Introduction
Worldwide, renal cell carcinoma (RCC) accounts for
~90% of all kidney neoplasms and 2–3% of adult malignant tumors,
and it is the ninth most common cancer (1–3). RCC
leads to a considerable mortality rate, which is ~100,000 patients
a year, and the morbidity and mortality rates are increasing at a
rate of 2–3% per decade (4). There
were ~63,920 newly diagnosed cases of RCC in the US in 2014
(5). The treatment of metastatic
RCC has improved in the past few decades. Although cytoreductive
nephrectomy is effective for the treatment of early and local RCC,
~30% of patients suffer from metastatic disease after surgery. In
addition, other therapeutic methods, such as immunotherapy have
been used for the treatment of RCC; however, the effectiveness is
limited (6). Therefore, the
identification of potential biomarkers and therapeutic targets for
RCC is urgently needed.
With the advancement of techniques used in
genome-wide platforms, long non-coding RNAs (lncRNAs) have been
identified and defined as RNA molecules of greater than 200
nucleotides in length and without apparent protein-coding potential
(7,8). Thousands of lncRNAs in both humans and
mice have been identified by the combined analysis of Chip-seq and
RNA-seq data; however, only a few have been assigned any function
(9–11). Accumulating evidence suggests that
lncRNAs play a vital role in various biological responses, such as
the regulation of epigenetic silencing by chromatin remodeling,
regulation of splicing, recruitment of transcription factors, and
regulation of mRNA stability. lncRNAs can function by regulating
the alternative splicing of pre-mRNAs and as competing endogenous
RNAs (ceRNAs) that competitively bind to microRNAs (miRNAs) to
suppress the inactivation of the target genes of miRNAs (12–14).
Considering the potential significance of lncRNAs in physiological
and pathological responses, many efforts have been made to
elucidate the function of lncRNAs in carcinogenesis and cancer
metastasis (15).
miRNAs are another class of non-coding RNAs with a
length of ~22 nucleotides that have been extensively studied
(16). miRNAs regulate the
expression of target genes by partial complementary binding to
miRNA response elements (MREs), which contain the corresponding
mRNA sequences (17). One miRNA is
able to regulate several mRNA transcripts, and one transcript may
be the target of several different miRNAs. The miRNA regulatory
network plays an important role in many biological responses,
including carcinogenesis and tumor metastasis (18,19).
ceRNAs are transcripts that cross-regulate each other by competing
for shared miRNAs (20,21). mRNAs, lncRNAs and other RNAs can
function as natural miRNA sponges by using shared MREs to suppress
miRNA function (20). Certain
lncRNAs have been identified and experimentally validated (22). H19 is an lncRNA that modulates the
let-7 miRNA family members and miR-106a availability by acting as a
molecular sponge (23,24). The muscle-specific lncRNA linc-MD1
can interact with miR-133 to regulate the expression of
muscle-specific gene expression (25). Such a RNA interaction could regulate
many physiological and pathological responses and thus may provide
new ideas for cancer therapy. Thus far, the lncRNA-miRNA-mRNA ceRNA
network in hepatocellular, breast and gastric cancer, has been
constructed and analyzed (26–29).
However, similar studies involving RCC are rare. Moreover,
genome-wide comprehensive analysis of RCC-associated miRNAs and
lncRNAs has been limited.
In the present study, we analyzed the mRNA, lncRNA,
and miRNA expression profiles from RCC-associated databases
(GSE66270, GSE40914 and GSE71302, respectively). We identified a
total of 2,378 aberrantly expressed RNAs; 2 upregulated and 3
downregulated RNAs were categorized as lncRNAs. Furthermore, we
identified differentially expressed miRNAs (DEmiRNAs) in patients
with RCC to elucidate the potential crosstalk between lncRNA, miRNA
and mRNA. Ultimately, a lncRNA-miRNA-mRNA ceRNA network was
constructed from 4 dysregulated lncRNAs, 17 mRNAs and 2 miRNAs by
bioinformatic analysis.
Materials and methods
Study materials
Three RNA datasets (mRNA, lncRNA and miRNA) were
selected in this study. In these datasets, GSE66270, GSE40914 and
GSE71302 were analyzed as they were associated with renal cancer
(30–32). GSE66270 (mRNA) of the Affymetrix
Human Genome U133 Plus 2.0 Array (GPL570) platform consisted of 28
samples including 14 renal cancer and 14 normal samples. GSE40914
(lncRNA) was composed of three sub-datasets, GSE40911, GSE40912 and
GSE40913. The GSE40911 dataset was selected for the extraction of
differentially expressed lncRNAs (DElncRNAs). GSE40911 was based on
the IQUSP_Human_intronic_4k_v2.0 (GPL3985) platform, which
consisted of 44 samples including 22 renal cancer and 22 normal
samples. GSE71302 (miRNA) was based on the Agilent-021827 Human
miRNA Microarray (V3) (miRBase release 12.0 miRNA ID version)
(GPL10850) platform, which consisted of 10 samples including 5
renal cancer and 5 normal samples.
Identification of differentially
expressed genes
The analysis and extraction of differentially
expressed genes were conducted with R language software, which
included background correction, normalization, and calculation of
the expression value of the original data and screening of the
differentially expressed genes. The Benjamini-Hochberg (BH) method
was used for statistical corrections. The online tool GEO2R was
used for the extraction of differentially expressed genes as
original data of the expression level for GSE40911 and GSE71302
were not available. In terms of the difference in expression
threshold, genes were considered as significant with
|log2FoldChange| >1 and corrected P<0.05.
After all genes with different expression levels
were collected, which were determined according to the previously
set threshold for GSE40911, the corresponding GPL3985 platform was
downloaded to annotate the data. The empty comment lines were
deleted, and all the information of the non-coding RNAs were
filtered out. Based on the information of the non-coding RNAs and
all the differentially expressed genes, DElncRNAs were
obtained.
Survival analysis
A total of 417 renal cancer samples in the database
were retrieved from the Cancer Genome Atlas data portal. The
survival package of R statistical programming software was used for
the survival analysis of filtered DElncRNAs and DEmiRNAs.
Functional enrichment analysis
When a sufficient number of differentially expressed
mRNAs (DEmRNAs) was obtained, enrichment analysis of the
differentially expressed genes was carried out using the cluster
Profile package of R software. The enrichKEGG and enrichGO
functions were used for KEGG and Gene Ontology (GO) enrichment
analysis, respectively. GO terms and KEGG pathways with
BH-corrected P<0.05 were filtered out to obtain the final
results. As for the pathway that ranked first in KEGG enrichment
analysis and the pathway relevant to renal cancer, the Pathview
package of R software was used for the mapping of related genes and
determination of the location of differentially expressed genes in
the entire pathway.
Construction of the ceRNA network and
interactions among DEmRNAs
lncRNA-miRNA interactions and miRNA-mRNA
interactions were predicted using miRcode (http://www.mircode.org/) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) (33,34).
The miRNA-mRNA pairs were validated by various methods including
qRT-PCR, western blotting, ChIP-seq and microarray. To obtain a
more reliable ceRNA network, high-correlation sets in the
miRTarBase were selected to predict the interactions between miRNAs
and mRNAs. The results of the lncRNA-miRNA-mRNA network were
visualized by Cytoscape 3.3.0 (35).
STRING database was used to screen interactions
among DEmRNAs with the threshold of combined score >0.4. The
renal cancer sub-network was imported into the Cytoscape database
to calculate the connectivity, and the top 10 genes with the
highest degree were selected as hub genes.
Real-time quantitative PCR
Total RNA was isolated from ACHN and HK-2 cells
using an RNeasy® Mini kit (Qiagen, Hilden, Germany)
according to the manufacturer's instructions. First-strand cDNA was
synthesized from 11 µg of total RNA using the Transcript or First
Strand cDNA Synthesis kit (Roche, Mannheim, Germany) as instructed
by the manufacturer. qRT-PCR reactions were performed on an Applied
Biosystems® 7500 real-time PCR system (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) using the following procedure:
95°C for 10 min, followed by 40 cycles of 95°C for 30 sec and 60°C
for 1 min. To create a qRT-PCR standard, glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) was used as an internal control. The
2−ΔΔCt method was used for data analysis.
Results
DEmRNAs and enrichment analysis in
renal cancer
According to the threshold value of absolute
log2 fold change >1 and corrected P<0.05, 2,318
mRNAs including 1,370 upregulated and 948 downregulated genes were
identified as DEmRNAs in the GSE66270 database. The top 100
upregulated and the top 100 downregulated genes were outlined
through the heatmap function in the gplots package of R software
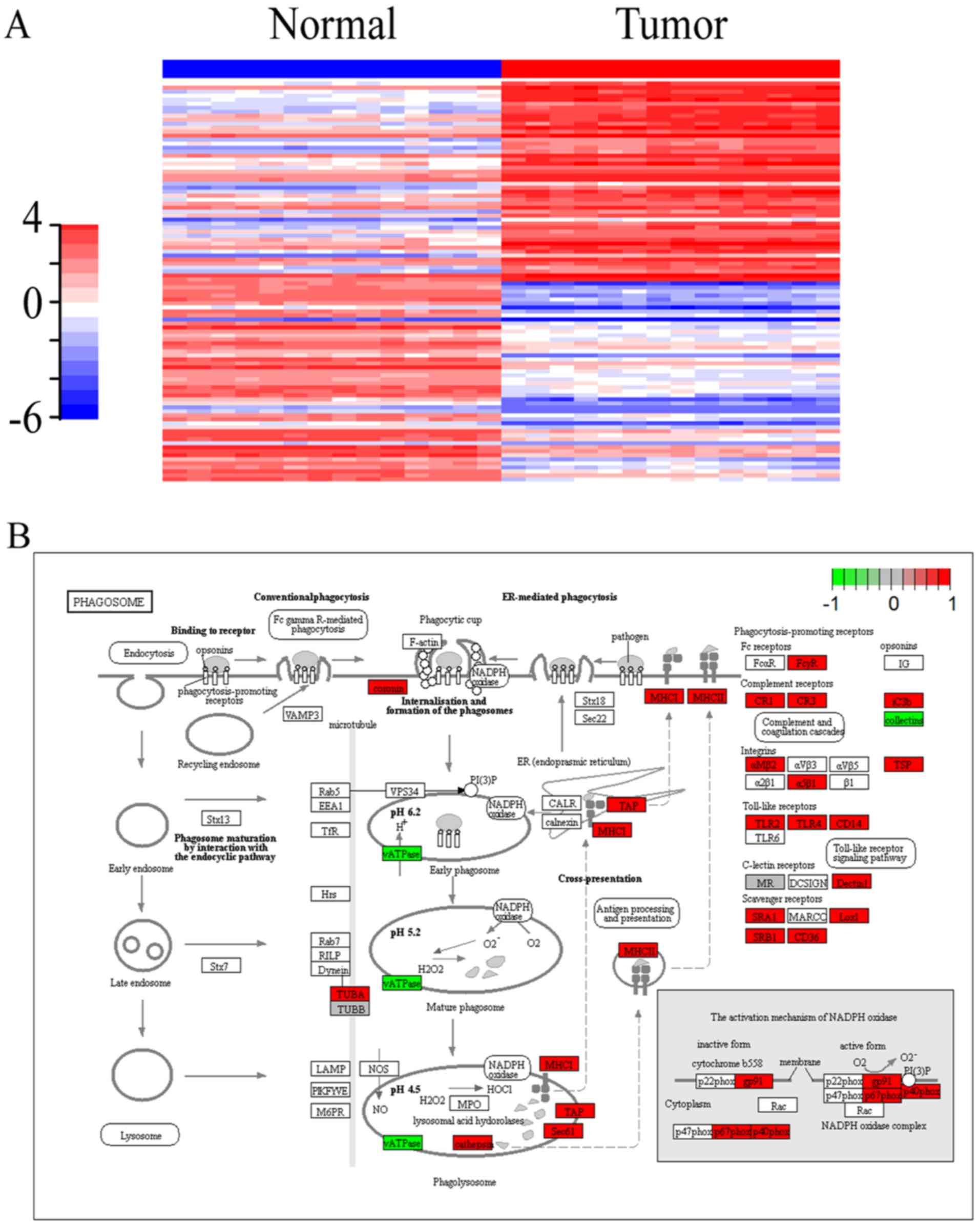
(Fig. 1A). The results demonstrated
that the expression of selected RNAs was significantly different
between renal cancer and normal tissues.
For a better understanding of the mechanisms of
these differentially expressed genes, which were implicated in the
tumorigenesis of renal cancer, functional enrichment analysis was
performed using the enrichKEGG function of R software based on
KOBAS 2.0. The phagosome pathway was identified as the pathway most
significantly related to cancer (P<0.01); thus, visualization of
the phagosome pathway was performed using the Pathview package. As
shown in Fig. 1B, 37 DEmRNAs (33
upregulated and 4 downregulated) were involved in the phagosome
pathway. Furthermore, the mapping of hsa05211 (the RCC pathway),
associated with the tumorigenesis of renal cancer, was also
performed. As shown in Fig. 1C, 15
DEmRNAs (12 upregulated and 3 downregulated) were involved in the
hsa05211 pathway. In addition, Fig.
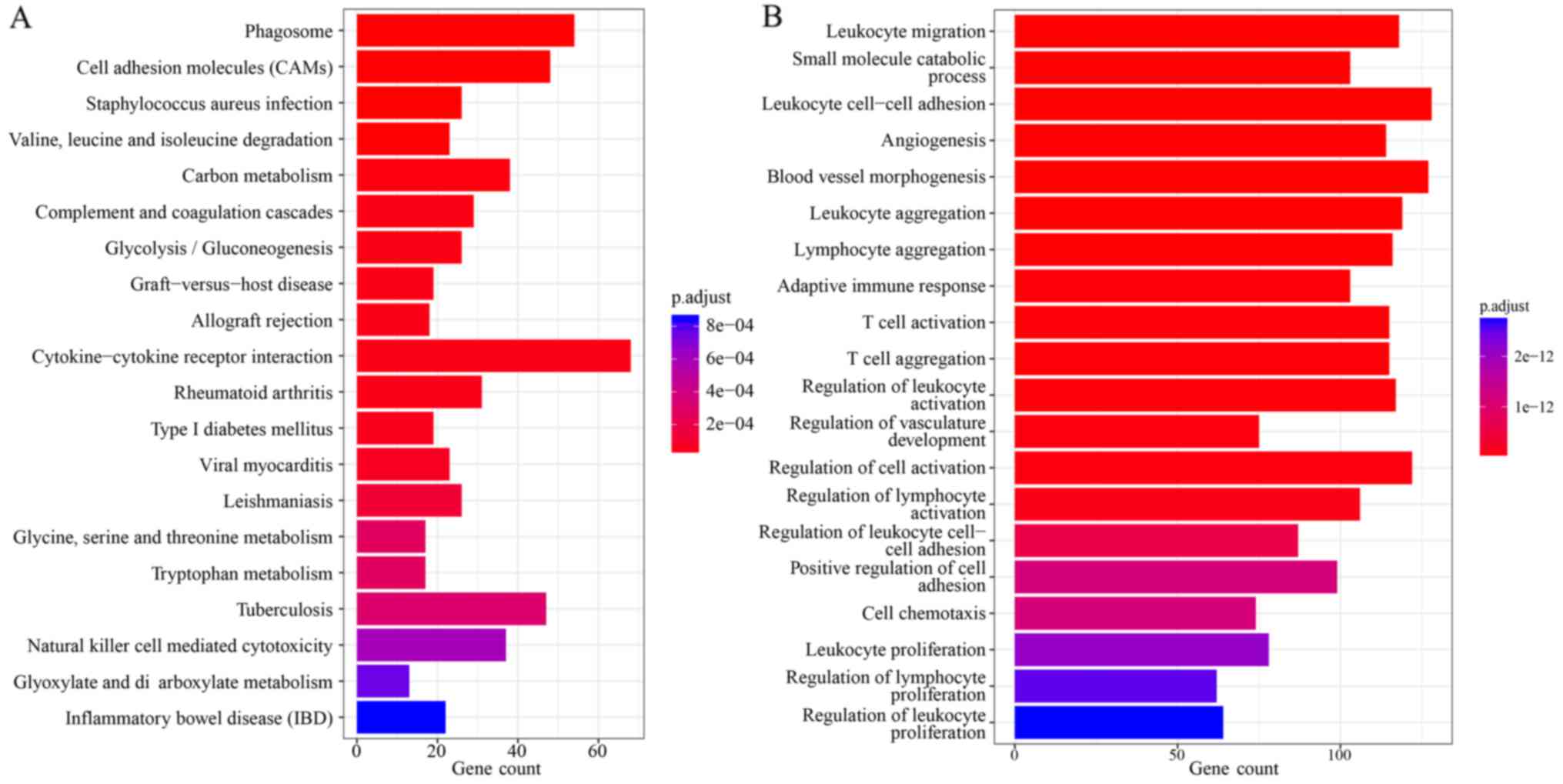
2 shows the top 20 significantly enriched GO terms (Fig. 2A) and KEGG pathways (Fig. 2B) of DEmRNAs.
DElncRNAs and DEmiRNAs in renal
cancer
According to the threshold value of absolute
log2 fold change >1 and corrected P<0.05, 5
lncRNAs, which included 2 upregulated and 3 downregulated lncRNAs
(Table I), were identified as
differentially expressed in the GSE40911 database. In addition, 55
miRNAs, which included 28 upregulated and 27 downregulated miRNAs,
were identified as differentially expressed in the GSE71302
database.
 | Table I.Differentially expressed lncRNAs in
renal cancer. |
Table I.
Differentially expressed lncRNAs in
renal cancer.
| GENE | logFC | adj.P.Val | P-value |
|---|
| ACTN4 | 1.005228 | 6.45E-08 | 1.43E-09 |
| CTHRC1 | −2.63837 | 1.56E-12 | 7.13E-15 |
| IGFBP7 | −2.29773 | 4.46E-10 | 5.25E-12 |
| RAB31 | −1.199 | 2.86E-10 | 3.27E-12 |
| RBPMS | 1.296871 | 1.56E-09 | 1.99E-11 |
The ceRNA network in renal cancer
For a better understanding of the characteristics of
DElncRNAs in renal cancer, a dysregulated lncRNA-miRNA-mRNA ceRNA
network was constructed by extracting the interaction pairs of
lncRNA-miRNA in miRcode and miRNA-mRNA in miRTarBase. In the ceRNA
network, 17 DEmRNAs (14 upregulated and 3 downregulated) and 5
DElncRNAs (2 upregulated and 3 downregulated) were identified.
Moreover, 2 upregulated miRNAs (hsa-miR-570 and hsa-miR-508-3p)
were identified in the network. The ceRNA network was divided into
two parts, and the miRNA was located at the center of each part.
The two parts were connected through 2 lncRNAs, which included the
upregulated RBPMS and the downregulated RAB31. The network is shown
in Fig. 3A.
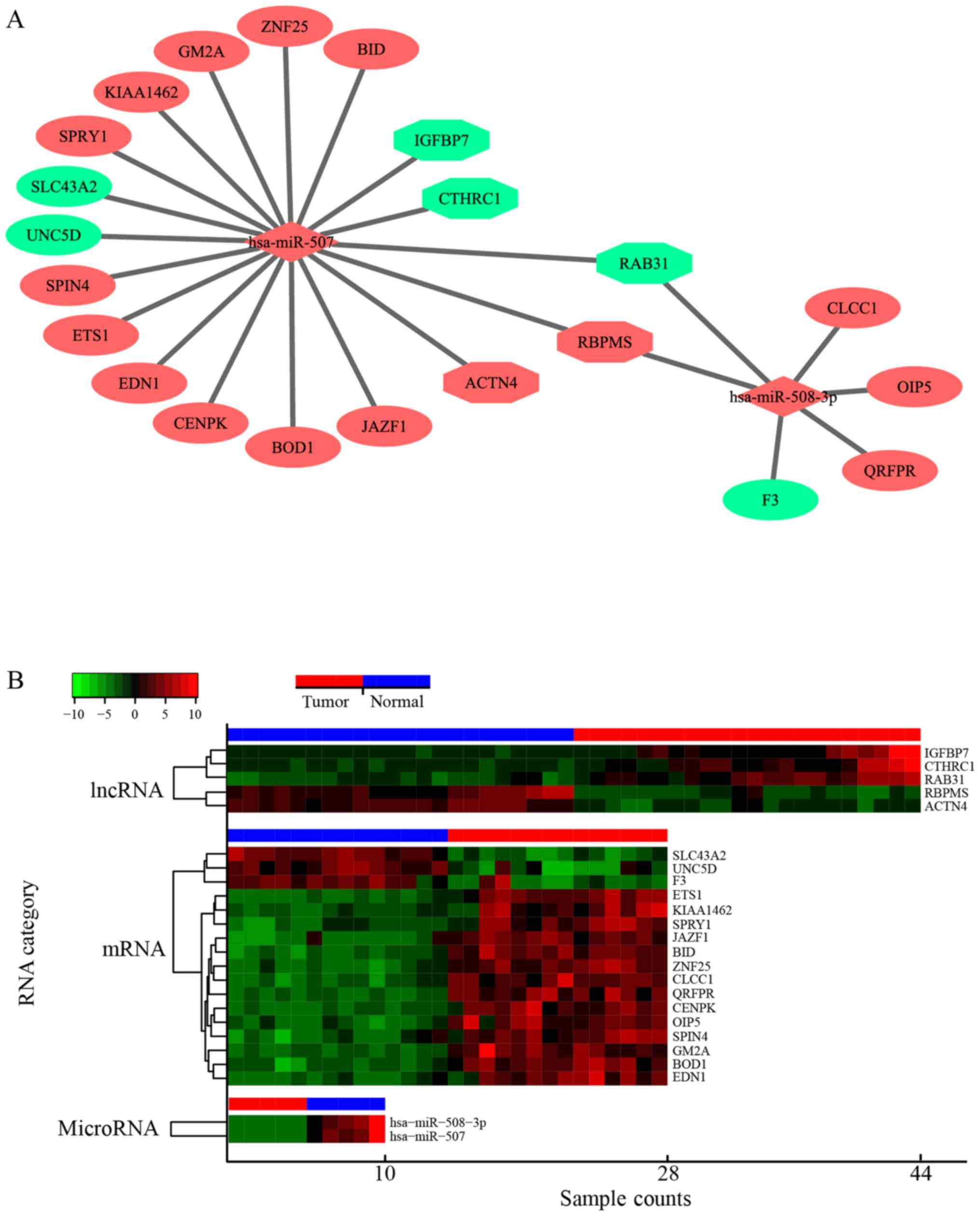 | Figure 3.The ceRNA network mediated by
DElncRNAs in renal cancer. (A) Global view of the lncRNA-miRNA-mRNA
network in renal cancer. Upregulated genes are indicated in red
color, and downregulated genes are indicated in green color.
lncRNAs, miRNAs, and mRNAs are represented as hexagon, rhombus, and
ellipse, respectively. (B) Heatmap of the expression profiles of
lncRNAs, miRNAs and mRNAs in the network. The horizontal axis
represents the sample capacity and the vertical axis represents the
types of RNA. The blue and red bars at the top of the heatmap
represent normal and renal cancer samples, respectively. ceRNA,
competing endogenous RNA; DEmRNAs, differentially expressed
miRNAs. |
Furthermore, the DEmRNAs involved in the ceRNA
network were analyzed by KOBAS 3.0 to uncover signaling pathways
that are regulated by DElncRNAs indirectly. As shown in Table II, 6 KEGG pathways were
significantly enriched (P<0.05), and the top KEGG pathway was
the diabetic complications pathway. Moreover, the other top 2 KEGG
pathways were renal cancer-related pathways.
 | Table II.KEGG pathways of DEmRNAs that were
involved in the ceRNA network. |
Table II.
KEGG pathways of DEmRNAs that were
involved in the ceRNA network.
| ID | Pathway
category | P-value |
|---|
|
hsa04933 | AGE-RAGE signaling
pathway in diabetic complications | 7.78E-04 |
|
hsa05211 | Renal cell
carcinoma | 2.70E-02 |
|
hsa04610 | Complement and
coagulation cascades | 3.17E-02 |
|
hsa04066 | HIF-1 signaling
pathway | 4.10E-02 |
|
hsa04668 | TNF signaling
pathway | 4.37E-02 |
|
hsa04142 | Lysosome | 4.87E-02 |
It is possible that DElncRNAs could indirectly
interact with DEmRNAs in renal cancer through miRNAs, such as
hsa-miR-570 and hsa-miR-508-3p, as demonstrated in the ceRNA
network. To explore the co-expression of each RNA in the network,
the heatmaps of the expression profiles of DElncRNAs, DEmRNAs and
DEmiRNAs were obtained (Fig.
3B).
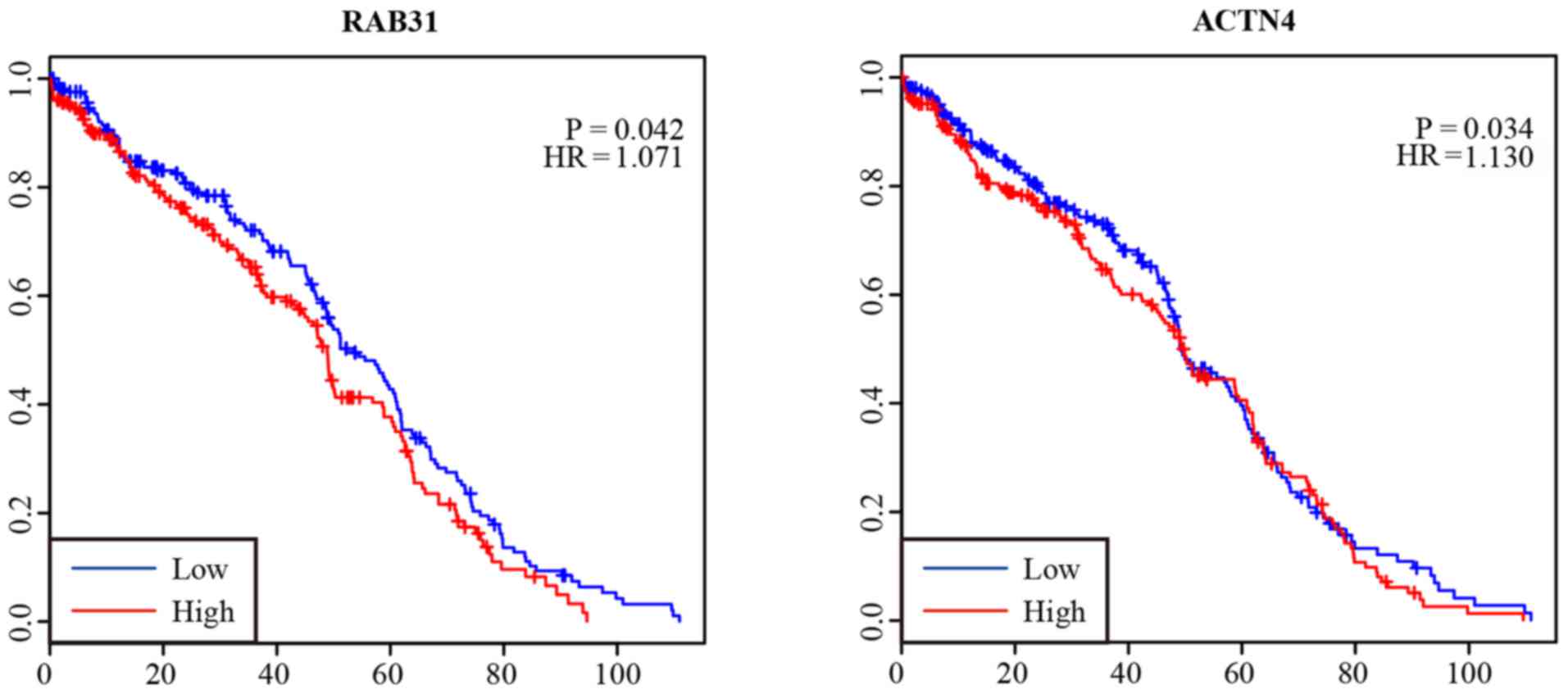
Survival analysis of lncRNAs
The overall survival analysis of the 5 DElncRNAs in
patients with renal cancer was performed by Kaplan-Meier curve
analysis. As shown in Fig. 4, the
expression levels of RAB31 (HR=1.017, 95% CI, 0.971–1.432) and
ACTN4 (HR=1.130, 95% CI, 1.011–1.659) were negatively correlated
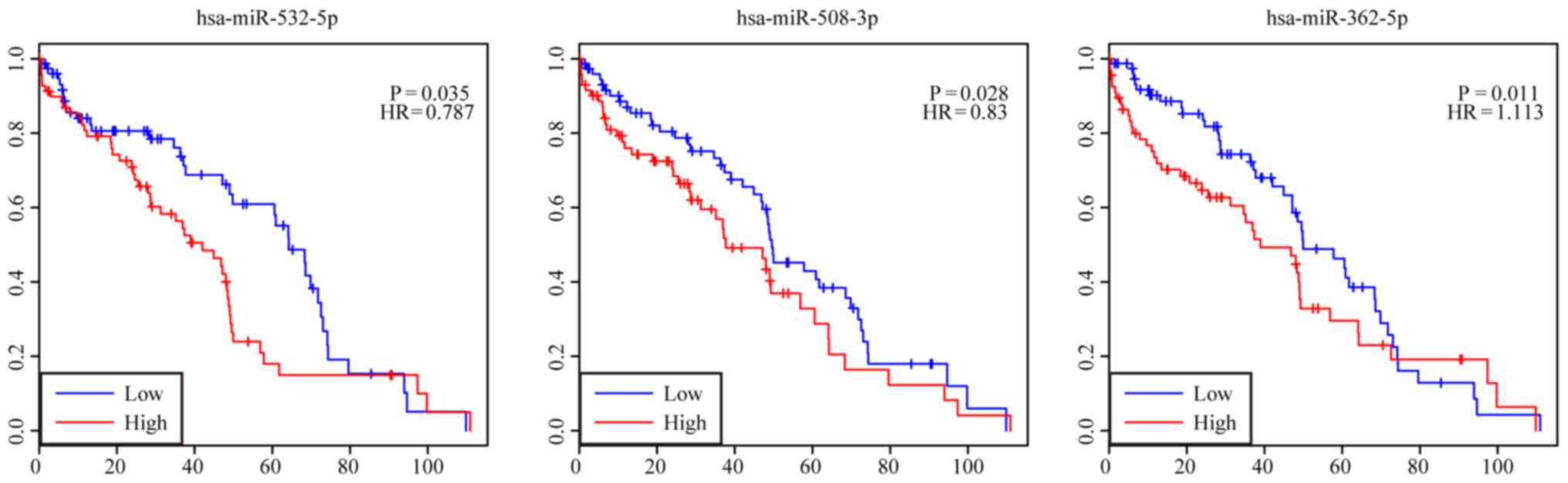
with the overall survival of patients (P<0.05). Survival
analysis of the DEmiRNAs showed that 3 highly expressed DEmiRNAs
were negatively correlated with the patient overall survival, which
were miR-362-5p (HR=1.113, 95% CI, 1.017–1.590), miR-508-3p
(HR=0.830, 95% CI, 0.613–0.932), and miR-532-5p (HR=0.787, 95% CI,
0.548–0.916). The results are presented in Fig. 5.
Screening of key genes
According to the threshold of combined score
>0.4, a total of 3,277 interaction pairs among the DEmRNAs were
obtained. The top 10 genes with the highest degree are shown in
Table III.
 | Table III.The top 10 DEmRNAs with highest
degree. |
Table III.
The top 10 DEmRNAs with highest
degree.
| Gene symbol | Gene name | Degree |
|---|
| FN1 | Fibronectin 1 | 98 |
| FBXO6 | F-box protein
6 | 82 |
| TP53 | Tumor protein
P53 | 72 |
| MYC | MYC
proto-oncogene | 66 |
| CDK1 | Cyclin-dependent
kinase 1 | 62 |
| JUN | Jun
proto-oncogene | 47 |
| LYN | LYN
proto-oncogene | 47 |
| SHC1 | SHC adaptor protein
1 | 46 |
| LCK | LCK
proto-oncogene | 45 |
| ITGA4 | Integrin subunit
α4 | 42 |
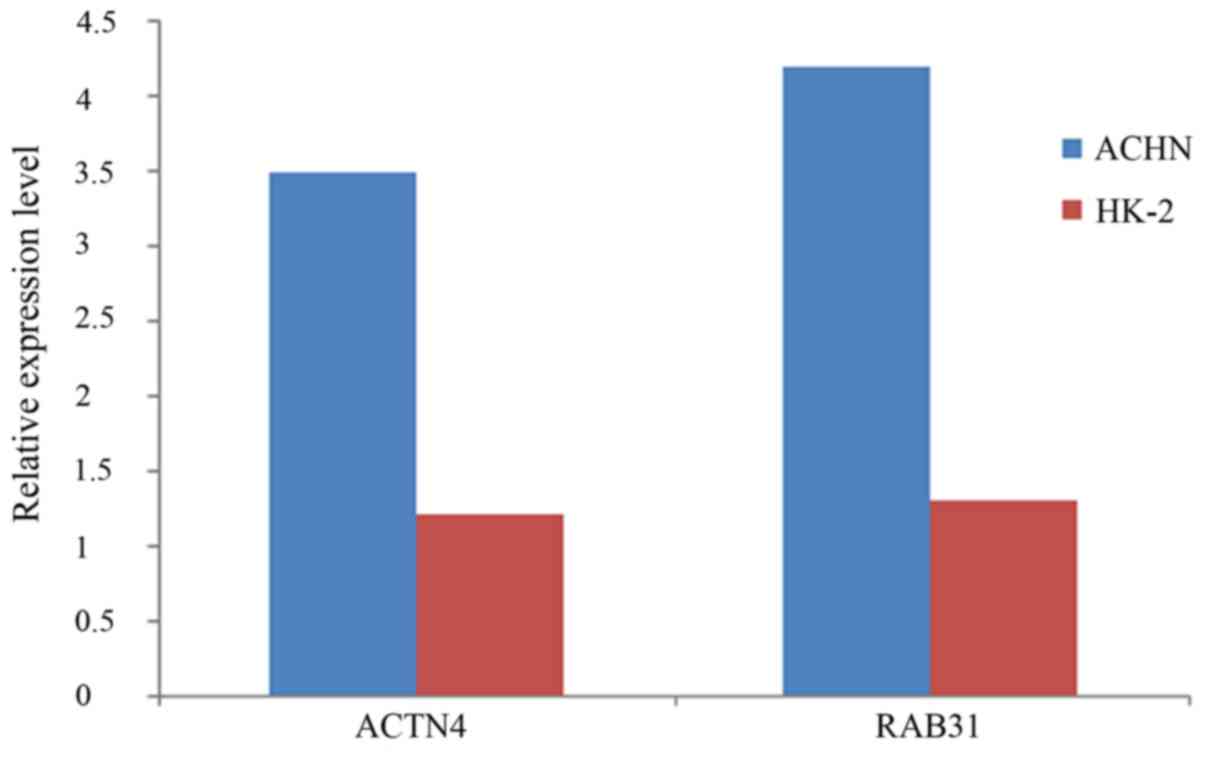
Real-time quantitative PCR
Expression differences of ACTN4 and RAB31 between
RCC cells ACHN and normal renal cells HK-2 were explored thorough
qRT-PCR. Primers used for ACTN4 were (5′→3′): forward primer,
5-ATGGTGGACTACCACGCGGCGAACC-3 and reverse primer,
5-TCACAGGTCGCTCTCGCCATACAAG-3; primers used for RAB31 were (5′→3′):
forward primer, 5-ATGATGGCGATACGGGAGCTCAAAG-3 and reverse primer,
5-TCAACAGCACCGGCGGCTGGCTTGC-3. Consistent with the results from the
microarray analysis, both RAB31 and ACTN4 were upregulated in RCC
cells compared with levels in the normal cells (Fig. 6).
Discussion
Increasing evidence suggests that lncRNAs are
crucial in human cancer. Various studies have revealed that the
differential expression of lncRNAs plays an important role in the
development of cancer (36,37). Recently, there have been efforts to
identify aberrantly expressed lncRNAs in muscle-invasive bladder
cancer. Based on a large sample size from the TCGA data portal,
several dysregulated lncRNAs have been identified (38). However, a limited number of studies
have identified RCC-specific lncRNAs.
Currently, lncRNAs closely associated with tumor
status are thought to be more suitable than mRNAs as diagnostic and
prognostic biomarkers (39). Some
lncRNAs that have been well studied, such as H19, are considered as
powerful predictors or potential targets in cancers (40). However, the relationship of lncRNAs
with RCC remains unclear. Therefore, the expression profiles of
lncRNAs, miRNAs and mRNAs in RCC were analyzed with a focus on the
clinical diagnostic significance of DElncRNAs. In the present
study, 5 lncRNAs with aberrant expression were identified in RCC
compared with normal samples. In addition, 2 DElncRNAs (ACTN4 and
RAB31) were included in the ceRNA network, which were significantly
correlated with the overall survival of patients with RCC; this
result strongly indicated that these DElncRNAs may function not
only as key oncogenes but also as prognostic markers in RCC
progression.
ACTN4 is a non-muscle type α-catenin that is only
expressed in non-muscle cells (41). In particular, ACTN4 is associated
with cell migration and cell adhesion, and it was first identified
in 1988 as a metastasis-related gene (42). In this study, the expression of
ACTN4 was negatively correlated with overall survival, which was
consistent with the results of a previous study reporting that
patients with ACTN4 amplification have significantly worse overall
survival in comparison with those without ACTN4 amplification
(43). Various studies have shown
that ACTN4 is expressed in several types of cancers, such as
invasive ductal adenocarcinoma and ovarian cancer, and it has been
used as a biomarker for therapeutic response (41,43,44).
RAB31 belongs to the Ras superfamily of small GTPase and was first
identified in human melanocytes (45,46).
There has been increased interest in the relationship between RAB31
and human cancer. Gene expression profiling analysis has revealed
the overexpression of RAB31 in estrogen receptor α-positive breast
carcinomas (47). Elevated
transcript levels of RAB31 were reported in breast cancer cells
expressing the urokinase-type plasminogen activator (uPA)-receptor
splice variant uPAR-del4/5, and high Rab31 levels were found to be
significantly associated with overall survival and distant
metastasis-free survival (48–50). A
meta-analysis of microarray data found that Rab31 is one of the 10
key genes in glioblastoma multiforme development (51). In this study, our results showed
that the expression of RAB31 was negatively associated with the
overall survival of patients, which was consistent with the
previously reported results for many types of cancers. These
lncRNAs were identified as prognostic predictors, and they may
contribute to the targeted therapy of RCC in the future.
Several miRNAs have been well characterized in human
diseases including lung cancer and bladder cancer (52,53).
However, only a few lncRNAs have been mechanically and functionally
characterized. In this study, we identified specific lncRNAs and
miRNAs of RCC in a database and constructed a ceRNA network, which
would be relevant for further investigations. Many protein-coding
genes in the ceRNA network, such as FN1, TP53 and MYC, are known as
oncogenes and/or tumor inhibitors related to RCC development and
progression, which may be potential therapeutic targets of cancer
(54–56). Six pathways of DEmRNAs were
significantly enriched in the ceRNA network, and the top KEGG
pathway was the diabetic complications pathway. This finding
indicated that the diabetic complications pathway may be involved
in the progression of RCC.
Although there has been increased interest in the
function of lncRNAs in recent years, the functional association of
lncRNAs and miRNAs remains unclear. The identification of DEmiRNAs
in RCC is necessary for determining the oncogenic pathways mediated
by miRNA. In this study, we identified miRNAs related to tumor
initiation in renal cells. Moreover, a lncRNA-miRNA-mRNA ceRNA
network was constructed to illustrate the interaction between
miRNAs, lncRNAs and coding genes. Two DElncRNAs (ACTN4 and RAB31)
were identified in this study, which may be potential prognostic
markers and oncogenes in RCC progression. In particular, the ceRNA
network built in this study would contribute to the elucidation of
the unknown ceRNA regulatory network in RCC.
In conclusion, we identified tumor
initiation-related miRNAs, and the lncRNA-miRNA-mRNA ceRNA network
in RCC was constructed. The interaction between miRNAs, lncRNAs and
coding genes was investigated. Further studies would help to
elucidate the primary mechanism of miRNAs and lncRNAs in RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81472682 and
81772756) and the Natural Science Foundation of Tianjin (nos.
17JCZDJC35300, 15JCZDJC35400 and 15JCYBJC27200).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HWG and XRC put forward the ideas of the research,
analyzed the data and written the manuscript. ZQS put forward the
ideas of the article and helped revising the manuscript. YJN
provided valuable instructions and suggestions on this thesis and
helped revising the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jonasch E, Gao J and Rathmell WK: Renal
cell carcinoma. BMJ. 349:g47972014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ljungberg B, Campbell SC, Choi HY, Jacqmin
D, Lee JE, Weikert S and Kiemeney LA: The epidemiology of renal
cell carcinoma. Eur Urol. 60:615–621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta S, Kang HC, Ganeshan DM, Bathala TK
and Kundra V: Diagnostic approach to hereditary renal cell
carcinoma. AJR Am J Roentgenol. 204:1031–1041. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bex A, Jonasch E, Kirkali Z, Mejean A,
Mulders P, Oudard S, Patard JJ, Powles T, van Poppel H and Wood CG:
Integrating surgery with targeted therapies for renal cell
carcinoma: Current evidence and ongoing trials. Eur Urol.
58:819–828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wyczólkowski M, Klima W, Bieda W and Walas
K: Spontaneous regression of hepatic metastases after nephrectomy
and metastasectomy of renal cell carcinoma. Urol Int. 66:119–120.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
International Human Genome Sequencing
Consortium: Finishing the euchromatic sequence of the human genome.
Nature. 431:931–945. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bergmann JH and Spector DL: Long
non-coding RNAs: Modulators of nuclear structure and function. Curr
Opin Cell Biol. 26:10–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guttman M, Garber M, Levin JZ, Donaghey J,
Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et
al: Ab initio reconstruction of cell type-specific transcriptomes
in mouse reveals the conserved multi-exonic structure of lincRNAs.
Nat Biotechnol. 28:503–510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cabili MN, Trapnell C, Goff L, Koziol M,
Tazon-Vega B, Regev A and Rinn JL: Integrative annotation of human
large intergenic noncoding RNAs reveals global properties and
specific subclasses. Genes Dev. 25:1915–1927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z,
Cai G, Li G, Yang BB and Zhang Y: The effect of central loops in
miRNA: MRE duplexes on the efficiency of miRNA-mediated gene
regulation. PLoS One. 3:e17192008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Amirkhah R, Schmitz U, Linnebacher M,
Wolkenhauer O and Farazmand A: MicroRNA-mRNA interactions in
colorectal cancer and their role in tumor progression. Genes
Chromosomes Cancer. 54:129–141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang C, Hu DZ and Liu JZ: Identification
of critical TF-miRNA-mRNA regulation loops for colorectal cancer
metastasis. Genet Mol Res. 14:5485–5495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Militello G, Weirick T, John D, Döring C,
Dimmeler S and Uchida S: Screening and validation of lncRNAs and
circRNAs as miRNA sponges. Brief Bioinform. 18:780–788.
2017.PubMed/NCBI
|
|
23
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Imig J, Brunschweiger A, Brümmer A,
Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M
and Hall J: miR-CLIP capture of a miRNA targetome uncovers a
lincRNA H19-miR-106a interaction. Nat Chem Biol. 11:107–114. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou X, Liu J and Wang W: Construction and
investigation of breast-cancer-specific ceRNA network based on the
mRNA and miRNA expression data. IET Syst Biol. 8:96–103. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
30
|
Wotschofsky Z, Gummlich L, Liep J, Stephan
C, Kilic E, Jung K, Billaud JN and Meyer HA: Integrated microRNA
and mRNA signature associated with the transition from the locally
confined to the metastasized clear cell renal cell carcinoma
exemplified by miR-146-5p. PLoS One. 11:e01487462016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fachel AA, Tahira AC, Vilella-Arias SA,
Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM and
Verjovski-Almeida S: Expression analysis and in silico
characterization of intronic long noncoding RNAs in renal cell
carcinoma: Emerging functional associations. Mol Cancer.
12:1402013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Chen X, Han W, Ruan A, Chen L,
Wang R, Xu Z, Xiao P, Lu X, Zhao Y, et al: miR-200c targets CDK2
and suppresses tumorigenesis in renal cell carcinoma. Mol Cancer
Res. 13:1567–1577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Walsh AL, Tuzova AV, Bolton EM, Lynch TH
and Perry AS: Long noncoding RNAs and prostate carcinogenesis: The
missing ‘linc’? Trends Mol Med. 20:428–436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang F, Zhang L and Zhang C: Long
noncoding RNAs and tumorigenesis: Genetic associations, molecular
mechanisms, and therapeutic strategies. Tumour Biol. 37:163–175.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang H, Niu L, Jiang S, Zhai J, Wang P,
Kong F and Jin X: Comprehensive analysis of aberrantly expressed
profiles of lncRNAs and miRNAs with associated ceRNA network in
muscle-invasive bladder cancer. Oncotarget. 7:86174–86185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang
J, Dong N, He J, Sun Q, Lv G, et al: Long noncoding RNA H19
indicates a poor prognosis of colorectal cancer and promotes tumor
growth by recruiting and binding to eIF4A3. Oncotarget.
7:22159–22173. 2016.PubMed/NCBI
|
|
41
|
Honda K: The biological role of actinin-4
(ACTN4) in malignant phenotypes of cancer. Cell Biosci. 5:412015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Honda K, Yamada T, Endo R, Ino Y, Gotoh M,
Tsuda H, Yamada Y, Chiba H and Hirohashi S: Actinin-4, a novel
actin-bundling protein associated with cell motility and cancer
invasion. J Cell Biol. 140:1383–1393. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yamamoto S, Tsuda H, Honda K, Onozato K,
Takano M, Tamai S, Imoto I, Inazawa J, Yamada T and Matsubara O:
Actinin-4 gene amplification in ovarian cancer: A candidate
oncogene associated with poor patient prognosis and tumor
chemoresistance. Mod Pathol. 22:499–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kikuchi S, Honda K, Tsuda H, Hiraoka N,
Imoto I, Kosuge T, Umaki T, Onozato K, Shitashige M, Yamaguchi U,
et al: Expression and gene amplification of actinin-4 in invasive
ductal carcinoma of the pancreas. Clin Cancer Res. 14:5348–5356.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rojas AM, Fuentes G, Rausell A and
Valencia A: The Ras protein superfamily: Evolutionary tree and role
of conserved amino acids. J Cell Biol. 196:189–201. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen D, Guo J, Miki T, Tachibana M and
Gahl WA: Molecular cloning of two novel rab genes from human
melanocytes. Gene. 174:129–134. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Abba MC, Hu Y, Sun H, Drake JA, Gaddis S,
Baggerly K, Sahin A and Aldaz CM: Gene expression signature of
estrogen receptor alpha status in breast cancer. BMC Genomics.
6:372005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Luther T, Kotzsch M, Meye A, Langerholc T,
Füssel S, Olbricht N, Albrecht S, Ockert D, Muehlenweg B, Friedrich
K, et al: Identification of a novel urokinase receptor splice
variant and its prognostic relevance in breast cancer. Thromb
Haemost. 89:705–717. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kotzsch M, Farthmann J, Meye A, Fuessel S,
Baretton G, Tjan-Heijnen VC, Schmitt M, Luther T, Sweep FC,
Magdolen V and Span PN: Prognostic relevance of uPAR-del4/5 and
TIMP-3 mRNA expression levels in breast cancer. Eur J Cancer.
41:2760–2768. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sato S, Kopitz C, Grismayer B, Beaufort N,
Reuning U, Schmitt M, Luther T, Kotzsch M, Krüger A and Magdolen V:
Overexpression of the urokinase receptor mRNA splice variant
uPAR-del4/5 affects tumor-associated processes of breast cancer
cells in vitro and in vivo. Breast Cancer Res Treat. 127:649–657.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kunkle BW, Yoo C and Roy D: Reverse
engineering of modified genes by Bayesian network analysis defines
molecular determinants critical to the development of glioblastoma.
PLoS One. 8:e641402013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Usó M, Jantus-Lewintre E, Sirera R,
Bremnes RM and Camps C: miRNA detection methods and clinical
implications in lung cancer. Future Oncol. 10:2279–2292. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kumra H and Reinhardt DP:
Fibronectin-targeted drug delivery in cancer. Adv Drug Deliv Rev.
97:101–110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Muller PA and Vousden KH: Mutant p53 in
cancer: New functions and therapeutic opportunities. Cancer Cell.
25:304–317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huang H, Weng H, Zhou H and Qu L:
Attacking c-Myc: Targeted and combined therapies for cancer. Curr
Pharm Des. 20:6543–6554. 2014. View Article : Google Scholar : PubMed/NCBI
|