Introduction
Oral squamous cell carcinoma (OSCC) is the one of
the most common cancer types (1).
With a propensity to lymph node metastasis, the 5-year survival
rate of OSCC is merely 40–50% (2).
However, advances in surgical techniques as well as novel
chemoradiation approaches remain less optimistic on the treatment
of advanced OSCC (3), due to lymph
node metastasis (4). Therefore,
basic researches on OSCC focusing on tumor metastases are required,
aiming to identify specific biomarkers that may offer novel
therapeutic directions and new insight (5).
X-box-binding protein 1 (XBP1) belongs to the basic
region/leucine zipper protein family, which is involved in unfolded
protein response (UPR) (6). Once
cleaved by inositol-requiring enzyme1ɑ (IRE1) by removing 26-bp
intron, and XBP1 mRNA fragments from the functional nuclear
transcriptional factor, XBP1-s (spliced XBP1) (7,8).
Previous findings showed that XBP1 is induced in various types of
cancer, and controls cell type- and tissue-specific transcriptional
regulatory networks in different cancer types (9–12). In
addition, XBP1 was identified as a survival factor in certain
malignant neoplasms (13,14). Mounting evidence supported a direct
role of XBP1 in tumor invasion, while loss of XBP1 was shown to
severely inhibit tumor metastasis in vitro and in
vivo (15). XBP1 also plays a
pivotal role in tumor invasion through the upregulation of MMP9
expression in esophageal squamous cell carcinoma and triggering
tumor EMT via promoting snail in breast cancer cells (16,17).
To the best of our knowledge, the role of XBP1 in OSCC metastasis
and prognosis has not been clearly elaborated. Therefore, exploring
the functional role and possible downstream signaling of XBP1 in
OSCC is crucial to elucidate the effect of XBP1 on cancer
development and progression.
The AXL receptor tyrosine kinase (AXL) is a member
of the TAM (TYRO3-AXL-MER) family of receptor tyrosine kinases
(18). A number of studies are
available to support AXL as a candidate in tumor metastasis
(18,19) and cancer progression (20,21).
It is reported that AXL regulates cancer invasion in breast
carcinoma (19,22) and hepatocellular carcinoma (23) via the PI3K/Akt signaling pathway, by
activating matrix metalloproteinase (MMPs) (24). Through genome-wide profiling, a
recent study demonstrated that the expression of AXL was decreased
when XBP1 expression was inhibited in XBP1-deficient breast cancer
cells (9). However, no further
research was performed to confirm the relationship between AXL and
XBP1. Therefore, the aim of the present study was to demonstrate
the role of XBP1 in OSCC invasion and prognosis. Moreover, the
relationship between XBP1 and AXL signaling was explored to gain
better understanding of the possible downstream signaling regulated
by XBP1 in OSCC.
Materials and methods
Cell culture
The Cal27 cell line (derived from a tongue SCC
patient) and the 293.2 sus cell line were obtained from the
American Type Culture Collection (ATCC, Manassas, VA, USA). The
head and neck squamous cell carcinoma cell line, UM-SCC-23, was a
gift of Dr Thomas E. Carey. The material transfer agreement of this
cell line was from the University of Michigan in June, 2012
(25). UM-SCC-23 and Cal-27 cells
were cultured in DMEM (Hyclone Laboratories, Logan, UT, USA), and
293.2 sus was cultured in RPMI-1640 medium (Hyclone Laboratories).
Cells were cultured as previously described (26).
Patients and tissue microarrays
The 96 OSCC samples for microarray were obtained
from patients that underwent primary surgery at the Stomatology
Hospital of Wuhan University (Hubei, China) between January 2002
and February 2009. All the adjacent normal epithelium samples were
obtained by local excision. Ethics approval from the Institutional
Review Board of School and Hospital of Stomatology, Wuhan
University was received for the examination of patient samples.
Specimens were fixed with formalin (Promoter Biotechnology, Hubei,
China) postoperatively, and embedded with paraffin before being
converted into tissue microarray slides. The histological types and
tumor grades were analyzed by two pathologists. There were 228
spots in all from 76 patients without lymph node metastasis: For
each patient, three spots were selected, respectively, from normal
adjacent epithelium, tumor center and tumor front, respectively.
The 228 spots were then spread evenly on 3 slides. In 20 patients
with lymph node metastasis, 4 spots were selected from normal
adjacent epithelium, tumor center, tumor front, and lymph node
metastases, respectively. Then, 80 spots in all were spread on one
slide. The diameter of each spot was 2 mm. Tissue microarrays were
produced by Outdo Biotech (Shanghai, China) after the design. Of
the 96 tumors, 47 (48.96%) were in grade I, 35 (36.46%) were in
grade II, and 14 (14.58%) were in grade III. The pathological grade
classification of the tumors was according to the 7th edition of
the cancer staging manual published by the American Joint Committee
on Cancer (27). Other information
related to the samples is listed in Table I.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Group | Cases (96) | Percentage % |
|---|
| Sex |
|
|
|
Male | 57 | 59 |
|
Female | 39 | 41 |
| Age |
|
|
|
<55 | 47 | 49 |
|
≥55 | 49 | 51 |
| Location |
|
|
|
Tongue | 34 | 35 |
|
Other | 62 | 65 |
| Lymph node
metastases |
|
|
|
With | 20 | 21 |
|
Without | 76 | 79 |
| Pathological
grades |
|
|
| I | 47 | 49 |
| II | 35 | 36 |
|
III | 14 | 15 |
| Clinical
stages |
|
|
| I | 31 | 32 |
| II | 30 | 31 |
|
III | 23 | 24 |
| IV | 12 | 13 |
Immunohistochemical staining
Paraffin-embedded specimens were sliced into 4-µm
sections. After deparaffinating, dehydrating and performing antigen
retrieval with high pressure, to quench the endogenous peroxidase
activity, the sections were incubated with 3% hydrogen superoxide
for 20 min followed by blocking non-specific binding in 10% normal
goat serum. Immunohistochemical staining was performed as follows:
The sections were incubated overnight at 4°C with polyclonal rabbit
anti-human XBP1 (1:300; cat. no. ab37152; Abcam, Cambridge, UK),
then labeled with HRP secondary antibody (Universal
streptavidin-peroxidase kit including endogenous peroxidase
blocking agent, goat serum, biotin-labeled goat anti-rabbit IgG,
and horseradish peroxidase-labeled streptavidin; cat. no. SP-9001;
Zhongshan, Beijing, China) followed by DAB (Maxim, Fuzhou, Fujian,
China) color reaction. The sections were then counterstained with
haematoxylin.
Scoring system, hierarchical
clustering and data visualization
The tissue microarray slices stained with XBP1
antibody were scanned using Aperio ScanScope CS scanner (Vista, CA,
USA), and Aperio ImageScope (Version 11.2) was used for nuclear
quantification. Four high-power fields of interests were selected
in each spot for quantifying the average optical density. Each
field was read and signed with four kinds of colors according to
color gradation (red for strong positive, orange for moderate
positive, yellow for weak positive, and blue for negative). The
mean histoscore of nuclear and cytoplasm staining was calculated
using the formula: [(intensity of strong positive) ×3+ (intensity
of moderate positive) ×2+ (intensity of weak positive)
×1)]/selected area/4 (28). The
hierarchical analysis was achieved by the HemI 1.0 with average
linkage based on Pearson's correlation coefficient (29).
Plasmids and stable transduction
For the gene knockdown study, the GV-248-XBP1-sh1,
2, 3 and GV-248-con lentiviral vectors were purchased from Shanghai
GeneChem Co., Ltd. (Shangai, China). The GV-248-XBP1-shs plasmids
were generated by inserting the oligonucleotide containing the
specific shRNA target sequences into the GV-248 vector and verified
by sequencing. Three shRNA sequence pairs are shown in Table II. A cocktail for each transduction
was produced by mixing the constructed, envelope, and packaging
plasmids with serum-free OPTI-MEM (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), which was added to 293.2 sus cells
dropwise. After 48 h of transduction, lentiviral particles were
harvested and stored in −80°C. To stably infect the target cells,
lentiviral particle solution with polybrene (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added to the 50–60% confluent target
cells. GFP fluorescence sorting was used to select transducted
cells.
 | Table II.Specific XBP1-shRNA target
sequences |
Table II.
Specific XBP1-shRNA target
sequences
| XBP1 | Sequences |
|---|
| −sh1 | A:
5′-CCGGGACCCAGTCATGTTCTTCAAACTCGAGTTTGAAGAACATGACTGGGTCTTTTTG-3′ |
|
| B:
5′-AATTCAAAAAGACCCAGTCATGTTCTTCAAACTCGAGTTTGAAGAACATGACTGGGTC-3′ |
| −sh2 | A:
5′-CCGGGCGGTATTGACTCTTCAGATTCTCGAGAATCTGAAGAGTCAATACCGCTTTTTG-3′ |
|
| B:
5′-AATTCAAAAAGCGGTATTGACTCTTCAGATTCTCGAGAATCTGAAGAGTCAATACCGC-3′ |
| −sh3 | A:
5′-CCGGGAACAGCAAGTGGTAGATTTACTCGAGTAAATCTACCACTTGCTGTTCTTTTTG-3′ |
|
| B:
5′-AATTCAAAAAGAACAGCAAGTGGTAGATTTACTCGAGTAAATCTACCACTTGCTGTTC-3′ |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the OSCC cell lines
using the HP total RNA isolation kit (Omega Biotek, Norcross, GA,
USA). RNA was transcribed into cDNA using the Takara® RT
reagent kit (Takara Biotechnology, Co., Ltd., Dalian, Japan), and
gene expression was quantified by Roche FastStart Essential DNA
Green Master (Roche, Basel, Switzerland). Primer sequences are
shown in Table III. The cycling
parameters used were 95°C for 15 min; followed by 40 cycles of 95°C
for 15 sec; 55°C for 30–40 sec and 72°C for 30 sec. The mRNA
expression was normalized to that of β-actin. The differential
expression of mRNA between transduced and control cells was deduced
from 2−ΔΔCq, where ΔΔCq=ΔCq transducted cells-ΔCq
control cells.
 | Table III.List of primers for RT-qPCR. |
Table III.
List of primers for RT-qPCR.
| Gene | Primer
sequence |
|---|
| β-actin | F:
5′-ACCAACTGGGACGACATGGAGAAA-3′ |
|
| R:
5′-TAGCACAGCCTGGATAGCAACGTA-3′ |
| XBP1 | F:
5′-CCTGGTTGCTGAAGAGGAGG-3′ |
|
| R:
5′-CCATGGGGAGATGTTCTGGAG-3′ |
| AXL | F:
5′-TCAAGGTGGCTGTGAAGACGATGA-3′ |
|
| R:
5′-AACCCTGGAAACAGACACCGATGA-3′ |
| E-cadherin | F:
5′-AAGTCAGTTCAGACTCCAGCC-3′ |
|
| R: 5′-
TGTAGCTCTCGGCGTCAAA-3′ |
| N-cadherin | F:
5′-TGAAACGGCGGGATAAAGAG-3′ |
|
| R:
5′-GGCTCCACAGTATCTGGTTG-3′ |
| Slug | F:
5′-TGGTTGCTTCAAGGACACAT −3′ |
|
| R:
5′-GTTGCAGTGAGGGCAAGAA-3′ |
| Snail1 | F:
5′-GCGAGCTGCAGGACTCTAAT-3′ |
|
| R:
5′-GGACAGAGTCCCAGATGAGC-3′ |
| Twist1 | F:
5′-GCCGGAGACCTAGATGTCATT-3′ |
|
| R:
5′-CACGCCCTGTTTCTTTGAAT-3′ |
| MMP1 | F:
5′-AAGGCCAGTATGCACAGCTT-3′ |
|
| R:
5′-GGGCCACTATTTCTCCGCTT-3′ |
| MMP2 | F:
5′-TGATGGCATCGCTCAGATCC-3′ |
|
| R:
5′-GGCCTCGTATACCGCATCAA-3′ |
| MMP3 | F:
5′-CACAGTTGGAGTTTGACCC-3′ |
|
| R:
5′-TAAGCAGCAGCCCATTTG −3′ |
| MMP9 | F:
5′-TTTGAGTCCGGTGGACGATG-3′ |
|
| R:
5′-GCTCCTCAAAGACCGAGTCC-3′ |
| MMP12 | F:
5′-TGCTGATGACATACGTGGCA-3′ |
|
| R:
5′-AGGATTTGGCAAGCGTTGG-3′ |
| LAMA1 | F:
5′-TCTGGGGAGAGATGTTGTGT-3′ |
|
| R:
5′-ACGTTTAAAAAGAGAGCCAGGG-3′ |
| PPFIBP1 | F:
5′-CAGGGAGGGAGGAGAGAAGG-3′ |
|
| R:
5′-GCCTGCACTACACCATGTCA-3′ |
| CXCL-1 | F:
5′-TCACAGTGTGTGGTCAACAT-3′ |
|
| R:
5′-AGCCCCTTTGTTCTAAGCCA-3′ |
| CXCL-2 | F:
5′-CACAGTGTGTGGTCAACATTTCT-3′ |
|
| R:
5′-TGCTCTAACACAGAGGGAAACA-3′ |
| MAX | F:
5′-TGCTCTAACACAGAGGGAAACA-3′ |
|
| R:
5′-CGGGATGCCTTCTCTCCTTG-3′ |
| Stat3 | F:
5′-ATCCTGGTGTCTCCACTGGT-3′ |
|
| R:
5′-CCTGGGTCAGCTTCAGGATG-3′ |
| JUN | F:
5′-GTGCCGAAAAAGGAAGCTGG-3′ |
|
| R:
5′-CTGCGTTAGCATGAGTTGGC-3′ |
| PI3K | F:
5′-TGCAGTTTTGGAAGCAGTCAC-3′ |
|
| R:
5′-CTGGAATAAGAACTATTCCTGCTCA-3′ |
| PPFIBP1 | F:
5′-CAGGGAGGGAGGAGAGAAGG-3′ |
|
| R:
5′-GCCTGCACTACACCATGTCA-3′ |
| C-MYC | F:
5′-TCCTCGGATTCTCTGCTCTC-3′ |
|
| R:
5′-CTCTGACCTTTTGCCAGGAG-3′ |
| IL-6 | F:
5′-TCAATATTAGAGTCTCAACCCCCA-3′ |
|
| R:
5′-GAGAAGGCAACTGGACCGAA-3′ |
Protein extraction and western blot
analysis
The total proteins were isolated from the OSCC cell
lines using RIPA buffer and western blot analysis was conducted as
previously described (26). The
membranes were then probed with anti-human XBP1 antibody (1:800;
cat. no. ab37152; Abcam) and anti-human AXL antibody (1:500; cat.
no. AF154; R&D Systems, Minnneapolis, MN, USA) overnight at
4°C. The membranes were also probed with anti-β-actin (Santa Cruz
Biotechnology, Dallas, TX, USA) to ensure equal amounts of protein.
Bound antibodies were detected using horseradish
peroxidase-conjugated secondary antibody (Santa Cruz
Biotechnology). Reactive protein was detected by ECL
chemiluminescence system (Advanstar, Santa Monica, CA, USA).
Western blot experiments were repeated in triplicate to confirm the
results. The protein amounts were estimated through densitometry as
the ratio detected protein/β-actin.
Cell invasion assay
Matrigel-coated chamber (BD Biosciences Inc. San
Jose, CA, USA) was prepared as previously described (26). Cells (5×105) were then
seeded onto the Matrigel-coated chamber. Cancer cells were seeded
using serum-free media to the upper chamber pairing with the lower
chamber filled with 20% serum DMEM media. At 24 h after allowing
cells to invade, cells on the lower side of the chamber were fixed
in ethanol and stained with crystal violet (Guge Biotechnology,
Wuhan, China). The total number of cells per high-power microscopic
field on the lower side of Matrigel-coated chamber were counted and
scored for invading cells. The mean number of cells in five
high-power microscopic fields was calculated with standard
deviations.
Thapsigargin stimulation and
semi-quantitative RT-PCR
ER stress was induced in epithelial cells by
exposure to thapsigargin. By the time of reaching near-confluence,
the cells were exposed to thapsigargin (50 nM; Sigma-Aldrich; Merck
KGaA) for 6 h (30). Dimethyl
sulfoxide (DMSO; Merck KGaA) served as a solvent control for
thapsigargin. To amplify the spliced and unspliced XBP1 mRNA,
semi-quantitative RT-PCR was performed and XBP1 primers are shown
in Table IV. PCR products were
electrophoresed on 2.5% agarose gel and β-actin was used as a
loading control. The size difference between the spliced and the
unspliced XBP1 was 26 nucleotides.
 | Table IV.List of primers for semi-quantitative
RT-PCR. |
Table IV.
List of primers for semi-quantitative
RT-PCR.
| Gene | Primer
sequence |
|---|
| β-actin | F:
5′-CCACCATGTACCCTGGCATT-3′ |
|
| R:
5′-CGCATCTCATATTTGGAATGACT-3′ |
| XBP1 | F:
5′-CCTTGTAGTTGAGAACCAGG-3′ |
|
| R:
5′-GGGGCTTGGTATATATGTGG-3′ |
Statistical analysis
Data analyses were performed using GraphPad Prism
5.0. One-way ANOVA followed by the Tukey's multiple comparison
tests were performed to analyze the differences in protein levels
and invasion cell number among groups. The χ2 test was
used to compare the dichotomous variables. The overall survival was
analyzed by the Kaplan-Meier method and log-rank test. Data were
presented as the mean ± SEM. P<0.05 was considered statistically
significant for all tests.
Results
XBP1 is increased in primary OSCC and
lymph node metastasis
XBP1 expression was determined in tissue microarray
using immunohistochemical staining. XBP1 protein mainly located in
nuclei and/or cytoplasm presented as yellow or brown (Fig. 1A). The weak nuclei staining of XBP1
was observed in the adjacent normal epithelial (109.2±9.91, mean ±
SEM). More intensive XBP1 expression was detected in tumor center
(TC: 205.1±12.70, mean ± SEM) and tumor front (TF: 271.3±15.9, mean
± SEM) (Fig. 1C). Statistical
analysis revealed that the relative expression level of XBP1 in the
tumor center and tumor front was significantly higher than that in
adjacent normal tissue (P<0.001). Moreover, compared with the
tumor center, the tumor front exhibited a stronger XBP1 expression
(P<0.05). In the samples with lymph node metastasis, the
expression level of XBP1 showed a gradual increasing tendency from
adjacent normal tissue, tumor center, and tumor front, to lymph
node metastasis. The heatmap was performed to reveal the difference
of XBP1 expression between normal adjacent epithelium, tumor
center, tumor front, and lymph node metastasis (Fig. 1B). Statistically, the mean staining
score of XBP1 in lymph node metastasis (404.9 ±52.09, mean ± SEM)
was 2.80-fold of normal adjacent epithelial, 2.50-fold of tumor
center and 1.83-fold of tumor front (P<0.001, Fig. 1C).
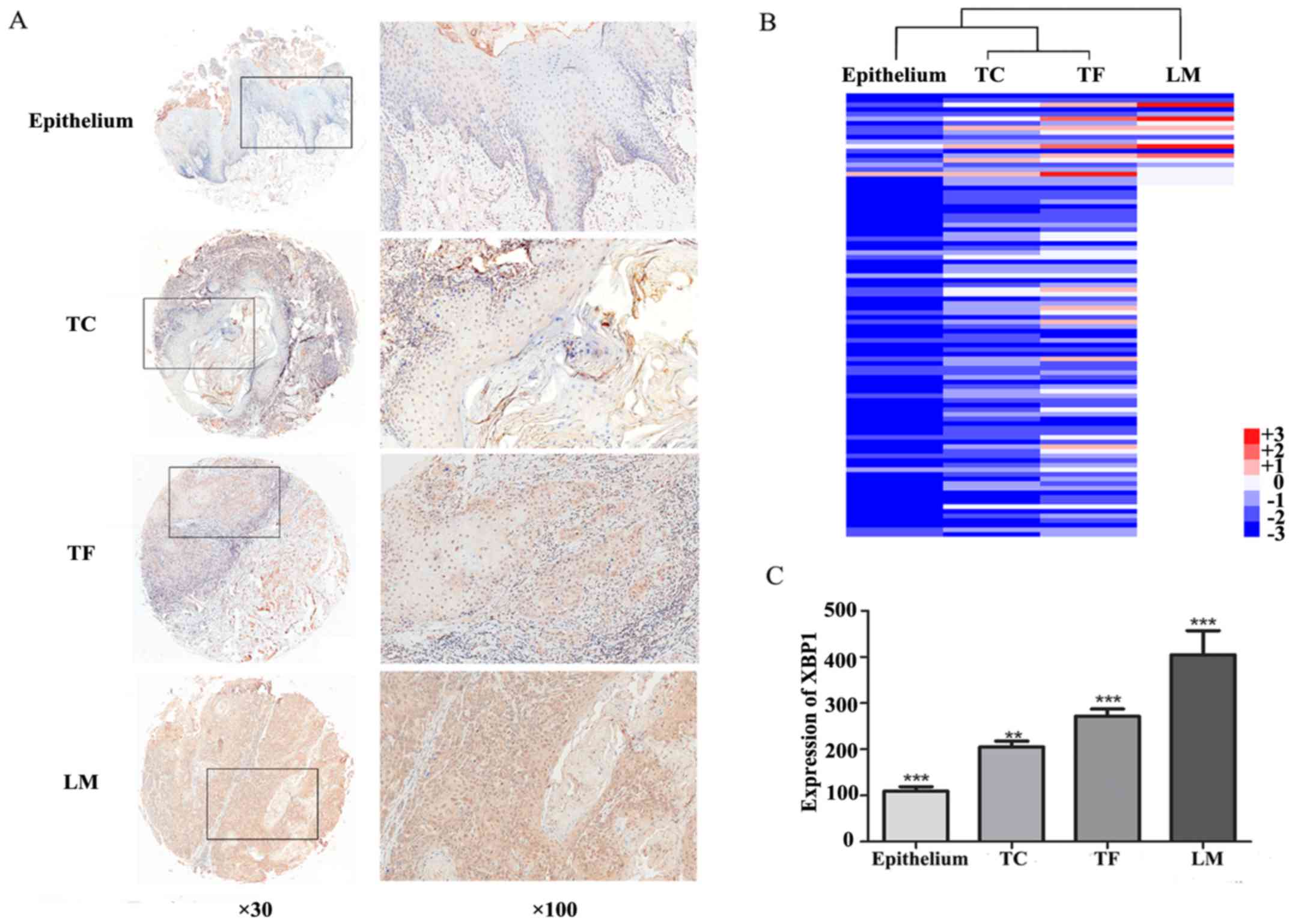 | Figure 1.Expression of XBP1 in adjacent normal
epithelium, tumor center, tumor front, and lymph node metastasis.
(A) Immunohistochemical staining of XBP1 in representative cores of
adjacent normal epithelium, tumor center (TC), tumor front (TF) and
lymph nodes metastases (LM). Magnification, cores, ×30; insets,
×100. (B) Heat map visualization of XBP1 expression in epithelium,
tumor center (TC), tumor front (TF) and lymph node metastasis (LM)
in 96 OSCC cases. Hierarchical clustering was analyzed based on the
results of IHC. Columns, cases; Rows, different areas of the
tissue; color key indicates XBP1 expression value; blue, lowest;
red, highest. (C) The histogram shows the expression of XBP1
presenting a significantly increasing trend in epithelium, tumor
center (TC), tumor front (TF) and lymph node metastasis (LM).
Significant differences were observed, respectively. ***P<0.001;
**P<0.05. |
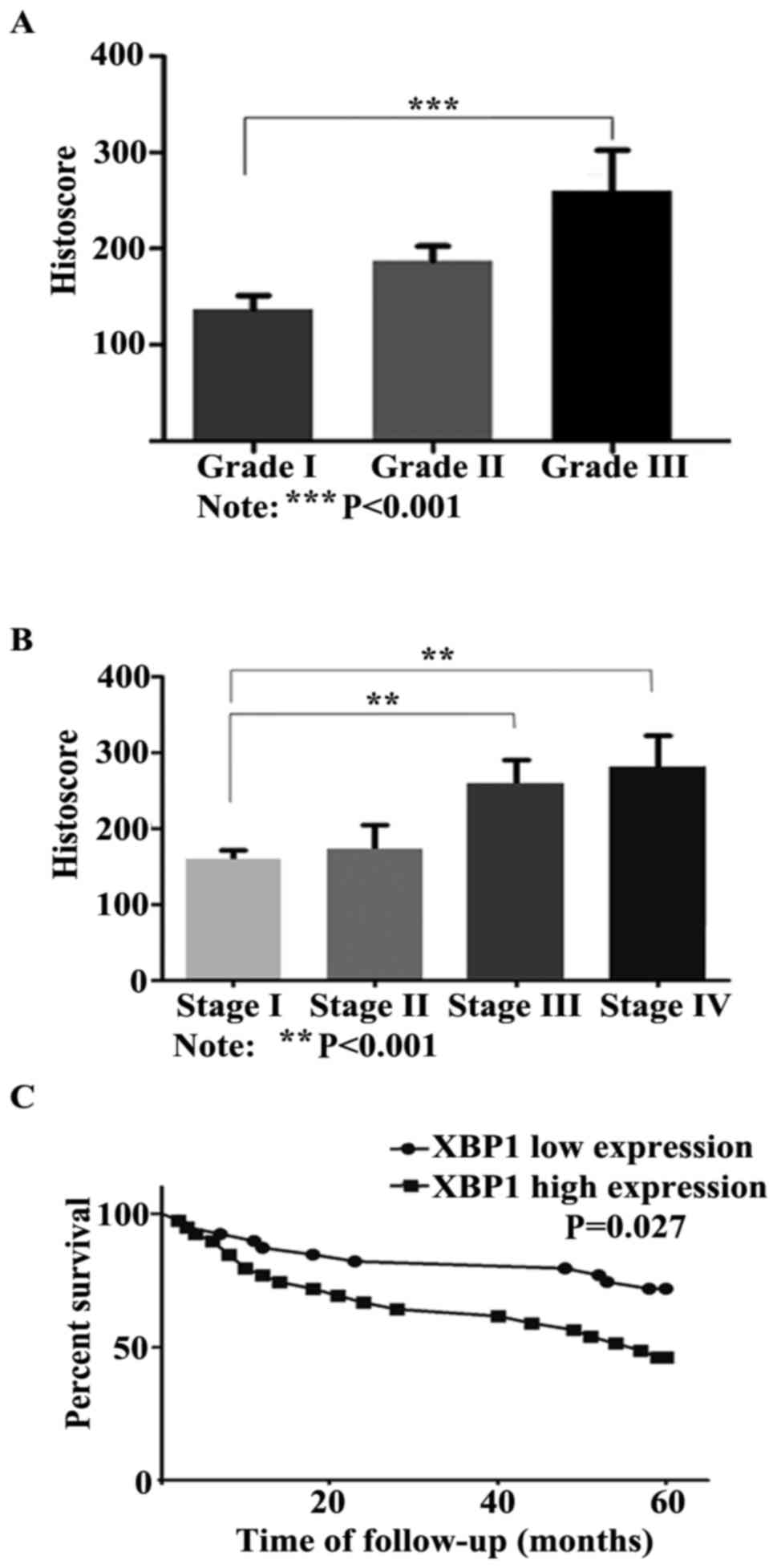
High XBP1 expression is associated
with poor clinicopathological characteristics in OSCC patients
To determine the association between XBP1 protein
expression level and the clinicopathological characteristics of
OSCC patients, we analyzed the correlation between XBP1 expression
and histological grades, clinical stages, sex and age. A high
nuclei XBP1 expression level was detected in histological grade II
(187.5±15.03, mean ± SEM) and III (260.0±42.67, mean ± SEM),
compared with histological grade I (137.2±13.84, mean ± SEM). The
expression level of XBP1 in grade III was 1.9-fold higher than that
in grade I (P<0.001, Fig. 2A).
Additionally, the statistical data demonstrated that XBP1
expression was significantly correlated with advanced clinical
stages (P<0.01, Fig. 2B).
Significantly stronger XBP1 staining was observed in stage III
(258.6±31.78, mean ± SEM) and stage IV (280.4±42.30, mean ± SEM)
than that in stage I (159.6±11.91, mean ± SEM). However, no
significant difference was detected between XBP1 expression and sex
as or age.
High XBP1 expression predicts poor
5-year survival
To elicit the correlation between XBP1 expression
and prognosis, the Kaplan-Meier survival analysis for XBP1
expression was performed. In total, 78 OSCC patients were followed
up until death or more than 5 years (range, 60–111 months), and 18
patients were lost. During the follow-up period, 32 patients
(41.0%) succumbed to the disease within 60 months, 1 patient (1.3%)
succumbed to the disease more than 60 months (77 months), and 45
patients (57.7%) were alive. Based on the result of
immunohistochemical staining, the samples with the tumor front
staining score ≥222.65 (median) were classified as high XBP1
expression, otherwise as low expression. The Kaplan-Meier survival
analysis and the log-rank test showed that patients with a high
level of XBP1 significantly were associated with unfavorable 5-year
survival (P=0.027, Fig. 2C).
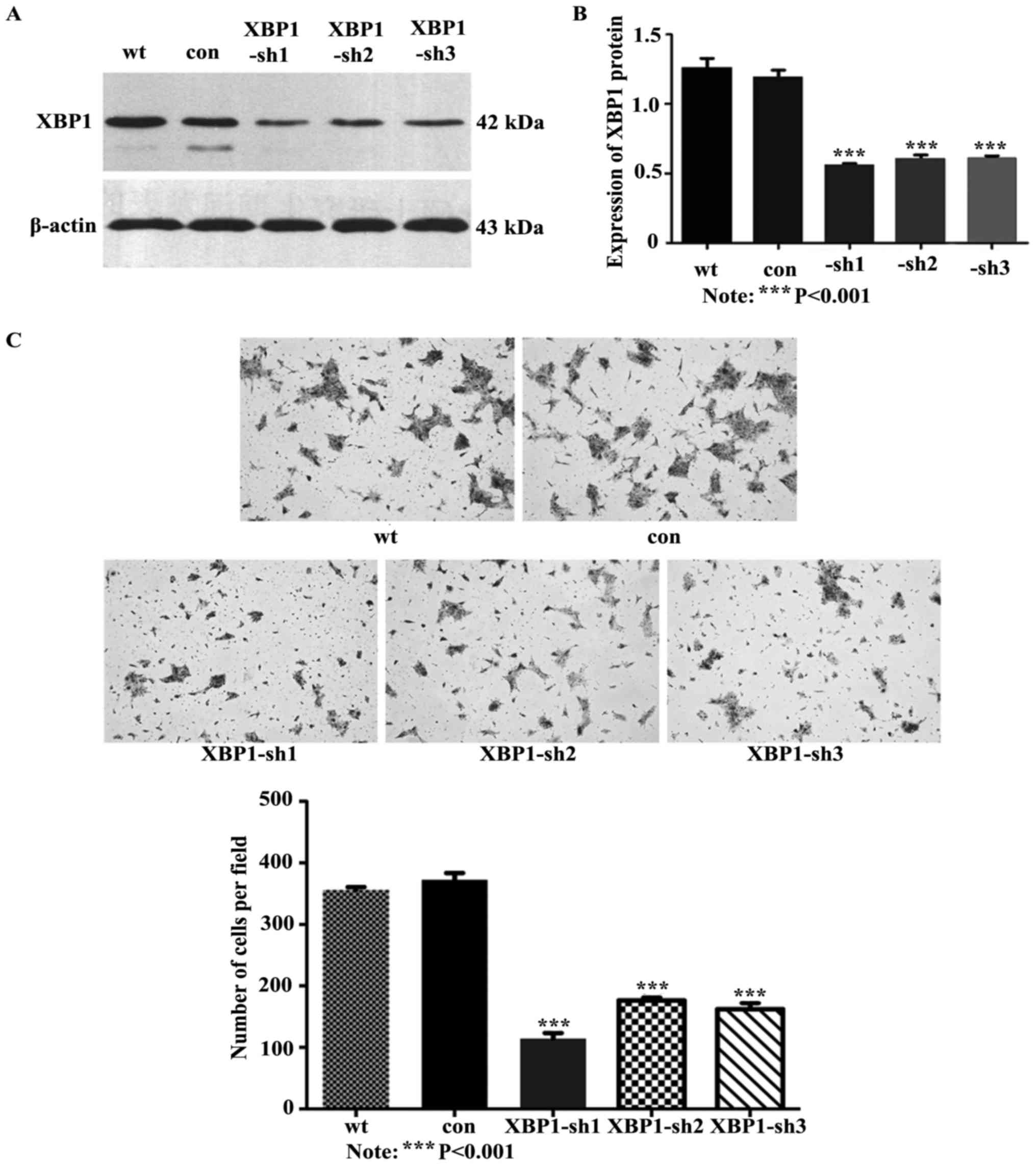
Inhibition of XBP1 suppresses cell
invasion in vitro
To further elucidate the function of XBP1 in the
metastatic process of OSCC, XBP1 were stably knocked down in SCC
cells using three constructed lentiviral vectors GV-248-XBP1-sh1,
2, 3. The XBP1 protein expression was decreased significantly in
XBP1-shs-transducted cells compared with the control (P<0.001,
Fig. 3A and B). The suppressed XBP1
expression caused a significant reduction of cell invasion
capability (Fig. 3C).
Statistically, the knockdown of XBP1 resulted in a 3.25-, 2.11- and
2.29-fold decrease of cell invasion, respectively, in
UM-SCC-23-XBP1-sh1, -sh2 and -sh3, compared with the cells
transduced with the control vector (P<0.001, Fig. 3C).
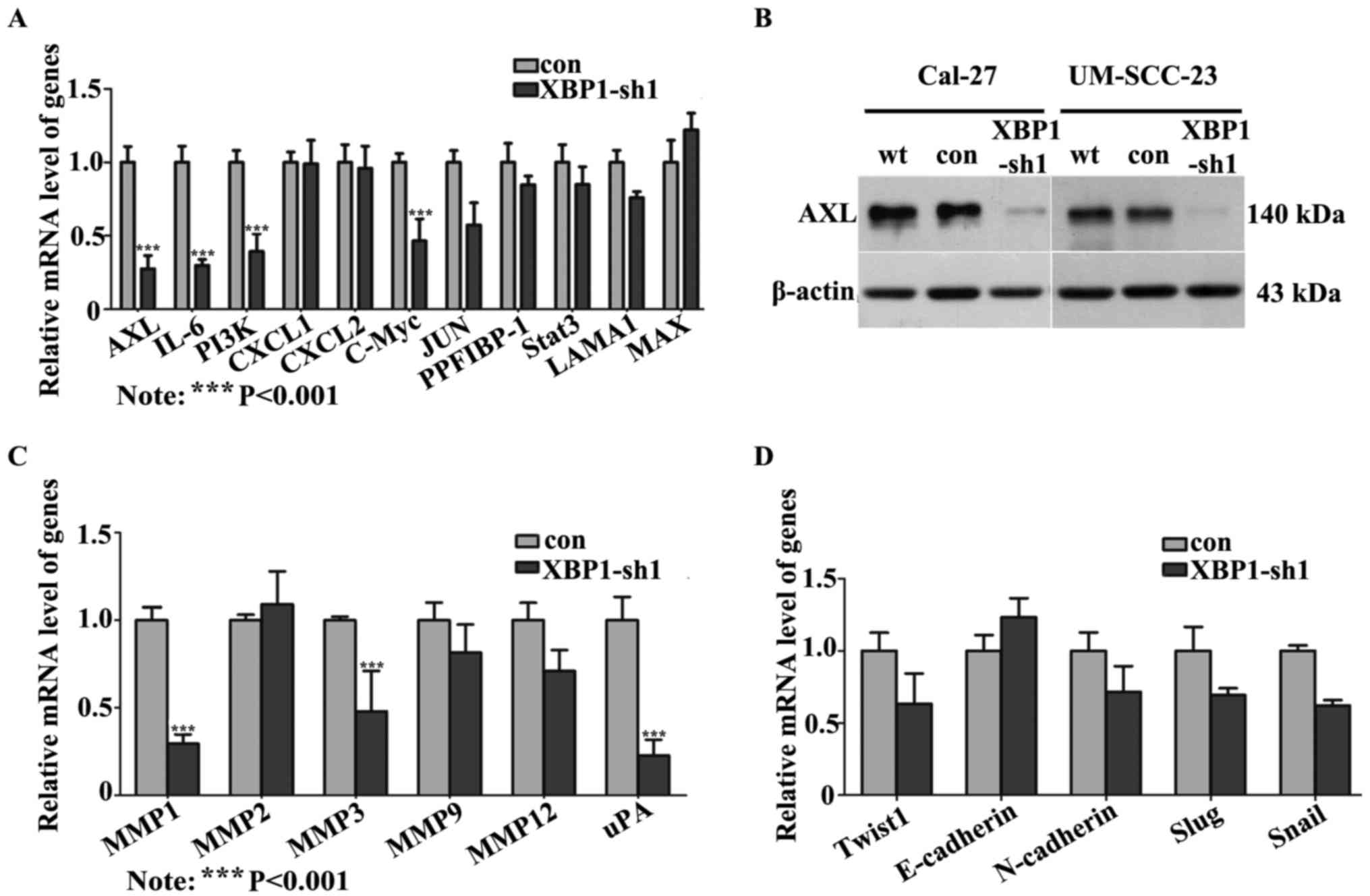
Suppression of XBP1 inhibits AXL
signaling
To explore the possible downstream molecular factor
of XBP1 involved in tumor metastasis, several invasion-related
genes putatively implicated in XBP1 signaling according to the
published data were investigated. The mRNA expression levels of 11
candidate genes were detected (Fig.
4A). Among these genes, the mRNA levels of AXL, PI3K, IL-6 and
C-MYC were significantly reduced by more than 2-fold in XBP1-sh1
cells. Furthermore, the decreased expression of AXL in XBP1-shs
cells was also confirmed in the protein level (Fig. 4B).
Recently, aberrant AXL was reported to be associated
with tumor invasion by inducing MMPs or uPA or triggering EMT
signaling (19,31). As a result, MMP1, 2, 3, 9, 12 and
uPA, and the EMT-associated molecules, such as twist1, slug, snail,
E-cadherin and N-cadherin were examined to evaluate the molecular
contribution of XBP1 in OSCC invasion through AXL signaling.
Significant reductions of MMP1, MMP3 and uPA were detected in
XBP1-sh1 cells compared to the control cells (Fig. 4C). However, the mRNA expression of
EMT markers showed no significant difference between cells (Cal-27)
transduced with lentivirus encoding XBP1-sh1 and scramble vector
(Fig. 4D).
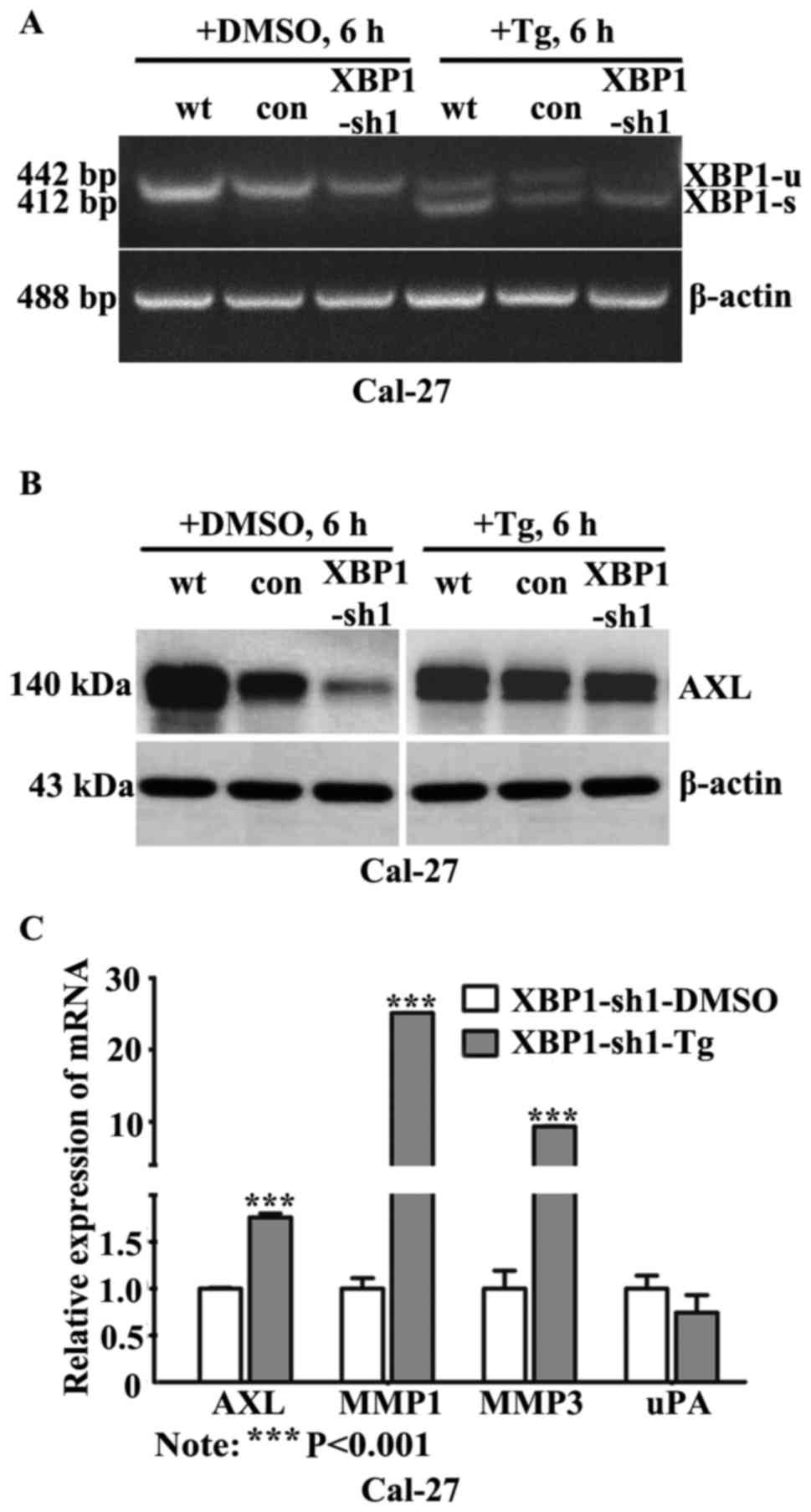
XBP1 activation rescued AXL expression
and promotes MMPs expression
Previous findings have demonstrated that XBP1 was
regulated through a novel mechanism of mRNA splicing initiated by
IRE1, an ER transmembrane kinase/endoribonuclease (32). To verify whether re-activation of
XBP1 could rescue the expression of AXL, the ER stress inducer
thapsigargin (Tg) was supplied to stimulate OSCC cells. After
treating the cells with 50 nM Tg for 6 h, the mRNA expression level
of XBP1-s was significantly induced in XBP1-sh1 cells, as well as
in control cells (Fig. 5A).
Simultaneously, the protein levels of AXL were restored obviously
when XBP1 was activated in XBP1-sh cells with Tg treatment
(Fig. 5B). In addition, the mRNA
expression levels of MMP1 and MMP3 were significantly increased in
XBP1-sh1 cells treated with 50 nM Tg for 6 h, compared with cells
treated with DMSO (Fig. 5C).
Aberrant AXL expression is correlated
with XBP1 overexpression in OSCC tissues
In order to evaluate the expression of AXL in OSCC
tissues, the IHC staining was performed on tissue microarrays
described before. Negative to weak cytoplasm staining of AXL was
detected in the adjacent normal epithelium, while moderate to
strong AXL staining was observed in the primary cancer cells and
lymph node metastases. The expression levels of AXL in adjacent
normal epithelium, primary cancer and lymph node metastasis showed
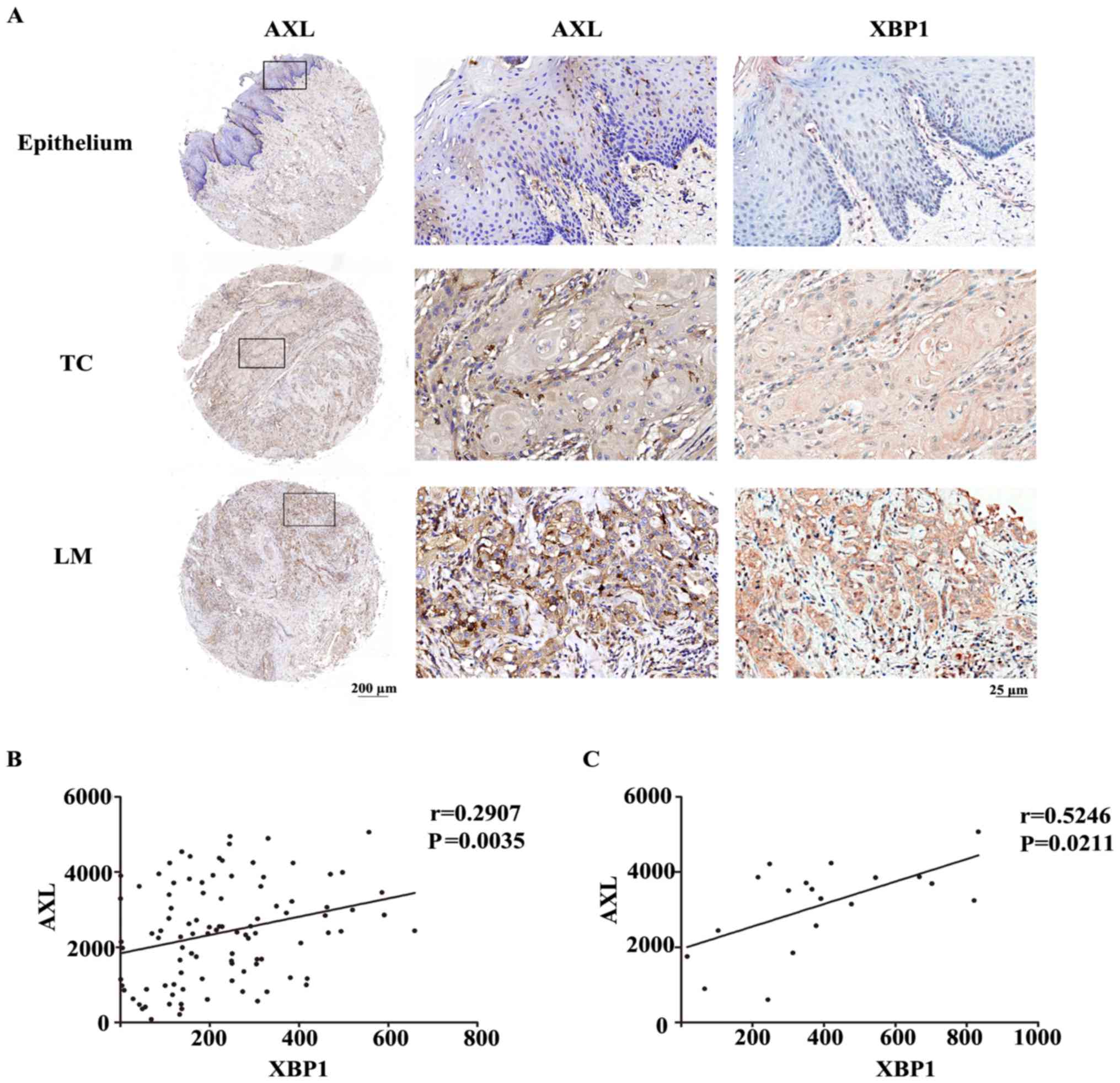
an increasing tendency, which was consistent with XBP1 (Fig. 6A). To compare the relationship
between the expression of AXL and XBP1 in OSCC, Spearman's
correlation method was used to quantify the degree of linear
association between two variables. The result showed that the
levels of AXL were positively correlated with XBP1 in primary
cancer (Fig. 6B, P=0.0035) and
lymph node metastases (Fig. 6C,
P=0.0211), respectively.
Discussion
XBP1, known as a candidate oncogenic gene, is
overexpressed in certain cancers (13). The expression pattern and oncogenic
function of XBP1 in OSCC is currently unclear. It was reported that
unspliced forms of XBP1 (XBP1-u) mainly existed in the cytoplasm,
and only the functional XBP1-s could translocate into the nucleus
and trigger transcriptional programs by regulating a subset of
genes involved in cancer progression (33). In the present study, XBP1 was
detected overexpressed in both cytoplasm and nuclear in OSCC tumor
cells compared with normal epithelial cells, with immunostaining
being much stronger in nucleus. This result is in accordance with
studies in esophageal SCC (16).
Since the XBP1 antibody we used in this study was able to recognize
both XBP1-s and XBP1-u, the location of XBP1 staining in the
nucleus may indicate that the abundant XBP1 forms were spliced and
activated in tumor cells and that its downstream transcriptional
program may be initiated.
Previous findings have shown that ectopic
overexpression of XBP1 resulted in metastasis in breast cancer
(6,9,17),
esophageal SCC (16), and
colorectal carcinoma (34). In the
present study, we characterized the metastatic function of XBP1 in
OSCC. Our findings demonstrated that XBP1 expression presented a
rising tendency from normal epithelial to cancer metastasis. In
lymph node metastasis, XBP1 showed the highest nuclear staining,
followed by adjacent tumor front, tumor center, and adjacent normal
epithelial. Moreover, transformed cells with XBP1 deficiency
dramatically decreased OSCC cell invasion ability in vitro.
These results suggest the effective function of XBP1 in promoting
OSCC invasion and lymph node metastasis.
Recognition of XBP1 target gene or downstream
signaling is of great importance to understand the molecular
mechanisms of XBP1-mediated tumorigenesis. It is demonstrated that
XBP1 controls specific transcriptional signaling in specific cancer
types. For example, XBP1 is reported to regulate the HIF1α pathway
in breast cancer (9), MMP9 in
esophageal squamous cell carcinoma (16), β-catenin in bladder cancer (11), and the PI3K/mTOR pathway in
osteosarcoma (13), which are
involved in cell proliferation, invasion and survival. The
relationship between XBP1 and AXL was previously reported in breast
cancer cells according to deep sequence data (9). However, to the best of our knowledge,
no study has clarified the mechanism as to how XBP1 could directly
control AXL expression. In our study, AXL was confirmed to be a
novel downstream signaling of XBP1 in OSCC. We provide evidence
that the expression of AXL gene decreased at mRNA and
protein levels when XBP1 expression was suppressed in OSCC cells.
Conversely, activation of XBP1 by Tg immediately rescued the
expression of AXL. Our result demonstrates AXL may be essentially
involved in XBP1-mediated transcriptional events in OSCC. In
addition, we demonstrated that the expression of AXL correlated
with XBP1 positively in OSCC tumor center and lymph node
metastasis. To better understand the downstream signal regulation
of XBP1, further efforts are required to elucidate more detailed
regulation mechanism between XBP1 and AXL.
AXL is detected as an essential biomarker in cancer
progression associated with metastasis (22,35,36),
overall survival (18), and
apoptosis (37). A crucial
mechanism reported in endometrial cancer (35) is that AXL induces MMPs and uPA
expression through the PI3K/AKT signaling pathway, thereby,
promoting tumor metastasis. Several MMP genes have been identified
to be regulated by AXL signaling which contribute to tumor invasion
and metastasis. MMP2 and MMP9 are by far the most commonly reported
tumor invasion-related genes associated with AXL pathway in the MMP
family (38,39). Besides these, MMP3 is activated by
AXL in metastatic head and neck cancer (24). Moreover, both MMP1 and MMP3 were
regulated by AXL, resulting endometrial cancer metastasis (35). uPA is another crucial factor
involved in extracellular matrix degradation through AXL signaling
(35), which plays a major role in
metastasis of cancer cells, by mediating directed extracellular
proteolysis on the surface of migrating or invading cells. In the
present study, we found that once XBP1 was suppressed in OSCC cell
lines, several downstream molecules of AXL signaling, such as PI3K,
MMP1, MMP3 and uPA, were significantly decreased, while MMP2 and
MMP9 remained unchanged. Moreover, once XBP1 was re-activated in
XBP1 knockdown cells, the expression levels of MMP1 and MMP3 were
increased significantly. Thus, AXL as well as the downstream
molecules PI3K, MMP1, MMP3 and uPA, may be closely associated with
XBP1-initiated OSCC metastasis. On the other hand, although AXL has
been reported to promote tumor metastasis via EMT in breast cancer
(18), we were not able to
demonstrate the effect of XBP1 on EMT-associated factors in XBP1
defection OSCC cell lines. This indicates that XBP1 is not capable
of significantly triggering tumor EMT phenotype in OSCC.
XBP1 was reported to be significantly correlated
with clinical outcome in various tumors, such as human osteosarcoma
(13), multiple myeloma (40), diffuse large B-cell lymphomas
(41) and breast carcinoma
(9). Therefore, the prognosis of
patients with OSCC as well as the clinical pathology factors was
taken into consideration in our study. After more than 5 years of
follow-up, the findings of this study have shown that a relatively
high XBP1 expression predicted unfavorable overall survival in OSCC
patients. Moreover, XBP1 tends to show a higher expression in
poorly differentiated tumor cells and advanced clinical stages.
Thus, XBP1 is a potential survival factor in predicting OSCC
patient outcome.
In summary, the above results demonstrate that XBP1
is a novel survival marker of OSCC, which was closely associated
with tumor lymph node metastasis, advanced clinical stages, poor
pathological differentiation and unfavorable prognosis. Aberrant
XBP1 expression has a powerful effect on tumor invasion and
apoptosis, and strongly modulates AXL signaling.
Acknowledgements
We would like to thank Shichun Xiong and Yuan Li of
Wuhan University for their technical assistance and Professor T.E
Carey of Michigan University for presenting UM-SCC-23 cell
lines.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81572664).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YS performed the construction of plasmid, verified
the knockdown efficiency, conducted the invasion assay and examined
the genes' expression downstream of XBP1; YS was a major
contributor in acquisition of data and writing the manuscript; FJ
participated in the construction of the TMA and performed IHC as
well as the patients follow-up; YP undertook the task of cell
culture; XC contributed to verify the histological types and tumor
grades of tumor sample; JC shared the work of construction of TMA
and patients follow-up; YW analyzed the data; XZ took part in the
image processing; JZ made substantial contributions to the
conception and design and to the acquisition of funding. JZ also
contributed to the writing of the manuscript as well as the TMA
construction. All authors read and approved the final manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Ethics approval from the Institutional Review Board
of School and Hospital of Stomatology, Wuhan University was
received for the examination of patient samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fan S, Tang QL, Lin YJ, Chen WL, Li JS,
Huang ZQ, Yang ZH, Wang YY, Zhang DM, Wang HJ, et al: A review of
clinical and histological parameters associated with contralateral
neck metastases in oral squamous cell carcinoma. Int J Oral Sci.
3:180–191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Russo D, Merolla F, Mascolo M, Ilardi G,
Romano S, Varricchio S, Napolitano V, Celetti A, Postiglione L, Di
Lorenzo PP, et al: FKBP51 Immunohistochemical expression: A new
prognostic biomarker for OSCC? Int J Mol Sci. 18:pii. E4432017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lingen MW, Kalmar JR, Karrison T and
Speight PM: Critical evaluation of diagnostic aids for the
detection of oral cancer. Oral Oncol. 44:10–22. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arellano-Garcia ME, Hu S, Wang J, Henson
B, Zhou H, Chia D and Wong DT: Multiplexed immunobead-based assay
for detection of oral cancer protein biomarkers in saliva. Oral
Dis. 14:705–712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Andres SA and Wittliff JL: Relationships
of ESR1 and XBP1 expression in human breast carcinoma and stromal
cells isolated by laser capture microdissection compared to intact
breast cancer tissue. Endocrine. 40:212–221. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bright MD, Itzhak DN, Wardell CP, Morgan
GJ and Davies FE: Cleavage of BLOC1S1 mRNA by IRE1 Is sequence
specific, temporally separate from XBP1 splicing, and dispensable
for cell viability under acute endoplasmic reticulum stress. Mol
Cell Biol. 35:2186–2202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Xing P, Cui W, Wang W, Cui Y, Ying
G, Wang X and Li B: Acute endoplasmic reticulum stress-independent
unconventional splicing of XBP1 mRNA in the nucleus of mammalian
cells. Int J Mol Sci. 16:13302–13321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen X, Iliopoulos D, Zhang Q, Tang Q,
Greenblatt MB, Hatziapostolou M, Lim E, Tam WL, Ni M, Chen Y, et
al: XBP1 promotes triple-negative breast cancer by controlling the
HIF1alpha pathway. Nature. 508:103–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu H, Abulimiti M, Liu H, Su XJ, Liu CH
and Pei HP: RITA enhances irradiation-induced apoptosis in
p53-defective cervical cancer cells via upregulation of
IRE1alpha/XBP1 signaling. Oncol Rep. 34:1279–1288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen W, Zhou J, Wu K, Huang J, Ding Y, Yun
EJ, Wang B, Ding C, Hernandez E, Santoyo J, et al: Targeting
XBP1-mediated β-catenin expression associated with bladder cancer
with newly synthetic Oridonin analogues. Oncotarget. 7:56842–56854.
2016.PubMed/NCBI
|
|
12
|
Bae J, Samur M, Munshi A, Hideshima T,
Keskin D, Kimmelman A, Lee AH, Dranoff G, Anderson KC and Munshi
NC: Heteroclitic XBP1 peptides evoke tumor-specific memory
cytotoxic T lymphocytes against breast cancer, colon cancer, and
pancreatic cancer cells. Oncoimmunology. 3:e9709142014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang J, Cheng D, Zhou S, Zhu B, Hu T and
Yang Q: Overexpression of X-Box binding protein 1 (XBP1) correlates
to poor prognosis and up-regulation of PI3K/mTOR in human
osteosarcoma. Int J Mol Sci. 16:28635–28646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Davies MP, Barraclough DL, Stewart C,
Joyce KA, Eccles RM, Barraclough R, Rudland PS and Sibson DR:
Expression and splicing of the unfolded protein response gene XBP-1
are significantly associated with clinical outcome of
endocrine-treated breast cancer. Int J Cancer. 123:85–88. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mhaidat NM, Alzoubi KH and Abushbak A:
X-box binding protein 1 (XBP-1) enhances colorectal cancer cell
invasion. J Chemotherapy. 27:167–173. 2015. View Article : Google Scholar
|
|
16
|
Xia T, Tong S, Fan K, Zhai W, Fang B, Wang
SH and Wang JJ: XBP1 induces MMP-9 expression to promote
proliferation and invasion in human esophageal squamous cell
carcinoma. Am J Cancer Res. 6:2031–2040. 2016.PubMed/NCBI
|
|
17
|
Li H, Chen X, Gao Y, Wu J, Zeng F and Song
F: XBP1 induces snail expression to promote
epithelial-to-mesenchymal transition and invasion of breast cancer
cells. Cell Signal. 27:82–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gjerdrum C, Tiron C, Høiby T, Stefansson
I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT,
et al: Axl is an essential epithelial-to-mesenchymal
transition-induced regulator of breast cancer metastasis and
patient survival. Proc Natl Acad Sci USA. 107:1124–1129. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Jia L, Ren D, Liu C, Gong Y, Wang N,
Zhang X and Zhao Y: Axl mediates tumor invasion and
chemosensitivity through PI3K/Akt signaling pathway and is
transcriptionally regulated by slug in breast carcinoma. IUBMB
Life. 66:507–518. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee CH, Liu SY, Chou KC, Yeh CT, Shiah SG,
Huang RY, Cheng JC, Yen CY and Shieh YS: Tumor-associated
macrophages promote oral cancer progression through activation of
the Axl signaling pathway. Ann Surg Oncol. 21:1031–1037. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rankin EB and Giaccia AJ: The receptor
tyrosine kinase AXL in cancer progression. Cancers (Basel). 8:pii:
E103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Jia L, Liu C, Gong Y, Ren D, Wang N,
Zhang X and Zhao Y: Axl as a downstream effector of TGF-β1 via
PI3K/Akt-PAK1 signaling pathway promotes tumor invasion and
chemoresistance in breast carcinoma. Tumor Biol. 36:1115–1127.
2015. View Article : Google Scholar
|
|
23
|
Xu J, Jia L, Ma H, Li Y, Ma Z and Zhao Y:
Axl gene knockdown inhibits the metastasis properties of
hepatocellular carcinoma via PI3K/Akt-PAK1 signal pathway. Tumor
Biol. 35:3809–3817. 2014. View Article : Google Scholar
|
|
24
|
Xu H, Chen X, Huang J, Deng W, Zhong Q,
Yue C, Wang P and Huang Z: Identification of GPR65, a novel
regulator of matrix metalloproteinases using high through-put
screening. Biochem Biophys Res Commun. 436:96–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brenner JC, Graham MP, Kumar B, Saunders
LM, Kupfer R, Lyons RH, Bradford CR and Carey TE: Genotyping of 73
UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck.
32:417–426. 2010.PubMed/NCBI
|
|
26
|
Zhang J, Wang Y, Chen X, Zhou Y, Jiang F,
Chen J, Wang L and Zhang WF: MiR-34a suppresses amphiregulin and
tumor metastatic potential of head and neck squamous cell carcinoma
(HNSCC). Oncotarget. 6:7454–7469. 2015.PubMed/NCBI
|
|
27
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma SR, Wang WM, Huang CF, Zhang WF and Sun
ZJ: Anterior gradient protein 2 expression in high grade head and
neck squamous cell carcinoma correlated with cancer stem cell and
epithelial mesenchymal transition. Oncotarget. 6:8807–8821. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng W, Wang Y, Liu Z, Cheng H and Xue Y:
HemI: A toolkit for illustrating heatmaps. PLoS One. 9:e1119882014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
van Schadewijk A, van't Wout EF, Stolk J
and Hiemstra PS: A quantitative method for detection of spliced
X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic
reticulum (ER) stress. Cell Stress Chaperones. 17:275–279. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Corno C, Gatti L, Lanzi C, Zaffaroni N,
Colombo D and Perego P: Role of the receptor tyrosine kinase Axl
and its targeting in cancer cells. Curr Med Chem. 23:1496–1512.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Castillo-Carranza DL, Zhang Y,
Guerrero-Muñoz MJ, Kayed R, Rincon-Limas DE and Fernandez-Funez P:
Differential activation of the ER stress factor XBP1 by oligomeric
assemblies. Neurochem Res. 37:1707–1717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoshida H, Oku M, Suzuki M and Mori K:
pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded
protein response activator pXBP1(S) in mammalian ER stress
response. J Cell Biol. 172:565–575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin C, Jin Z, Chen NZ, Lu M, Liu CB, Hu WL
and Zheng CG: Activation of IRE1α-XBP1 pathway induces cell
proliferation and invasion in colorectal carcinoma. Biochem Bioph
Res Commun. 470:75–81. 2016. View Article : Google Scholar
|
|
35
|
Divine LM, Nguyen MR, Meller E, Desai RA,
Arif B, Rankin EB, Bligard KH, Meyerson C, Hagemann IS, Massad M,
et al: AXL modulates extracellular matrix protein expression and is
essential for invasion and metastasis in endometrial cancer.
Oncotarget. 7:77291–77305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mudduluru G, Vajkoczy P and Allgayer H:
Myeloid zinc finger 1 induces migration, invasion, and in vivo
metastasis through Axl gene expression in solid cancer. Mol Cancer
Res. 8:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cho CY, Huang JS, Shiah SG, Chung SY, Lay
JD, Yang YY, Lai GM, Cheng AL, Chen LT and Chuang SE: Negative
feedback regulation of AXL by miR-34a modulates apoptosis in lung
cancer cells. RNA. 22:303–315. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han J, Tian R, Yong B, Luo C, Tan P, Shen
J and Peng T: Gas6/Axl mediates tumor cell apoptosis, migration and
invasion and predicts the clinical outcome of osteosarcoma
patients. Biochem Bioph Res Commun. 435:493–500. 2013. View Article : Google Scholar
|
|
39
|
Chiu KC, Lee CH, Liu SY, Yeh CT, Huang RY,
Yuh DY, Cheng JC, Chou YT and Shieh YS: Protumoral effect of
macrophage through Axl activation on mucoepidermoid carcinoma. J
Oral Pathol Med. 43:538–544. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen L, Li Q, She T, Li H, Yue Y, Gao S,
Yan T, Liu S, Ma J and Wang Y: IRE1α-XBP1 signaling pathway, a
potential therapeutic target in multiple myeloma. Leuk Res.
49:7–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Balague O, Mozos A, Martinez D, Hernandez
L, Colomo L, Mate JL, Teruya-Feldstein J, Lin O, Campo E,
Lopez-Guillermo A, et al: Activation of the endoplasmic reticulum
stress-associated transcription factor × box-binding protein-1
occurs in a subset of normal germinal-center B cells and in
aggressive B-cell lymphomas with prognostic implications. Am J
Pathol. 174:2337–2346. 2009. View Article : Google Scholar : PubMed/NCBI
|