Introduction
Colorectal cancer (CRC) is the third most common
type of cancer and the second leading cause of cancer-related
deaths worldwide (1). CRC can
develop sporadically or in the background of several colonic
disorders. Inflammatory bowel disease (IBD), including ulcerative
colitis (UC) and Crohn's disease (CD), is characterized by the
idiopathic, chronic and recurrent inflammatory conditions of the
human bowel and is an important risk factor for developing CRC,
thus called colitis-associated colorectal cancer (CAC) (2,3). UC
patients with refractory and long-standing disease duration are
reported to have a significantly increased cancer risk which
increases with the earlier age of disease onset, the duration of
the disease and the extent of colonic involvement (4,5). Eaden
et al (6) estimated the
cumulative cancer incidence for developing CAC as 2% at 10 years,
8% at 20 years and 18% at 30 years in patients affected by chronic
colitis using a widely cited meta-analysis of 116 studies with age
stratified data. A more recent study reported that the risk of CRC
was 60% higher with extensive UC than in a without colitis-matched
cohort of people (7). In contrast
to UC, the role of CD in CRC risk remains controversial, and the
majority of studies could not detect different incidence rates in
comparison with the general population (8). Relative to sporadic CRC, CAC is
usually linked to an earlier age of onset, a fast progressing
course and a higher rate of mortality (9). Approximately 59% of CAC patients were
deceased at the 5-year follow-up according to the statistics of
Ording et al (10). These
previous studies demonstrated that repeated colonic inflammation is
a major contributing factor for CRC development.
Studies have revealed that the pathogenesis of CAC
differs from the sporadic form. Though they share some disease
mechanisms, such as mutations in oncogenes or tumor suppressors and
genetic instabilities, differences in morphology and biological
behaviors, timing and the sequence of genetic and epigenetic
alterations are present (11).
While most sporadic CRC demonstrates an ‘adenoma-to-carcinoma’
profile, CAC arises through an ‘inflammation-associated
dysplasia-to-carcinoma’ sequence, which, highlights important
differences in their carcinogenic pathways (12). In either situation, the cancer
develops from acquiring hallmark molecular alterations in the
epithelium of the colon and rectum. Regrettably, 20 to 50% of UC
patients with CAC were diagnosed with dysplasia only in the
preoperative pathological diagnosis (13). This indicates the difficulty of
surveillance for UC patients using colonoscopy. Thus, novel tumor
markers for early clinical risk assessment and differential
diagnosis are urgently required.
MicroRNAs (miRNAs) are endogenous small non-coding
RNAs of 18 to 25 nucleotides in length that regulate gene
expression through binding to the 3′-untranslated region (3′-UTR)
of target messenger RNAs, either targeting the transcripts for
degradation or blocking their translation. miRNAs are expressed in
the human body in a cell- and tissue-specific manner and are
crucial regulators of development and disease (14,15).
The role of miRNAs in human sporadic CRC has been intensively
studied in recent years. However, far less is known about their
implications in UC-CAC development. Considering that the etiology
of the disease is unclear, researchers are pursuing multiple
independent approaches to identify sensitive, as well as specific
biomarkers for early CAC (16).
Though there are currently many biomarkers in development to
discriminate IBD subtypes, there are few biomarkers for
differentiating IBD progressors from non-progressors (17). As post-transcriptional modulators of
intracellular signaling pathways, miRNAs are frequently found
upregulated or downregulated during different stages of chronic
inflammatory diseases. In the present study, we identified that
miR-449a, which is a member of the miR-34/449 superfamily, was
differentially expressed during the progression to carcinogenesis
from UC, and may thus hold significant diagnostic potential in
CAC.
Materials and methods
Patient information and tissue
samples
Human colon mucosa samples were obtained from
patients who received endoscopic biopsy or surgery at The Shanghai
General Hospital and The Shanghai East Hospital between January
2007 and July 2017. In total, 41 cases of CAC tissues and paired
adjacent non-cancerous tissues (ANTs) were collected from patients
with pathologically diagnosed CAC (28 males, 13 females; mean age,
59.72±10.05 years) who had clear histories of chronic colitis
suffering. Twenty-eight dysplasia-associated lesion or mass (DALM)
samples (16 males, 12 females; mean age, 51.72±13.05 years) were
collected and the dysplasia was defined according to the modified
Vienna criteria (18). Sixty-three
UC samples (33 males, 30 females; mean age, 45.58±10.96 years) and
49 CD samples (24 males, 25 females; mean age, 42.81±12.17 years)
were from patients in the active stage of disease. The diagnosis of
IBD was based on clinical, endoscopic and pathohistological
criteria as previously described (19,20).
Another 40 normal colorectal tissues obtained from people free of
intestinal diseases (20 males, 20 females; mean age, 45.23±9.56
years) were used as a control. All biopsy specimens were fixed in
liquid nitrogen or 10% formalin for analyses.
The present study was approved by the Ethics
Committee of The Shanghai General Hospital and the Ethics Committee
of The Shanghai East Hospital. The clinical study protocol strictly
conformed to the ethical guidelines of the 1975 Declaration of
Helsinki. Written informed consent was obtained from all
patients.
Cell lines and cell culture
The human CRC cell lines SW620 and SW480 were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA) and grown in complete growth medium as recommended by the
manufacturer. The cancer cells were maintained in a humidified 5%
CO2 atmosphere at 37°C. Both cell lines were regularly
authenticated by checking their morphology and confirming the
absence of mycoplasma contamination (MycoAlert Mycoplasma Detection
kit; Lonza, Rockland, ME, USA).
CAC animal model establishment
A cohort of 40 pathogen-free male Balb/c mice (6
weeks old, 18–20 g) was obtained from the Shanghai Institute of
Materia Medica, Chinese Academy of Sciences (Shanghai, China).
Every 5 mice were housed in an individual ventilated cage under
controlled conditions of humidity (50±10%), lighting (12-h
light/dark cycle) and temperature (25±2°C) with pure water and a
freely accessible pelleted basal diet. All protocols concerning
laboratory animal use were submitted to, and validated by, the
Animal Care Ethics Committee of Shanghai General Hospital.
Colitis-associated neoplasm was induced as
previously described (21). In
brief, animals were randomly divided into two groups (n=20). The 20
mice in the experimental group received intraperitoneally a
single-dose injection of azoxymethane (AOM; Sigma-Aldrich, Munich,
Germany) (10 mg/kg) on the first day. Dextran sodium sulfate (DSS;
MP Biomedicals, Aurora, OH, USA) (4%) was dissolved in distilled
water and administered in the first, third, fifth and seventh week.
The same procedure was performed with intraperitoneal normal saline
and drinking distilled water instead of the AOM/DSS treatment in
the control group.
On the first day and at the end of weeks 1/7/9, 5
mice in each group were euthanized by decapitation and colon
tissues were harvested and stored in 10% buffered formalin or at
−80°C in liquid nitrogen. The weight loss, stool consistency and
bleeding were recorded daily to monitor the disease activity.
Histological change was evaluated by hematoxylin and eosin
(H&E) staining. The formalin-fixed distal colon tissues were
embedded in paraffin blocks. Sliced sections (4 µm) were
deparaffinized and rehydrated by a xylene-ethanol-water gradient
system. H&E staining was then performed followed by a
dehydration process.
In situ hybridization (ISH) and
staining assessment
ISH analysis was performed using double-digoxigenin
(DIG) labeled 2′O-methyl locked nucleic acid (LNA)-ZEN probes
(Boster Biological Technology Co., Ltd., Wuhan, China)
complimentary to miR-449a (5′-ACCAGCTAACAATACACTGCCA-3′) along with
a scrambled negative control probe (5′-GUAUUAUAGCCGAUUAACG-3′).
Hybridization, washing, and scanning were performed according to
previous studies (22).
A staining evaluation system incorporating the
intensity and percentage of positive cells was conducted to assess
the miR-449a staining degree. The staining intensity was classified
into four grades: 0, no staining; 1, weak; 2, moderate; 3, strong.
The percentage of cells stained was graded as follows: 0, no
staining; 1, <10%; 2, 10–50%; and 3, >50% cells. The final
score was calculated by multiplying the grade for percentage
staining by the grade for intensity. Scores of ≥4 were defined as
positive staining and high expression. Each sample was evaluated by
two experienced pathologists who were not aware of the experimental
protocols.
Vectors and cell transfection
The DNA fragment of hsa-miR-449a was amplified from
genomic DNA and inserted into the BamHI/HindIII site
of lentivirus expression vector pGCL (Shanghai GenePharma, Co.,
Ltd., Shanghai, China). An empty pGCL vector was used as a negative
control. The Notch-1 expression vector was generated as previously
described. In brief, the coding sequence of Notch-1 was amplified
from cDNA and was then subcloned into a pcDNA3.1 plasmid (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Cell transfection was
performed with the Opti-MEM medium (Gibco; Thermo Fisher
Scientific, Inc.) and Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions.
Dual-luciferase reporter assay
Luciferase constructs were generated by ligating
oligonucleotides containing the wild-type or mutant putative target
site of the Notch-1 3′-UTR into the Psi-CHECK2 vector (Promega
Corp., Madison, WI, USA) downstream of the luciferase gene. Human
CRC cells were transfected with miR-449a expression vectors or the
controls in combination with each individual psiCHECK2 luciferase
vector. Cells were collected at 24 h after transfection, and
firefly and Renilla luciferase activities were assessed
using the Dual-Luciferase assay kit (Promega Corp.) (23).
Quantitative real-time RT-PCR
Total RNA was extracted from cells or tissues using
TRIzol reagent (Invitrogen) following the manufacturer's
instructions. The expression of miRNAs was quantified by two-step
quantitative RT-PCR, beginning with first-strand cDNA synthesis
using the One-step PrimeScript miRNA cDNA Synthesis kit (Takara,
Kyoto, Japan), followed by quantitative real-time PCR amplification
with the miScript SYBR-Green PCR kit (Takara). The quantitative
SYBR-Green PCR kit (Qiagen, Hilden, Germany) was used to quantify
the mRNA expression levels of genes in the Notch-1 signaling
pathway. All reactions were run in triplicate on the MasterCycler
Real-Time PCR Detection system (Eppendorf, Hamburg, Germany). The
primer sequences used in the study are listed in Table I. The expression of miRNA was
normalized to small nuclear RNA U6, while the other genes were
normalized using the GAPDH gene. The fold change of
miRNA/mRNA expression was calculated using the 2−ΔΔCq
method (24).
 | Table I.Primers used in the study. |
Table I.
Primers used in the study.
| Oligo | Sequence |
|---|
| Notch-1 | F:
TCCTTCTACTGCGAGTGTCC |
|
| R:
TCGTTACAGGGGTTGCTGAT |
| Jagged-1 | F:
CTTCAATCTCAAGGCCAGCC |
|
| R:
CAGGCGAAACTGAAAGGCAG |
| Dll-4 | F:
TGTGGCAAACAGCAAAACCA |
|
| R:
CCGACACTCTGGCTTTTCACT |
| Hes-1 | F:
TTTGGATGCTCTGAAGAAAGATAGC |
|
| R:
CGGTACTTCCCCAGCACACTT |
| Hey-1 | F:
CGAGCTGGACGAGACCAT |
|
| R:
CTAGAGCCGAACTCAAGTTTCC |
| GAPDH | F:
AGCGAGAACAATTACCCTGGGCAC |
|
| R:
ATTCTTGCCCTTCGCCTCTT |
Western blot analysis
Total protein was extracted from the cells and
measured with the bicinchoninic acid assay (BCA) kit (Beyotime
Institute of Biotechnology, Shanghai, China). Whole protein lysates
per lane (30 µg) were used to detect the indicated protein. The
cell lysate was separated by 8% SDS-PAGE and then transferred to a
olyvinylidene difluoride (PVDF) membrane at 320 mA for 2 h at 4°C.
The protein on the PVDF membrane was blocked with 5% fat-free milk
in the PBST solution, cultured in primary anti-Notch1 antibody
(1:1,000; cat. no. ab65297; Abcam, Cambridge, UK) and then
incubated in the secondary rabbit anti-goat IgG-HRP antibody
(1:2,000; cat. no. sc-2768; Santa Cruz Biotechnology, Dallas, TX,
USA) for ~1-2 h at room temperature. PVDF membranes were washed and
the signals of protein bands were visualized using ECL reagent
(Promega Corp.). The densitometry scan of the western blotting was
performed by Image-Pro Plus 6.0 (Media Cybernetics, Inc.,
Rockville, MD, USA). The resulting values of the proteins of
interest were normalized to GAPDH.
Cell proliferation, migration and
invasion assays
Cells (3×103) were plated in 96-well
plates and allowed to grow for 24–120 h. Subsequently, the cells in
each well were incubated with 10 µl of Cell Counting Kit-8 (CCK-8;
Dojindo Laboratories, Kumamoto, Japan) at 37°C for 4 h. The
absorbance was assessed at 450 nm using a microplate reader. Each
variant of these tests was performed in triplicate.
Cell migration was assessed with a wound-healing
assay. Briefly, the cells were plated in a 6-well plate, and 24 h
later, a scratch was made through the confluent cell monolayer
using a pipette tip. The spread of the wound closure was
photographed after 48 h.
For the invasion assays, cells (1×105)
suspended in serum-free medium were plated in the upper chamber
(8-µm pore size) coated with Matrigel (BD Biosciences, Flanklin
Lakes, NJ, USA) and 600 µl of DMEM containing 10% FBS was added to
the lower chamber to serve as a chemoattractant. Following 48 h,
the non-invaded cells were gently removed with a cotton swab, while
the cells located on the lower side of the chamber were fixed with
4% paraformaldehyde, stained with 0.1% crystal violet, air dried,
photographed and quantified by counting them in five random fields
using a light microscope (Leica Microsystems, Wetzalr, Germany).
Each experiment was repeated at least three times.
Bioinformatics analysis
Integrated analysis of three miRNA target gene
predication databases, including TargetScan (http://www.targetscan.org/vert_72/), Microcosm
(https://www.microcosm.com/) and miRanda
(http://www.microrna.org) was conducted to
identify the number of miRNA-regulated target gene pairs. Only
miRNAs shown to bind to the same region of the target 3′-UTR
sequence in all the three programs were selected.
Statistical analysis
All data are presented as the mean ± standard
deviation (SD) from at least three independent experiments with
similar results. Statistical analyses were undertaken using SPSS
version 19.0 software (IBM Corp., Armonk, NY, USA). Student's
t-test was used to evaluate differences between two groups. One-way
ANOVA test and Bonferroni post hoc test were used for evaluating
differences among multiple groups. A value of P<0.05 was
considered to indicate a statistically significant difference.
Results
The expression level of miR-449a is
decreased in CAC tissues compared with the ANTs
Since the role of the miR-449 family members, which
share the same seed region with the miR-34 family, has not been
intensively studied in IBD or CAC, we first examined the expression
of miR-449a, miR-449b and miR-449c in 41 pairs of CAC tissues and
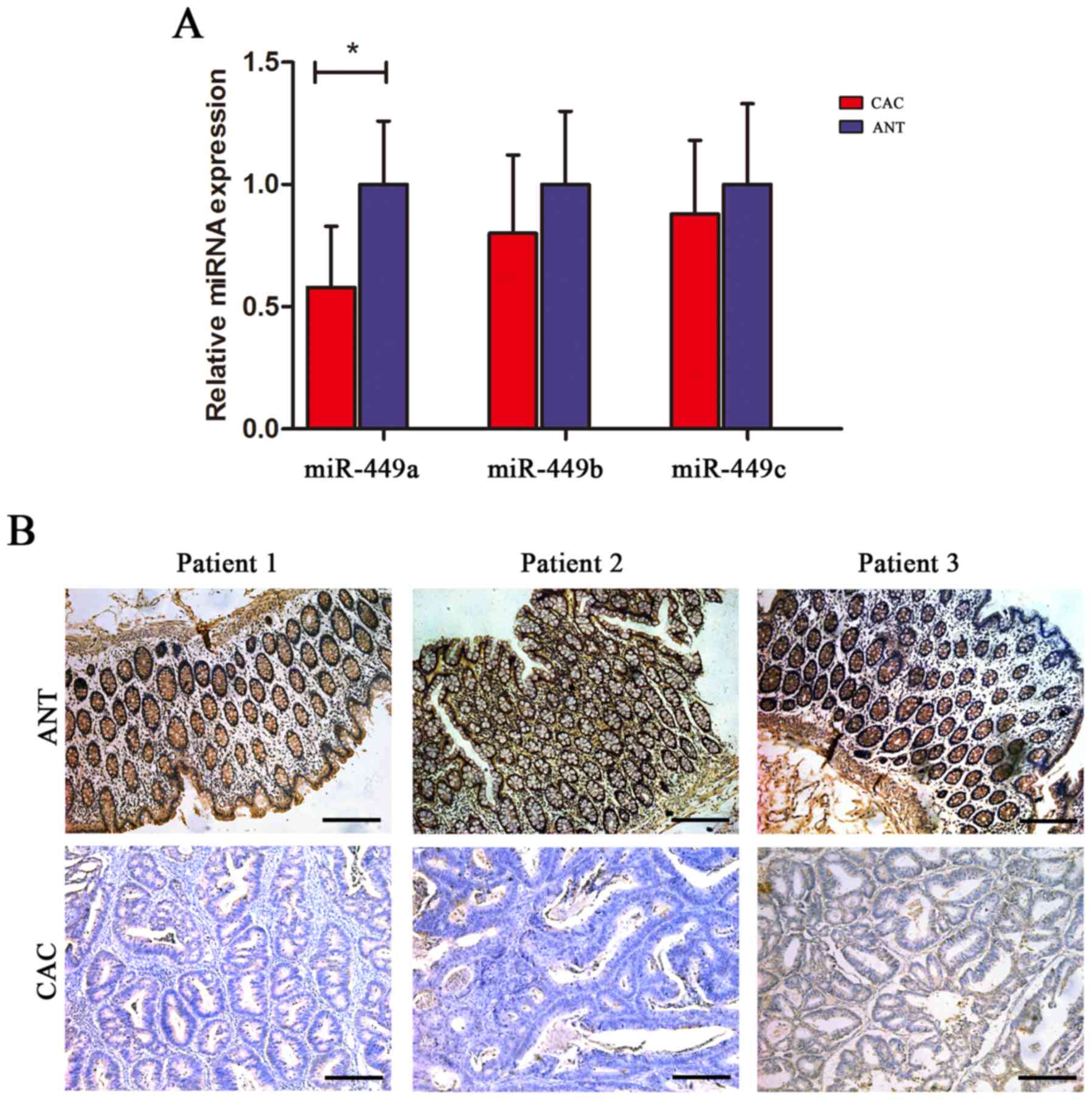
ANTs. Using RT-qPCR analysis, we found that miR-449a expression was
significantly decreased in CAC tissues compared with the ANTs,
whereas the levels of miR-449b and miR-449c did not exhibit a
statistical difference between the CAC and ANTs (Fig. 1A). Furthermore, we performed an ISH
analysis of miR-449a and confirmed that miR-449a was extensively
expressed in adjacent mucosa, either with active or inactive
inflammation, but not in malignant epithelial tissues (Fig. 1B).
Decreased miR-449a level is associated
with CAC development
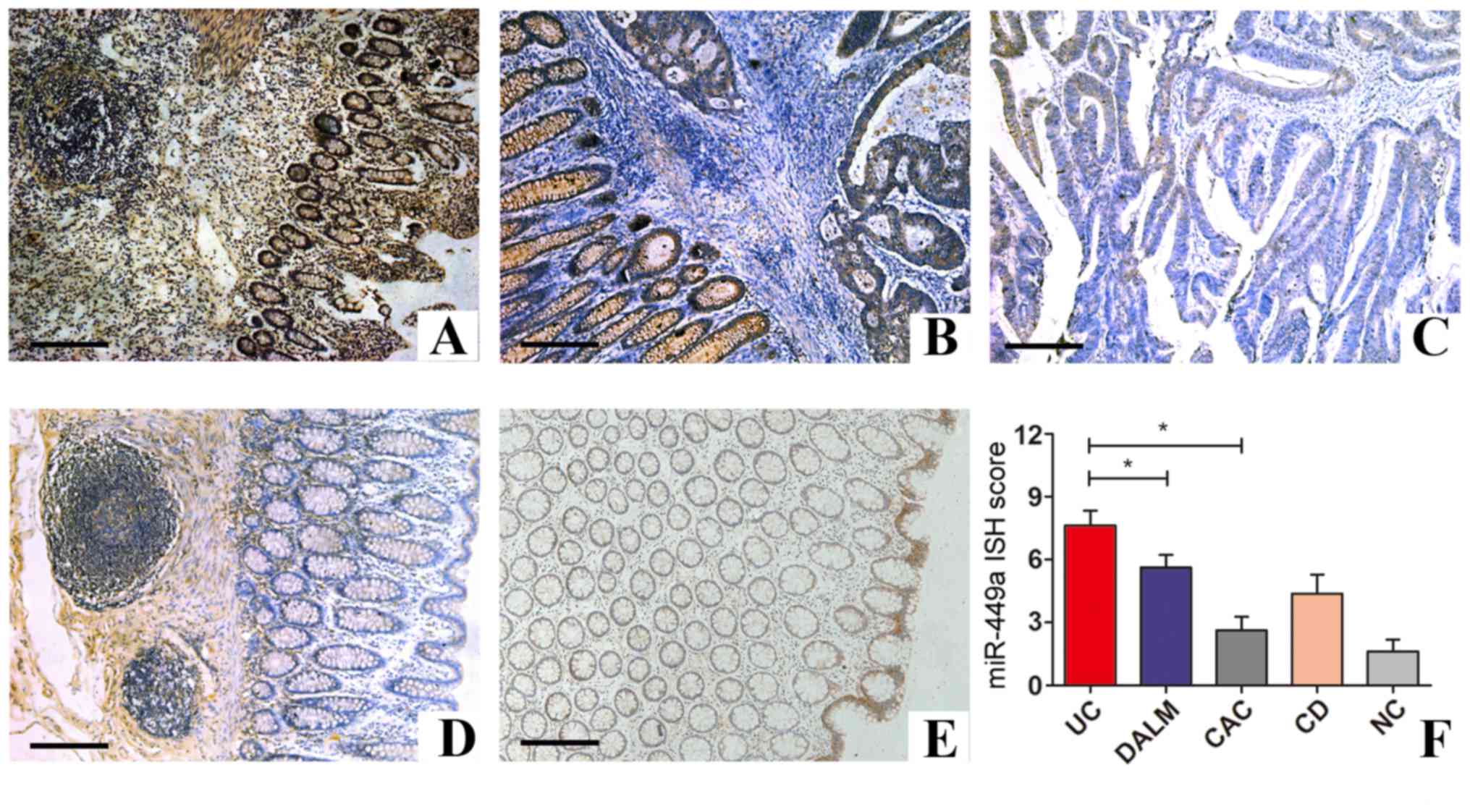
Subsequently, we investigated the expression of
miR-449a in different stages of CAC progression and in CD and
control samples by ISH staining. As shown in Table II, positive miR-449a expression was
detected in 54/63 (85.7%) cases of UC samples and in 17/28 (60.7%)
of DALM specimens, whereas CACs were predominately negative (75.6%)
for miR-449a staining. In addition, positive miR-449a staining was
observed in 26/49 (53.1%) cases of CD and only 6/40 (15.0%) cases
of normal controls free of intestinal diseases. Collectively, these
data indicated that miR-449a was mostly inactive in normal mucosa,
but was expressed at a high level during active intestinal
inflammation. Notably, the expression of miR-449a was gradually
decreased with advancing stages of UC-CAC development (Fig. 2).
 | Table II.Expression of miR-449a in different
gastrointestinal diseases. |
Table II.
Expression of miR-449a in different
gastrointestinal diseases.
|
|
| miR-449a
expression |
|
|---|
|
|
|
|
|
|---|
| Gastrointestinal
diseases | No. of
patients | Low n (%) | High n (%) | aP-value |
|---|
| Ulcerative
colitis | 63 | 9
(14.3) | 54 (85.7) |
|
| DALM | 28 | 11 (39.3) | 17 (60.7) |
|
| CAC | 41 | 31 (75.6) | 10 (24.4) | 0.002a |
| Crohn's
disease | 49 | 23 (46.9) | 26 (53.1) |
|
| Normal control | 40 | 34 (85.0) | 6
(15.0) |
|
Furthermore, we explored miR-449a expression in the
41 pairs of CAC samples to determine its clinical and pathological
relevance. ISH analysis demonstrated that low expression of
miR-449a was observed in 31/41 (75.6%) cases of CAC patients,
whereas high expression of miR-449a was observed in only 10/41
(24.4%) of the patients. Clinicopathological investigations
revealed that decreased expression level of miR-449a was associated
with local invasion (P=0.001), lymph node metastasis (P=0.024),
AJCC stage (P=0.021) and histological grade (P=0.017) of the
colitis-associated tumors, but not with patient age (P=0.232), sex
(P=0.564) or tumor location (P=0.191) (Table III). These results indicated that
suppression of miR-449a was closely linked with the aggressive
parameters of CAC and may thus represent a potential predictive
factor of CAC progression.
 | Table III.Correlation between
clinicopathological characteristics and the expression of miR-449a
in patients with CAC. |
Table III.
Correlation between
clinicopathological characteristics and the expression of miR-449a
in patients with CAC.
|
|
| miR-449a
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | No. of patients
(%) | High n=10 (%) | Low n=31 (%) | aP-value |
|---|
| Age, years |
|
<55 | 17 (41.5) | 4 (40.0) | 13 (41.9) | 0.232 |
|
≥55 | 24 (58.5) | 6 (60.0) | 18 (58.1) |
|
| Sex |
|
Male | 28 (68.3) | 7 (70.0) | 21 (67.7) | 0.564 |
|
Female | 13 (31.7) | 3 (30.0) | 10 (32.3) |
|
| Tumor location |
|
Left | 30 (73.2) | 7 (70.0) | 23 (74.2) | 0.191 |
|
Right | 11 (26.8) | 3 (30.0) | 8
(25.8) |
|
| pT status |
|
T1/T2 | 14 (34.1) | 8 (80.0) | 6
(19.4) | 0.001a |
|
T3/T4 | 27 (65.9) | 2 (20.0) | 25 (80.6) |
|
| pN status |
|
Absent | 25 (61.0) | 8 (80.0) | 17 (54.8) | 0.024a |
|
Present | 16 (39.0) | 2 (20.0) | 14 (45.2) |
|
| AJCC stage |
|
I/II | 24 (58.5) | 8 (80.0) | 16 (51.6) | 0.021a |
|
III/IV | 17 (41.5) | 2 (20.0) | 15 (48.4) |
|
| Histology |
|
Well/moderate | 31 (75.6) | 9 (90.0) | 22 (71.0) | 0.017a |
|
Poor | 10 (24.4) | 1 (10.0) | 9
(29.0) |
|
Colonic expression of miR-449a is also
decreased during CAC tumorigenesis in AOM/DSS-treated mice
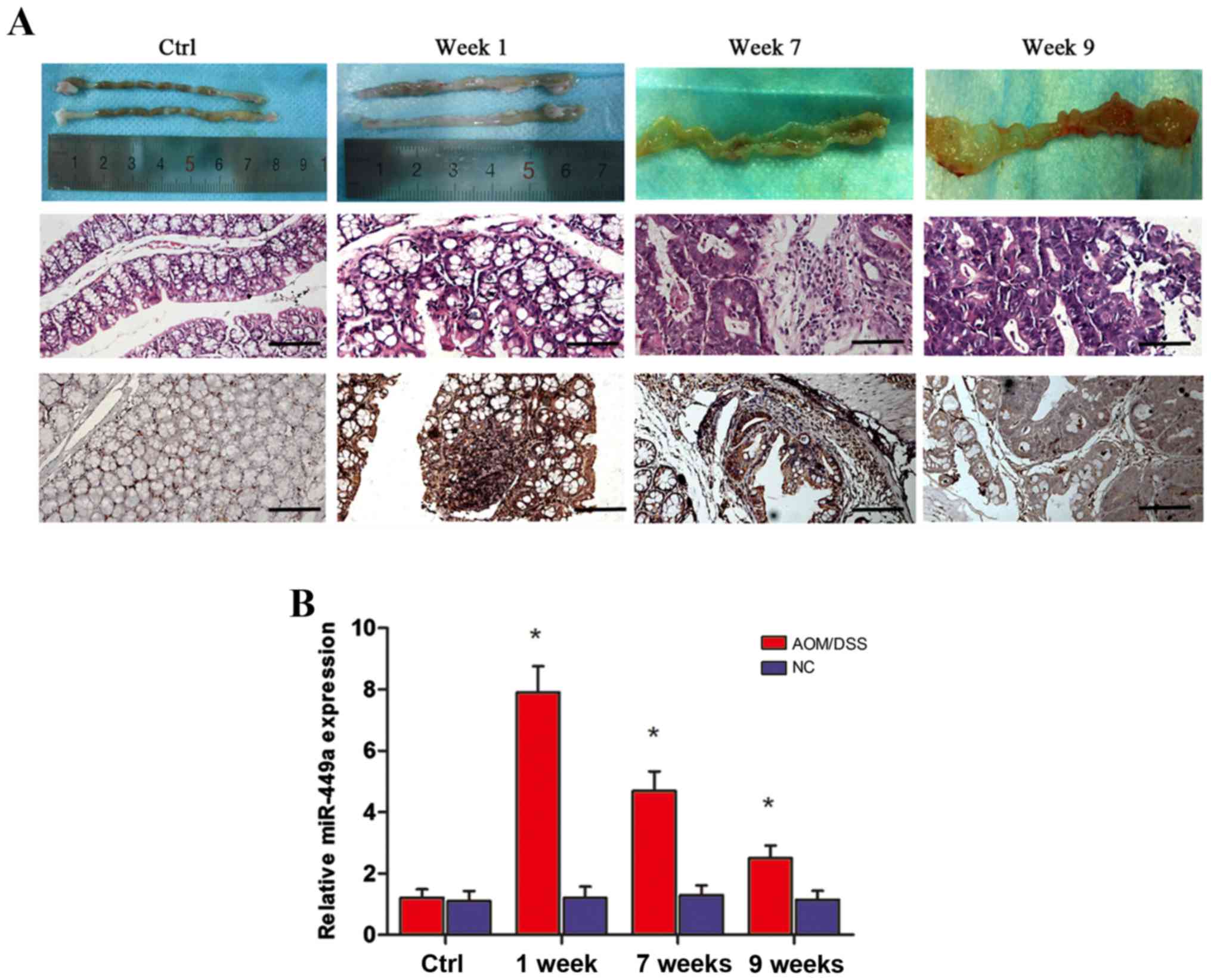
For further validation, an AOM/DSS-induced CAC mouse
model was established and RNA samples were extracted from distal
colorectal mucosa on the first day (negative control) and at the
end of week 1 (colitis), week 7 (dysplasia) and week 9 (cancer).
Morphological and histological evaluation revealed that there was
significant colonic inflammatory infiltration and active colitis by
the end of the first week; while at the 7th week time-point,
neoplastic transformation was observed in the distal colon. Lastly,
by the end of week 9, high grade dysplasia and carcinoma were
observed in the entire intestinal tract (Fig. 3A, upper and middle panels).
Subsequently, RT-qPCR and ISH analyses were used to assess the
expression level of miR-449a. As anticipated, miR-449a was markedly
increased in the inflammatory mucosa compared with the normal
colon, and then gradually declined during the course of
colitis-to-colorectal neoplasm (Fig.
3B). miR-449a exhibited strong to moderate expression in
inflammatory tissues and weak or negative staining in CAC tissues
(Fig. 3A, lower panel). Thus, these
data demonstrated dynamic expression of miR-449a during the
tumorigenic course of CAC formation in a rodent model, which, is in
line with the clinical observations.
miR-449a inhibits CAC growth and
invasion by targeting Notch-1
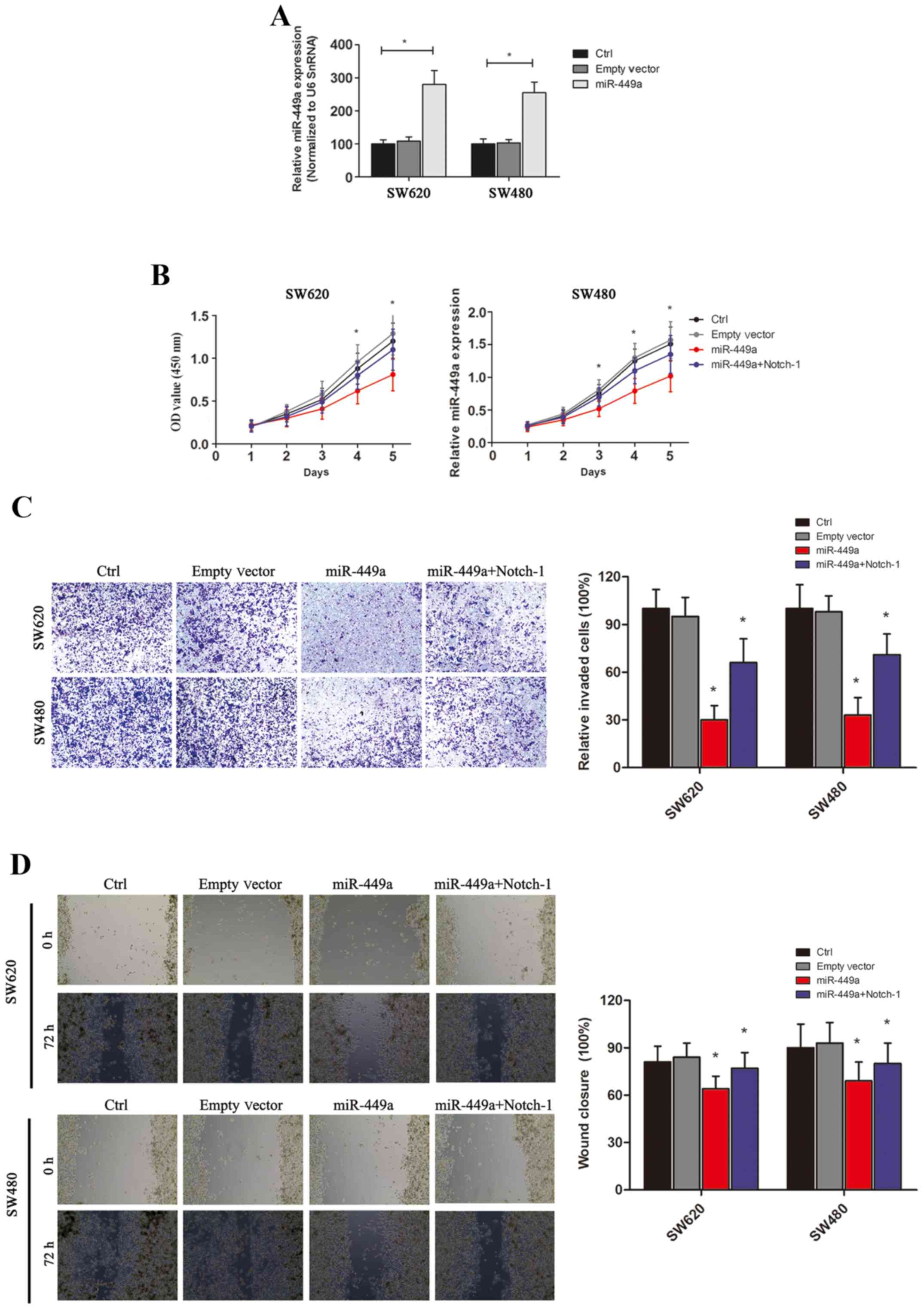
To establish the role of miR-449a in colon cancer,
we first constructed miR-449a expression vectors as well as the
empty control vectors and transfected them into the human CRC cell
lines SW620 and SW480 (Fig. 4A
displays the transfection efficiency). Subsequently, we tested the
effects of miR-449a on CRC cell proliferation by CCK-8 assay.
Overexpression of miR-449a in two CRC cell lines significantly
inhibited the proliferation rate of tumor cells in vitro,
whereas transfection of the empty controls revealed no significant
effects on cell proliferation (Fig.
4B). We next investigated the influence of miR-449a on CRC cell
migration and invasion. Using a Transwell assay, we observed that
significantly less miR-449a-overexpressed SW480 or SW620 cells
invaded through the Matrigel and migrated to the lower surface of
the filter, compared with the respective controls (Fig. 4C), indicating that miR-449a
effectively reduced the invasive capability of CRC cells.
Furthermore, the wound-healing assay revealed that miR-449a
significantly inhibited cell migration (Fig. 4D). Collectively, these results
demonstrated that miR-449a may act as an important tumor suppressor
in CRC, as well as CAC.
Subsequently, we explored the mechanism by which
suppression of miR-449a during CAC development promoted tumor
growth. After searching three miRNA prediction databases
(TargetScan, Microcosm and miRanda), a putative miR-449a binding
site in the 3′-UTR of Notch-1, a well-characterized oncogene in
human CRC, was identified. Previous evidence has demonstrated that
activated Notch signaling is maintained throughout CRC
tumorigenesis (25). In addition,
enhanced levels of Notch-1 have been associated with progression,
tumor grade and metastasis (26).
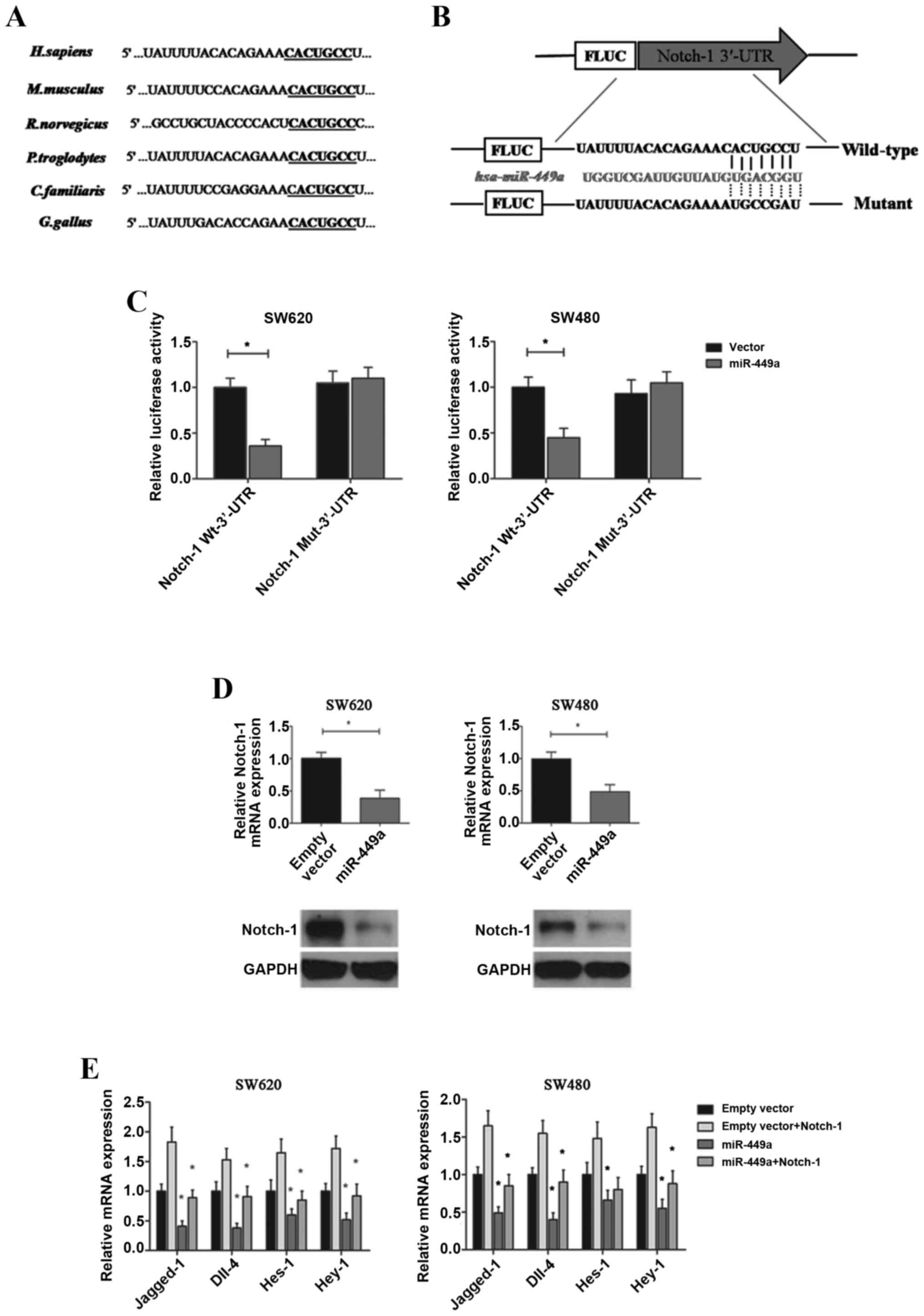
Using the miRNA target prediction programs, we found that the
3′-UTRs of Notch-1 mRNAs contain conserved miR-449a binding sites,
which are highly conserved among human, mouse, rat, chimpanzee, dog
and chicken species (Fig. 5A). We
thus conducted a dual-luciferase reporter assay by constructing
luciferase reporter constructs containing the wild-type or mutant
Notch-1 3′-UTR (Fig. 5B) and
co-transfected them with miR-449a expression vectors or the empty
controls into target cells. We found that overexpression of
miR-449a markedly reduced the luciferase activity of the wild-type
3′-UTR of Notch-1, but not the mutant reporter constructs, in both
SW620 and SW480 cells (Fig. 5C).
Further PCR and western blot verifications confirmed the direct
inhibition of miR-449a on the gene and protein levels of Notch-1
expression (Fig. 5D). Additionally,
we examined the influence of miR-449a on intracellular Notch
signaling. It was observed that Jagged-1, Dll-4, Hes-1 and Hey-1,
which are key factors of the canonical Notch pathway and are
critical regulators of CRC/CAC metastasis and angiogenesis
(27), were all significantly
decreased after miR-449a overexpression in SW620 and SW480 cells.
However, through the restoration of Notch-1 expression, these
effects were partially reversed (Fig.
5E). Lastly, we performed in vitro studies and confirmed
that Nocth-1 restoration could also reverse the inhibition of
miR-449a on CRC cell proliferation, migration and invasion
(Fig. 4B-D). Therefore, our
findings indicated that miR-449a inhibited the tumorigenesis of CAC
by directly targeting Notch-1 and its downstream oncogenic
effectors, and the decreased miR-449a level was supposed to be an
important factor for sustained activation of the Notch signaling in
CAC progression.
Discussion
It is widely accepted that perpetuated intestinal
inflammation markedly increases the risk of cancer. However, the
detailed mechanisms concerning the biological and genetic
alterations that endow malignant cells with growth and motility
advantages over normal epithelial cells have not been
comprehensively elucidated. In the present study, we identified a
novel miRNA, miR-449a, that was expressed in a decreased pattern
during the neoplastic transformation of CAC. In addition, we
demonstrated that miR-449a served its tumor-suppressor functions by
inhibiting the Notch-1 signaling pathway. These findings may aid in
uncovering the molecular events that drive CAC progression, and may
indicate the broad clinical prospects for miR-449a in the
management of human CAC.
It is known that miRNA genomic clusters are often
co-transcribed as related miRNAs, which share the same seed region
or belong to functionally related miRNA families. A typical example
is given by the miR-34/449 superfamily, which exhibits extensive
genomic redundancy by encompassing six homologous miRNAs (miR-34a,
miR-34b/c and miR-449a/b/c) located on three distinct genomic loci
(28). While the tumor suppressor
role of miR-34 has been largely established, the effects of the
miR-449 family members have not been addressed in human CRC, as
well as CAC (29,30). Therefore, we first determined the
expression of miR-449a, miR-449b and miR-449c in colon samples
obtained from CAC patients with a clear history of chronic colitis,
and we determined that only miR-449a was significantly lower in CAC
tissues than in ANTs. Further ISH staining demonstrated that
miR-449a was markedly activated in inflamed UC tissues, and
expressed increasingly less in DALM and CAC samples. Notably, high
expression of miR-449a was detected in most UC tissues (85.7%),
however, half the CD samples expressed miR-449a at low levels
(46.9%), which is probably due to the different immunoreaction
types and inflammatory activities between the two subtypes of IBD
(31,32). Specifically, these findings also
identified miR-449a as a molecular candidate for the discrimination
of IBD subtypes.
Long-term UC patients are known to have a high risk
for developing CRC compared with the general population. The
current standard of care for surveillance of chronic UC patients
largely depends on routine colonoscopy examination (33,34),
it is therefore important to identify and develop biomarkers for
stratifying the patients who undergo repeated colorectal
inflammation and have a relatively high cancer risk. Since CAC
progresses in a step-wise fashion, which is usually characterized
as colitis negative for dysplasia (NEG), indefinite for dysplasia
(IND), low-grade dysplasia (LGD) and high-grade dysplasia (HGD)
cancer, new molecular markers detecting the premalignant lesions to
discriminate UC progressors from non-progressors are useful for CAC
prevention (35,36). An ideal biomarker for diagnosis
should be highly specific, sensitive and less invasive. Previous
research has demonstrated that miRNAs exhibit unique expression
profiles in different tumor types and are critical for the
initiation and progression of human cancers (37). The crucial functions of miRNAs in
tumorigenesis have promoted intensive research into miRNA-based
diagnostic and therapeutic applications. In the present study, for
the first time, we identified miR-449a as a novel CAC-associated
miRNA that was expressed in close relevance with patient disease
status during UC-CAC progression. Notably, a CAC rodent model
established by AOM/DSS induction further confirmed that miR-449a
followed a linear pathway of downregulation in CAC carcinogenesis.
Furthermore, our clinicopathological investigation on a cohort of
41 CAC patients revealed that decreased miR-449a expression was
also associated with advanced T or N status, later clinical stage,
and poor histological differentiation of tumors. These data
collectively provided strong evidence for the translational
potential of miR-449a in the prediction, diagnosis and prognosis of
human CAC.
Cancer formation can be attributed to the
dysfunction of cellular signaling transduction. The Notch pathway,
which is highly conserved across evolution, is one of the master
regulators of colonic epithelium development, differentiation and
cell proliferation and apoptosis (38). Abnormal Notch signaling along with
genetic or epigenetic mutations has been highly noted in a variety
of gastrointestinal pathologies, including mucosal inflammation and
tumorigenesis (25,39). Modulation of Notch signaling
therefore, offers an approach for CRC treatment (40). In the present study, we revealed
that miR-449a is a novel Notch-targeting miRNA, by directly binding
to the 3′-UTR of Notch-1, a major receptor of signaling; miR-449a
markedly suppressed Notch-1 expression at the gene and protein
levels. It has been documented that Notch-1 elevation was
positively associated with tumor growth, metastasis and poor
clinical outcomes in human CRC, which could be related to
inhibition of apoptosis promoted by Notch-1 (41). In the present study, we demonstrated
that the key effectors of the canonical Notch pathway, such as
Jagged-1, Dll-4, Hes-1 and Hey-1, were all significantly suppressed
by miR-449a overexpression and were partially reversed after
Notch-1 restoration. Thus, miR-449a suppresses CAC via the specific
regulation of the Notch signaling cascade.
In conclusion, the present study has characterized
miR-449a as a novel tumor suppressor miRNA by targeting the Notch
pathway. The expression of miR-449a was gradually decreased during
the progression of CAC, which, renders it a promising predictive
and diagnostic factor for UC-CAC patients. Further large-scale
clinical investigations and long-term prognostic analyses are still
needed for the verification of the clinical and therapeutic
application of miR-449a in human CAC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81270518 and
81470858).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YF, YWD, YNS and JHX performed the experiments, LGL
designed the study, XYG and WLJ prepared and wrote the study. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shanghai General Hospital and the Ethics Committee of Shanghai East
Hospital. The clinical study protocol strictly conformed to the
ethical guidelines of the 1975 Declaration of Helsinki. Written
informed consent was obtained from all of the patients. All
protocols concerning laboratory animal use were validated by the
Animal Care Ethics Committee of Shanghai General Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
CRC
|
colorectal cancer
|
|
CAC
|
colitis-associated colorectal
cancer
|
|
3′-UTR
|
3′-untranslated region
|
|
IBD
|
inflammatory bowel disease
|
|
UC
|
ulcerative colitis
|
|
CD
|
Crohn's disease
|
|
DALM
|
dysplasia-associated lesion or
mass
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abraham C and Cho JH: Inflammatory bowel
disease. N Engl J Med. 361:2066–2078. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kaplan GG and Ng SC: Globalisation of
inflammatory bowel disease: Perspectives from the evolution of
inflammatory bowel disease in the UK and China. Lancet
Gastroenterol Hepatol. 1:307–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fumery M, Dulai PS, Gupta S, Prokop LJ,
Ramamoorthy S, Sandborn WJ and Singh S: Incidence, risk factors,
and outcomes of colorectal cancer in patients with ulcerative
colitis with low-grade dysplasia: A systematic review and
meta-analysis. Clin Gastroenterol Hepatol. 15:665–674.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rogler G: Chronic ulcerative colitis and
colorectal cancer. Cancer Lett. 345:235–241. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eaden JA, Abrams KR and Mayberry JF: The
risk of colorectal cancer in ulcerative colitis: A meta-analysis.
Gut. 48:526–535. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herrinton LJ, Liu L, Levin TR, Allison JE,
Lewis JD and Velayos F: Incidence and mortality of colorectal
adenocarcinoma in persons with inflammatory bowel disease from 1998
to 2010. Gastroenterology. 143:382–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sebastian S, Hernández V, Myrelid P, Kariv
R, Tsianos E, Toruner M, Marti-Gallostra M, Spinelli A, van der
Meulen-de Jong AE, Yuksel ES, et al: Colorectal cancer in
inflammatory bowel disease: Results of the 3rd ECCO pathogenesis
scientific workshop (I). J Crohns Colitis. 8:5–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yashiro M: Ulcerative colitis-associated
colorectal cancer. World J Gastroenterol. 20:16389–16397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ording AG, Horváth-Puhó E, Erichsen R,
Long MD, Baron JA, Lash TL and Sørensen HT: Five-year mortality in
colorectal cancer patients with ulcerative colitis or Crohn's
disease: A nationwide population-based cohort study. Inflamm Bowel
Dis. 19:800–805. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Itzkowitz SH and Yio X: Inflammation and
cancer IV. Colorectal cancer in inflammatory bowel disease: The
role of inflammation. Am J Physiol Gastrointest Liver Physiol.
287:G7–G17. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo C and Zhang H: The role of
proinflammatory pathways in the pathogenesis of colitis-associated
colorectal cancer. Mediators Inflamm. 2017:51260482017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kinugasa T and Akagi Y: Status of
colitis-associated cancer in ulcerative colitis. World J
Gastrointest Oncol. 8:351–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ambros V: MicroRNAs and developmental
timing. Curr Opin Genet Dev. 21:511–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saraggi D, Fassan M, Mescoli C, Scarpa M,
Valeri N, Michielan A, D'Incá R and Rugge M: The molecular
landscape of colitis-associated carcinogenesis. Dig Liver Dis.
49:326–330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Romano M, De Francesco F, Zarantonello L,
Ruffolo C, Ferraro GA, Zanus G, Giordano A, Bassi N and Cillo U:
From inflammation to cancer in inflammatory bowel disease:
Molecular perspectives. Anticancer Res. 36:1447–1460.
2016.PubMed/NCBI
|
|
18
|
Schlemper RJ, Riddell RH, Kato Y, Borchard
F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preiser CM, Fléjou JF,
Geboes K, et al: The Vienna classification of gastrointestinal
epithelial neoplasia. Gut. 47:251–255. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ungaro R, Mehandru S, Allen PB,
Peyrin-Biroulet L and Colombel JF: Ulcerative colitis. Lancet.
389:1756–1770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Torres J, Mehandru S, Colombel JF and
Peyrin-Biroulet L: Crohn's disease. Lancet. 389:1741–1755. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bader JE, Enos RT, Velázquez KT, Carson
MS, Nagarkatti M, Nagarkatti PS, Chatzistamou I, Davis JM, Carson
JA, Robinson CM, et al: Macrophage depletion using clodronate
liposomes decreases tumorigenesis and alters gut microbiota in the
AOM/DSS mouse model of colon cancer. Am J Physiol Gastrointest
Liver Physiol. 314:G22–G31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y,
Zhang T, Khaliq J and Li Y: miR-140-5p suppresses the
proliferation, migration and invasion of gastric cancer by
regulating YES1. Mol Cancer. 16:1392017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen J, Zhang S, Li Y, Zhang W, Chen J,
Zhang M, Wang T, Jiang L, Zou X, et al: p14ARF inhibits the
functions of adenovirus E1A oncoprotein. Biochem J. 434:275–285.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vinson KE, George DC, Fender AW, Bertrand
FE and Sigounas G: The Notch pathway in colorectal cancer. Int J
Cancer. 138:1835–1842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li G, Zhou Z, Zhou H, Zhao L, Chen D, Chen
H, Zou H, Qi Y, Jia W and Pang L: The expression profile and
clinicopathological significance of Notch1 in patients with
colorectal cancer: A meta-analysis. Future Oncol. 13:2103–2118.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamy M, Ferreira A, Dias JS, Braga S,
Silva G and Barbas A: Notch-out for breast cancer therapies. N
Biotechnol. 39:215–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mercey O, Popa A, Cavard A, Paquet A,
Chevalier B, Pons N, Magnone V, Zangari J, Brest P, Zaragosi LE, et
al: Characterizing isomiR variants within the microRNA-34/449
family. FEBS Lett. 591:693–705. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang LG, Ni Y, Su BH, Mu XR, Shen HC and
Du JJ: MicroRNA-34b functions as a tumor suppressor and acts as a
nodal point in the feedback loop with Met. Int J Oncol. 42:957–962.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang L and Hermeking H: miR-34a and
miR-34b/c suppress intestinal tumorigenesis. Cancer Res.
77:2746–2758. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wallace KL, Zheng LB, Kanazawa Y and Shih
DQ: Immunopathology of inflammatory bowel disease. World J
Gastroenterol. 20:6–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Peyrin-Biroulet L, Panés J, Sandborn WJ,
Vermeire S, Danese S, Feagan BG, Colombel JF, Hanauer SB and
Rycroft B: Defining disease severity in inflammatory bowel
diseases: Current and future directions. Clin Gastroenterol
Hepatol. 14:348–354.e7. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hata K, Kishikawa J, Anzai H, Shinagawa T,
Kazama S, Ishii H, Nozawa H, Kawai K, Kiyomatsu T, Tanaka J, et al:
Surveillance colonoscopy for colitis-associated dysplasia and
cancer in ulcerative colitis patients. Dig Endosc. 28:260–265.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feuerstein JD and Cheifetz AS: Ulcerative
colitis: Epidemiology, diagnosis, and management. Mayo Clinic Proc.
89:1553–1563. 2014. View Article : Google Scholar
|
|
35
|
Bressenot A, Cahn V, Danese S and
Peyrin-Biroulet L: Microscopic features of colorectal neoplasia in
inflammatory bowel diseases. World J Gastroenterol. 20:3164–3172.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen R, Lai LA, Brentnall TA and Pan S:
Biomarkers for colitis-associated colorectal cancer. World J
Gastroenterol. 22:7882–7891. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
de Santa Barbara P, van den Brink GR and
Roberts DJ: Development and differentiation of the intestinal
epithelium. Cell Mol Life Sci. 60:1322–1332. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fazio C and Ricciardiello L: Inflammation
and Notch signaling: A crosstalk with opposite effects on
tumorigenesis. Cell Death Dis. 7:e25152016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Takebe N, Nguyen D and Yang SX: Targeting
notch signaling pathway in cancer: Clinical development advances
and challenges. Pharmacol Ther. 141:140–149. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chu D, Zhang Z, Zhou Y, Wang W, Li Y,
Zhang H, Dong G, Zhao Q and Ji G: Notch1 and Notch2 have opposite
prognostic effects on patients with colorectal cancer. Ann Oncol.
22:2440–2447. 2011. View Article : Google Scholar : PubMed/NCBI
|