Introduction
Liver cancer, including hepatocellular carcinoma
(HCC) and cholangiocarcinoma (CCA) in adults, and hepatoblastoma
(HB) mainly in children, is one of the most malignant tumor types
with poor response to drugs used at present against cancer
(1–4). Traditional treatments for liver cancer
include surgery, radiofrequency ablation and chemotherapy. However,
liver cancer is usually diagnosed at an advanced stage and the
patients therefore miss the opportunity for surgical resection.
Systemic chemotherapy via or trans-arterial chemoembolization is
the second line of treatment, but the overall response rate is
rather low due to the high chemoresistance of liver cancer cells
(5,6). Therefore, dissecting the underlying
mechanisms of liver cancer development and progression is essential
for developing new therapeutic drugs and strategies.
HMBOX1 (homeobox containing 1), a novel human
homeobox gene, was first isolated from the human pancreatic cDNA
library. HMBOX1 has the atypical homeobox domain with 78 amino
acids and a putative HNF1-N domain, and is classified into the HNF
homeobox class of the Hmbox family (7–9).
Recently, several studies have reported the possible biological
functions of HMBOX1. Su et al demonstrated that HMBOX1 was
the key factor in the differentiation of bone marrow stromal cells
(BMSCs) to endothelial cells (ECs) by regulating IP-10 and Ets-1
(10). Ma et al reported
that HMBOX1 regulated intracellular free zinc levels by interacting
with MT2A, inhibiting apoptosis and promoting autophagy of VECs
(11). In addition, HMBOX1 could
directly bind to telomeric double-stranded DNA and helped in
telomere maintenance in cells with ALT (alternative lengthening of
telomeres) (12).
In our previous study, we found that HMBOX1 acted as
a transcriptional repressor of interferon γ (IFN-γ) in natural
killer (NK) cells (13,14). Furthermore, HMBOX1 was localized in
both the cytoplasm and nucleus, and distributed widely in many
tissues, including the liver. Notably, HMBOX1 was expressed in
significantly lower levels in hepatic carcinoma tissues compared to
adjacent healthy tissues (8,15).
Therefore, HMBOX1 may play a role in the progression of liver
cancer, although the exact biological function of HMBOX1 in liver
cancer is still unknown.
In this study, we demonstrated that the expression
level of HMBOX1 was negatively associated to the differentiation
degree and clinical stage of liver cancer. Furthermore, we also
revealed that HMBOX1 could significantly enhance autophagy and
downregulate the ‘stemness’ genes in liver cancer cells. Notably,
HMBOX1 overexpression increased the sensitivity of tumor cells to
NK cell cytolysis and increased NK function. These results
indicated that HMBOX1 may be a novel therapeutic target for human
liver cancer that is worth investigating.
Materials and methods
Cell lines and cell culture
The human natural killer cell line NK-92 was
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA) and maintained in a-MEM supplemented with 12.5%
horse serum (both from Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 12.5% fetal bovine serum (FBS), 100 U/ml rhIL-2,
0.1 mM P-mercaptoethanol and 0.02 mM folic acid. Human liver cancer
cell line HepG2 (HCC/HB) (16),
human hepatoma cell line PLC/PRF/5, human immortalized liver cell
line HL7702 and murine hepatoma cell line Hepa1-6 were purchased
from the Cell Bank of Type Culture Collection of Chinese Academy of
Sciences (Shanghai, China) and maintained in RPMI-1640 medium
supplemented with 10% FBS. All these cell lines were conserved in
our laboratory.
Liver cancer specimens
Paraffin sections from 14 liver specimens of liver
cancer patients, including 10 males and 4 females with the age of
54.6±4.9 years old, were procured from Qilu Hospital of Shandong
University (Jinan, China) and written informed consents were
obtained from all patients. The samples were collected from July
2014 to May 2015, and all of them were processed for routine
histological examination and were classified by their degree of
differentiation (poorly, moderately and well-differentiated) and
clinical stage (I to III). The use of the liver specimens was
approved by the Ethics Committee of Qilu Hospital of Shandong
University and was consistent with the standards established by the
Declaration of Helsinki (as revised in Fortaleza, Brazil, October
2013).
Animal model
Dimethylnitrosamine (DEN, 25 mg/kg) were injected
intraperitoneally into 2-week-old male C57BL/6 mice (n=10, 6.0–6.5
g) once a week for three times total, followed by 500 µl/kg Carbon
tetrachloride (CCL4) once a week for 12 times total. Mice at 42
weeks old were anesthetized by lidocaine (100 mg/kg)
intraperitoneally. Mice with visible tumors (≥0.5 mm) on the
surface of the liver were used in this study. Tumor tissues and
age-matched healthy liver tissues were collected and either fixed
in 4% paraformaldehyde for further analysis.
Animal experiment protocols were approved by the
Ethics Committee of Shandong University. Mice were housed in a
temperature-controlled pathogen-free environment on a 12-h
light/12-h dark cycle, and had access to food and water ad
libitum.
Immunohistochemistry
After preconditioning, the sections were incubated
overnight with a primary antibody in a humidified chamber at 4°C.
Anti-HMBOX1 mAb (dilution 1:100; cat. no. 2A5F4 mAb) as previously
described (8) was used as the
primary HMBOX1 antibody. After washing with PBS, the sections were
incubated with anti-mouse secondary antibody with no dilution (cat.
no. SP-9002; Zsbio, Beijing, China) for 30 min at 37°C. The
sections were washed again with phosphate-buffered saline (PBS) and
stained with DAB solution (Zsbio) according to the manufacturer's
protocol, and the nuclei were dyed with hematoxylin. PBS was used
in place of the primary antibody as a negative control.
Immunofluorescence
HepG2 cells transfected with pEGFP-N1-HMBOX1 or
siRNA-HMBOX1 were cultured in 96-well plates for 6 h. After a
30-min fixation with 1% paraformaldehyde, the cells were blocked
with 5% BSA at room temperature and incubated overnight with an
LC3B antibody (dilution 1:500; cat. no. 2775; Cell Signaling
Technology, Danvers, MA, USA) in a humidified chamber at 4°C. The
cells were then washed with PBS and incubated with an anti-rabbit
secondary antibody (dilution 1:200; cat. no. 8889; Cell Signaling
Technology) for 1 h at 37°C in the dark. The nuclei were
counterstained with DAPI and fluorescently-labeled cells were
visualized at the requisite excitation wavelength (LC3B: 594 nm;
DAPI: 340 nm) under a fluorescence microscope (Olympus TH4-200;
Olympus Corp., Tokyo, Japan) at ×400 magnification
Transfection and RNA interference
Cells were transfected with either specific siRNA
against HMBOX1 or the negative control using Lipofectamine™ 2000
(Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The siRNAs (Target
sequence: GGAAGTTCATATGGGAATA) were designed and synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Cytotoxicity assay
The cytotoxic activity of NK-92 cells against HepG2
cells was assessed by MTT assay. HepG2 cells transfected with
different plasmids were seeded in 96 well plates at
1×104 cells/well 24 h after transfection. NK92 cells
were added at a ratio of 10:1, 5:1 or 2.5:1, and incubated for an
additional 6 h. MTT (Sigma-Aldrich, Merck; St. Louis, MO, USA)
solution was prepared at 10 mg/ml and 20 µl was added to the
medium. The cells were incubated for another 4 h and the absorbance
was determined at 570/630 nm using a multifunctional microplate
reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
percentage of cytotoxicity was calculated by the formula: lysis (%)
= 1 - (ODE+T - ODE)/ODT × 100%; E,
effector cell group; T, target cell group; E+T, effector cell group
and target cell group.
Quantitative real-time PCR
analysis
Total RNA was extracted from cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
used to synthesize cDNA using M-MLV Reverse Transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol.
The specific transcripts were detected via Real-time
PCR (qRT-PCR) with SYBR Green Master Mix (Toyobo, Osaka, Japan)
using an iCycleriQ Real-Time PCR system (Bio-Rad Laboratories,
Inc.). The expression of specific transcripts was normalized to
GAPDH levels. The qRT-PCR began with a step of denaturation (95°C
for 25 sec), annealing (60°C for 20 sec) and extension (72°C for 30
sec) by 35 cycles. For the conventional RT-PCR, it began with a
step of denaturation (95°C for 30 sec), annealing (60°C for 30 sec)
and extension (72°C for 60 sec) by 35 cycles, and the production
was separated by Gel electrophoresis (1.5%) with ethidium bromide.
ImageJ software (version 1.4.3.67; National Institutes of Health,
Bethesda, MD, USA) was used for densitometry analysis. Forward and
reverse primers were shown in Table
1.
 | Table I.Sequences of forward and reverse
primers. |
Table I.
Sequences of forward and reverse
primers.
| Gene | Squence of primers
(5′-3′) |
|---|
| HMBOX1 | Forward:
AACCCTGGCGCTACACTAAG |
|
| Reverse:
TCCTTTCTCCAGGTAAATCGAC |
| CD133 | Forward:
ACATGAAAAGACCTGGGGG |
|
| Reverse:
GATCTGGTGTCCAGCATG |
| Klf4 | Forward:
GCGGCAAAACCTACACAAAG |
|
| Reverse:
CCCCGTGTGTTTACGGTAGT |
| NANOG | Forward:
TTTGTGGGCCTGAAGAAAACT |
|
| Reverse:
AGGGCTGTCCTGAATAAGCAG |
| Sox2 | Forward:
GCGAACCATCTCTGTGGTCT |
|
| Reverse:
GGAAAGTTGGGATCGAACAA |
| ESG | Forward:
GCGCAGTATCACAGCCTTAAA |
|
| Reverse:
TCAATCTCTTGGCGATTTCA |
| APOA1BP | Forward:
TTCAGCGTGGACCAACTTATG |
|
| Reverse:
GGCCTTTTGGGGTAATAGATGG |
| CYP2W1 | Forward:
CCGATTTGACTACCGGGACC |
|
| Reverse:
CGTCCACATAGCTGCACAC |
| CYP46A1 | Forward:
GTGCGCTCCAGACTGTGTTT |
|
| Reverse:
CCAGGTCTATGACTCTCCGCT |
| CYP26B1 | Forward:
TGGACCTCCTCATTGAGAGCA |
|
| Reverse:
GGCATAGGCCGCAAAGATCA |
| HPD | Forward:
GAACCTCTAGCCTACAGGGG |
|
| Reverse:
TCTTTGTTCCAGGGGTTGAGC |
| Fas | Forward:
TCTGGTTCTTACGTCTGTTGC |
|
| Reverse:
CTGTGCAGTCCCTAGCTTTCC |
| GAPDH | Forward:
GAAGGTGAAGGTCGGAGT |
|
| Reverse:
CATGGGTGGAATCATATTGGAA |
Western blotting
Total protein was extracted from cells lysed with a
RIPA lysis buffer [50 mM Tris-HCl (pH 8.0), 1% NP-40, 0.5% sodium
deoxycholate, 150 mM NaCl and 1 mM PMSF], and the concentrations
were determined with BSA methods. The proteins (30 µg/lane) were
separated by SDS-PAGE on a 10% polyacrylamide gel and then
transferred onto polyvinylidene difluoride membranes (PVDF) (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 5%
non-fat milk in TBS/0.1% Tween-20 for 1 h at room temperature. Then
the protein were probed with specific antibodies (1:1,000):
anti-HMBOX1 (cat. no. ab97643; Abcam, Cambridge, UK), anti-p38 and
anti-p-p38 (cat. nos. 8690 and 4511; Cell Signaling Technology),
anti-AKT and anti-p-AKT (cat. nos. 4685 and 4060; Cell Signaling
Technology), anti-mTOR and anti-p-mTOR (cat. nos. ab32028 and
ab109268; Abcam), anti-LC3 (cat. no. 2775; Cell Signaling
Technology) and β-actin (cat. no. sc-58673; Santa Cruz
Biotechnology). The protein were incubated with secondary antibody
(dilution 1:1,000; cat. nos. A0216 and A0208; Beyotime Institute of
Biotechnology, Shanghai, China) for 1 h at room temperature and the
bands were visualized using Immobilon Western Chemiluminescent HRP
Substrate (EMD Millipore) and analyzed with Alpha Ease FC software
(Bio-Rad Laboratories, Inc.).
Flow cytometry
The cells were phenotypically analyzed by flow
cytometry with the following antibodies (1:10): PE-cy5-labeled
anti-Fas (cat. no. 556641; BD Pharmingen BD Biosciences, San Diego,
CA, USA), PE-conjugated anti-NKG2A (cat. no. FAB1059P; R&D
Systems, Minneapolis, MN, USA), FITC-conjugated anti-NKG2D and
PE-conjugated anti-PD-L1 (cat. nos. 11-5878-73 and 12-5983-42;
eBiosciences, San Diego, CA, USA). The cells were stained with a
saturating amount of the antibodies for 1 h at 4°C. After washing
with PBS, the stained cells were acquired using a FACS Calibur
system (BD Biosciences, San Jose, CA, USA) and analyzed with WinMDI
2.0 software (Scripps Research Institute, La Jolla, CA, USA).
Statistical analyses
All data are presented as the mean ± SD of three or
more independent experiments. Statistical significance was
calculated using Kruskal Wallis test, post hoc tests (Dunn's test,
Mann Whitney U with Bonferroni's correction applied) and Student's
t-test. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
HMBOX1 expression is correlated to the
differentiation of liver cancer
Our previous study (8) revealed that the expression of HMBOX1
in liver cancer was lower than that in adjacent non-cancerous
tissues. To elucidate whether HMBOX1 expression was associated with
the development of liver cancer, tissue specimens with varying
degrees of differentiation and of different clinical stages were
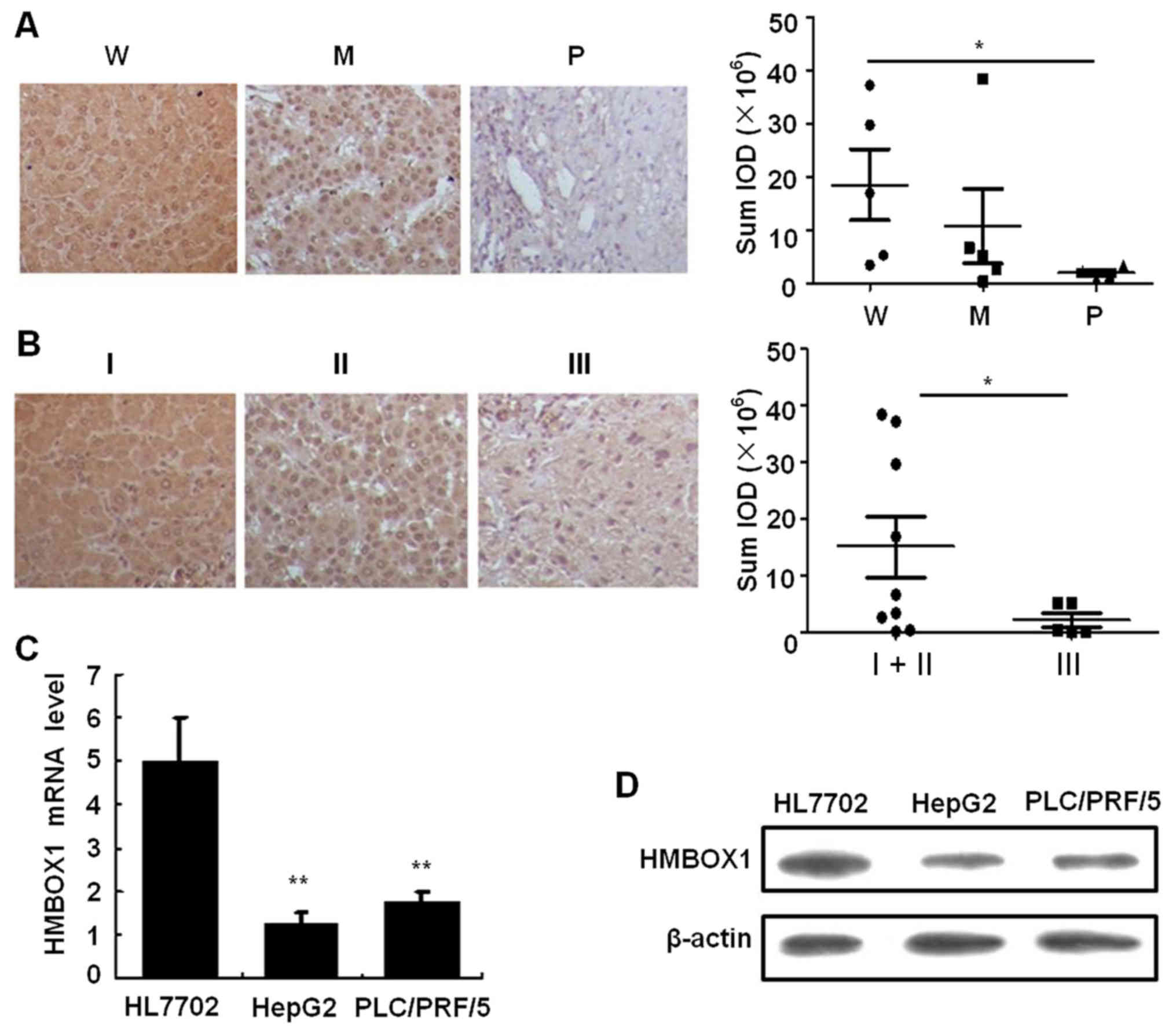
analyzed. As shown in Fig. 1A,
HMBOX1 expression revealed a significant negative association with
the degree of tumor differentiation (P<0.05) as well as the
clinical stage (Fig. 1B).
Furthermore, both mRNA and protein levels of HMBOX1 were suppressed
in the HepG2 and PLC/PRF/5 cell lines compared to the immortalized
HL7702 hepatocyte cell line (Fig. 1C
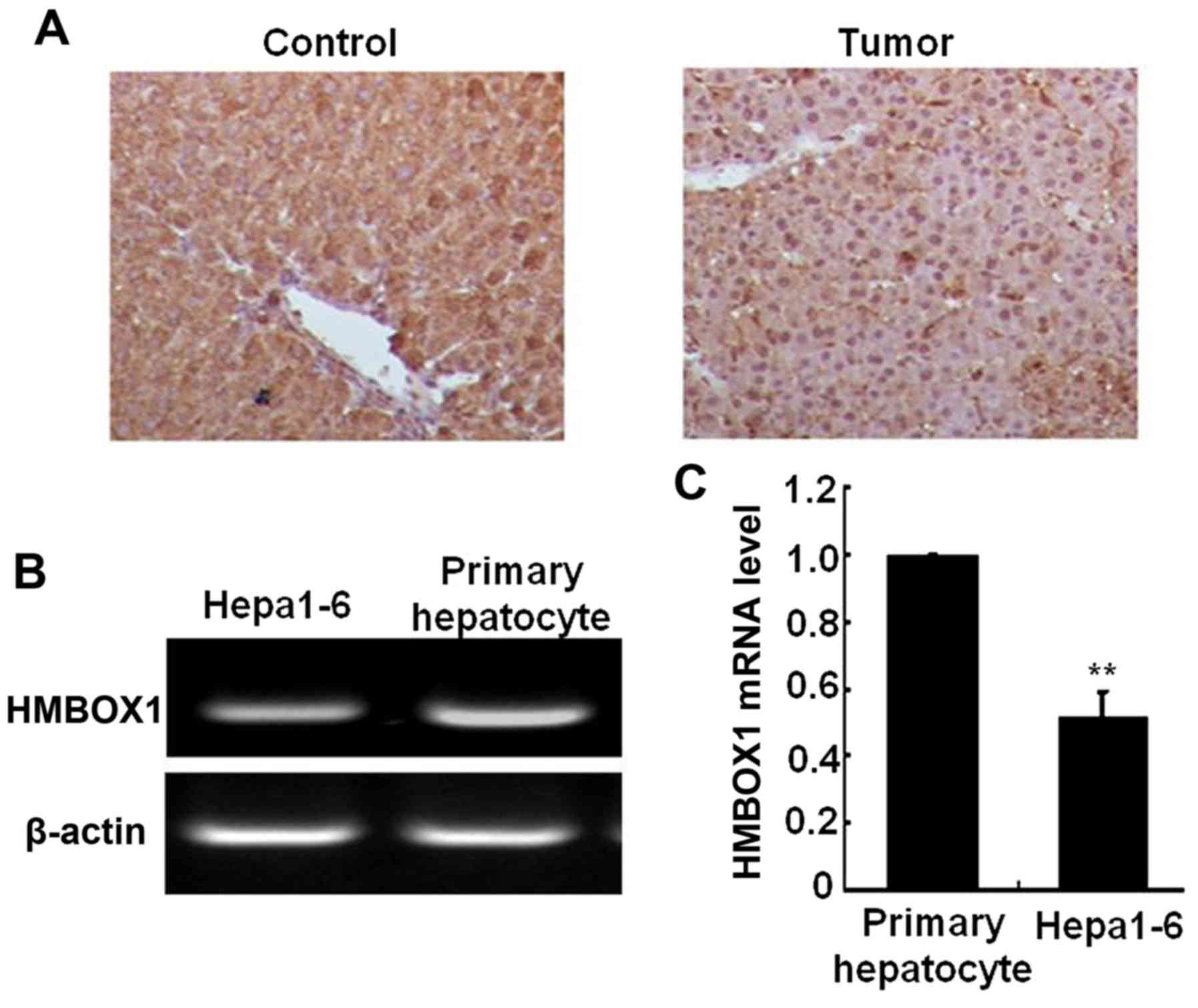
and D). In the mouse liver cancer model induced by DEN and
CCL4, liver HMBOX1 expression level was also
significantly decreased in cancer tissue compared with normal
tissue (Fig. 2A). HMBOX1 expression
in the murine Hepa1-6 cell line was also lower than in primary
murine hepatocytes (Fig. 2B and C).
All these results indicated an important role of HMBOX1 in
hepatocarcinogenesis.
HMBOX1 promotes autophagy in liver
cancer cells
Autophagy is a common tumor suppression mechanism
that prevents cellular damage and maintains cellular homeostasis.
To elucidate the function of HMBOX1 on cell autophagy, HepG2 cells
were transfected with either HMBOX1 siRNA or pEGFP-N1-HMBOX1
plasmids to suppress or overexpress HMBOX1 respectively. After 48 h
of transfection, autophagy was detected by assessing the levels of
autophagic protein, microtubule-associated protein 1 light chain 3B
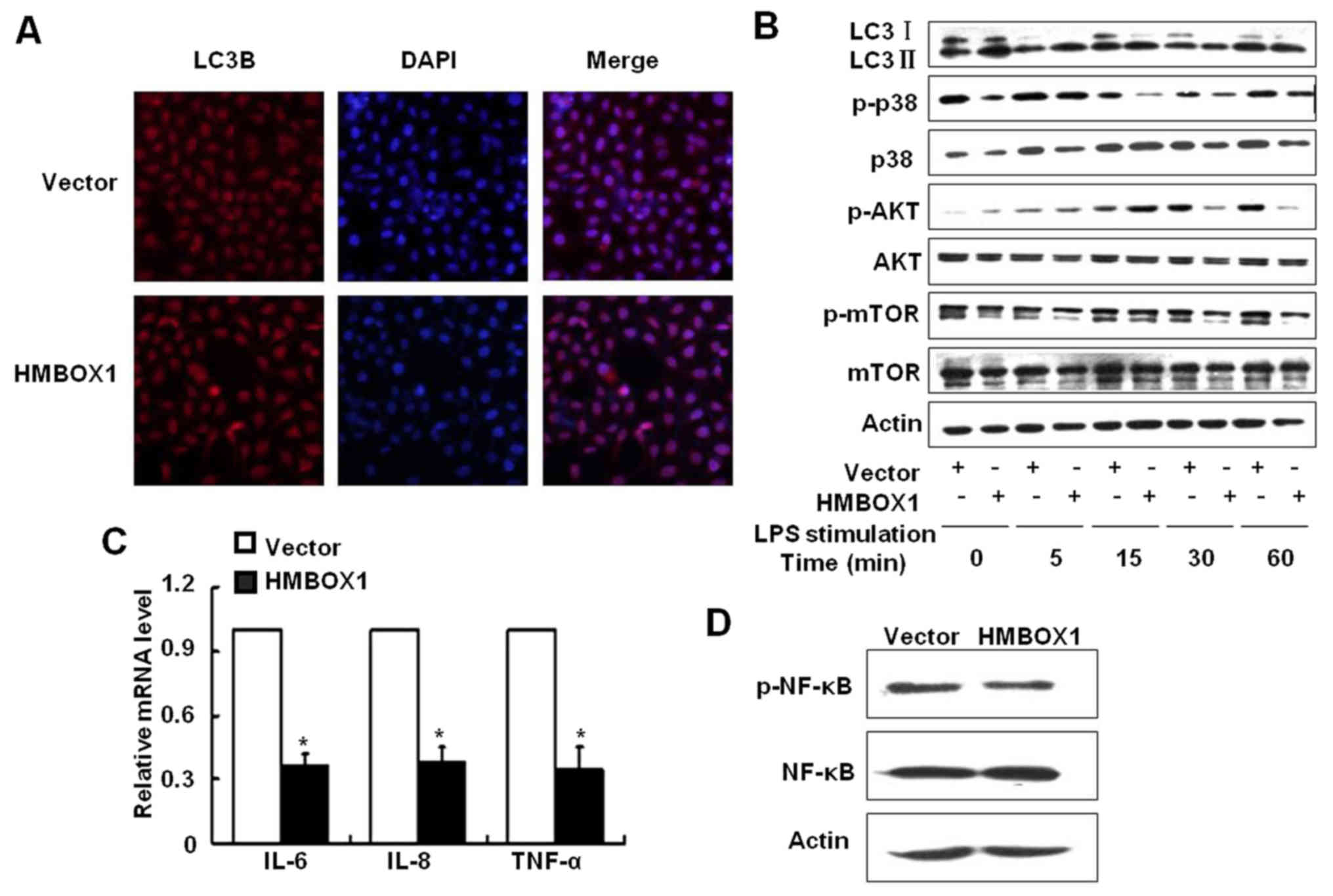
(LC3B). As shown in Fig. 3A, the
expression of LC3B was increased in HepG2 cells overexpressing
HMBOX1. To further validate the effect of HMBOX1 on autophagic
flux, HepG2 cells were stimulated with 10 µg/ml lipopolysaccharide
(LPS) and the LC3 II/LC3 I ratio was analyzed by western blotting.
As shown in Fig. 3B, the LC3 II/LC3
I ratio was upregulated in HMBOX1 overexpressing HepG2 cells,
clearly indicating that HMBOX1 promoted autophagy of HepG2
cells.
Mammalian target of rapamycin (mTOR) prevents
mammalian cell autophagy (17,18),
and is activated by the PI3K/protein kinase B (Akt) pathway kinases
and the p38 mitogen-activated protein kinase (MAPK). HMBOX1
overexpression inhibited the phosphorylation of mTOR as well its
upstream activators p38MAPK and AKT, indicating a possible
underlying mechanism (Fig. 3B).
HMBOX1 overexpression also suppressed the levels of
pro-inflammatory IL-6, IL-8 and TNF-α (Fig. 3C), along with inhibiting NF-κB
activation (Fig. 3D). These results
indicated that HMBOX1 could suppress cancer development by
upregulating autophagy, which inhibited cancer-associated
inflammation.
HMBOX1 inhibits expression of
‘stemness’ genes in liver cancer cells
Considering that HMBOX1 expression is negatively
associated to the differentiation of liver cancer, we hypothesized
that HMBOX1 may affect the expression of genes regulating stemness
and differentiation in liver cancer cells. Cancer stem cells (CSCs)
are a subpopulation of tumor cells which resemble normal stem cells
with respect to their ability to self-renew and differentiate into
multiple cell types (19). CD133, a
CSC marker, is related to tumor initiation and progression, as well
as colony formation ability and differentiation potential of the
cells (13,14). Bahnassy et al reported that
increased expression of CD133 in the liver tumor microenvironment
promoted liver cancer progression (20). CSCs are also known to endogenously
express stemness-related genes like OCT3/4, SOX2, NANOG and KLF4 in
many cancers (21–23). To investigate whether HMBOX1
contributed to the regulation of stemness-related genes, their
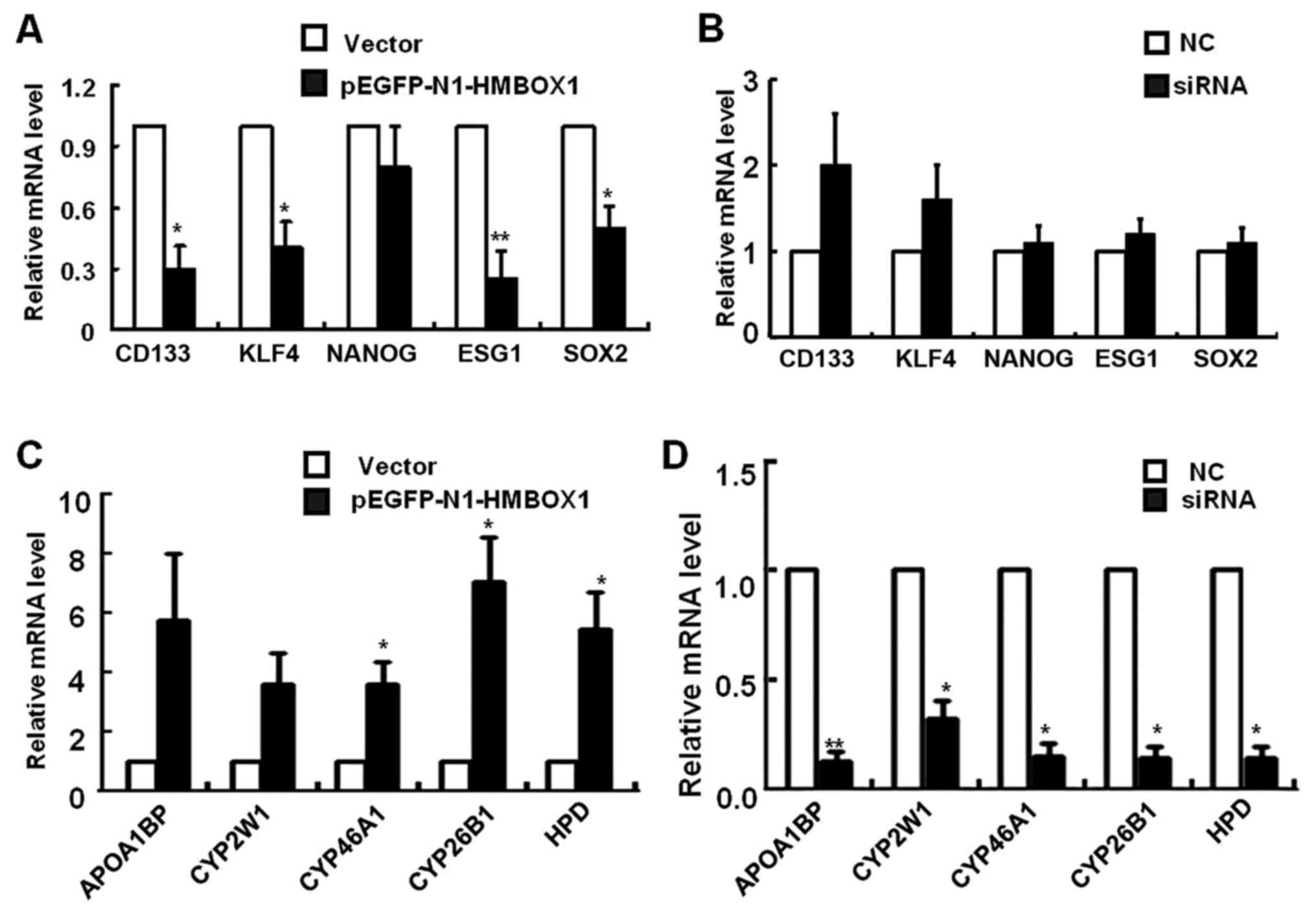
expression was analyzed by real-time PCR. As shown in Fig. 4A, the mRNA levels of CD133, KLF4,
ESG1 and SOX2 were significantly reduced in HepG2 cells 3 days
after pEGFP-N1-HMBOX1 transfection. Contrasting results were
observed after HMBOXI silencing, with an increasing tendency of the
aforementioned genes level, however, no statistical difference was
observed (Fig. 4B).
To gain deeper insights into the regulatory function
of HMBOX1 in liver CSCs, mRNA microarray analysis was performed in
HepG2 cells overexpressing HMBOX1. A cluster of liver
metabolism-related genes was upregulated, including apolipoprotein
A-1 binding protein (APOA1BP), cytochrome P450 proteins (CYP2W1,
CYP46A1 and CYP26B1) and 4-hydroxyphenylpyruvate dioxygenase (HPD)
(Fig. 4C). As anticipated, the mRNA
levels of these genes were significantly downregulated upon HMBOX1
silencing (Fig. 4D). These results
indicated a potential function of HMBOX1 in reversing the phenotype
of liver CSCs to that of normal hepatocytes.
HMBOX1 increases the sensitivity of
liver cancer cells to NK cell-mediated cytolysis
NK cells, the major cellular component of innate
immunity, predominantly reside in the liver (24–28).
Several studies have revealed significantly reduced cytotoxicity of
NK cells obtained from liver perfusates of liver cancer patients
(29), indicating that a functional
defect of NK cells is responsible for the failure of antitumor
immune responses (30). To
elucidate whether HMBOX1 is involved in cancer-induced immune
system escape, the human NK-92 cell line was used as the effector
cell population. After co-incubation with the target HMBOX1
overexpressing or silencing HepG2 cells, NK cell cytotoxicity was
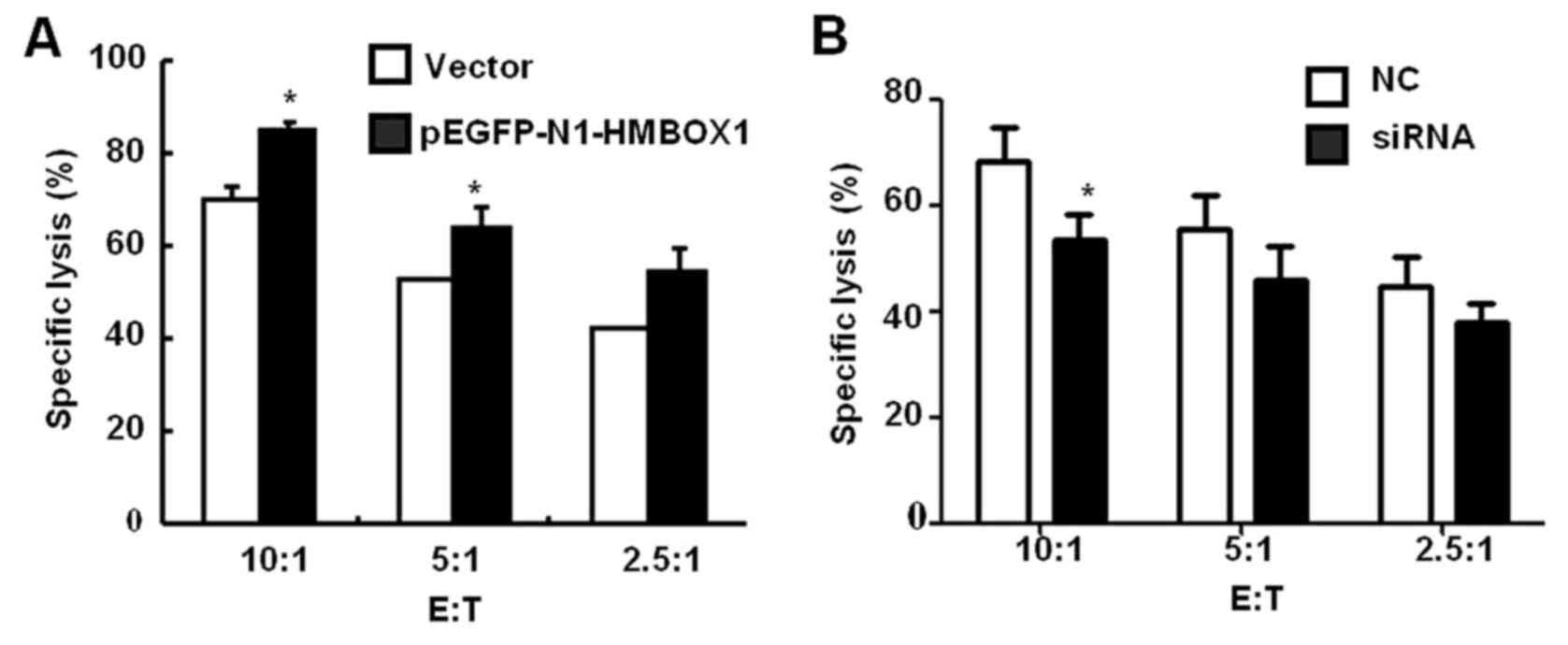
detected by MTT assay. As shown in Fig.
5A, HMBOX1 overexpression increased the sensitivity of HepG2
cells to NK cell cytolysis by 15–25%, while HMBOX1 knockdown
aggravated the resistance of HepG2 cells to NK cell cytolysis
(Fig. 5B). These findings indicated
that low expression of HMBOX1 would help liver cancer cells escape
NK cell-mediated immune surveillance.
HMBOX1 regulates the expression of
genes associated with NK-cell cytolytic activity
The interaction between active/inactive NK cell
receptors and cancer cell surface ligands is the first step of NK
cell-mediated target cell killing. Therefore, we determined whether
HMBOX1 could influence the expression of factors involved in NK
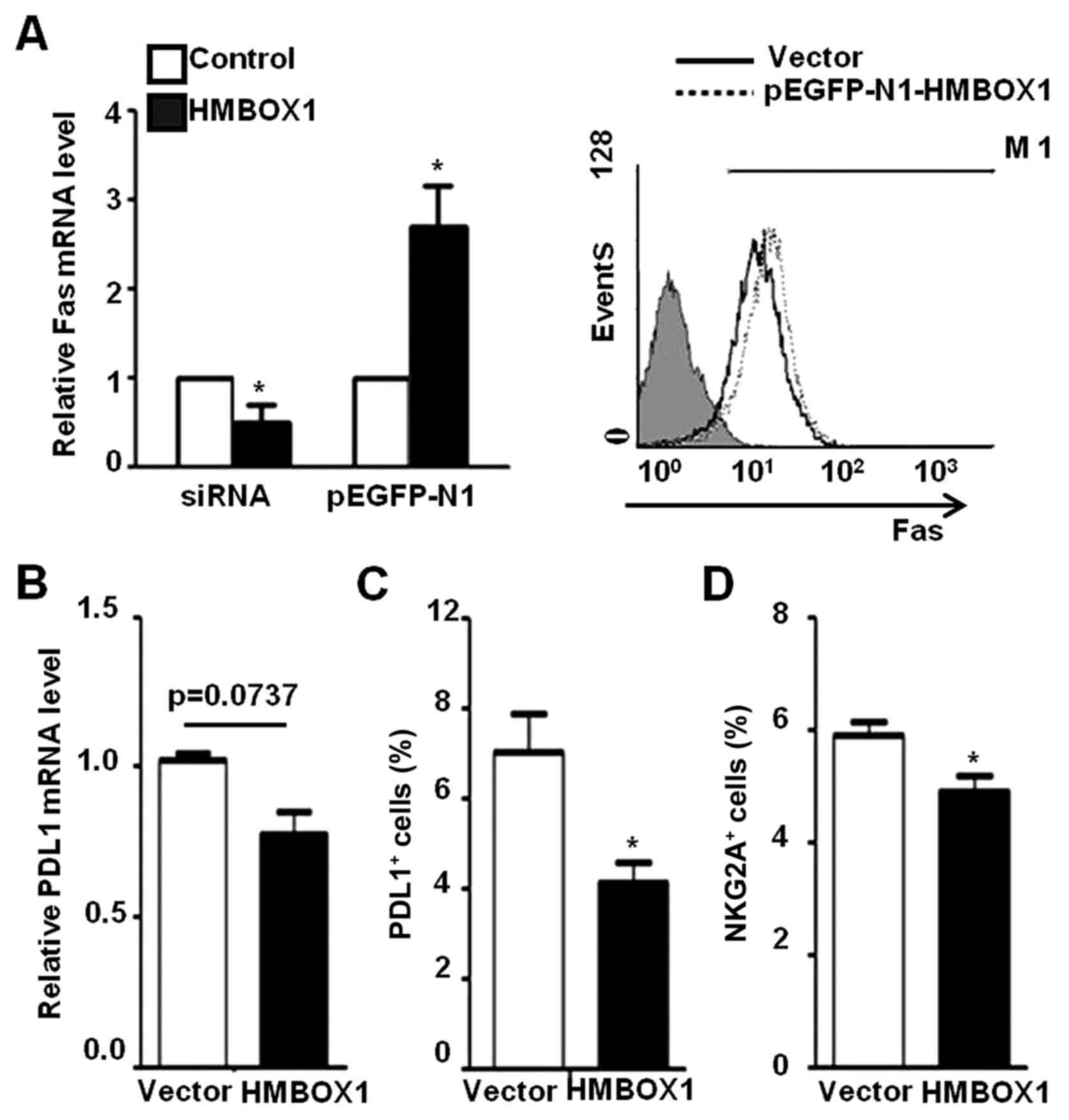
cell-mediated cytotoxicity. As shown in Fig. 6A, Fas expression was increased in
HepG2 cells transfected with pEGFP-N1-HMBOX1, and decreased upon
HMBOX1 silencing by siRNA. In addition, the expression of PD-L1 was
decreased in HepG2 cells transfected with pEGFP-N1-HMBOX1 (Fig. 6B and C). However, no differences
were observed in the expression levels of ULBP2 and HLA-A/B/C, as
well as anti-inflammatory factors IL-10 and TGF-β (data not shown).
In addition, the inhibitory NKG2A receptor expressed on NK-92 cells
was suppressed after co-incubation with HepG2 cells transfected
with pEGFP-HMBOX1 (Fig. 6D) while
no significant changes were observed with the activating receptor
NKG2D (data not shown). These results indicated a protective role
of HMBOX1 against liver cancer development via promotion of NK cell
surveillance.
Discussion
HMBOX1 is a novel transcription repressor whose
structure and function have not been fully characterized. In a
previous study, we found that the expression of HMBOX1 was lower in
liver carcinoma cells (8,15), although the biological significance
was unclear. In the present study, we found a significant negative
association between high HMBOX1 expression and differentiation
degree of liver cancer (Fig. 1),
indicating a role of HMBOX1 in liver cancer development.
Autophagy is a normal cellular process that degrades
dysfunctional and unnecessary components. Studies have revealed a
role of autophagy in various human liver diseases (31–33).
Earlier research demonstrated that autophagy exerts a suppressive
function on tumors. During initial carcinogenesis, autophagy
limited inflammation, p62 accumulation and oxidative stress
response, thereby inhibiting genomic instability by maintaining
cellular metabolic homeostasis (34). In addition, autophagy is known to
inhibit inflammasome activation by removing endogenous sources of
inflammasome agonists (35,36). Autophagy-deficient macrophages have
insufficient microbial clearing capacity, resulting in increased
bacillary burden and excessive pulmonary inflammation characterized
by neutrophil infiltration, IL-17 response and increased IL-1α
levels (37). In the present study,
we found that HMBOX1 overexpression inhibited inflammation and
promoted autophagy via the p38MAPK/AKT/mTOR signaling pathway in
HepG2 cells (Fig. 3). This suggests
that downregulated HMBOX1 would reduce autophagy, thereby
destroying the protective role of the latter in inflammatory
regulation and ultimately leading to the progression of liver
cancer.
Many studies have suggested a hierarchical
organization of heterogeneous cancer cells with a rare subset of
cancer cells with stem cell features, known as cancer stem cells
(CSCs), at the apex. CSCs are defined as a group of cells with high
tumorigenicity, metastasis, and resistance to chemotherapy and
radiation. Furthermore, CSCs have been implicated in tumor relapse
after therapy (38–40). CSCs have also been detected in liver
cancer tissues, and the role of CD133 as a CSC marker in liver
cancer has been confirmed in several studies. CD133-expressing
cancer cells are responsible for tumor initiation or progression,
and display stem-cell-like properties such as colony-forming
ability and multi-potent differentiation potential (41,42).
Notably, stem cell-related genes like CD133, KLF4, ESG1 and SOX2
were inhibited with increased HMBOX1 expression (Fig. 4). Concomitantly, liver
metabolism-related genes like HPD and cytochrome p450 were
upregulated by HMBOX1 overexpression. These results indicated that
HMBOX1 suppressed the stemness of CSCs and maintained the normal
metabolic function of hepatocytes, which is indispensable for
physiological functions of the liver.
The liver is an important organ of the immune system
as it harbors a large number of innate immune cells. Studies have
increasingly revealed that NK cells play a critical role in tumor
immuno-surveillance and act as the first line of the defense
against carcinoma cells. The recognition between the NK cell
receptors and cancer cell surface ligands is the first step in NK
cell-mediated cell killing. Liver cancer can inhibit antitumor
immune responses in the host through various mechanisms (43). We observed that HMBOX1
overexpression improved NK cell-mediated antitumor immune responses
(Fig. 5).
HMBOX1 mediated PD-L1 expression could be the
underlying molecular mechanism which attenuates the
immunosuppressive effect induced by PD-1/PD-L1. In addition, HMBOX1
also increased Fas expression in cancer cells, and consequently
Fas/FasL-mediated apoptosis. Lastly, co-incubation of
HMBOX1-overexpressing HepG2 cells and NK-92 cells downregulated
NKG2A expression of the latter, which indirectly triggered NK
cell-mediated antitumor response (Fig.
6).
Collectively, to the best of our knowledge, we
revealed for the first time that HMBOX1 expression level was
negatively associated with the development of liver cancer. HMBOX1
possesses several protective roles, which include promoting cell
autophagy, downregulating stemness-related genes, increasing HepG2
sensitivity to NK cell cytolysis and enhancing the function of NK
cells, all of which suppress the development of liver cancer. These
findings indicated that the targeted use of HMBOX1 agonists may
enhance the therapeutic effects on liver cancer, and presents a new
perspective on the mechanism of development and clinical therapy of
liver cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (no. 81373222).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and are available from the
corresponding author upon reasonable request.
Authors' contributions
HZ performed the research on cell autophagy,
stemness gene analysis and NK cell cytolytic activity, and was a
major contributor in writing the manuscript. HJ and QH analyzed and
interpreted the liver cancer specimen data and NK cell-mediated
cytolysis. JZ performed the final verification of the manuscript
and was involved in the conception of the study. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The study was performed according to a protocol
approved by the Ethics Committee of Qilu Hospital of Shandong
University and was consistent with the standards established by the
Declaration of Helsinki (as revised in Fortaleza, Brazil, October
2013). Written informed consents were obtained from all patients.
Animal experiment protocols were approved by the Ethics Committee
of Shandong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schütte K, Bornschein J and Malfertheiner
P: Hepatocellular carcinoma-epidemiological trends and risk
factors. Dig Dis. 27:80–92. 2009. View Article : Google Scholar
|
|
2
|
Lv B, Zhang L, Miao R, Xiang X, Dong S,
Lin T, Li K and Kai Q: Comprehensive analysis and experimental
verification of LINC01314 as a tumor suppressor in hepatoblastoma.
Biomed Pharmacother. 98:783–792. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Marin JJG, Briz O, Herraez E, Lozano E,
Asensio M, Di Giacomo S, Romero MR, Osorio-Padilla LM,
Santos-Llamas AI, Serrano MA, et al: Molecular bases of the poor
response of liver cancer to chemotherapy. Clin Res Hepatol
Gastroenterol. 42:182–192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng Z, Li X and Ding J: Characteristics
of liver cancer stem cells and clinical correlations. Cancer Lett.
379:230–238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Llovet JM and Bruix J: Systematic review
of randomized trials for unresectable hepatocellular carcinoma:
Chemoembolization improves survival. Hepatology. 37:429–442. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen S, Saiyin H, Zeng X, Xi J, Liu X, Li
X and Yu L: Isolation and functional analysis of human HMBOX1, a
homeobox containing protein with transcriptional repressor
activity. Cytogenet Genome Res. 114:131–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dai J, Wu L, Zhang C, Zheng X, Tian Z and
Zhang J: Recombinant expression of a novel human transcriptional
repressor HMBOX1 and preparation of anti-HMBOX1 monoclonal
antibody. Cell Mol Immunol. 6:261–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Holland PW, Booth HA and Bruford EA:
Classification and nomenclature of all human homeobox genes. BMC
Biol. 5:472007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su L, Zhao H, Sun C, Zhao B, Zhao J, Zhang
S, Su H and Miao J: The role of Hmbox1 in endothelial
differentiation of bone-marrow stromal cells by a small molecule.
ACS Chem Biol. 5:1035–1043. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma H, Su L, Yue H, Yin X, Zhao J, Zhang S,
Kung H, Xu Z and Miao J: HMBOX1 interacts with MT2A to regulate
autophagy and apoptosis in vascular endothelial cells. Sci Rep.
12:151212015. View Article : Google Scholar
|
|
12
|
Feng X, Luo Z, Jiang S, Li F, Han X, Hu Y,
Wang D, Zhao Y, Ma W, Liu D, et al: The telomere-associated
homeobox-containing protein TAH1/HMBOX1 participates in telomere
maintenance in ALT cells. J Cell Sci. 126:3982–3989. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu L, Zhang C, Zheng X, Tian Z and Zhang
J: HMBOX1, homeobox transcription factor, negatively regulates
interferon-γ production in natural killer cells. Int
Immunopharmacol. 11:1895–1900. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu L, Zhang C and Zhang J: HMBOX1
negatively regulates NK cell functions by suppressing the
NKG2D/DAP10 signaling pathway. Cell Mol Immunol. 8:433–440. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dai J, Zhang C, Tian Z and Zhang J:
Expression profile of HMBOX1, a novel transcription factor, in
human cancers using highly specific monoclonal antibodies. Exp Ther
Med. 2:487–490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
17
|
Hou L, Li Y, Song H, Zhang Z, Sun Y, Zhang
X and Wu K: Protective macroautophagy is involved in vitamin E
succinate effects on human gastric carcinoma cell line SGC-7901 by
inhibiting mTOR axis phosphorylation. PLoS One. 10:e01328292015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang C, Jia X, Wang K, Bao J, Li P, Chen
M, Wan JB, Su H, Mei Z and He C: Polyphyllin VII induces an
autophagic cell death by activation of the JNK pathway and
inhibition of PI3K/AKT/mTOR pathway in HepG2 cells. PLoS One.
11:e01474052016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghoshal S, Fuchs BC and Tanabe KK: STAT3
is a key transcriptional regulator of cancer stem cell marker CD133
in HCC. Hepatobiliary Surg Nutr. 5:201–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bahnassy AA, Fawzy M, El-Wakil M, Zekri
AR, Abdel-Sayed A and Sheta M: Aberrant expression of cancer stem
cell markers (CD44, CD90, and CD133) contributes to disease
progression and reduced survival in hepatoblastoma patients: 4-year
survival data. Transl Res. 165:396–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh U, Quintanilla RH, Grecian S, Gee
KR, Rao MS and Lakshmipathy U: Novel live alkaline phosphatase
substrate for identification of pluripotent stem cells. Stem Cell
Rev. 8:1021–1029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, Li A, Glas M, Lal B, Ying M, Sang Y,
Xia S, Trageser D, Guerrero-Cázares H, Eberhart CG, et al: c-Met
signaling induces a reprogramming network and supports the
glioblastoma stem-like phenotype. Proc Nat Acad Sci USA.
108:9951–9956. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noguchi K, Eguchi H, Konno M, Kawamoto K,
Nishida N, Koseki J, Wada H, Marubashi S, Nagano H, Doki Y, et al:
Susceptibility of pancreatic cancer stem cells to reprogramming.
Cancer Sci. 106:1182–1187. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawasaki A, Shinkai Y, Yagita H and
Okumura K: Expression of perforin in murine natural killer cells
and cytotoxic T lymphocytes in vivo. Eur J Immunol. 22:1215–1219.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Subleski JJ, Hall VL, Back TC, Ortaldo JR
and Wiltrout RH: Enhanced antitumor response by divergent
modulation of natural killer and natural killer T cells in the
liver. Cancer Res. 66:11005–11012. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bai X, Wang S, Tomiyama-Miyaji C, Shen J,
Taniguchi T, Izumi N, Li C, Bakir HY, Nagura T, Takahashi S, et al:
Transient appearance of hepatic natural killer cells with high
cytotoxicity and unique phenotype in very young mice. Scand J
Immunol. 63:275–281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Han Q, Hou Z, Zhang C, Tian Z and
Zhang J: Exosomes mediate hepatitis B virus (HBV) transmission and
NK-cell dysfunction. Cell Mol Immunol. 14:465–475. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu D, Han Q, Hou Z, Zhang C and Zhang J:
miR-146a negatively regulates NK cell functions via STAT1
signaling. Cell Mol Immunol. 14:712–720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishiyama K, Ohdan H, Ohira M, Mitsuta H,
Arihiro K and Asahara T: Difference in cytotoxicity against
hepatocellular carcinoma between liver periphery natural killer
cells in humans. Hepatology. 43:362–372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sung MW, Johnson JT, Van Dongen G and
Whiteside TL: Protective effects of interferon-gamma on
squamous-cell carcinoma of head and neck targets in
antibody-dependent cellular cytotoxicity mediated by human natural
killer cells. Int J Cancer. 66:393–399. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rautou PE, Mansouri A, Lebrec D, Durand F,
Valla D and Moreau R: Autophagy in liver diseases. J Hepatol.
53:1123–1134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Czaja MJ: Functions of autophagy in
hepatic and pancreatic physiology and disease. Gastroenterology.
140:1895–1908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kwanten WJ, Martinet W, Michielsen PP and
Francque SM: Role of autophagy in the pathophysiology of
nonalcoholic fatty liver disease: A controversial issue. World J
Gastroenterol. 20:7325–7338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun K, Guo XL, Zhao QD, Jing YY, Kou XR,
Xie XQ, Zhou Y, Cai N, Gao L, Zhao X, et al: Paradoxical role of
autophagy in the dysplastic and tumor-forming stages of
hepatocarcinoma development in rats. Cell Death Dis. 4:e5012013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou R, Yazdi AS, Menu P and Tschopp J: A
role for mitochondria in NLRP3 inflammasome activation. Nature.
469:221–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakahira K, Haspel JA, Rathinam VA, Lee
SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim
HP, et al: Autophagy proteins regulate innate immune responses by
inhibiting the release of mitochondrial DNA mediated by the NALP3
inflammasome. Nat Immunol. 12:222–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Castillo EF, Dekonenko A, Arko-Mensah J,
Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS,
Bhattacharya D, Yang H, et al: Autophagy protects against active
tuberculosis by suppressing bacterial burden and inflammation. Proc
Nat Acad Sci USA. 109:E3168–E3176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jordan CT, Guzman ML and Noble M: Cancer
stem cells. N Engl J Med. 355:1253–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Magee JA, Piskounova E and Morrison SJ:
Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer
Cell. 21:283–296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma S: Biology and clinical implications of
CD133+ liver cancer stem cells. Exp Cell Res.
319:126–132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO,
Zheng BJ and Guan XY: Identification and characterization of
tumorigenic liver cancer stem/progenitor cells. Gastroenterology.
132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rountree CB, Senadheera S, Mato JM, Crooks
GM and Lu SC: Expansion of liver cancer stem cells during aging in
methionine adenosyltransferase 1A-deficientmice. Hepatology.
47:1288–1297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sui Q, Zhang J, Sun X, Zhang C, Han Q and
Tian Z: NK cells are the crucial antitumor mediators when
STAT3-mediated immunosuppression is blocked in hepatocellular
carcinoma. J Immunol. 193:2016–2023. 2014. View Article : Google Scholar : PubMed/NCBI
|