Introduction
Endometrial cancer (EC) is the most common
gynecologic tumor in developed countries, with an increasing
prevalence (1). It is estimated
that 61,380 new cases and 10,920 deaths resulting from EC occurred
in 2017 (2). Adenocarcinoma of the
endometrium accounts for over 70% of all uterine cancers. Most
patients (80%) are diagnosed with early disease and can be
surgically cured. However, the outcome of advanced or recurrent
cases remains far worse, and the adjuvant treatment options are
quite limited (1). Discovery of
novel targets is warranted to better understand the EC pathogenesis
and to develop new therapeutic approaches for this disease. It is
well known that tumor progression and metastasis are often linked
to epithelial-mesenchymal transition (EMT). During this process, a
more invasive cell phenotype is established, accompanied by
alterations in the expression of many core molecules, particularly
E-cadherin (3). Thus, it is
worthwhile to explore the regulatory mechanism of EMT in the tumor
biology of EC.
S100 calcium binding protein A4 (S100A4) has been
shown to be involved in biological functions that contribute to
malignant tumors, such as proliferation, apoptosis, metastasis,
angiogenesis and immune evasion (4). More importantly, S100A4 plays pivotal
roles in tumor invasion by triggering EMT, mechanically serves as a
downstream target gene of the wnt/β-catenin pathway, modulates
membrane integrin signaling, and directly promotes cell motility
through interaction with cytoskeletal proteins such as myosin,
actin and tropomyosin (5–7). Elevated S100A4 expression has been
found in many types of tumors and is closely related to poor
outcome in tumor patients (8). Our
previous research revealed that S100A4 is highly expressed in EC
cells, and knockdown of S100A4 expression resulted in suppression
of the migration and invasion capability of EC cells, which may
partially occur via EMT-related modifications (9). However, the regulatory mechanisms of
S100A4 expression in EC remain to be elucidated.
Estrogen-related receptors (ERRs; ERRα, ERRβ and
ERRγ) comprise a subgroup of orphan nuclear receptors that share
highly homologous DNA-binding domains with estrogen receptors.
However, ERRs do not bind to endogenous estrogens or their
derivatives, and thus are designated orphan receptors (10). High ERRγ expression levels are often
associated with high metabolic demand in human tissues, such as
skeletal muscle, heart and brown adipose tissue. Accumulating
evidence indicates a central role of ERRγ in metabolic genes and
cellular energy metabolism regulation (11). Apart from metabolic disease, recent
studies have revealed the clinical significance of ERRγ in several
cancer types, including EC. In breast cancer, ERRγ is generally
overexpressed and related to lymph node status and upregulated
during tamoxifen resistance acquisition, indicating a cancer
promoting role of ERRγ (12,13).
The role of ERRγ in EC remains unclear. Overexpression of ERRγ is
correlated with increased clinical stage, deeper myometrium
invasion and positive lymph node status (14,15).
In addition, ERRγ mediates estrogen-induced proliferation of EC
cells (16). However, the effect of
ERRγ on EC cell migration and metastasis has never been
explored.
In the present study, augmented expression of ERRγ
was found, and for the first time, a correlation between ERRγ and
S100A4 expression was identified in clinical EC tissues via
experimental techniques and public database mining. Furthermore,
ERRγ directly facilitates S100A4 transcription through promoter
activation, thus promoting migration and invasion of EC cells both
in vitro and in vivo, demonstrating the emerging
roles of ERRγ in EC progression through transcriptional regulation
of S100A4.
Materials and methods
Patients and specimens
The present study was performed in accordance with
the Declaration of Helsinki, and approval to conduct the present
study was obtained from the Ethics Committee of Tongji Medical
College, Huazhong University of Science and Technology (IORG no.
IORG0003571). Tissues were collected after receiving informed
consent from the patients. Formalin-fixed paraffin-embedded
specimens were obtained from 20 primary EC patients [age (mean ±
SD), 51.5±12.1 years] who had undergone surgical resection at WuHan
Union Hospital between September 2015 and December 2016. Those that
had already received adjuvant therapy, such as chemotherapy,
hormone therapy or radiotherapy, were excluded. Fresh specimens
from the above-mentioned EC patients and another 20 normal
endometrium cases (age, 49.6±9.4 years) were collected for protein
and total RNA extraction and stored in liquid nitrogen until
use.
Immunohistochemical staining
Fresh specimens were fixed in 10% formaldehyde for
at least 24 h and embedded in paraffin. Tissue slides with
4-µm-thick sections were constructed and dewaxed in xylene and
rehydrated in a graded alcohol series. Antigen retrieval was
conducted by heating slides in 0.01 M of sodium citrate buffer for
20 min. Endogenous non-specific peroxidase activity was blocked
with 3% H2O2 for 15 min, and non-specific
staining was blocked by incubation with 10% normal goat serum for
30 min. Then, the samples were incubated with 200 µl of primary
antibodies against ERRγ (1:400; cat. no. ab49129; Abcam, Cambridge,
MA, USA), S100A4 (1:400; cat. no. 13018S; Cell Signaling
Technology, Inc., Beverly, MA, USA) and Ki-67 (1:200; cat. no.
sc-23900; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at
4°C overnight. After being washed with phosphate-buffered saline
(PBS), the slides were incubated with EnVision/HRP, rabbit
secondary antibody (1:1,000; cat. no. GB23303; Servicebio
Technology, Co., Ltd., Wuhan, China) for 30 min. Diaminobenzidine
substrate was used for visualization, followed by counterstaining
with hematoxylin. Finally, the slides were dehydrated and mounted.
The immunohistochemistry scoring strategy was performed as
previously described (9).
Western blot analysis
Collected fresh EC tissues or cultured EC cells were
washed with ice-cold PBS 3 times and lysed with
radio-immunoprecipitation assay buffer containing protease
inhibitors. The protein concentration was determined using a
bicinchoninic acid (BCA) protein assay kit. Equal amounts of
proteins (30 µg) were added to 12% SDS-PAGE gels and transferred
onto nitrocellulose membranes. After being blocked with 5% skim
milk in 1X TBS buffer containing 0.1% Tween-20 at room temperature
for 1 h, the membranes were incubated with primary antibodies
against ERRγ (1:800; cat. no. ab49129; Abcam), S100A4 (1:800; cat.
no. 13018S; Cell Signaling Technology), GAPDH (1:1,000; cat. no.
sc-66163; Santa Cruz Biotechnology) and E-cadherin (1:200; cat. no.
sc-52327; Santa Cruz Biotechnology) at 4°C overnight. The target
proteins were visualized using enhanced chemiluminescence reagents
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) after incubation
with goat anti-rabbit secondary antibody (1:5,000; cat. no.
sc-2004; Santa Cruz Biotechnology). Enhanced chemiluminescence
reagents (Thermo Fisher Scientific) were used for bands detection.
The optical density was quantified using Bio-Rad Image Lab™ v4.1
software.
Real-time quantitative RT-PCR
Total RNA from EC tissues and cultured EC cells was
isolated with RNAiso Plus (Takara Bio, Co., Ltd., Otsu, Japan)
following the manufacturer's protocol. The reverse transcription
reactions were carried out using a PrimeScript RT reagent kit
(Takara Bio). The primers involved were as follows: ERRγ (102 bp),
5′-CCCGACAGTGACATCAAAGCC-3′ (sense) and
5′-CGTGGAGAAGCCTGGAATATGC-3′ (antisense); S100A4 (251 bp),
5′-TACTCGGGCAAAGAGGGTGA-3′ (sense) and 5′-CATTTCTTCCTGGGCTGCTTA-3′
(antisense); E-cadherin (162 bp), 5′-GAGAACGCATTGCCACATACAC-3′
(sense) and 5′-GAGCACCTTCCATGACAGACCC-3′ (antisense); GAPDH (255
bp), 5′-ACTTTGGTATCGTGGAAGGACTAT-3′ (sense) and
5′-GTTTTTCTAGACGGCAGGTCAGG-3′ (antisense). Real-time PCR was
conducted with Premix Ex Taq (Takara Bio), as previously described
(9). The 2−ΔΔCt method
was employed for relative transcript abundance determination. Each
experiment was performed in triplicate at least 3 times.
Cell culture and transfection
The human EC cell lines Ishikawa, AN3CA, HEC-1A and
HEC-1B were purchased from the American Type Culture Collection
(ATCC, Manassas, VA, USA). Cells were cultured in appropriate
medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml of
penicillin and 100 µg/ml of streptomycin. All the cells were
incubated at 37°C in a humidified atmosphere containing 5%
CO2.
Plasmid/construct transfection
Oligonucleotides encoding short hairpin RNAs
(shRNAs) for ERRγ (sh-ERRγ), S100A4 (sh-S100A4), scramble shRNA
(sh-Scb), and S100A4 expression cDNA (GV230-S100A4) were purchased
from Shanghai GeneChem (Shanghai, China). Invitrogen™ Lipofectamine
2000 (Thermo Fisher Scientific) was used for transfection. The
target sequences for synthetic oligonucleotide primers are listed
in Table I. A lentiviral vector
carrying ERRγ gene fragments was constructed to upregulate ERRγ
expression in HEC-1A and AN3CA cells. The empty vector served as
the control. Stable cell lines were selected by administration of
geneticin or puromycin (Sigma-Aldrich; Merck KGaA).
 | Table I.Oligonucleotide sets used for
constructs and short hairpin RNAs. |
Table I.
Oligonucleotide sets used for
constructs and short hairpin RNAs.
| Oligo set | Sequences |
|---|
| sh-Scb |
5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′
(sense) |
|
|
5′-AATTCAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAA-3′
(antisense) |
| sh-ERRγ-1 |
5′-CCGGCCTCACTACACTGTGTGACTTCTCGAGAAGTCACACAGTGTAGTGAGGTTTTTG-3′
(sense) |
|
|
5′-AATTCAAAAACCTCACTACACTGTGTGACTTCTCGAGAAGTCACACAGTGTAGTGAGG-3′
(antisense) |
| sh-ERRγ-2 |
5′-CCGGCGAATGAATGTGAAATCACAACTCGAGTTGTGATTTCACATTCATTCGTTTTTG-3′
(sense) |
|
|
5′-AATTCAAAAACGAATGAATGTGAAATCACAACTCGAGTTGTGATTTCACATTCATTCG-3′
(antisense) |
| sh-S100A4 |
5′-CCGGCGCCATGATGTGTAACGAATTCTCGAGAATTCGTTACACATCATGGCGTTTTTG-3′
(sense) |
|
|
5′-AATTCAAAAACGCCATGATGTGTAACGAATTCTCGAGAATTCGTTACACATCATGGCG-3′
(antisense) |
Luciferase reporter assay
Sequences containing different S100A4 promoter
sections were cloned and sub-cloned to pcDNA3.0 basic vectors. EC
cells were plated on 24-well plates overnight and co-transfected
with luciferase reporter vectors and the internal control plasmid
pRL-SV40 (Promega Corp., Madison, WI, USA) carrying the
Renilla luciferase gene for 24 h. Firefly and Renilla
luciferase activities were measured with a dual-luciferase assay
system kit (Promega Corp.). Experiments performed for the analysis
were repeated at least 3 times.
Transwell migration and invasion
assays
Migration assays were performed using Transwell
inserts with 8.0-µm pore membrane filters (Corning Costar,
Tewksbury, MA, USA). For invasion assays, the microfilters were
precoated with 50 µl of Matrigel matrix (BD Biosciences, Sparks,
MD, USA). Then, 5×104 homogeneous single cells were
plated in the upper chambers of each well with low-serum medium (1%
FBS), and a chemo-attractant (medium containing 10% FBS) was added
to the lower chamber, followed by a 24-h incubation. Afterwards,
the membranes were fixed with 4% paraformaldehyde and stained with
0.5% crystal violet. The numbers of migrating and invading cells
were determined in 5 random fields of each membrane using CX23
Olympus light microscopy (Olympus Optical Co., Ltd., Tokyo, Japan).
Three replicates were performed for this analysis.
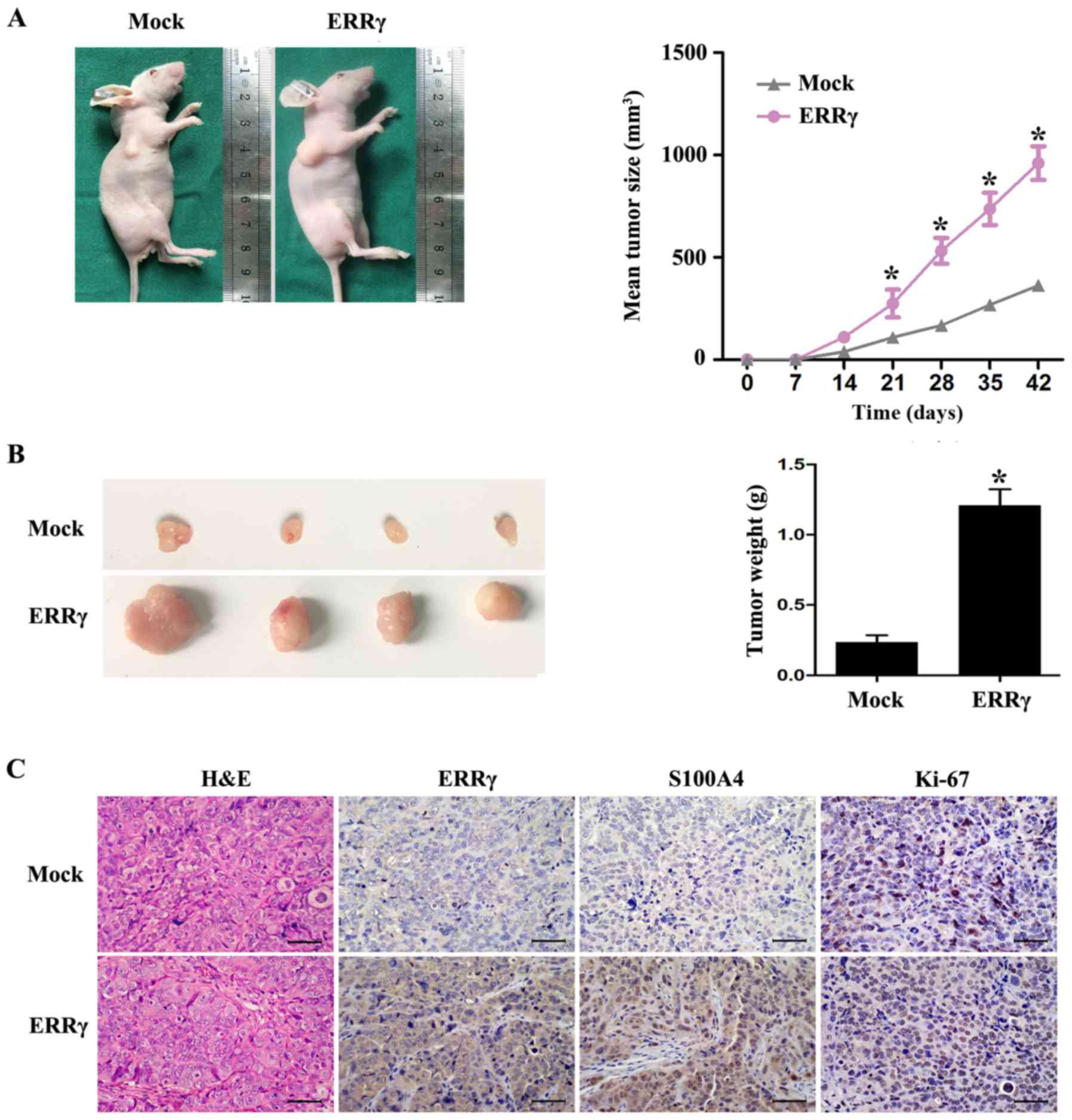
In vivo tumor growth assay
All animal experiments were approved by the Animal
Care Committee of Tongji Medical College. Female 4- to 6-week-old
and weight ~20 g BALB/c nude mice were purchased from Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). The
animals were housed at 23±1°C under a standard 12h/12h light-dark
cycle with free access to food and water. Mice were injected
subcutaneously in the right flank with 5×106 HEC-1A
cells stably transfected with ERRγ or empty vectors as indicated
(n=5 per group). Tumor volume (V) was measured every 4 days and
calculated via the formula: V = length × width2/2. At 42
days after injection, the mice were sacrificed, and subcutaneous
tumors were weighed and photographed using a camera. The protein
levels of ERRγ, S100A4 and Ki-67 were analyzed in tumor tissues via
immunohistochemistry.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism v5.0 statistical software (GraphPad Software, Inc., La Jolla,
CA, USA). All results are expressed as the mean ± standard error of
the mean (SEM). Differences among groups were determined with
Student's t-test or one-way analysis of variance (ANOVA). The post
hoc test was performed for intergroup comparison of parametric
data. Pearson's coefficient correlation was applied to investigate
the association between ERRγ and S100A4 expression levels. P-values
<0.05 were statistically significant.
Results
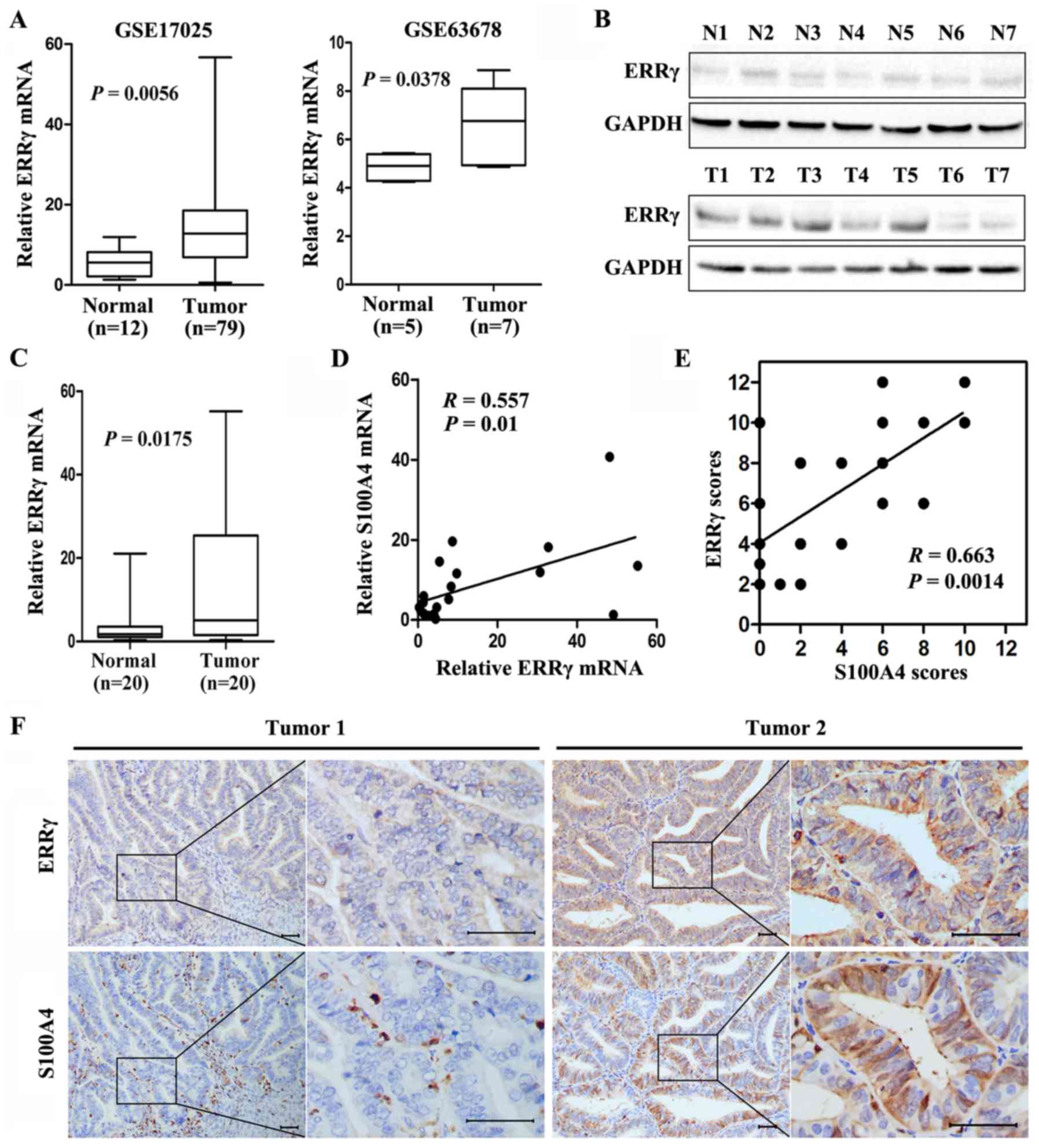
ERRγ is highly expressed and
positively correlated with S100A4 expression in EC tissues
Mining the publicly available databases Gene
Expression Omnibus website (GEO; http://www.ncbi.nlm.nih.gov/geo/) and R2, microarray
analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) revealed
increased ERRγ transcription levels in EC compared with benign
endometrium (Fig. 1A), and an
inverse correlation between ERRγ and S100A4 transcription levels
was observed in several types of cancers, including non-small cell
lung and colon cancer, squamous cell carcinoma of the tongue, oral
cavity cancer, and Wilms' tumors (data not shown). To investigate
ERRγ expression in EC, fresh tissues from 20 well established
primary EC patients and 20 normal endometrium cases were collected.
Real-time quantitative RT-PCR and western blotting showed higher
expression levels of ERRγ in EC specimens than the levels noted in
the benign endometrium (Fig. 1B and
C). In addition, a positive correlation between ERRγ and S100A4
transcription levels in EC tissues was verified with real-time
quantitative RT-PCR (correlation coefficient R=0.557, P=0.01,
Fig. 1D) and immunohistochemical
staining (correlation coefficient R=0.663, P=0.0014, Fig. 1E and F). These results indicated
that ERRγ is overexpressed and positively correlated with S100A4 in
EC patients.
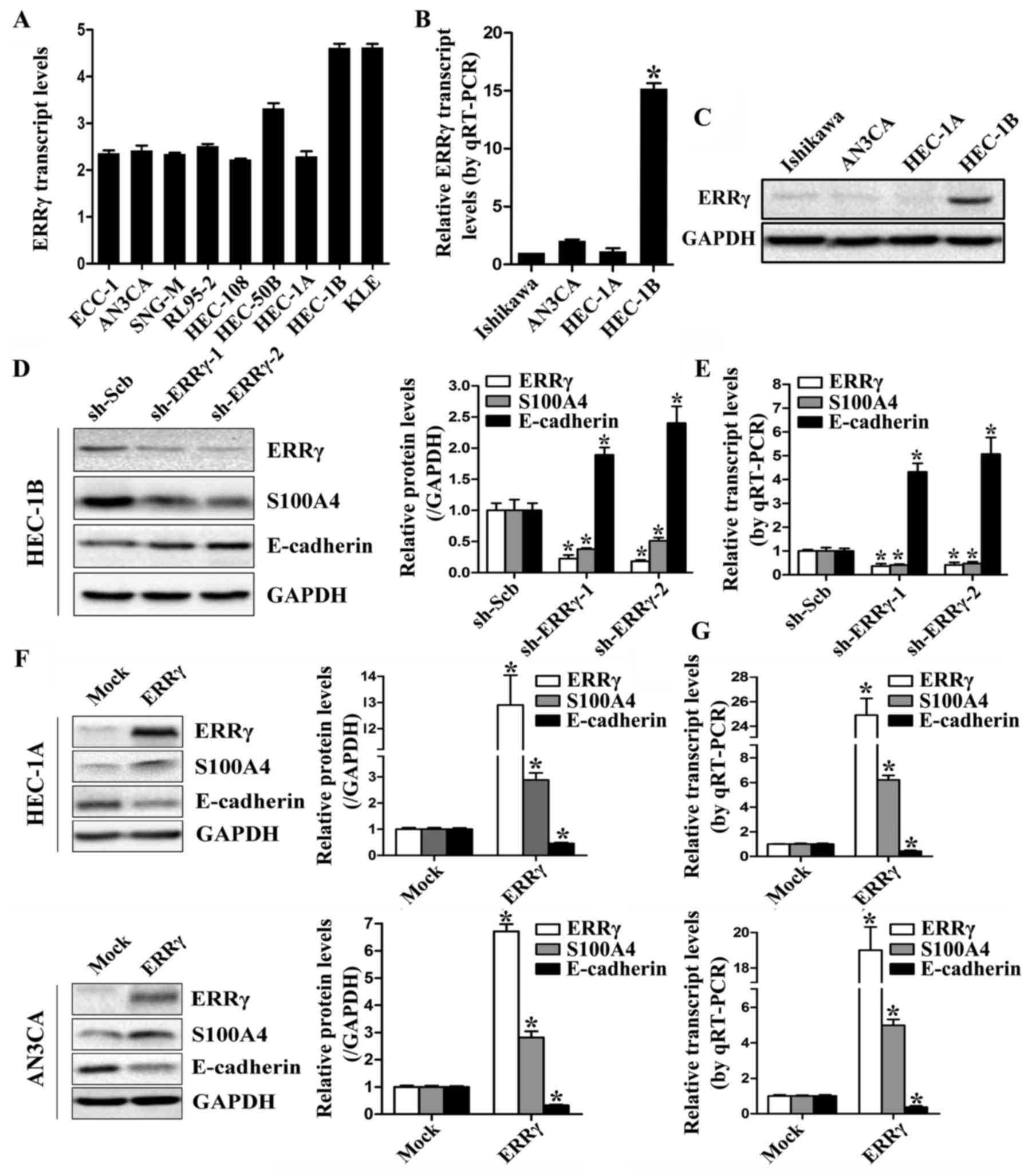
ERRγ transcriptionally regulates the
expression of S100A4 in cultured EC cells
The transcription levels of ERRγ are low in most EC
cell lines, as indicated by the Cancer Cell Line Encyclopedia
(CCLE) program (http://www.broadinstitute.org/ccle; Fig. 2A), and this finding was validated by
western blotting and real-time quantitative RT-PCR in 4
representative EC cell lines (Fig. 2B
and C). Expression of ERRγ was relatively high in HEC-1B cells
but almost undetectable in Ishikawa, HEC-1A and AN3CA EC cells. To
explore the hypothesis that ERRγ may modulate the expression of
S100A4 in EC, HEC-1B cells were stably transfected with sh-ERRγ,
leading to decreased protein and transcription levels of ERRγ and
S100A4 compared to those in cells transfected with sh-Scb (Fig. 2D and E). Additionally, expression of
the S100A4 downstream gene E-cadherin was significantly upregulated
in ERRγ-silenced EC cells. As restoration of S100A4 partially
abolished the impact of ERRγ on E-cadherin expression (data not
shown), we believed that ERRγ regulated E-cadherin expression
through S100A4 in EC cells. Inversely, stable transfection of
HEC-1A and AN3CA EC cells with ERRγ notably upregulated and
downregulated the expression of S100A4 and E-cadherin,
respectively, compared with cells transfected with the empty vector
(Fig. 2F and G). These results
demonstrated that ERRγ could modulate the expression of S100A4 in
EC cells.
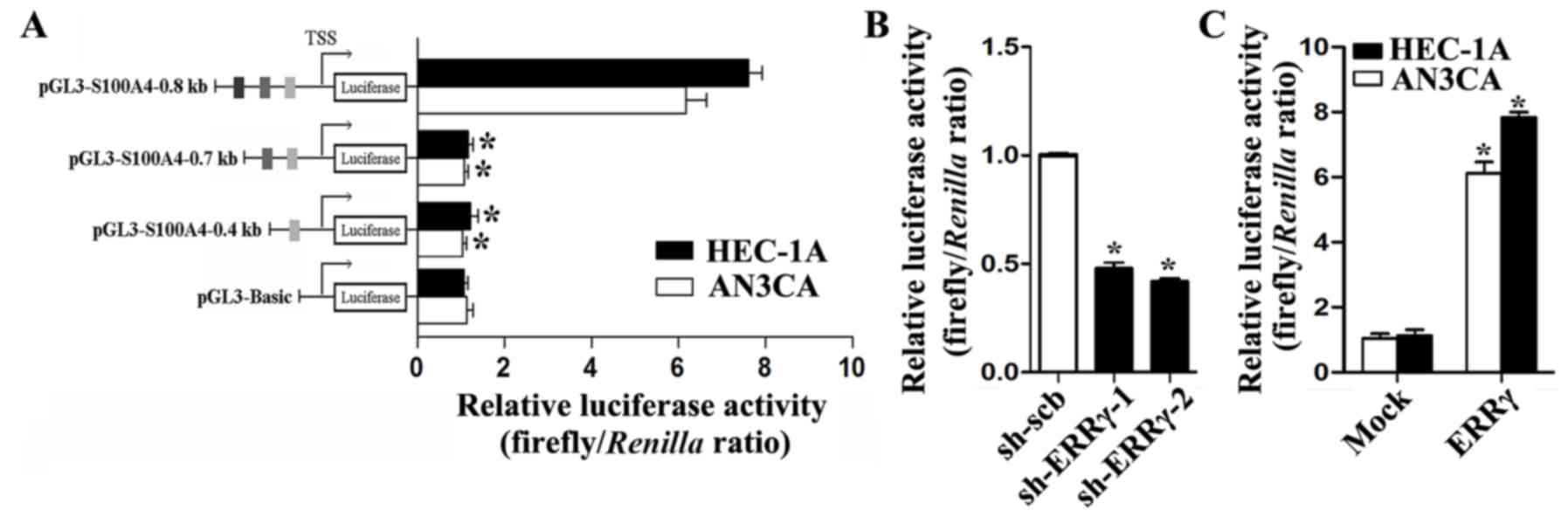
To investigate whether ERRγ could transcriptionally
increase S100A4 expression, computational assessment from the
JASPAR CORE Database (http://jaspar.genereg.net) revealed three potential
binding sites of ERRγ within the S100A4 promoter, located 368–377,
639–648 and 731–740 bp upstream of the transcription start site
(TSS). An S100A4 promoter luciferase reporter and its truncation
vectors were constructed and used to transfect EC cells. A
dual-luciferase assay revealed that the region −656/-784 bp
relative to the TSS was essential for S100A4 promoter activities,
and deletion of this region resulted in remarkably decreased S100A4
promoter activities in cultured HEC-1A and AN3CA cells (Fig. 3A). Moreover, knockdown of ERRγ in
cultured HEC-1B cells attenuated the promoter activities of S100A4,
and ectopic expression of ERRγ enhanced the S100A4 promoter
activities in HEC-1A and AN3CA cells (Fig. 3B and C). These results indicated
that ERRγ could trigger S100A4 transcription through promoter
activation.
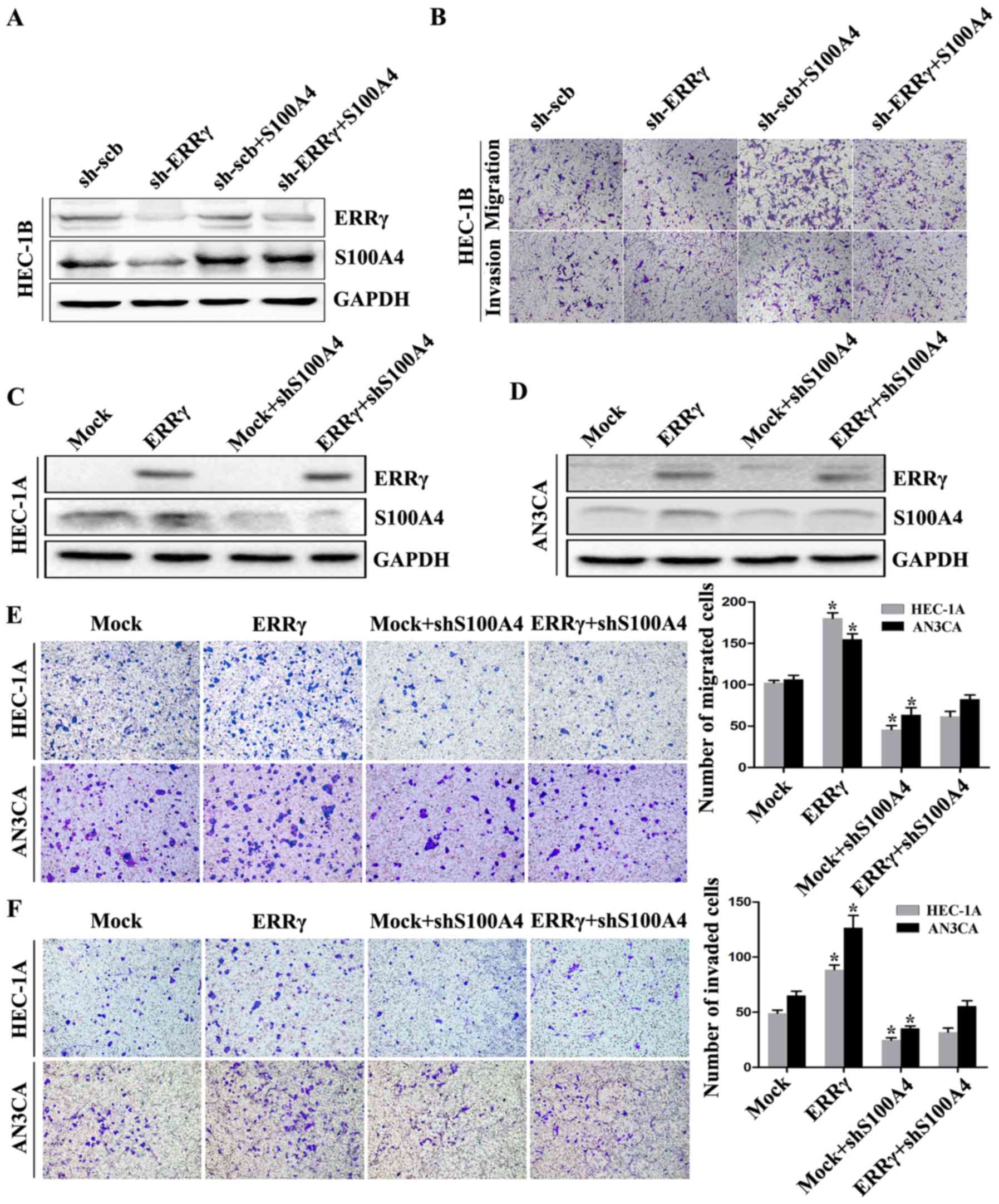
ERRγ modulates the migration and
invasion capability of EC cells through S100A4 in vitro
We first explored the effects of ERRγ knockdown and
S100A4 restoration on the migration and invasion capacity of EC
cells. ERRγ knockdown decreased the expression of S100A4, and
ectopic expression of S100A4 restored the ERRγ knockdown-induced
S100A4 downregulation in HEC-1B cells (Fig. 4A). In Transwell migration assays,
ERRγ knockdown inhibited the migration capability of HEC-1B cells
compared to that of cells transfected with sh-Scb. Matrigel
invasion assays showed that HEC-1B cells stably transfected with
sh-ERRγ presented an impaired invasion capacity compared to sh-Scb
group cells. In addition, restoration of S100A4 expression rescued
the EC cells from the defects in migration and invasion
capabilities induced by ERRγ downregulation (Fig. 4B). These results revealed that
S100A4 was involved in ERRγ knockdown-induced EC cell migration and
invasion inhibition.
The impacts of ERRγ overexpression and S100A4
restoration on cultured EC cells were further studied. Transfection
of HEC-1A and AN3CA cells with sh-S100A4 resulted in reduced S100A4
protein levels and restored the upregulation of S100A4 induced by
ERRγ (Fig. 4C and D). In Transwell
migration assays, ectopic ERRγ expression increased the migration
capability of HEC-1A and AN3CA cells compared with cells
transfected with empty vector (mock) (Fig. 4E). Matrigel invasion assays revealed
that EC cells stably transfected with ERRγ exhibited an enhanced
invasion capacity compared with mock group cells (Fig. 4F). Moreover, restoration of S100A4
expression prevented the enhanced migration and invasion capacity
in EC cells induced by stable overexpression of ERRγ (Fig. 4E and F). These findings suggest that
S100A4 could, at least in part, mediate ERRγ-induced promotion of
EC cell aggressiveness.
ERRγ promotes the growth of EC cells
in vivo
The efficacy of ERRγ overexpression on tumor growth
in vivo was further investigated. HEC-1A cells with fixed
ERRγ expression were subcutaneously injected into nude mice,
leading to an increased proliferative index and tumor weight
compared with tumors formed from cells transfected with the empty
vector (Fig. 5A and B).
Immunohistochemical analysis also showed that the expression of
ERRγ and its downstream gene S100A4 were increased by stable
transfection with ERRγ. Notably, the cell proliferation marker
Ki-67 was also upregulated in HEC-1A cells (Fig. 5C). These results revealed an
oncogenic role of ERRγ in EC and were consistent with the in
vitro studies.
ERRγ and S100A4 are upregulated in
estrogen-treated HEC-1A cells
Since estrogen is a major factor in EC pathogenesis
and progression, we hypothesized that estrogen may affect the
expression of ERRγ and S100A4 in EC cells. We first manipulated
HEC-1A cells with increasing estrogen concentrations (0, 10, 20, 50
and 100 nM) for 24 h. In real-time quantitative RT-PCR assays, ERRγ
mRNA was significantly increased when estrogen concentration
reached 50 nM, and no differences were detected between the 50- and
100-nM groups (Fig. 6A). We next
treated HEC-1A cells with 50 nM of estrogen for increasing
durations (0, 10, 20, 30, 60, 120 and 240 min). Western blot
analysis revealed that there was a time-dependent diversification
in the protein levels of both ERRγ and S100A4, and both reached a
peak at 30 min (Fig. 6B).
Furthermore, S100A4 levels increased almost coincidently with ERRγ,
and a positive correlation was detected (correlation coefficient
R=0.886, P=0.0079). Mining the public GEO database (GSE11869)
revealed that a positive correlation between ERRγ and S100A4
expression has also been found elsewhere in several types of EC
cells after estrogen stimulation (correlation coefficient R=0.448,
P<0.0001). The above findings suggest that estrogen may be an
upstream regulator of ERRγ and S100A4 expression in EC.
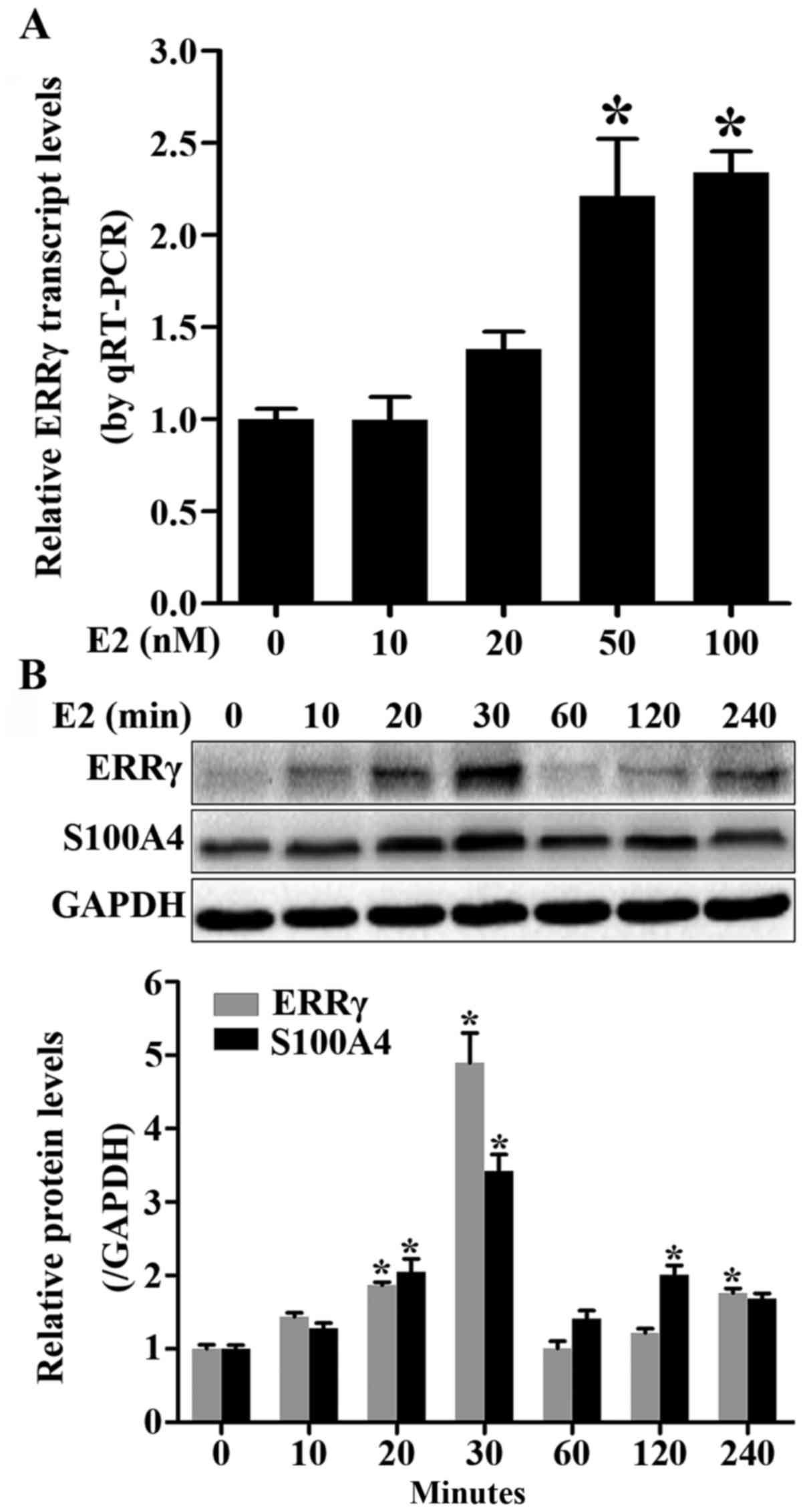 | Figure 6.Upregulation of ERRγ and S100A4 in
estrogen-treated HEC-1A cells. (A) Transcription levels of ERRγ in
HEC-1A cells treated with 0, 10, 20, 50 and 100 nM of estrogen for
24 h. (B) Time course of ERRγ and S100A4 protein and mRNA levels in
HEC-1A cells treated with 50 nM of estrogen for 0, 10, 20, 30, 60,
120 and 240 min. *P<0.01. S100A4, S100 calcium binding
protein A4; ERRγ, estrogen-related receptor γ. |
Discussion
It is generally accepted that S100A4 has profound
impacts on numerous types of cancers, including EC, and S100A4
upregulation results in tumor progression and aggressiveness.
Additionally, overexpression of S100A4 is a predictive indicator of
metastasis and poor survival of cancer patients (17–20).
Our previous studies demonstrated that S100A4 promoted endometrial
cancer (EC) cell aggressiveness via EMT-related modifications
(9). However, the regulatory
mechanisms essential for S100A4 expression in EC remain largely
unknown. Studies have suggested that tyrosine-protein kinase erbB 2
(ERBB2) receptor signaling and integrin signaling regulate S100A4
expression in human medulloblastoma and breast cancer cells
(21,22). More importantly, S100A4 gene
expression can be regulated at the transcriptional level, because
its promoter contains several putative regulatory elements for
transcription factors. In colorectal cancer (CRC), functionally
active β-catenin is indispensable for induction of S100A4
expression and results in enhanced S100A4-induced migration and
invasion (23). An electrophoretic
mobility shift assay (EMSA) and chromatin immunoprecipitation
(ChIP) assay further confirmed binding of β-catenin to the S100A4
promoter (7). In the present study,
synthetic approaches were employed to analyze transcription
profiling of several cancer specimens and transcription factor
binding reported in public databases and identified ERRγ as a
crucial modulator facilitating S100A4 expression in EC. Notably,
ERRγ is highly expressed and positively correlated with S100A4
levels in several types of cancers, including EC specimens.
Early works regarding ERRγ and malignancy mainly
focused on the potential crosstalk of ERRγ with the classical
estrogen pathway and excavating its master regulation role in
energy metabolism (24–26). ERRγ may exert oncogenic or tumor
suppressive functions with tumor specificity. High ERRγ expression
is correlated with more favorable clinical outcomes in ovarian,
breast, and prostate cancer, indicating its tumor-suppressing
function in these cancers (13,27,28).
Conversely, ERRγ-positive staining in hepatocellular carcinoma
(HCC) specimens was remarkably higher than that in adjacent
non-tumor liver tissues and was associated with advanced clinical
stage and pathological grade, and knocking down ERRγ inhibited HCC
cell proliferation and induced G1-phase arrest (29). In human EC, ERRγ is expressed in
~31.3% of EC tissues, and its immunoreactivity was correlated with
worse progression-free survival and overall survival.
Interestingly, the opposite EC cell responsiveness was observed
under forced ERRγ expression or estrogen stimulation with ERα
status dependence (15). In
addition, the transcription levels of ERRγ in EC were increased
with clinical staging, myometrial invasion, and metastatic lymph
nodes. Inhibition of ERRγ activity attenuated estrogen-induced
proliferation of EC cells through AKT and ERK1/2 signaling
abolition (16). However, the exact
biological functions of ERRγ in EC have never been explored. By
means of gain- and loss-of-function studies, we demonstrated that
ERRγ facilitated migration and invasion of EC cells in vitro
and promoted tumor growth in vivo, suggesting an oncogenic
role of ERRγ during EC progression.
Strict binding site specificity experiments
indicated that the 3 members of ERRs preferentially recognize
almost identical DNA elements, distinct from the traditional
estrogen receptor element (ERE), referred to as the
estrogen-related receptor response element (ERRE; TnAAGGTCA)
(10). Subsequent studies
identified widespread distribution of ERRγ targets, and
transcriptionally active ERRγ forms a heterodimer or homodimer that
binds to the promoter of target genes, while ligand is unnecessary
for ERRγ activity (30,31). Studies have confirmed that ERRγ
regulates target genes mainly involved in cellular metabolism,
including tricarboxylic acid (TCA) cycle genes, fatty acid
β-oxidation (FAO) genes, and electron transport chain (ETC) genes
(32). Ectopic expression of ERRγ
enhanced oxidative phosphorylation in breast cancer cells, and the
shift to oxidative metabolism attenuated breast cancer cell
proliferation and tumor growth in vitro and in vivo
(33). In addition, ERRγ can also
directly bind to genes involved in cell growth, such as p21 and
p27. Consistent with the favorable role of ERRγ in breast cancer,
ERRγ reprograms the genetic profiles of breast cancer cells in a
manner characteristic of mesenchymal-to-epithelial transition, in
which E-cadherin was activated by ERRγ directly (34). The target genes of ERRγ involved in
initiation and aggressiveness of EC still warrant investigation. In
the present study, we showed that ERRγ facilitated transcription of
S100A4 in EC cells via S100A4 promotor activation. Furthermore,
since restoration of S100A4 expression rescued EC cells from
ERRγ-induced phenotype changes in aggressiveness, ERRγ may exert
its oncogenic functions by activating S100A4 transcription in
EC.
Estrogen is not a natural ligand for ERRγ, as
indicated by ligand binding studies and transfection experiments
with reporter genes. However, ERRγ stimulates ERE-mediated
transcription and functions as an estrogen responsive gene in
breast cancer cells. Estrogen exposure resulted in ERRγ
overexpression or translocation from the cytoplasm to the nucleus
in breast cancer and EC, and ERRγ further mediates the cell
proliferation promotion effects induced by estrogen (16,35).
Apart from the crucial role of estrogen in cell growth, emerging
evidence has indicated the involvement of estrogen in cell
aggressiveness in certain types of cancers, such as breast, ovarian
and EC, partially through cell stemness, motility and EMT promotion
(36,37). In the present study, we found that
ERRγ expression is stimulated dose- and time-dependently by
estrogen in HEC-1A EC cells. In addition, ERRγ expression is
unexpectedly correlated with S100A4 after different times of
estrogen exposure. With consistent data from public datasets, we
suspect that ERRγ may mediate estrogen signaling in EC progression
by modulating S100A4 expression, which warrants further
investigation.
In conclusion, for the first time, this study
demonstrates that ERRγ is upregulated and positively related to
S100A4 expression in EC. Additionally, ERRγ facilitates S100A4
transcription through promoter activation and promotes the
migration and invasion capability of EC cell lines. Furthermore,
the expression of both ERRγ and S100A4 could be regulated by
estrogen stimulation. These findings extend our current knowledge
of the mechanism of S100A4 regulation by transcription factors and
suggest that ERRγ could be a potential novel therapeutic target in
human EC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81601260).
Availability of data and materials
The datasets used during the present study are
available from the authors upon reasonable request.
Authors' contributions
HBW, YCZ and TH conceived and designed the study.
TH, XXW, SQC and YL performed the experiments. DLF and YCZ were
involved in data analysis. TH and XXW wrote the paper. HBW, YCZ and
DLF reviewed and edited the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki, and approval to conduct the present
study was obtained from the Ethics Committee of Tongji Medical
College, Huazhong University of Science and Technology (IORG no:
IORG0003571). Tissues were collected after receiving informed
consent from the patients. All animal experiments were approved by
the Animal Care Committee of Tongji Medical College.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Makker A and Goel MM: Tumor progression,
metastasis, and modulators of epithelial-mesenchymal transition in
endometrioid endometrial carcinoma: An update. Endocr Relat Cancer.
23:R85–R111. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Donato R, Cannon BR, Sorci G, Riuzzi F,
Hsu K, Weber DJ and Geczy CL: Functions of S100 proteins. Curr Mol
Med. 13:24–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart RL and O'Connor KL: Clinical
significance of the integrin α6β4 in human malignancies. Lab
Invest. 95:976–986. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kiss B, Kalmar L, Nyitray L and Pal G:
Structural determinants governing S100A4-induced isoform-selective
disassembly of nonmuscle myosin II filaments. FEBS J.
283:2164–2180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dahlmann M, Kobelt D, Walther W, Mudduluru
G and Stein U: S100A4 in cancer metastasis: Wnt signaling-driven
interventions for metastasis restriction. Cancers. 8:pii: E59.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hua T, Liu S, Xin X, Cai L, Shi R, Chi S,
Feng D and Wang H: S100A4 promotes endometrial cancer progress
through epithelial-mesenchymal transition regulation. Oncol Rep.
35:3419–3426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deblois G and Giguere V: Functional and
physiological genomics of estrogen-related receptors (ERRs) in
health and disease. Biochim Biophys Acta. 1812:1032–1040. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ranhotra HS: The orphan estrogen-related
receptor alpha and metabolic regulation: New frontiers. J Recept
Signal Transduct Res. 35:565–568. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Madhavan S, Gusev Y, Singh S and Riggins
RB: ERRgamma target genes are poor prognostic factors in
Tamoxifen-treated breast cancer. J Exp Clin Cancer Res. 34:452015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Misawa A and Inoue S: Estrogen-Related
Receptors in Breast Cancer and Prostate Cancer. Front Endocrinol.
6:832015. View Article : Google Scholar
|
|
14
|
Gao M, Sun P, Wang J, Zhao D and Wei L:
Expression of estrogen receptor-related receptor isoforms and
clinical significance in endometrial adenocarcinoma. Int J Gynecol
Cancer. 16:827–833. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto T, Mori T, Sawada M, Kuroboshi H,
Tatsumi H, Yoshioka T, Matsushima H, Iwasaku K and Kitawaki J:
Estrogen-related receptor-gamma regulates estrogen receptor-alpha
responsiveness in uterine endometrial cancer. Int J Gynecol Cancer.
22:1509–1516. 2012.PubMed/NCBI
|
|
16
|
Sun Y, Wang C, Yang H and Ma X: The effect
of estrogen on the proliferation of endometrial cancer cells is
mediated by ERRgamma through AKT and ERK1/2. Eur J Cancer Prev.
23:418–424. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang S, Zheng J, Huang Y, Song L, Yin Y,
Ou D, He S, Chen X and Ouyang X: Impact of S100A4 expression on
clinicopathological characteristics and prognosis in pancreatic
cancer: A meta-analysis. Dis Markers. 2016:81373782016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling Z and Li R: Clinicopathological and
prognostic value of S100A4 expression in gastric cancer: A
meta-analysis. Int J Biol Markers. 29:e99–e111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stewart RL, Carpenter BL, West DS, Knifley
T, Liu L, Wang C, Weiss HL, Gal TS, Durbin EB, Arnold SM, et al:
S100A4 drives non-small cell lung cancer invasion, associates with
poor prognosis, and is effectively targeted by the FDA-approved
anti-helminthic agent niclosamide. Oncotarget. 7:34630–34642. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q,
Yang W, Zheng X, Tu K and Liu Q: miR-187-3p inhibits the metastasis
and epithelial-mesenchymal transition of hepatocellular carcinoma
by targeting S100A4. Cancer Lett. 381:380–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hernan R, Fasheh R, Calabrese C, Frank AJ,
Maclean KH, Allard D, Barraclough R and Gilbertson RJ: ERBB2
up-regulates S100A4 and several other prometastatic genes in
medulloblastoma. Cancer Res. 63:140–148. 2003.PubMed/NCBI
|
|
22
|
Kim TH, Kim HI, Soung YH, Shaw LA and
Chung J: Integrin (alpha6beta4) signals through Src to increase
expression of S100A4, a metastasis-promoting factor: Implications
for cancer cell invasion. Mol Cancer Res. 7:1605–1612. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang
F, Shao Z, Ding Y and Zhao L: LIM and SH3 protein 1 induces
TGFbeta-mediated epithelial-mesenchymal transition in human
colorectal cancer by regulating S100A4 expression. Clin Cancer Res.
20:5835–5847. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huss JM, Garbacz WG and Xie W:
Constitutive activities of estrogen-related receptors:
Transcriptional regulation of metabolism by the ERR pathways in
health and disease. Biochim Biophys Acta. 1852:1912–1927. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deblois G, St-Pierre J and Giguere V: The
PGC-1/ERR signaling axis in cancer. Oncogene. 32:3483–3490. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Giguere V: To ERR in the estrogen pathway.
Trends Endocrinol Metab. 13:220–225. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun P, Sehouli J, Denkert C, Mustea A,
Könsgen D, Koch I, Wei L and Lichtenegger W: Expression of estrogen
receptor-related receptors, a subfamily of orphan nuclear
receptors, as new tumor biomarkers in ovarian cancer cells. J Mol
Med. 83:457–467. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Audet-Walsh E, Yee T, McGuirk S, Vernier
M, Ouellet C, St-Pierre J and Giguère V: Androgen-dependent
repression of errgamma reprograms metabolism in prostate cancer.
Cancer Res. 77:378–389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim JH, Choi YK, Byun JK, Kim MK, Kang YN,
Kim SH, Lee S, Jang BK and Park KG: Estrogen-related receptor gamma
is upregulated in liver cancer and its inhibition suppresses liver
cancer cell proliferation via induction of p21 and p27. Exp Mol
Med. 48:e2132016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huppunen J and Aarnisalo P: Dimerization
modulates the activity of the orphan nuclear receptor ERRgamma.
Biochem Biophys Res Commun. 314:964–970. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Greschik H, Wurtz JM, Sanglier S, Bourguet
W, van Dorsselaer A, Moras D and Renaud JP: Structural and
functional evidence for ligand-independent transcriptional
activation by the estrogen-related receptor 3. Mol Cell. 9:303–313.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Audet-Walsh E and Giguere V: The multiple
universes of estrogen-related receptor α and γ in metabolic control
and related diseases. Acta Pharmacol Sin. 36:51–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eichner LJ, Perry MC, Dufour CR, Bertos N,
Park M, St-Pierre J and Giguère V: miR-378* mediates metabolic
shift in breast cancer cells via the PGC-1β/ERRγ transcriptional
pathway. Cell Metab. 12:352–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tiraby C, Hazen BC, Gantner ML and Kralli
A: Estrogen-related receptor gamma promotes
mesenchymal-to-epithelial transition and suppresses breast tumor
growth. Cancer Res. 71:2518–2528. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ijichi N, Shigekawa T, Ikeda K,
Horie-Inoue K, Fujimura T, Tsuda H, Osaki A, Saeki T and Inoue S:
Estrogen-related receptor γ modulates cell proliferation and
estrogen signaling in breast cancer. J Steroid Biochem Mol Biol.
123:1–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jeon SY, Hwang KA and Choi KC: Effect of
steroid hormones, estrogen and progesterone, on epithelial
mesenchymal transition in ovarian cancer development. J Steroid
Biochem Mol Biol. 158:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun Y, Wang Y, Fan C, Gao P, Wang X, Wei G
and Wei J: Estrogen promotes stemness and invasiveness of
ER-positive breast cancer cells through Gli1 activation. Mol
Cancer. 13:1372014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Day RS, McDade KK, Chandran UR, Lisovich
A, Conrads TP, Hood BL, Kolli VS, Kirchner D, Litzi T and Maxwell
GL: Identifier mapping performance for integrating transcriptomics
and proteomics experimental results. BMC Bioinformatics.
12:2132011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pappa KI, Polyzos A, Jacob-Hirsch J,
Amariglio N, Vlachos GD, Loutradis D and Anagnou NP: Profiling of
discrete gynecological cancers reveals novel transcriptional
modules and common features shared by other cancer types and
embryonic stem cells. PLoS One. 10:e01422292015. View Article : Google Scholar : PubMed/NCBI
|