Introduction
Laminins are a family of extracellular matrix (ECM)
glycoproteins, which are the major non-collagenous constituent of
basement membranes and serve an important role in cell
differentiation, migration and adhesion (1,2). In
gastric cancer, certain laminin family members are dysregulated and
are associated with malignant phenotypes. For example, laminin γ2
upregulation may constitute an adaptive stimulus that allows
E-cadherin-defective cells to survive and invade, which contributes
toward the subsequent cancer progression (3). Co-expression of laminin β3 and γ2 is
significantly correlated with the depth of invasion and advanced
tumor stage (4). Epigenetic
silencing of laminin β3 chain may reduce cancer cell invasion
(4).
The a4 subunit [laminin a4 (LAMA4)] is a component
of laminin-8 and laminin-9, which is present in tissues of
mesenchymal origin, in endothelial basement membranes and in
certain epithelial basement membranes (5). Recent studies have reported that
aberrant LAMA4 expression is associated with enhanced cell
migration and metastasis of certain types of cancer, including
hepatocellular (6) and breast
cancer (7), and renal carcinoma
(8). However, the effect of
LAMA4 dysregulation on gastric cancer is poorly
understood.
Zinc finger E-box-binding homeobox (ZEB) 1 is an
E-box binding transcription factor and is one of the key
epithelial-mesenchymal transition (EMT)-inducible genes in multiple
types of cancer (9–11). Previous studies have demonstrated
that ZEB1 is an independent factor for peritoneal dissemination in
patients with gastric cancer (12,13).
Knockdown of ZEB1 can significantly reduce Vimentin
expression and increase E-cadherin expression in gastric cancer
cells (14), and can also decrease
the invasive potential of the cancer cells (15,16).
As a transcriptional factor, ZEB1 can act as a transcriptional
activator (via binding to CtBP co-repressors) and repressor (via
binding to chromatin remodeling ATPase BRG1, histone
acetyl-transferase TIP60 and histone deacetylase SIRT1) (17), depending on specific genes and cells
(18,19). In gastric cancer, its downstream
regulation remains to be fully elucidated.
The present study investigated the prognostic value
and functional role of LAMA4 in gastric cancer and further
investigated the association between the expression of ZEB1 and
LAMA4.
Materials and methods
Bioinformatic analysis
The clinicopathological data of patients with
primary gastric cancer, the mRNA expression of LAMA4, MMP2,
MMP9 and ZEB1, and their associations in The Cancer
Genome Atlas-stomach adenocarcinoma (TCGA-STAD) were analyzed using
UCSC Xena Browser (http://xena.ucsc.edu/). The genes co-upregulated with
LAMA4 in TCGA-STAD were also identified using the UCSC
browser. LAMA4 protein expression in gastric cancer tissues and in
normal gastric tissues was reviewed using immunohistochemistry
(IHC) images from the Human Protein Atlas (http://www.proteinatlas.org/) (20), via www.proteinatlas.org/ENSG00000112769-LAMA4/tissue/stomach
and http://www.proteinatlas.org/ENSG00000112769-LAMA4/pathology/tissue/stomach+cancer.
The association between LAMA4 expression and
overall survival (OS) of patients with gastric cancer was examined
using data in TCGA-STAD and by data mining in Kaplan-Meier plotter
(http://kmplot.com/analysis/), an online
database containing gene expression data and survival information
of 1,065 patients with gastric cancer (21). The patients were divided into two
groups by setting the best performing threshold of LAMA4
expression as the cut-off. The hazard ratio (HR), 95% confidence
intervals (CI) and log-rank P-values were calculated. The
number-at-risk was indicated below the survival curves.
Cell culture
Human gastric cancer HGC-27 and SGC-7901 cell lines
were obtained from the Institute of Basic Medical Sciences of the
Chinese Academy of Medical Sciences (Beijing, China). The cells
were cultured with Dulbecco's modified Eagle's medium (DMEM)/high
glucose (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc.), 100 IU/ml penicillin G and 100 µg/ml
streptomycin at 37°C in a humidified 5% CO2
incubator.
Lentiviral LAMA4 shRNA particles (SHCLNV-NM_
002290) (pLOK.1-CMV-tGFP, with the sequence for shLAMA4-1,
5′-CCGGCGTCTATAATTTGGGAACTAACTCGAGTTAGTTCCCAAATTATAGACGTTTTTG-3′
and shLAMA4-2,
5′-CCGGGAACACCACTGACCGAATTTACTCGAGTAAATTCGGTCAGTGGTGTTCTTTTTG-3′)
and the corresponding negative control (empty pLOK.1-CMV-tGFP) were
purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). ZEB1
lentiviral particles and the corresponding negative controls were
purchased from GeneCopoeia (Rockville, MD, USA). The total
transducing units needed (TU) for infection was calculated by
(total number of cells per well ×3). The cancer cells were infected
with the lentiviral particles in the presence of Polybrene (8
µg/ml; Sigma-Aldrich; Merck KGaA) according to the manufacturer's
protocols and were subjected to analysis 48 h later.
Western blot analysis
Conventional western blotting was performed to
detect protein band signals. Cells were lysed using a
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) for protein extraction. Protein
concentration was determined using a bicinchoninic acid protein
assay kit (Beyotime Institute of Biotechnology, Haimen, China). The
proteins (25 µg protein/lane) were separated by 10% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes. The membranes
were blocked with 5% skimmed milk for 1 h at room temperature and
then incubated with primary antibodies overnight at 4°C. The
primary antibodies used were as follows: Anti-LAMA4 (1:1,000; cat.
no. ab209675; Abcam, Cambridge, UK), anti-MMP2 (1:1,000; cat. no.
ab37150; Abcam), anti-MMP9 (1:1,000; cat. no. ab38898; Abcam),
anti-ZEB1 (1:2,000; cat. no. ab180905; Abcam) and anti-β-actin
(1:2,000; cat. no. ab3280; Abcam). Following incubation with
horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG H&L
(1:5,000; cat. no. ab205719; Abcam) or HRP-conjugated goat
anti-rabbit IgG H&L (1:10,000; cat. no. ab205718; Abcam)
secondary antibody for 1 h in TBST with 5% skimmed milk at room
temperature, protein band signals were developed using the enhanced
chemiluminescence Plus kit (Amersham, Piscataway, NJ, USA). Band
densitometry was performed using ImageJ software (v2.1.4.6;
National Institutes of Health, Bethesda, MD, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cell samples using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and were used as the template for reverse
transcription with the ProtoScript First Strand cDNA Synthesis kit
(New England Biolabs, Ipswich, MA, USA). In brief, RNA was
denatured for 5 min at 70°C. Next, cDNA synthesis reaction was
conducted at 42°C for 1 h. Finally, the enzyme was inactivated at
80°C for 5 min. Subsequently, qPCR was performed to detect the
expression of LAMA4 mRNA using the SYBR® Select
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) in
an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The primer sequences used were as
follows: LAMA4 forward, 5′-GGAAAATAAGCGAGGCACCG-3′ and
reverse, 5′-AGCCACAGAGGCAGAACCGA-3′; and GAPDH forward,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′). The relative expression of
LAMA4 mRNA was calculated using the 2−ΔΔCq method
(22).
Wound healing assay
In brief, HGC-27 and SGC-7901 cells were cultured in
6-well plates and were infected with lentiviral LAMA4 shRNA
particles or negative controls. A total of 24 h later, confluent
cell monolayers were manually wounded by scraping the cells with a
200 µl pipette tip. Wound images were taken at 0 and 24 h after the
scratch under an inverted microscope (IX73; Olympus Corporation,
Tokyo, Japan), at a magnification of ×10. The wound areas were
measured using ImageJ software (v2.1.4.7; n=3).
Transwell assay
A Transwell assay was conducted using a Matrigel
invasion chamber (BD Biosciences, San Jose, CA, USA) in a 24-well
cell culture plate according to the manufacturer's protocols.
Briefly, 3×104 HGC-27 and SGC-7901 cells infected with
LAMA4 shRNA particles or the negative controls were seeded
into the upper chamber inserts containing an 8-µm pore size
membrane with a thin layer Matrigel matrix, with 500 µl serum-free
DMEM. The lower chamber of the well was filled with 700 µl DMEM
with 20% FBS. A total of 48 h later, cells that had invaded the
lower surface of the membrane were fixed with 70% methanol at room
temperature for 10 min while the non-invading cells on the upper
surface were removed. The invaded cells were stained with 0.1%
crystal violet for 30 min at room temperature, and the number was
then determined for 3 independent fields under an inverted
microscope (IX73; Olympus Corporation), at a magnification of
×100.
Dual-luciferase reporter assay
The promoter sequence of LAMA4 was obtained
from GeneCopoeia (>HPRM34295 NM_001105206). The possible ZEB1
binding sites in the LAMA4 promoter region were predicted
using the JASPAR database (http://jaspar.genereg.net/). LAMA4 promoter
fragments (−1,351 to +219 and −700 to +219) were PCR amplified from
the promoter clone (>HPRM34295). The fragments were then
inserted into the sites between XhoI-HindIII of
pGL3-basic plasmids (Promega Corporation, Madison, WI, USA). 293
cells cultured in 12-well plates were initially infected with
lentiviral ZEB1 expression particles or the empty control. A total
of 24 h later, the cells were co-transfected with 1.5 µg luciferase
construct plasmids (Promega Corporation) or the empty reporter
vector DNA and 0.05 µg phRL-TK (Promega Corporation) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). A total of 24 h after transfection, the cells
were lysed. The luciferase activity of the lysate was measured
using the dual-luciferase reporter assay system with a luminometer
and was normalized to that of Renilla luciferase activity
(Promega Corporation, Madison, WI, USA).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
All assays were performed in triplicate and data are reported as
the mean ± standard deviation. The group difference was examined by
two-tailed Student's t-tests or one-way analysis of variance with
Student-Newman-Keuls test as a post hoc test. The association
between LAMA4 RNA expression and the clinicopathological
features was assessed using χ2 tests. Receiver operating
characteristic (ROC) curves for mortality were constructed and the
optimal cut-off value of LAMA4 expression was determined
based on the Youden index. Log-rank tests were performed to assess
the difference between the survival curves. Prognostic values were
analyzed by univariate and multivariate Cox regression models.
P<0.05 was considered to indicate a statistically significant
difference.
Results
LAMA4 upregulation is associated with
higher grade tumors in gastric cancer
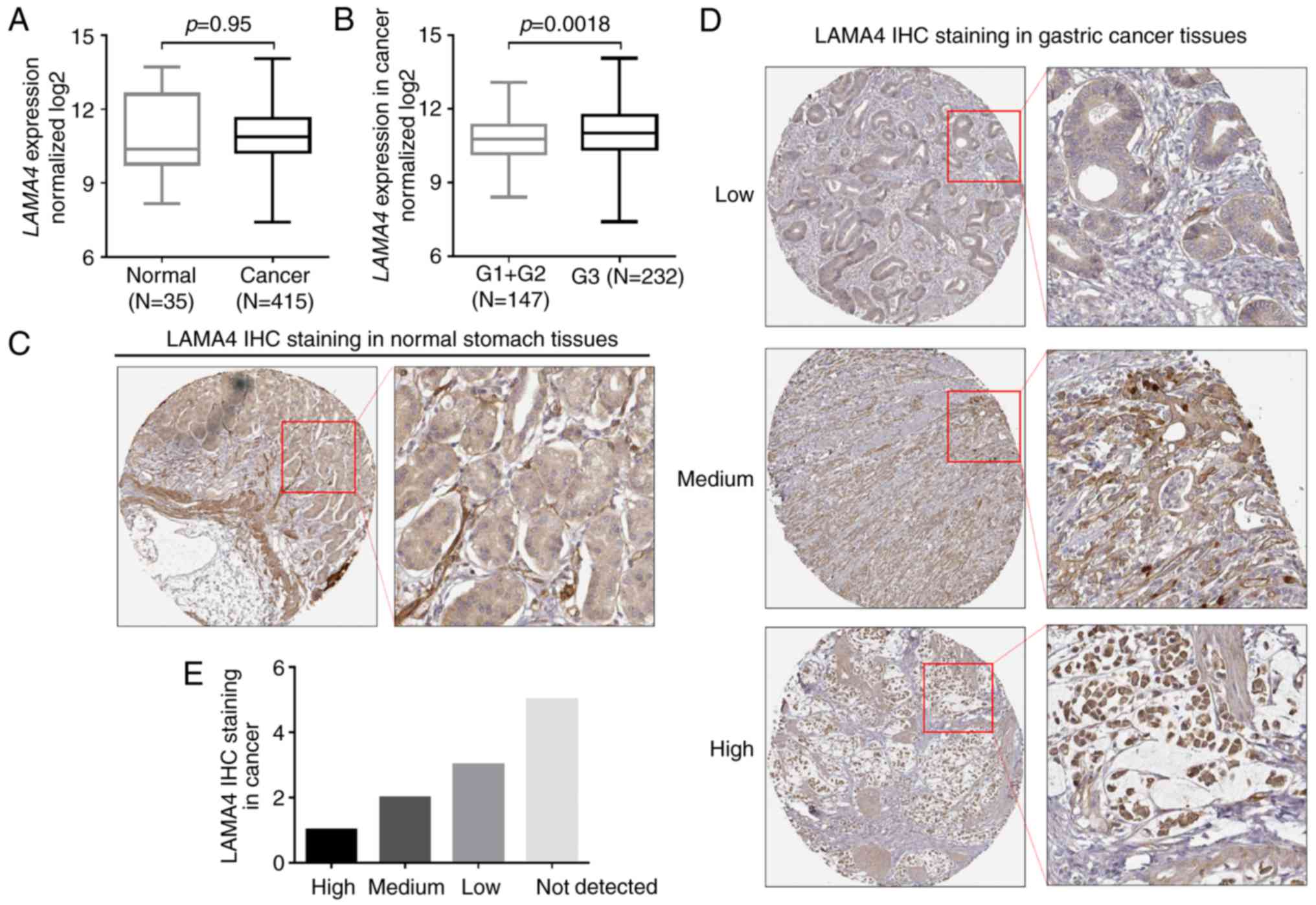
Using RNA-seq data in TCGA-STAD, it was revealed
that LAMA4 RNA expression was not altered in gastric cancer
tissues compared with normal stomach tissues (Fig. 1A). However, in the cancer cases, the
grade 3 tumors had significantly higher LAMA4 expression
than the grade 1/2 tumors (P=0.0018; Fig. 1B). By reviewing LAMA4 IHC images in
Human Protein Atlas, it was revealed that the LAMA4 staining was
usually low in the glandular cells in normal tissues (Fig. 1C). However, the intensity of LAMA4
staining varied significantly in different gastric cancer cases
(Fig. 1D). Among 11 gastric cancer
tissues, 3 cases had moderate/high LAMA4 staining, while 3 cases
had low LAMA4 staining (Fig.
1D).
Knockdown of LAMA4 impaired the
migration and invasion of gastric cancer cells
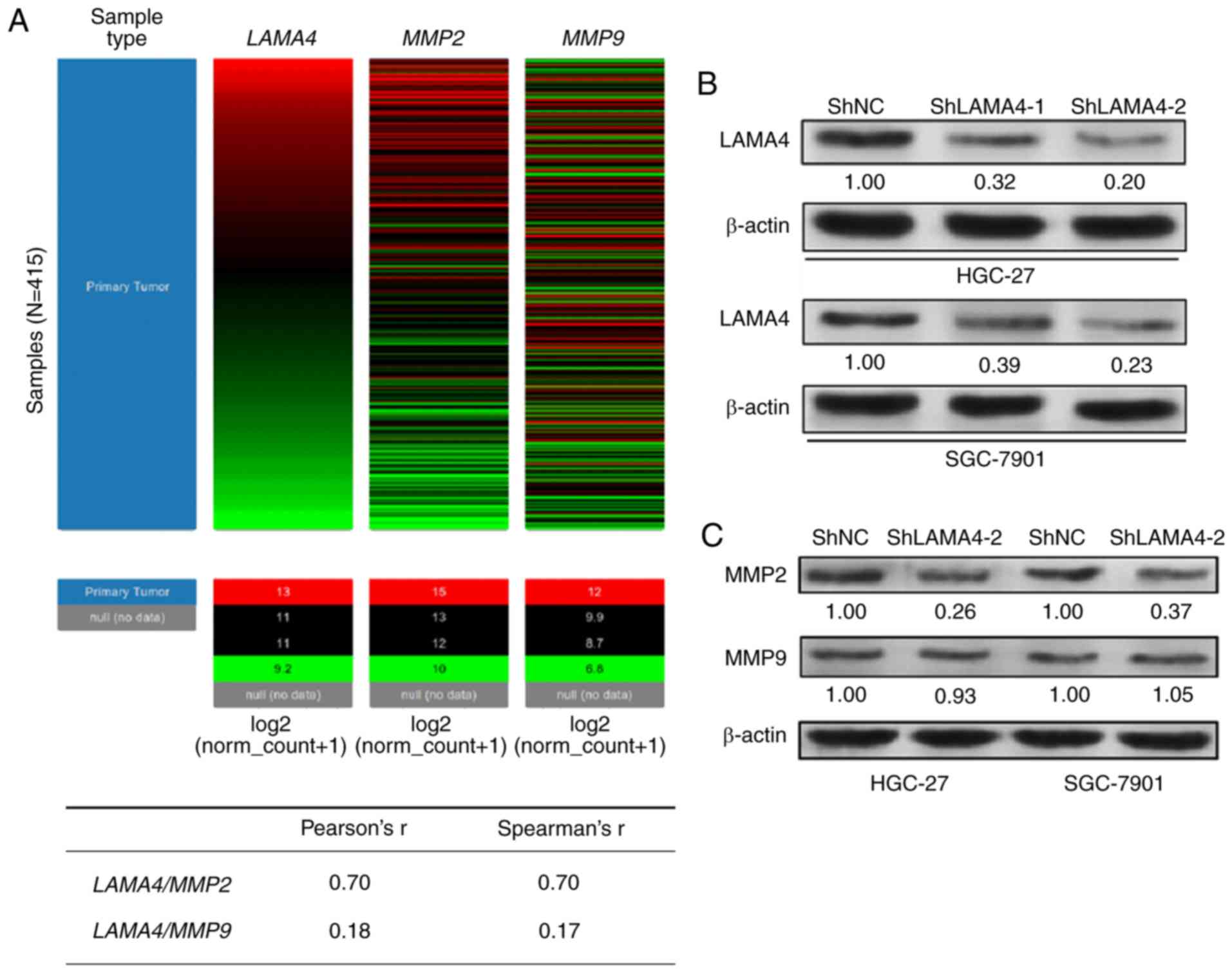
One previous study reported that LAMA4 could promote
trophoblast cell invasion and migration via upregulating MMP2 and
MMP9, two enzymes facilitating invasion by degrading the ECM
(5). By data mining in TCGA-STAD,
we the co-expression trend between LAMA4 and MMP2 or
MMP9 was characterized (Fig.
2A). Heat-map and subsequent regression analysis demonstrated
that LAMA4 was significantly co-upregulated with MMP2
(Pearson's r=0.70), but not with MMP9 (Pearson's r=0.18) among the
415 patients with gastric cancer (Fig.
2A). To investigate the functional role of LAMA4 in gastric
cancer, HGC-27 and SGC-7901 cells were infected with LAMA4
shRNA for knockdown (Fig. 2B). In
these two cell lines, LAMA4-knockdown significantly reduced
MMP2 expression, but had little influence on MMP9 expression
(Fig. 2C). Wound healing and
Transwell assays demonstrated that LAMA4 inhibition impaired
the speed of wound healing (Fig.
2D-E) and reduced the invasive capability of the cancer cells
(Fig. 2F-G).
ZEB1 directly increases LAMA4
expression via binding to its promoter
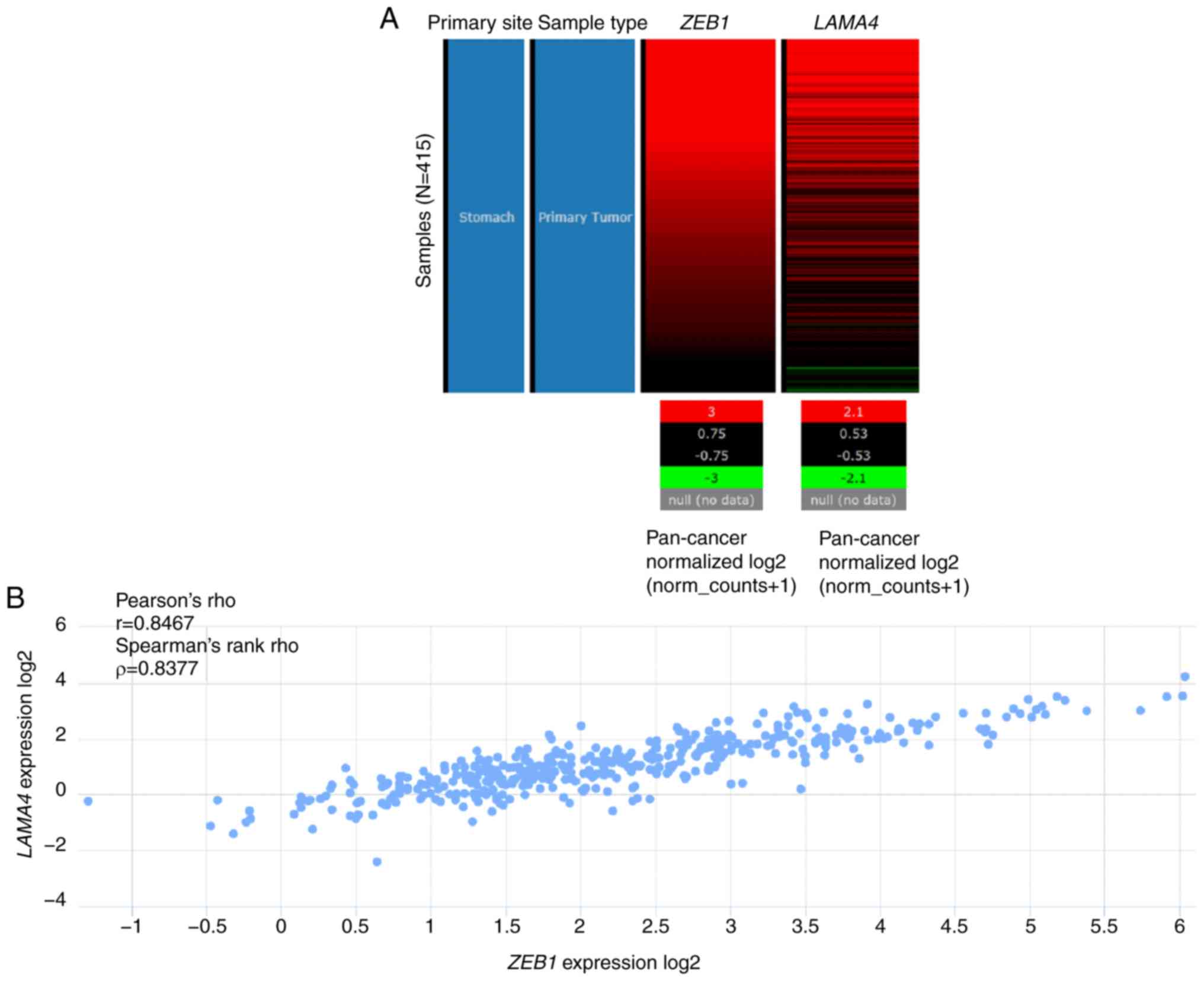
By screening the genes co-upregulated with
LAMA4 in TCGA-STAD, it was revealed that ZEB1 was
correlated with LAMA4 in gastric cancer (Pearson's r=0.85;
Fig. 3A). In fact, ZEB1
upregulation has well-characterized oncogenic effects on gastric
cancer (13,23). By using the UCSC Xena browser
(Fig. 3A and B) and the cBioPortal
for Cancer Genomics (Fig. 3C), two
online tools to analyze data in TCGA-STAD, a strong correlation
between the expression of ZEB1 and that of LAMA4 was
confirmed (Fig. 3A-C). To further
investigate the effect of ZEB1 on LAMA4 expression, HGC-27 and
SGC-7901 cells were infected with lentiviral LAMA4
expression particles for overexpression (Fig. 3D). Enforced ZEB1 expression
significantly elevated LAMA4 expression at the mRNA and
protein levels (Fig. 3D and E). By
promoter scanning, two possible and close ZEB1 binding sites were
identified in the promoter of LAMA4 (Fig. 3F). pGL3-basic-based luciferase
reporter plasmids carrying the intact LAMA4 promoter
sequence or truncated sequence were generated (Fig. 3G). A luciferase assay revealed that
ZEB1 overexpression significantly increased the luciferase activity
of the reporter with the intact LAMA4 promoter sequence
(Fig. 3H). By comparison,
ZEB1 overexpression had little influence on the luciferase
activity of the reporter with the truncated LAMA4 promoter
sequence (Fig. 3H).
High LAMA4 expression independently
predicts a poor OS in patients with primary gastric cancer
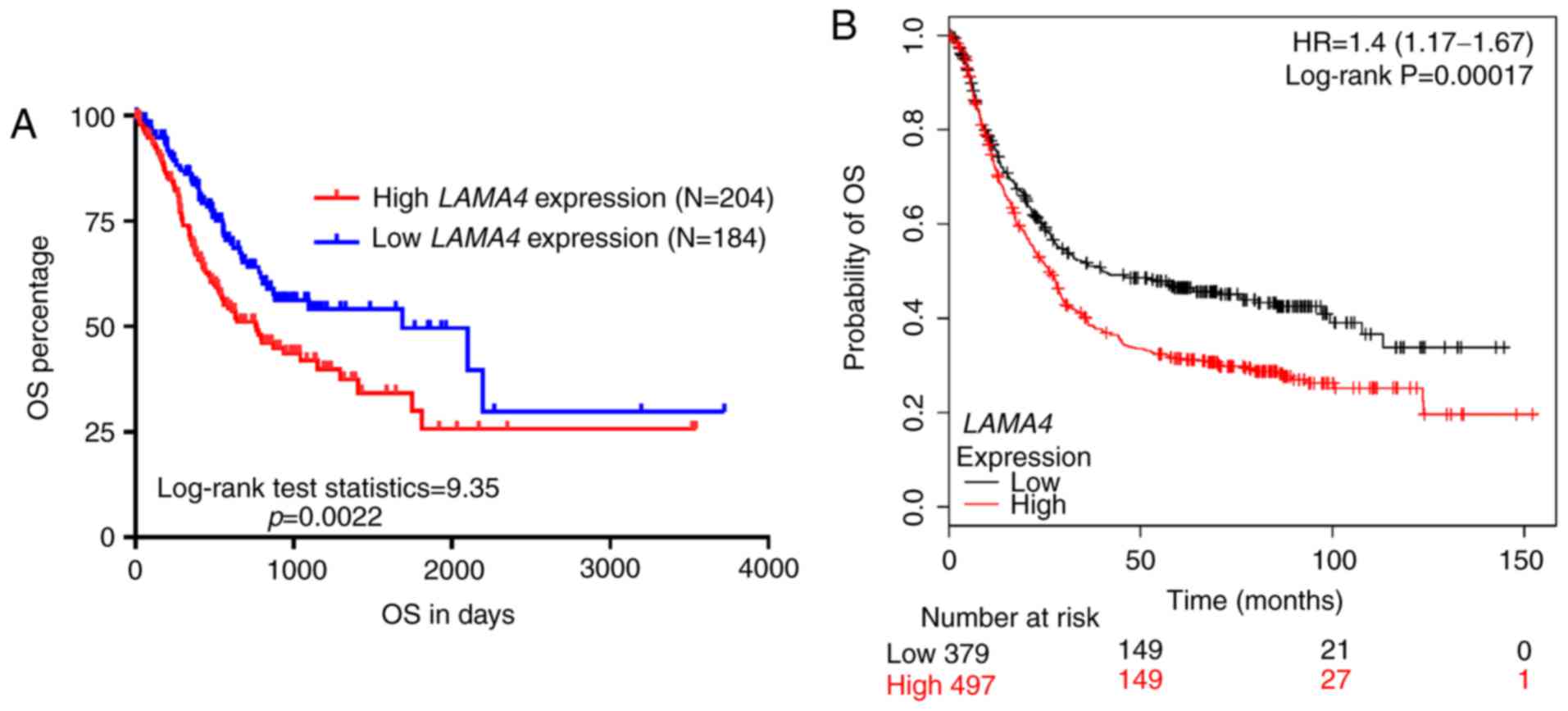
In order to investigate the prognostic value of
LAMA4 in gastric cancer, the association between
LAMA4 expression and OS was further assessed based on data
in TCGA-STAD and by data mining in Kaplan-Meier plotter. The
associations between LAMA4 expression and the
clinicopathological parameters in patients with primary gastric
cancer in TCGA were summarized in Table
I. The high LAMA4 expression group had significantly
higher ratios of grade 3 (G3) tumors (134/199, 67.3%) and mortality
(98/204, 48.0%) than the low LAMA4 expression group (G3,
98/180, 54.4%; mortality, 59/184, 32.1%; Table I). Kaplan-Meier curves demonstrated
that the high LAMA4 expression group (n=204) had
significantly poorer OS rates than the low LAMA4 expression
group (n=184; P=0.0022; Fig. 4A).
Data mining in Kaplan-Meier plotter also confirmed this association
(HR, 1.4; 95% CI, 1.17–1.67; P<0.001; Fig. 4B). In univariate analysis, it was
revealed that high age (>65), high grade (G3/G4), nodal
invasion, metastasis, advanced disease stage (III/IV) and high
LAMA4 expression were associated with significantly shorter
OS times (Table II). Multivariate
analysis revealed that the high LAMA4 expression could
independently predict a poor OS (HR, 1.614; 95% CI, 1.155–2.256;
P=0.005; Table II).
 | Table I.The association between LAMA4
expression and the clinicopathological parameters of patients with
primary gastric cancer in TCGA-STAD. |
Table I.
The association between LAMA4
expression and the clinicopathological parameters of patients with
primary gastric cancer in TCGA-STAD.
|
| LAMA4
expression |
|
|
|---|
|
|
|
|
|
|---|
| Parameter | High (n=204) | Low (n=184) | χ2 | P-value |
|---|
| Age, mean ± SD | 65.30±10.68 | 65.30±10.62 |
| 1.00 |
| Sex |
|
Female | 67 | 69 | 0.92 | 0.34 |
|
Male | 137 | 115 |
|
|
| Histological
grade |
|
G1/G2 | 65 | 82 | 6.62 | 0.01 |
| G3 | 134 | 98 |
|
|
| GX | 5 | 4 |
|
|
| Nodal status |
| N0 | 56 | 60 | 0.99 | 0.32 |
| N+ | 141 | 121 |
|
|
|
Null | 7 | 3 |
|
|
| Metastasis
status |
| M0 | 181 | 166 | 0.14 | 0.71 |
| M1 | 14 | 11 |
|
|
| MX | 9 | 7 |
|
|
| Clinical stage |
|
I/II | 81 | 91 | 3.01 | 0.080 |
|
III/IV | 114 | 89 |
|
|
|
Discrepancy + Null | 9 | 4 |
|
|
| Status |
|
Alive | 106 | 125 | 10.25 | 0.0014 |
|
Deceased | 98 | 59 |
|
|
 | Table II.Univariate and multivariate analyses
of overall survival in patients with primary gastric cancer in
TCGA-STAD. |
Table II.
Univariate and multivariate analyses
of overall survival in patients with primary gastric cancer in
TCGA-STAD.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Parameter | P-value | HR | 95% CI
(lower/upper) | P-value | HR | 95% CI
(lower/upper) |
|---|
| Age |
| >65
vs. ≤65 | 0.006 | 1.572 | 1.139 | 2.171 | 0.002 | 2.001 | 1.421 | 2.816 |
| Sex |
| Female
vs. male | 0.296 | 0.835 | 0.596 | 1.171 |
|
|
|
|
| Grade |
| G3/G4
vs. G1/G2 | 0.022 | 1.479 | 1.057 | 2.069 | 0.039 | 1.463 | 1.020 | 2.098 |
| Nodal status |
| N1+ vs.
N0 | 0.001 | 2.042 | 1.362 | 3.061 | 0.136 | 1.526 | 0.876 | 2.661 |
| Metastasis
status |
| M1 vs.
M0 | 0.002 | 2.334 | 1.367 | 3.986 | 0.002 | 2.429 | 1.371 | 4.305 |
| Clinical stage |
| III/IV
vs. I/II | <0.001 | 2.063 | 1.460 | 2.916 | 0.173 | 1.398 | 0.863 | 2.265 |
| LAMA4
expression |
| High
vs. low | 0.002 | 1.664 | 1.205 | 2.300 | 0.005 | 1.614 | 1.155 | 2.256 |
Discussion
In the present study, the results of bioinformatic
analysis indicated that LAMA4 upregulation was associated
with higher grades of gastric cancer. LAMA4 upregulation is
associated with enhanced invasion and metastasis of cancer cells.
In hepatocellular carcinoma, LAMA4 has specific in vivo
distribution in the tumor basement membrane and its upregulation is
correlated with tumor invasion and metastasis (6). In renal cell carcinoma, LAMA4
is upregulated in locally advanced tumors and in primary tumor and
secondary metastases (8).
LAMA4 upregulation may also predict poor survival in
patients with renal cell carcinoma (8). One recent study reported that LAMA4
could promote trophoblast cell invasion and migration via
upregulating MMP2 and MMP9 (5).
MMP2 and MMP9 are two critical enzymes degrading ECM, thereby
supporting cancer cell migration and invasion (24,25).
In fact, trophoblast research over the past decades revealed that
placental cells have high levels of similarities in proliferative,
migratory and invasive properties to those of cancer cells
(26). As LAMA4 dysregulation may
be associated with tumor grade in gastric cancer, the present study
investigated its regulative effect on the migration and invasion of
gastric cancer cells. In HGC-27 and SGC-7901 cells,
LAMA4-knockdown significantly reduced MMP2 expression, but
had little influence on MMP9 expression. Functional assays revealed
that LAMA4 inhibition impaired the speed of wound healing
and also reduced the invasive capability of the cancer cells.
To investigate the mechanism of LAMA4 dysregulation
in gastric cancer, we identified the genes significantly
co-expressed with LAMA4 in TCGA-STAD and observed that
ZEB1 is correlated with LAMA4 expression. The
oncogenic effects of aberrant ZEB1 expression in gastric cancer
have been widely reported (13,15,27).
As a transcription factor, ZEB1 can modulate the expression of a
series of genes in different types of cancer. In breast cancer,
ZEB1 can upregulate VEGF expression and promote angiogenesis
(28). Additionally, ZEB1 can
reduce NGN3 transcription via forming a ZEB1/DNA
methyltransferase (DNMT)3B/histone deacetylase 1 (HDAC1) complex on
the NGN3 promoter (29). In
tongue cancer cells, ZEB1 can bind to the CA9 promoter and
positively regulate its expression, thereby leading to enhanced
chemoresistance (30). In
gallbladder cancer cells, ZEB1 can repress T-cadherin expression
via binding to the promoter, thereby increasing their invasive
capability (31). These results
suggested that ZEB1 can be either an epigenetic activator or
repressor, depending on specific gene and cancer types. However,
the regulative effect of ZEB1 in gastric cancer is not yet fully
understood. The present study revealed that ZEB1 can directly
increase LAMA4 expression via binding to its promoter in
gastric cancer cells. This finding revealed a novel regulative
effect of ZEB1 in gastric cancer.
Based on data mining in two large databases,
including TCGA-STAD and Kaplan-Meier plotter, it was revealed that
LAMA4 upregulation is associated with unfavorable OS rates
in patients with gastric cancer. Univariate and multivariate
analysis demonstrated that the high LAMA4 expression could
independently predict a poor OS rate (HR, 1.614; 95% CI,
1.155–2.256; P=0.005), suggesting that LAMA4 expression may
be a valuable biomarker in gastric cancer.
Based on the aforementioned results, we hypothesized
that ZEB1 could epigenetically activate LAMA4 expression via
binding to its promoter in gastric cancer cells, while high
LAMA4 expression was an independent indicator for a poor OS
in patients with gastric cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XW and QH performed cellular studies and conducted
data analysis and interpretation. XW and XZ collected and analyzed
data from databases. All authors participated in the manuscript
preparation and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TCGA
|
The Cancer Genome Atlas
|
|
STAD
|
stomach adenocarcinoma
|
|
OS
|
overall survival
|
|
LAMA4
|
laminin subunit α4
|
|
ZEB1
|
zinc finger E-box-binding homeobox
1
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ECM
|
extracellular matrix
|
References
|
1
|
Cheng YS, Champliaud MF, Burgeson RE,
Marinkovich MP and Yurchenco PD: Self-assembly of laminin isoforms.
J Biol Chem. 272:31525–31532. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lugassy C, Torres-Munoz JE, Kleinman HK,
Ghanem G, Vernon S and Barnhill RL: Overexpression of
malignancy-associated laminins and laminin receptors by angiotropic
human melanoma cells in a chick chorioallantoic membrane model. J
Cutan Pathol. 36:1237–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Caldeira J, Figueiredo J, Bras-Pereira C,
Carneiro P, Moreira AM, Pinto MT, Relvas JB, Carneiro F, Barbosa M,
Casares F, et al: E-cadherin-defective gastric cancer cells depend
on Laminin to survive and invade. Hum Mol Genet. 24:5891–5900.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ii M, Yamamoto H, Taniguchi H, Adachi Y,
Nakazawa M, Ohashi H, Tanuma T, Sukawa Y, Suzuki H, Sasaki S, et
al: Co-expression of laminin beta3 and gamma2 chains and epigenetic
inactivation of laminin alpha3 chain in gastric cancer. Int J
Oncol. 39:593–599. 2011.PubMed/NCBI
|
|
5
|
Shan N, Zhang X, Xiao X, Zhang H, Tong C,
Luo X, Chen Y, Liu X, Yin N, Deng Q and Qi H: Laminin alpha4
(LAMA4) expression promotes trophoblast cell invasion, migration,
and angiogenesis, and is lowered in preeclamptic placentas.
Placenta. 36:809–820. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang X, Ji G, Wu Y, Wan B and Yu L:
LAMA4, highly expressed in human hepatocellular carcinoma from
Chinese patients, is a novel marker of tumor invasion and
metastasis. J Cancer Res Clin Oncol. 134:705–714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ross JB, Huh D, Noble LB and Tavazoie SF:
Identification of molecular determinants of primary and metastatic
tumour re-initiation in breast cancer. Nat Cell Biol. 17:651–664.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wragg JW, Finnity JP, Anderson JA,
Ferguson HJ, Porfiri E, Bhatt RI, Murray PG, Heath VL and Bicknell
R: MCAM and LAMA4 are highly enriched in tumor blood vessels of
fenal cell carcinoma and predict patient outcome. Cancer Res.
76:2314–2326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spaderna S, Schmalhofer O, Wahlbuhl M,
Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D and Eger
A: The transcriptional repressor ZEB1 promotes metastasis and loss
of cell polarity in cancer. Cancer Res. 68:537–544. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wellner U, Schubert J, Burk UC,
Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D,
zur Hausen A, et al: The EMT-activator ZEB1 promotes tumorigenicity
by repressing stemness-inhibiting microRNAs. Nat Cell Biol.
11:1487–1495. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo E, Wang Z and Wang S: MiR-200c and
miR-141 inhibit ZEB1 synergistically and suppress glioma cell
growth and migration. Eur Rev Med Pharmacol Sci. 20:3385–3391.
2016.PubMed/NCBI
|
|
12
|
Okugawa Y, Toiyama Y, Tanaka K, Matsusita
K, Fujikawa H, Saigusa S, Ohi M, Inoue Y, Mohri Y, Uchida K and
Kusunoki M: Clinical significance of Zinc finger E-box Binding
homeobox 1 (ZEB1) in human gastric cancer. J Surg Oncol.
106:280–285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia B, Liu H, Kong Q and Li B:
Overexpression of ZEB1 associated with metastasis and invasion in
patients with gastric carcinoma. Mol Cell Biochem. 366:223–229.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou X, Wang Y, Shan B, Han J, Zhu H, Lv
Y, Fan X, Sang M, Liu XD and Liu W: The downregulation of
miR-200c/141 promotes ZEB1/2 expression and gastric cancer
progression. Med Oncol. 32:4282015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yanaka Y, Muramatsu T, Uetake H, Kozaki K
and Inazawa J: miR-544a induces epithelial-mesenchymal transition
through the activation of WNT signaling pathway in gastric cancer.
Carcinogenesis. 36:1363–1371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sanchez-Tillo E, Liu Y, de Barrios O,
Siles L, Fanlo L, Cuatrecasas M, Darling DS, Dean DC, Castells A
and Postigo A: EMT-activating transcription factors in cancer:
Beyond EMT and tumor invasiveness. Cell Mol Life Sci. 69:3429–3456.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Postigo AA and Dean DC: ZEB represses
transcription through interaction with the corepressor CtBP. Proc
Natl Acad Sci USA. 96:6683–6688. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Larsen JE, Nathan V, Osborne JK, Farrow
RK, Deb D, Sullivan JP, Dospoy PD, Augustyn A, Hight SK and Sato M:
ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J
Clin Invest. 126:3219–3235. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lindskog C: The potential clinical impact
of the tissue-based map of the human proteome. Expert Rev
Proteomics. 12:213–215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szász AM, Lánczky A, Nagy Á, Förster S,
Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A and Győrffy B:
Cross-validation of survival associated biomarkers in gastric
cancer using transcriptomic data of 1,065 patients. Oncotarget.
7:49322–49333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yabusaki N, Yamada S, Murai T, Kanda M,
Kobayashi D, Tanaka C, Fujii T, Nakayama G, Sugimoto H, Koike M, et
al: Clinical significance of zinc-finger E-box binding homeobox 1
mRNA levels in peritoneal washing for gastric cancer. Mol Clin
Oncol. 3:435–441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sato H and Seiki M: Membrane-type matrix
metalloproteinases (MT-MMPs) in tumor metastasis. J Biochem.
119:209–215. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yasui W, Oue N, Aung PP, Matsumura S,
Shutoh M and Nakayama H: Molecular-pathological prognostic factors
of gastric cancer: A review. Gastric Cancer. 8:86–94. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ferretti C, Bruni L, Dangles-Marie V,
Pecking AP and Bellet D: Molecular circuits shared by placental and
cancer cells, and their implications in the proliferative, invasive
and migratory capacities of trophoblasts. Hum Reprod Update.
13:121–141. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Miao L, Xiong X, Lin Y, Cheng Y, Lu J,
Zhang J and Cheng N: Down-regulation of FoxM1 leads to the
inhibition of the epithelial-mesenchymal transition in gastric
cancer cells. Cancer Genet. 207:75–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Tong Q, Liu S, Cui J, Zhang Q, Sun
W and Yang S: ZEB1 upregulates VEGF expression and stimulates
angiogenesis in breast cancer. PLoS One. 11:e01487742016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou C, Jiang H, Zhang Z, Zhang G, Wang H,
Zhang Q, Sun P, Xiang R and Yang S: ZEB1 confers stem cell-like
properties in breast cancer by targeting neurogenin-3. Oncotarget.
8:54388–54401. 2017.PubMed/NCBI
|
|
30
|
Zheng G, Peng C, Jia X, Gu Y, Zhang Z,
Deng Y, Wang C, Li N, Yin J, Liu X, et al: ZEB1 transcriptionally
regulated carbonic anhydrase 9 mediates the chemoresistance of
tongue cancer via maintaining intracellular pH. Mol Cancer.
14:842015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Adachi Y, Takeuchi T, Nagayama T, Ohtsuki
Y and Furihata M: Zeb1-mediated T-cadherin repression increases the
invasive potential of gallbladder cancer. FEBS Lett. 583:430–436.
2009. View Article : Google Scholar : PubMed/NCBI
|