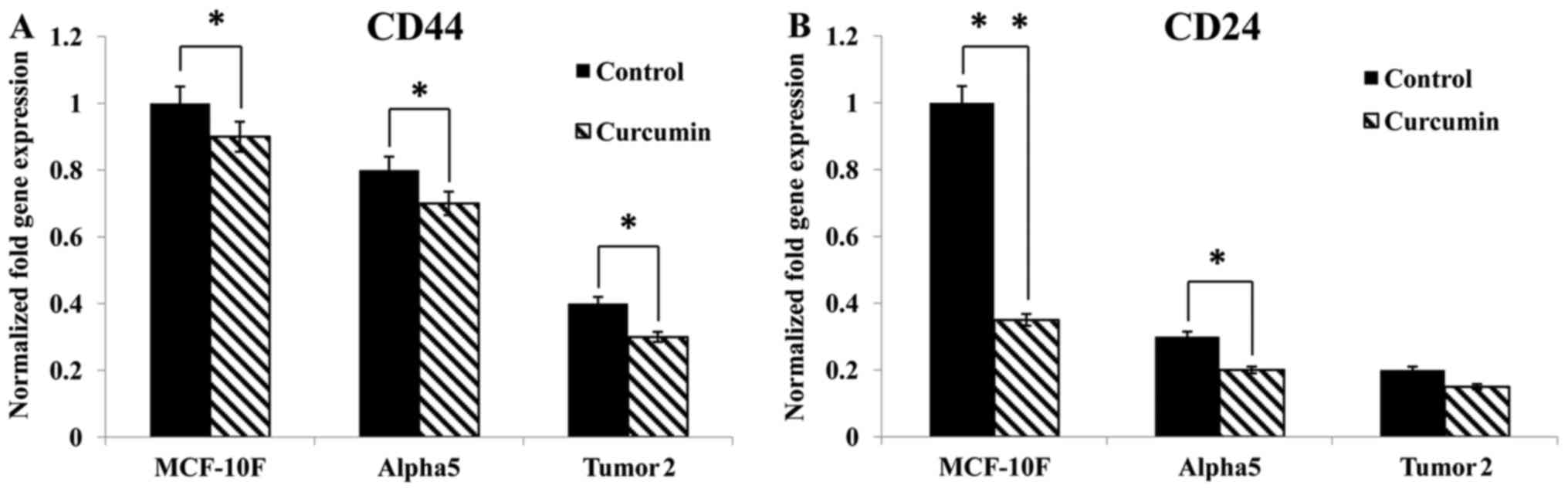
The results revealed that curcumin significantly
(P<0.05) inhibited CD44 and CD24 gene expression in MCF-10F,
Alpha5 and Tumor2 cell lines (Fig.
1), compared with the corresponding control cells. Notably,
CD44 and CD24 surface markers were lower in MCF-10F cells than in
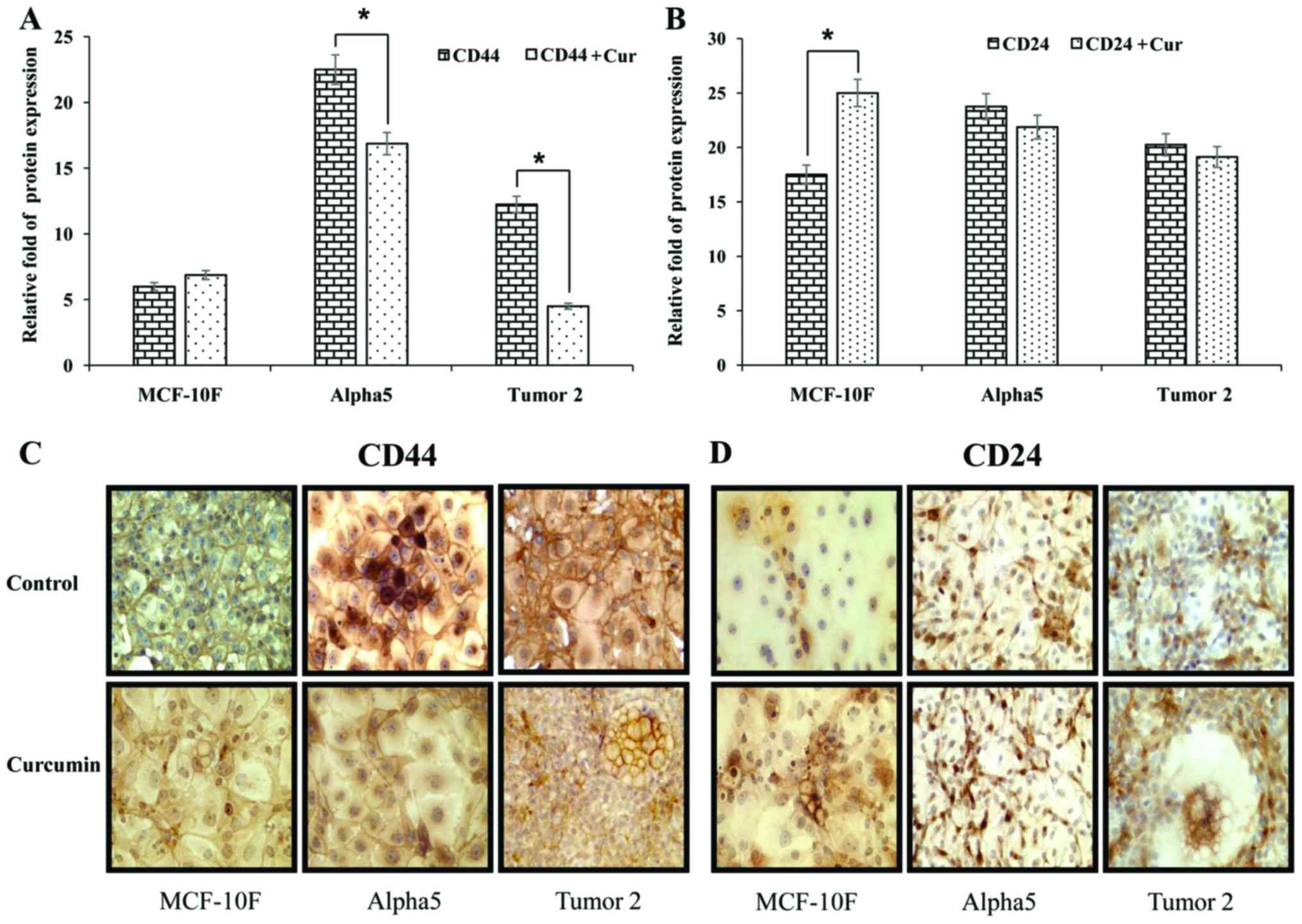
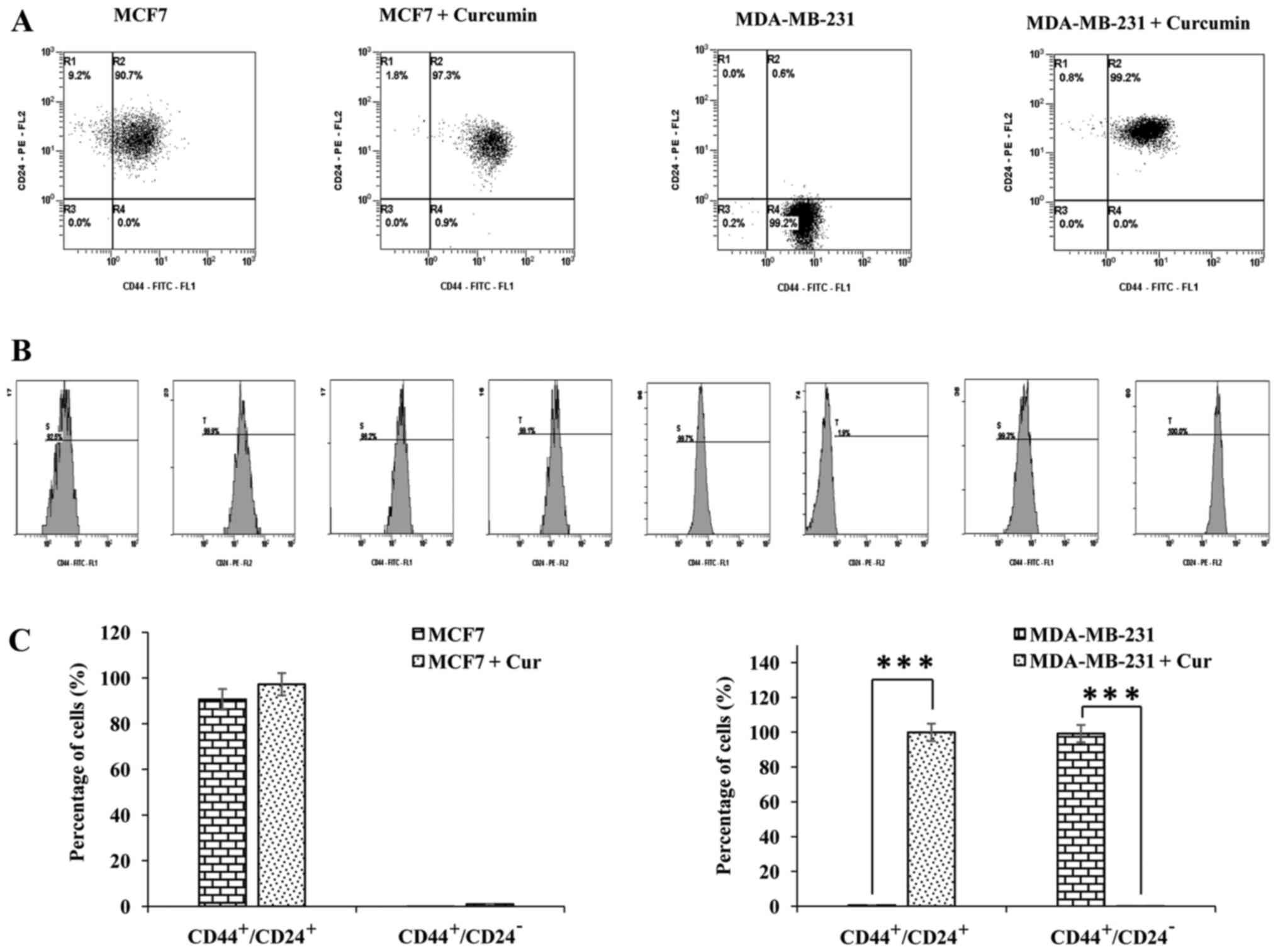
the premalignant and malignant cell lines. As displayed in Fig. 2A and B MCF-10F cells had lower CD44
and CD24 protein expression than Alpha5 and Tumor2 cell lines.
Curcumin decreased such expression in the malignant cell lines.
Protein expression was consistent with gene expression with regard
to the effect of curcumin on Alpha5 and Tumor2 cells.
Representative images of the effect of curcumin on CD44 and CD24
protein expression in these three cell lines are displayed in
Fig. 2C and D.
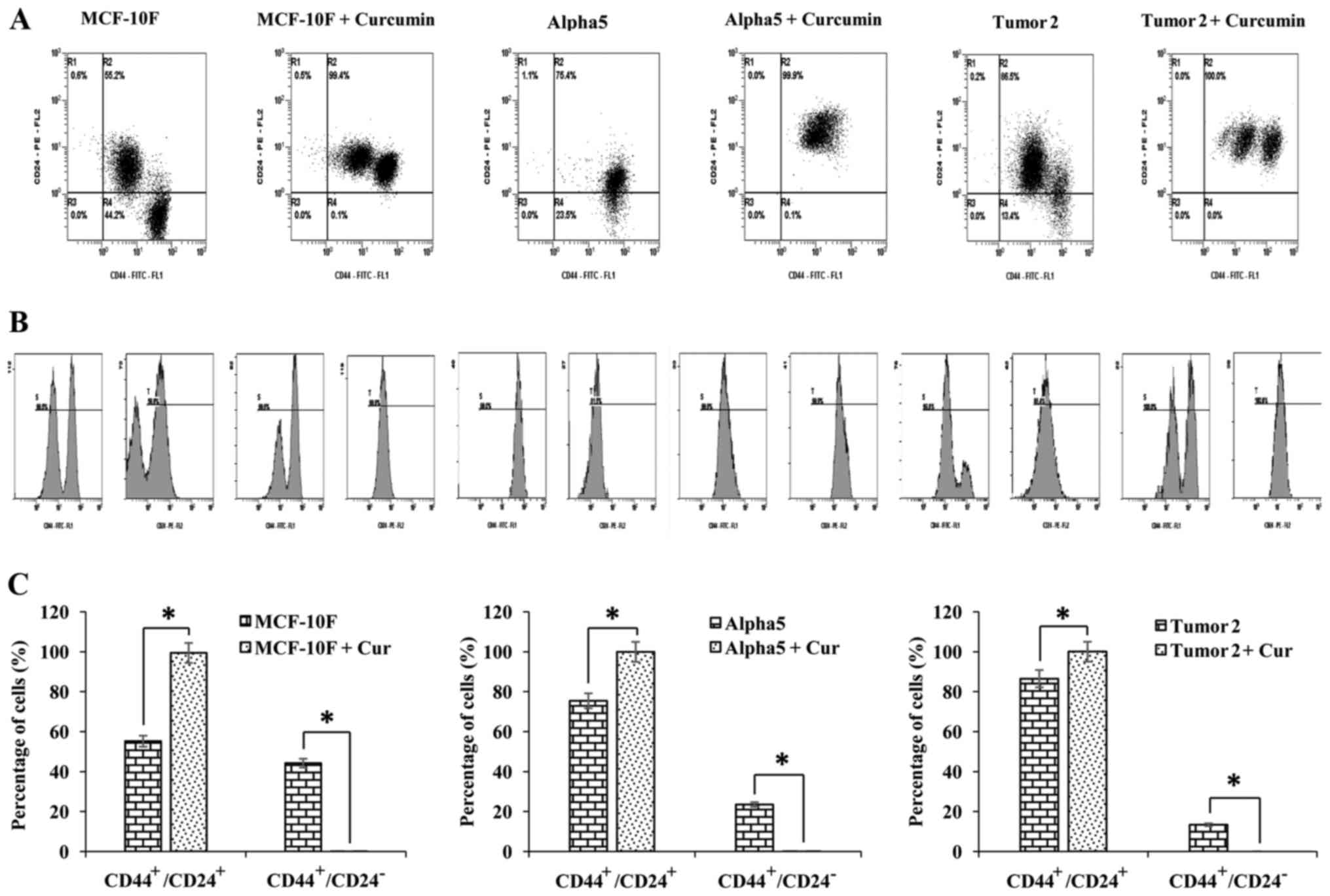
The effect of curcumin on CD44 and CD24 was also
analyzed by flow cytometry in MCF-10F, Alpha5 and Tumor2 cell
lines. The percentages of CD44+/CD24+ and
CD44+/CD24− subpopulations of cells
determined by dot-plots are illustrated in Fig. 3A. The histogram of the expression of
CD44 and CD24 is displayed in Fig.
3B and the graphs in Fig. 3C
depict the percentages of cells with the
CD44+/CD24+ and
CD44+/CD24− phenotypes. The
CD44+/CD24+ subpopulation was significantly
larger than the CD44+/CD24− subpopulation in
MCF-10F, Alpha5 and Tumor2 cells. Notably, curcumin decreased
CD44+/CD24− and increased
CD44+/CD24+ subpopulations in normal MCF-10F
and pre-tumorigenic Alpha5 cells, whereas there was no significant
effect in Tumor2 cells compared with the control cells of the same
type. Table II summarizes the
effects of curcumin on the percentage of CD44 and CD24 in the
MCF-10F, Alpha5, Tumor2, MCF-7 and MDA-MB-231 breast cell lines, as
determined by flow cytometry.
 | Table II.Percentage of CD44/CD24 cell surface
markers in breast cell lines untreated and treated with
curcumin. |
Table II.
Percentage of CD44/CD24 cell surface
markers in breast cell lines untreated and treated with
curcumin.
| Breast cell
lines |
CD44+/CD24+ (%) |
CD44+/CD24− (%) |
CD44−/CD24+ (%) |
CD44−/CD24− (%) |
|---|
| MCF-10F | 55.2 | 44.2 | 0.6 | 0.0 |
| MCF-10F + Cur | 99.4 | 0.1 | 0.5 | 0.0 |
| Alpha5 | 75.4 | 23.5 | 1.1 | 0.0 |
| Alpha5 + Cur | 99.9 | 0.1 | 0 | 0.0 |
| Tumor2 | 86.5 | 13.4 | 0.2 | 0.0 |
| Tumor2 + Cur | 100.0 | 0.0 | 0.0 | 0.0 |
| MCF7 | 90.7 | 0.0 | 9.2 | 0.0 |
| MCF7 + Cur | 97.3 | 0.9 | 1.8 | 0.0 |
| MDA-MB-231 | 0.6 | 99.2 | 0.0 | 0.2 |
| MDA-MB-231 +
Cur | 99.2 | 0.0 | 0.8 | 0.0 |
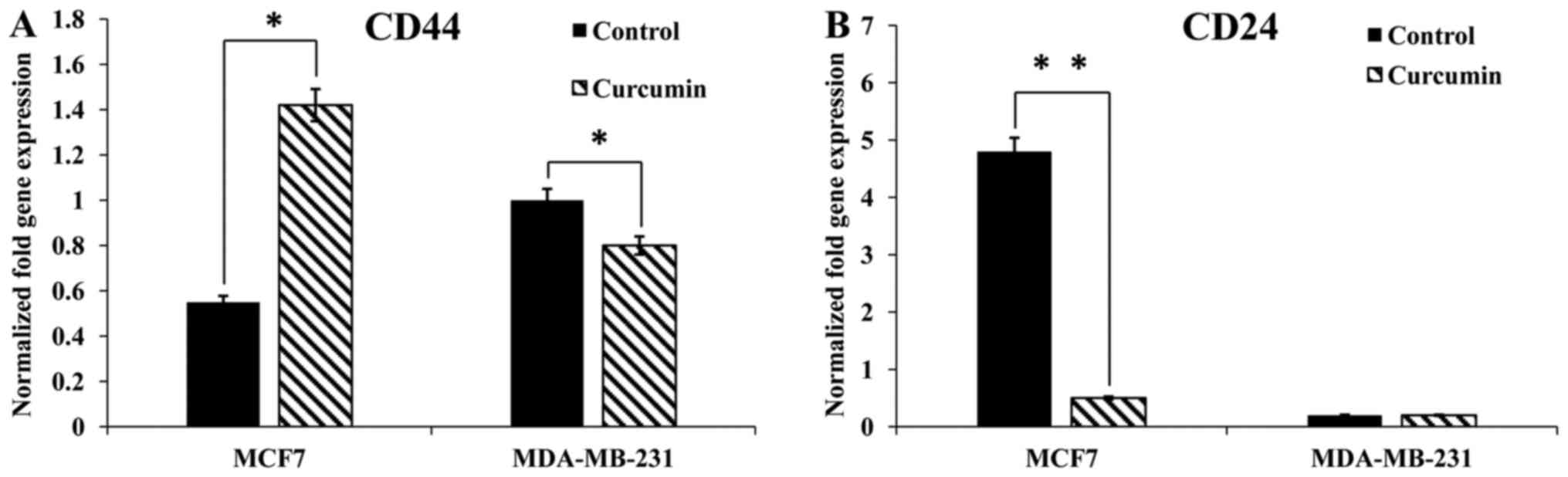
As displayed in Fig.
4, curcumin increased CD44 and decreased CD24 gene expression
in MCF-7 cells. Conversely, curcumin decreased CD44 gene expression
in the MDA-MB-231 cell line. Notably, CD24 was not present in
MDA-MB-231 cells. The effect of curcumin on CD44 and CD24 was
analyzed via flow cytometry in MCF-7 and MDA-MB-231 cells (Fig. 5A-C). Curcumin did not affect the
CD44+/CD24+ or the
CD44+/CD24− subpopulations in the MCF-7 cell
line. However, the CD44+/CD24+ subpopulation
was increased and the CD44+/CD24−
subpopulation was decreased in MDA-MB-231 cells following treatment
with curcumin. The percentages of CD44+/CD24+
and CD44+/CD24− cells are indicated in
Fig. 5C.
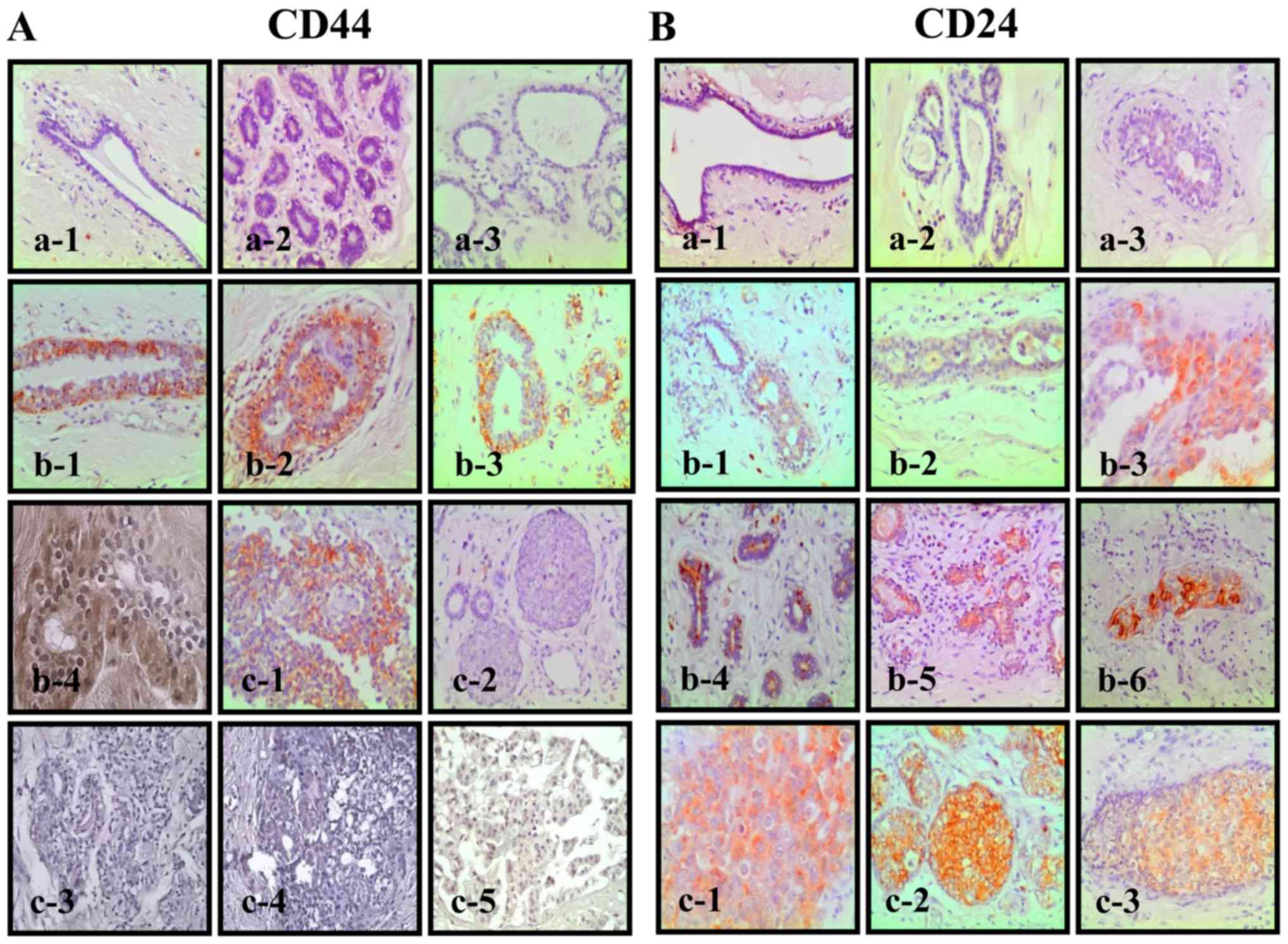
Biopsy specimens obtained from 33 patients diagnosed
with benign and breast cancer lesions were studied. Among them were
three normal tissues, which were negative for CD44 and CD24
(Fig. 6A-a and B-a), 8 dysplastic
tissues positive for CD44 and CD24 (Fig. 6A-b and B-b), 9 positive ductal
carcinomas, of which some were positive for CD44 (Fig. 6A-c1-5) and all of which were
positive for CD24 (Fig. 6B-c).
Benign specimens contained atypia, subnormal ductules and atypical
ductal hyperplasia. The 13 samples from reduction mammoplasties
were all negative for both markers.
Discussion
The present study was conducted to analyze the
effect of curcumin on the cell surface markers and adhesion
molecules CD44 and CD24 in breast cancer cell lines. The results
demonstrated that curcumin decreased CD44 and CD24 gene expression
levels in the Alpha model (MCF-10F, Alpha5 and Tumor2 cell lines).
Peroxidase staining results corroborated that CD44 and CD24 protein
expression were also decreased in Alpha5 and Tumor2 cells. However,
in MCF-10F cells, there were increases in both markers.
Furthermore, curcumin increased CD44 and decreased CD24 gene
expression in MCF-7 cells, but decreased CD44 expression in
MDA-MB-231 cells, while having no significant effect on CD24.
Certain previous studies (27–29)
have indicated that CD44 has several roles, including its activity
as an adhesion molecule and as a signal regulator. It has been
reported that CD44-positive cells exhibit a more mesenchymal-like
profile, are usually enriched for genes involved in cell motility,
proliferation and angiogenesis and form tumors in mice (27).
CD24-positive cells usually express genes implicated
in carbohydrate metabolism and RNA splicing (27). CD24 is a mucin-like adhesion
molecule that increases the metastatic potential of malignant cells
and is associated with poor clinical outcomes in breast carcinomas
(27).
In the present study, the effects of curcumin were
examined via flow cytometry to determine the percentages of two
subpopulations of cells, CD44+/CD24+ and
CD44+/CD24−, among MCF-10F, Alpha5 and Tumor2
cell lines. It was found that the CD44+/CD24+
and CD44+/CD24− subpopulations were present
in all three cell lines. When curcumin treatment was applied, the
CD44+/CD24+ subpopulation increased and the
CD44+/CD24− subpopulation decreased in the
normal MCF-10F and the pre-tumorigenic Alpha5 cells. Such an effect
was not observed in Tumor2 cells in comparison to the corresponding
control cells of the same type.
When subpopulations were analyzed by flow cytometry
it was observed that curcumin increased the
CD44+/CD24+ and decreased the
CD44+/CD24− subpopulations of MDA-MB-231
cells. However, curcumin did not affect the
CD44+/CD24+ or the
CD44+/CD24− subpopulations of MCF-7 cells.
The prognostic value of these markers in breast cancer remains
controversial and requires further research (30,31).
A pioneering study (32) in human breast cancer cells, such as
MCF-7 and MDA-MB-231, demonstrated that
CD44+/CD24− cells can be recognized as
prospective cancer stem cells. Later, these findings were supported
by those of Sheridan et al (33). Evidence indicates that such
phenotypes are not present in all breast cancers, only in breast
cancer stem cells (30,34,35).
It is important to point out that the
CD44+/CD24+ phenotype Is highly expressed in
differentiated epithelial cell types (30) and CD44+/CD24−
cells exhibit undifferentiated basal/mesenchymal cell properties
(33). It can be suggested that
curcumin can be used to improve the ratio of
CD44+/CD24+ cells and decrease the
CD44+/CD24− subpopulation, as well as the
cancerous types of breast cells. Such results have some kind of
significance to be taken into consideration concerning the use of
curcumin for breast cancer treatment. Evidence of migration, colony
formation and invasion in CD44+ MDA-MB-468 cells
supports the concept of regulation of phenotypes (36). In similar studies (37), evidence indicated that there can be
inter-conversion between the phenotypes and that epithelial-like
CD44+/CD24+ cells can readily give rise to
CD44+/CD24− cells during tumor initiation
(38).
In summary, the present study investigated the
expression of CD44 and CD24 in epithelial lesions derived from
biopsy specimens obtained from the archived tissues of patients
with benign and breast cancer lesions. The results indicated that
normal tissues were negative for CD44 and CD24 expression. However,
benign lesions were positive for both markers. Malignant tissues
were negative for CD44 and positive for CD24 in most of the cases.
In conclusion, these results indicated that a natural substance
such as curcumin may be used to improve the ratio of
CD44+/CD24+ cells and to decrease
CD44+/CD24− subpopulations, which have the
characteristics of undifferentiated basal/mesenchymal cells. Our
next study will be conducted with the aim to validate this
conclusion and further explore it in animal models where we will
study the effects of curcumin on the expression of CD44 and CD24
in vivo by delivering it orally in nude mouse xenografts of
Alpha5 and Tumor2 cells. Furthermore, we will consider the effects
of curcumin on malignant behavior, such as the proliferation or
motility of breast cancer cells used in the present study in
association with studies on the expression of CD44 and CD24.
Acknowledgements
The technical support of Georgina Vargas, Guiliana
Rojas and Leodán A. Crispin are greatly appreciated.
Funding
The present study was supported by UTA FIAC 1117
(GMC).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
GMC and JAQ conceived and designed the study. GMC,
RPC and JAQ performed the experiments. GMC and RPC wrote the the
manuscript. GMC, RPC and JAQ reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee School of Medicine, Saint-Luc Hospital, IMAG Unit (IREC)
of University of Louvain (Brussels, Belgium). All patients had
provided written informed consent prior to obtaining the
samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han JS and Crowe DL: Tumor initiating
cancer stem cells from human breast cancer cell lines. Int J Oncol.
34:1449–1453. 2009.PubMed/NCBI
|
|
2
|
Gudjonsson T, Villadsen R, Nielsen HL,
Rønnov-Jessen L, Bissell MJ and Petersen OW: Isolation,
immortalization, and characterization of a human breast epithelial
cell line with stem cell properties. Genes Dev. 16:693–706. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vesuna F, Lisok A, Kimble B and Raman V:
Twist modulates breast cancer stem cells by transcriptional
regulation of CD24 expression. Neoplasia. 11:1318–1328. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fang X, Zheng P, Tang J and Liu Y: CD24:
from A to Z. Cell Mol Immunol. 7:100–103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sleeman KE, Kendrick H, Ashworth A, Isacke
CM and Smalley MJ: CD24 staining of mouse mammary gland cells
defines luminal epithelial, myoepithelial/basal and non-epithelial
cells. Breast Cancer Res. 8:R72006. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fogel M, Friederichs J, Zeller Y, Husar M,
Smirnov A, Roitman L, Altevogt P and Sthoeger ZM: CD24 is a marker
for human breast carcinoma. Cancer Lett. 143:87–94. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kristiansen G, Winzer KJ, Mayordomo E,
Bellach J, Schlüns K, Denkert C, Dahl E, Pilarsky C, Altevogt P,
Guski H, et al: CD24 expression is a new prognostic marker in
breast cancer. Clin Cancer Res. 9:4906–4913. 2003.PubMed/NCBI
|
|
8
|
Lim SC: CD24 and human carcinoma: Tumor
biological aspects. Biomed Pharmacother. 59 Suppl 2:S351–S354.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jaggupilli A and Elkord E: Significance of
CD44 and CD24 as cancer stem cell markers: An enduring ambiguity.
Clin Dev Immunol. 2012:7080362012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ponta H, Sherman L and Herrlich PA: CD44:
From adhesion molecules to signalling regulators. Nat Rev Mol Cell
Biol. 4:33–45. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Orian-Rousseau V: CD44, a therapeutic
target for metastasising tumours. Eur J Cancer. 46:1271–1277. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marhaba R and Zöller M: CD44 in cancer
progression: Adhesion, migration and growth regulation. J Mol
Histol. 35:211–231. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Günthert U, Hofmann M, Rudy W, Reber S,
Zöller M, Haussmann I, Matzku S, Wenzel A, Ponta H and Herrlich P:
A new variant of glycoprotein CD44 confers metastatic potential to
rat carcinoma cells. Cell. 65:13–24. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kristiansen G, Sammar M and Altevogt P:
Tumour biological aspects of CD24, a mucin-like adhesion molecule.
J Mol Histol. 35:255–262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JH, Kim SH, Lee ES and Kim YS: CD24
overexpression in cancer development and progression: A
meta-analysis. Oncol Rep. 22:1149–1156. 2009.PubMed/NCBI
|
|
17
|
Kristiansen G, Machado E, Bretz N, Rupp C,
Winzer KJ, König AK, Moldenhauer G, Marmé F, Costa J and Altevogt
P: Molecular and clinical dissection of CD24 antibody specificity
by a comprehensive comparative analysis. Lab Invest. 90:1102–1116.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Darwish NS, Kim MA, Chang MS, Lee HS, Lee
BL, Kim YI and Kim WH: Prognostic significance of CD24 expression
in gastric carcinoma. Cancer Res Treat. 36:298–302. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choi YL, Kim SH, Shin YK, Hong YC, Lee SJ,
Kang SY and Ahn G: Cytoplasmic CD24 expression in advanced ovarian
serous borderline tumors. Gynecol Oncol. 97:379–386. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smith SC, Oxford G, Wu Z, Nitz MD, Conaway
M, Frierson HF, Hampton G and Theodorescu D: The
metastasis-associated gene CD24 is regulated by Ral GTPase
and is a mediator of cell proliferation and survival in human
cancer. Cancer Res. 66:1917–1922. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bretz N, Noske A, Keller S, Erbe-Hofmann
N, Schlange T, Salnikov AV, Moldenhauer G, Kristiansen G and
Altevogt P: CD24 promotes tumor cell invasion by suppressing tissue
factor pathway inhibitor-2 (TFPI-2) in a c-Src-dependent fashion.
Clin Exp Metastasis. 29:27–38. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baumann P, Cremers N, Kroese F, Orend G,
Chiquet-Ehrismann R, Uede T, Yagita H and Sleeman JP: CD24
expression causes the acquisition of multiple cellular properties
associated with tumor growth and metastasis. Cancer Res.
65:10783–10793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aggarwal BB, Sundaram C, Malani N and
Ichikawa H: Curcumin: The Indian solid gold. Adv Exp Med Biol.
595:1–75. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Labbozzetta M, Notarbartolo M, Poma P,
Maurici A, Inguglia L, Marchetti P, Rizzi M, Baruchello R, Simoni D
and D'Alessandro N: Curcumin as a possible lead compound against
hormone-independent, multidrug-resistant breast cancer. Ann NY Acad
Sci. 1155:278–283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO Classification of Tumors of the Breast.
4. 4th edition. International Agency for Research on Cancer; Lyon,
France: 2012
|
|
27
|
Dontu G, Al-Hajj M, Abdallah WM, Clarke MF
and Wicha MS: Stem cells in normal breast development and breast
cancer. Cell Prolif. 36 Suppl 1:59–72. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dontu G, El-Ashry D and Wicha MS: Breast
cancer, stem/progenitor cells and the estrogen receptor. Trends
Endocrinol Metab. 15:193–197. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kordon EC and Smith GH: An entire
functional mammary gland may comprise the progeny from a single
cell. Development. 125:1921–1930. 1998.PubMed/NCBI
|
|
30
|
Ricardo S, Vieira AF, Gerhard R, Leitão D,
Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F and Paredes
J: Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression
distribution within intrinsic molecular subtype. J Clin Pathol.
64:937–946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou L, Jiang Y, Yan T, Di G, Shen Z, Shao
Z and Lu J: The prognostic role of cancer stem cells in breast
cancer: A meta-analysis of published literatures. Breast Cancer Res
Treat. 122:795–801. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sheridan C, Kishimoto H, Fuchs RK,
Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet R Jr, Badve S and
Nakshatri H: CD44+/CD24− breast cancer cells
exhibit enhanced invasive properties: An early step necessary for
metastasis. Breast Cancer Res. 8:R592006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Honeth G, Bendahl PO, Ringnér M, Saal LH,
Gruvberger-Saal SK, Lövgren K, Grabau D, Fernö M, Borg A and
Hegardt C: The CD44+/CD24− phenotype Is
enriched in basal-like breast tumors. Breast Cancer Res.
10:R532008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mylona E, Giannopoulou I, Fasomytakis E,
Nomikos A, Magkou C, Bakarakos P and Nakopoulou L: The
clinicopathologic and prognostic significance of
CD44+/CD24−/low and
CD44−/CD24+ tumor cells in invasive breast
carcinomas. Hum Pathol. 39:1096–1102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Croker AK, Goodale D, Chu J, Postenka C,
Hedley BD, Hess DA and Allan AL: High aldehyde dehydrogenase and
expression of cancer stem cell markers selects for breast cancer
cells with enhanced malignant and metastatic ability. J Cell Mol
Med. 13:2236–2252. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meyer MJ, Fleming JM, Ali MA, Pesesky MW,
Ginsburg E and Vonderhaar BK: Dynamic regulation of CD24 and the
invasive, CD44posCD24neg phenotype In breast
cancer cell lines. Breast Cancer Res. 11:R822009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|