Introduction
Colorectal cancer (CRC) is the third most malignant
tumor and common cause of death in both men and women worldwidely
(1). Radiotherapy (RT),
chemotherapy and surgical resection are the conventional treatment
options of colorectal cancer (2).
In recent years, radiotherapy has generally become a routine
treatment for colorectal cancer (3). Compared to standardized total
mesorectal excision (TME) alone, preoperative radiotherapy can
produce a higher survival rate (4).
However, radiation resistance can lead to tumor recurrence and poor
prognosis. Consequently, overcoming tumor radioresistance and
promoting radiosensitivity are of vital importance for the
treatment and prognosis of CRC.
Non-coding RNAs (ncRNAs) are functional RNA
molecules that cannot be translated into proteins, compared to
protein-coding RNAs (5). These
ncRNAs are divided into short ncRNAs and long non-coding RNAs
(lncRNAs) according to their length (6). As a member of ncRNA family, lncRNAs
are defined as an RNA transcript of more than 200 nucleotides (nt)
(7,8). In the last decade, a number of lncRNAs
have been revealed to play an important role in the regulation of
tumors. Recently, it was revealed that MALAT1 promoted CRC tumor
development via its target protein AKAP-9 (9). Upregulation of HOTAIR drove malignancy
in gastrointestinal stromal tumors (10). High expression of lncRNA DANCR
(anti-differentiation ncRNA) was associated with poor prognosis for
both OS and DFS in CRC (11).
lncRNA-UCA1 has been demonstrated to play an oncogenic role in
breast cancer in part through the suppression of p27 and by
influencing cell proliferation, apoptosis and cell cycle
distribution of CRC cells (12,13).
In addition, UCA1 had been reported in many other cancers such as
tongue squamous cell carcinomas, bladder cancer, melanoma and
non-small cell lung cancer (14–19).
Εpithelial-mesenchymal transition (EMT) is a process
characterized by loss of cell-cell adhesion and gain of migratory
traits (20). The EMT process has
been reported in many types of cancer such as colorectal cancer
(20,21). EMT is controlled by many
transcriptional regulators and non-coding RNAs (22). lncRNA H19 modulated EMT by acting as
a competing endogenous RNA in CRC (20) knockdown of SPRY4-IT1 and MALAT1
inhibited EMT phenotype in glioma cells and cervical cancer cells
(23,24). To date, there is no study concerning
the influence of UCA1 on EMT.
Although UCA1 has been revealed to play an important
role in tumor regulation, whether it modulated the radiosensitivity
of CRC cells was still unknown. In the present study, we aimed to
explore the impact of UCA1 silencing on EMT and radiosensitivity of
CRC.
Materials and methods
Collection of clinical samples
CRC tissues and adjacent normal tissues from 32 CRC
patients were collected immediately after surgical resection from
The Second Affiliated Hospital of Soochow University (Suzhou,
China). Fresh colorectal cancer tissues and the paired adjacent
normal tissues with a distance of 10 cm from the border of the
tumor tissues were stored at −80°C. None of the patients had
undergone any treatment before surgical resection. In addition,
four pairs of CRC tissues before and after radiotherapy were
collected from The Second Affiliated Hospital of Soochow. The
present study was conducted in accordance with the Declaration of
Helsinki and approved by the Ethics Committee of Soochow
University. Signed informed consent was obtained from all
participants. The diagnosis of CRC was based on the World Health
Organization criteria. The clinical stage of CRC was evaluated
based on the TNM classification system. The data included age, sex,
tumor size, stage, differentiation levels, invasion level such as
lymphatic invasion, perineural invasion and vascular invasion.
Tables I and II reveal the clinical characteristics of
these patients. In addition, the radiotherapy effect was evaluated
according to the WHO standards: CR, tumor completely disappeared;
PR, >50% tumor shrinkage; SD, not >50% tumor shrinkage; PD,
tumor volume increased.
 | Table I.Clinical characteristics of the CRC
patients. |
Table I.
Clinical characteristics of the CRC
patients.
| Clinical
parameters | Numbers |
|---|
| Cases | 32 |
| Age (years) |
|
<65 | 17 |
|
≥65 | 15 |
| Tumor size
(cm) |
| Small
size (<5) | 14 |
| Large
size (≥5) | 18 |
| Sex |
|
Male | 16 |
|
Female | 16 |
| Invasion
levels |
|
Mucosa | 2 |
|
Submucosa | 3 |
|
Muscle | 6 |
|
Serosa | 21 |
| TNM stage |
| Stage
1–2 | 10 |
| Stage
3–4 | 22 |
| Lymph node
metastasis |
|
Positive | 15 |
|
Negative | 17 |
| Grade of
tumors |
| Low
grade | 8 |
|
Intermediate grade | 22 |
| High
grade | 2 |
| Vascular
invasion |
|
Positive | 12 |
|
Negative | 20 |
| Perineural
invasion |
|
Positive | 11 |
|
Negative | 21 |
 | Table II.Clinical characteristics of the CRC
patients who received radiotherapy. |
Table II.
Clinical characteristics of the CRC
patients who received radiotherapy.
| Sample number | Treatment
protocols | Radiotherapy
effect | Distant
metastasis |
|---|
| 1 | 6MvX-ray IMRT
DT5000cGy/200cGy/25f mFOLFOX6 | PR | NO |
| 2 | 6MvX-ray 3DCRT
Dt4500cGy/180Gy/25f FOLFOX | PR | NO |
| 3 | 6MvX-ray IMRT
DT5000cGy/200cGy/25f XELOX | SD | NO |
| 4 | 6MvX-ray 3DCRT
Dt4500cGy/180Gy/25f XELOX | SD | NO |
Cell culture and treatment
Human CRC-derived cell lines HCT116, CCL244, SW480,
LoVo and normal epithelial cells FHC were purchased from the
Shanghai Institute of Cell Biology (Shanghai, China) and stored in
the laboratory of the School for Radiological and Interdisciplinary
Sciences, Soochow University (Suzhou, China). All the cells were
cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM;
HyClone Laboratories; GE Healthcare, Chicago, IL, USA) supplemented
with 10% fetal bovine serum (FBS; Biological Industries, Kibbutz
Beit Haemek, Israel), 100 U/ml penicillin G and 100 mg/ml
streptomycin (HyClone Laboratories; GE Healthcare) and maintained
at 37°C in a humidified atmosphere with 5% CO2. Cells
were exposed to a single dose of X-ray irradiation from a linear
accelerator (Rad Source Technologies, Inc., Suwanee, GA, USA) at a
dose rate of 1.15 Gy/min or sham-irradiated.
Construction and transfection of
siRNAs
The siRNA sequences silencing human UCA1 (si-UCA1)
were designed and synthesized by Shanghai GenePharma, Co., Ltd.
(Shanghai, China). The negative control siRNA was also provided by
Shanghai GenePharma Co., Ltd. The siRNA sequences are presented in
Table III. CCL244 cells were
transiently transfected using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). After
transfection for 4 h, the serum-free medium was replaced by a
medium containing 10% FBS. After transfection for 48 h, the cells
were harvested for further experiments. Quantitative real-time PCR
was used to determine the efficiency of transfection.
 | Table III.Target sequences of UCA1 siRNAs and
negative control (NC) siRNA. |
Table III.
Target sequences of UCA1 siRNAs and
negative control (NC) siRNA.
| siRNAs | Target
sequences |
|---|
| si-UCA1-1
sense |
5′-GAGCCGAUCAGACAAACAATT-3′ |
| si-UCA1-1
antisense |
5′-UUGUUUGUCUGAUCGGCUCTT-3′ |
| si-UCA1-2
sense |
5′-GGGCUUGGGACAUUUCACUTT-3′ |
| si-UCA1-2
antisense |
5′-AGUGAAAUGUCCCAAGCCCTT-3′ |
| si-UCA1-3
sense |
5′-GGGAAUACUAUUCGUAUGATT-3′ |
| si-UCA1-3
antisense |
5′-UCAUACGAAUAGUAUUCCCTT-3′ |
| NC sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| NC antisense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
RNA extraction and real-time
RT-PCR
Total RNA was extracted from cells and tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
reverse transcription of mRNAs was performed using a Reverse
Transcription kit (Roche, Basel, Switzerland). The real-time PCR
assay was carried out with a SYBR-Green kit (Tiangen Biotech Co.,
Beijing, China) on an ABI 7500 system (Applied Biosystems, Foster
City, CA, USA). The thermocycling conditions were as follows:
Initial denaturation: At 94°C for 3 min 30 cycles: At 94°C for 30
sec, at 55°C for 30 sec, and at 72°C for 1 min; final extension: At
72°C for 5 min. The GAPDH genes were used for the detection of
normalization of each sample. The relative expression of UCA1 was
calculated using the 2−ΔΔCt method. The primers of UCA1
and GAPDH were designed by Shanghai GenePharma Co., Ltd. The primer
sequences are presented in Table
IV.
 | Table IV.Primer sequences for qRT-PCR
analysis. |
Table IV.
Primer sequences for qRT-PCR
analysis.
| Gene | Forward
sequence | Reverse
sequence |
|---|
| UCA1 |
5′-GCCCCTTGGACCATCACA-3′ |
5′-GACGGCAGTTGGTGTGCTAT-3′ |
| GAPDH |
5′-CATGAGAAGTATGACAACAGCCT-3′ |
5′-AGTCCTTCCACGATACCAAAGT-3′ |
Cell proliferation assays
An MTT assay was used to examine cell proliferation.
After transfection, the cells were seeded in a 96-well plate at
2,000 cells/well and cultured in normal medium. The cells were
subjected to 8-Gy irradiation after 24 h. After 48 and 72 h, the
cells were incubated in 0.5 mg/ml MTT at 37°C for 4 h and lysed in
dimethyl sulfoxide (DMSO) at room temperature for 10 min. Then,
viability was assessed by obtaining the optical density (OD) values
using a microplate reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) at a wavelength of 490 nm.
Colony-formation assay
After transfection, the cells were inoculated into
6-well plates at 300, 500, 3,000 and 8,000 cells/well and exposed
to X-rays with different irradiation doses of 0, 2, 4, 6 and 8 Gy
after 24 h. Following 14 days of incubation, the colonies were
fixed with 4% paraformaldehyde and stained with Giemsa. Colonies
with a minimum of 50 cells were counted. The radiation sensitivity
enhancement ratio (SER) was measured according to the multi-target
single hit model.
Cell apoptosis assays
Apoptosis was assessed using Annexin V/propidium
iodide (PI) double-staining following the manufacturer's
instructions (BD Biosciences, San Jose, CA, USA). After
transfection with siRNA and NC-siRNA for 24 h, the cells were
treated with 8 Gy X-ray irradiation. Cells were washed twice with
cold PBS and resuspended in 1× binding buffer at a concentration of
1×106 cells/ml. Then, 100 µl of the solution
(1×105 cells) was transferred into a 2-ml culture tube.
Next, the cells were incubated for 15 min in the dark following the
addition of 5 µl FITC-Annexin V and 5 µl of PI. Subsequently, 400
µl of 1× binding buffer was added into each tube. Flow cytometric
analysis was performed with the BD FACSDiva™ software
(BD Bioscienes, San Jose, CA, USA) within 1 h.
Cell cycle distribution analysis
After transfection, the cells were subjected to 6-Gy
irradiation. Twenty-four hours later, the cells were harvested by
trypsin digestion, pelleted by centrifugation, washed with ice-cold
PBS, and then fixed with 75% cold ethanol overnight. The staining
solution containing PI (50 mg/ml) (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and DNA-free RNAs (20 mg/ml) were added 30 min
before the detection. The fraction of the population in each phase
of the cell cycle was determined as a function of the DNA content
using a coulter flow cytometer (Beckman Coulter, Inc., Brea, CA,
USA).
Cell wound healing assay
lncRNA-UCA1-silenced and negative control cells with
or without 6-Gy irradiation were seeded in 6-well plates and
incubated overnight. Wounds were created by scratching cell
monolayers with a sterile 200-µl plastic pipette tip. The cells
were further incubated in a hypoxic environment for 24 h and images
were monitored using phase-contrast microscopy (Nikon Corp., Tokyo,
Japan). The analysis of the cell migration assay was performed
using the ImageJ software (National Institutes of Health, Bethesda,
MD, USA).
Protein extraction and western blot
analysis
Cells (1×105 cells/dish) in 6-well plates
were incubated in DMEM containing 10% FBS to 70% confluence. The
cells were transfected with siRNA for 24 h and then were
sham-irradiated or exposed to 6-Gy X-ray irradiation. The cells
were then harvested in lysis buffer with 1 mM phenylmethylsulfonyl
fluoride (PMSF). Subsequently, the cells were allowed to swell on
ice for 30 min. The homogenates were centrifuged for 15 min at
12,000 × g and the supernatant was used as the cytosolic extract.
Proteins were separated using 10% sodium dodecyl sulfate
poly-acrylamide gel electrophoresis (SDS-PAGE) and transferred onto
polyvinylidene difluoride (PVDF) membranes The membranes were
blocked with 5% non-fat dried milk in TBS containing 1% Tween-20
(TBST) for 2 h at room temperature, followed by incubation with the
appropriate primary antibodies caspase-3 (cat. no. sc-7272), Bcl-2
(sc-509; both from Santa Cruz Biotechnology, Inc., Santa Cruz, CA),
MMP2 (cat. no. ab92536), MMP9 (cat. no. ab76003), ZEB1 (cat. no.
ab124512) and vimentin (cat. no. ab137321; all from Abcam,
Cambridge, MA, USA) at a 1:1,000 dilution for 2 h, and either
horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse
antibodies (cat. nos. A0208 and A0216, respectively; both from
Beyotime Institute of Biotechnology, Nantong, China) for 1 h.
β-actin (Beyotime Institute of Biotechnology) was used as the
loading control. Antibody-bound proteins were detected using an ECL
Stable Peroxide solution (PointBio, Shanghai, China). All protein
bands were visualized using a FluroChem MI imaging system (Alpha
Innotech, Santa Clara, CA, USA) at the room temperature.
Statistical analysis
All experiments were performed at least three times
(n=3). Data were expressed as the means ± SEM. Student's t-test was
used to analyze the data when only two groups were compared or
one-way analysis of variance (ANOVA) when more than two groups were
compared. Differences were considered statistically significant
when P<0.05. SPSS Statistical software (version 19.0; IBM Corp.,
Armonk, NY, USA) was utilized for statistical analysis.
Results
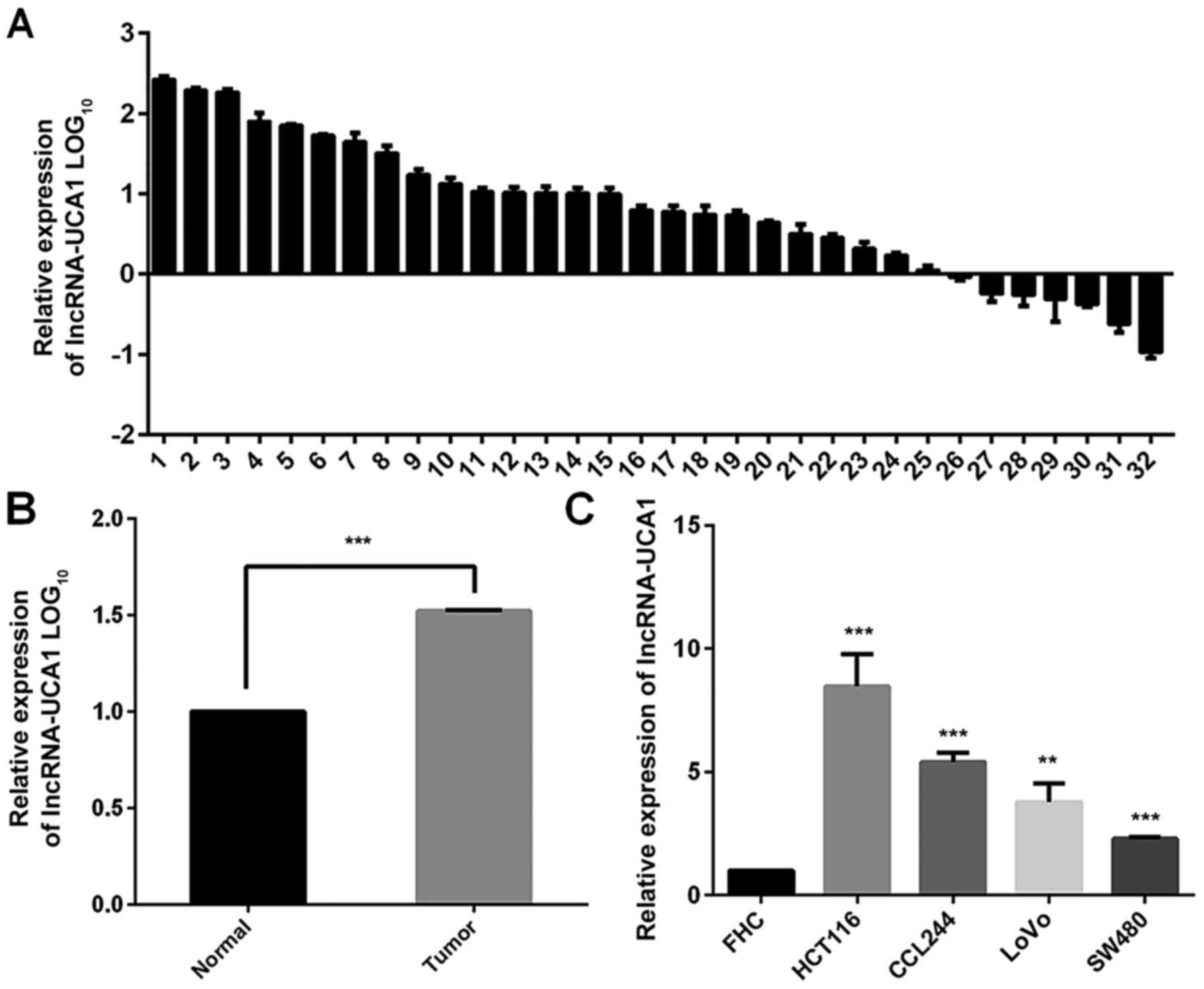
UCA1 levels are upregulated in CRC
tissues and cell lines
Thirty-two pairs of human CRC and adjacent normal
tissues were collected to investigate the expression level of UCA1
using real-time PCR. As shown in Fig.
1A and C, UCA1 was overexpressed in CRC tissues compared to
paired adjacent normal tissues (P<0.001). In addition, our
results demonstrated that UCA1 was highly expressed in human
CRC-derived cell lines HCT116, CCL244, SW480 and LoVo, in contrast
with the normal epithelial cells FHC (Fig. 1B). These results indicated that UCA1
was significantly upregulated in CRC tumor tissues and cell lines.
CCL244 cells were selected to perform the following experiments
since the ascending order of these four CRC cell lines in terms of
radiosensitivity was demonstrated to be CCL244, SW480, LoVo and
HCT116 (25).
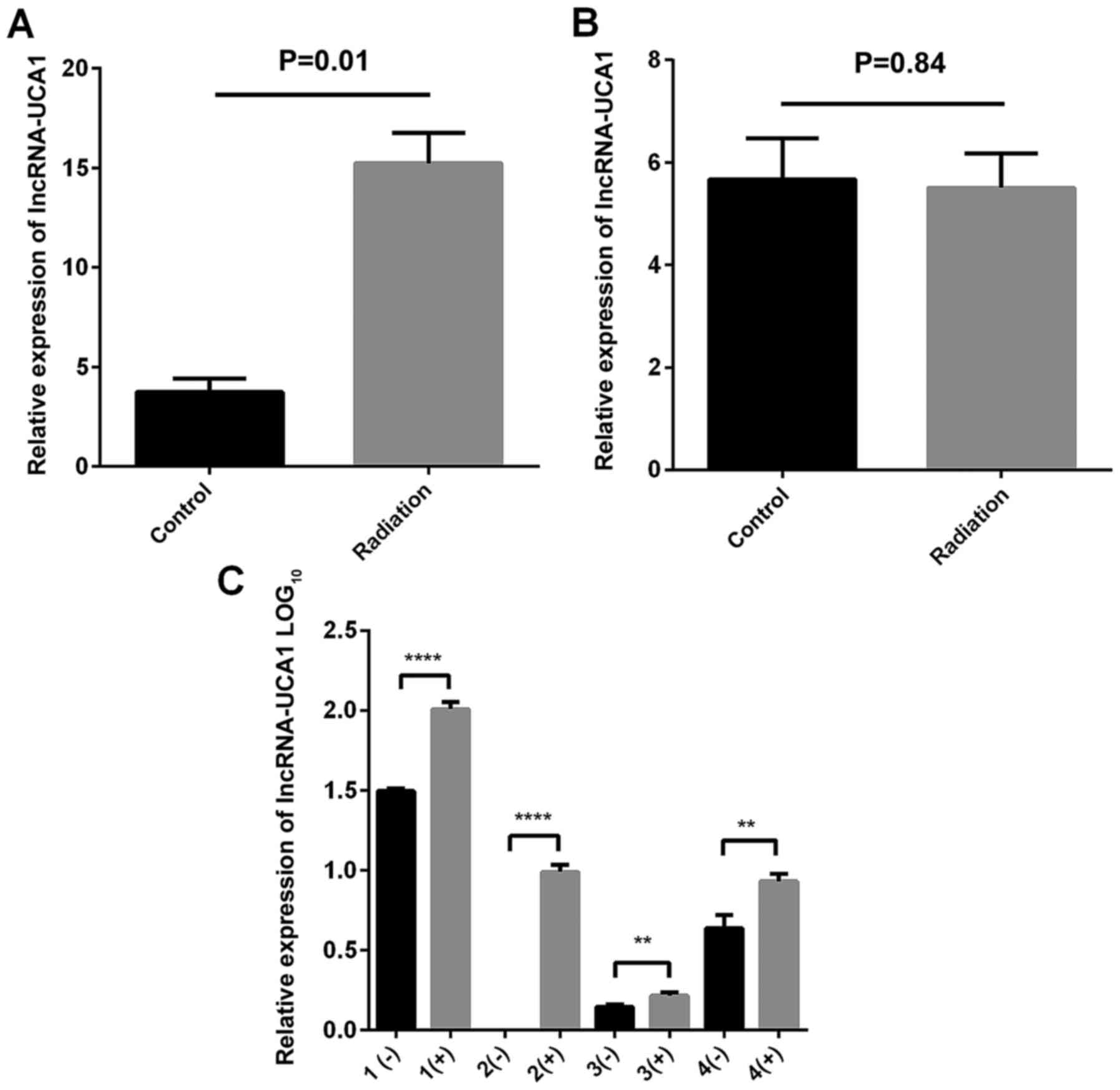
Expression of UCA1 in two CRC cell
lines and four clinical samples after irradiation compared to
unirradiated cells
CCL244 and HCT116 cells were seeded in 6-well plates
and then exposed to 8-Gy X-ray irradiation. After 48 h, total RNA
was extracted and then a real-time PCR assay was used to detect the
expression of UCA1. As shown in Fig. 2A
and B, the expression level was higher in the CCL244 cells with
irradiation than the cells without irradiation (P=0.01). However,
there was no statistically significant difference in HCT116 cells
(P=0.84). Similarly, Fig. 2C
indicated that the expression of UCA1 was upregulated in four
clinical samples after radiotherapy.
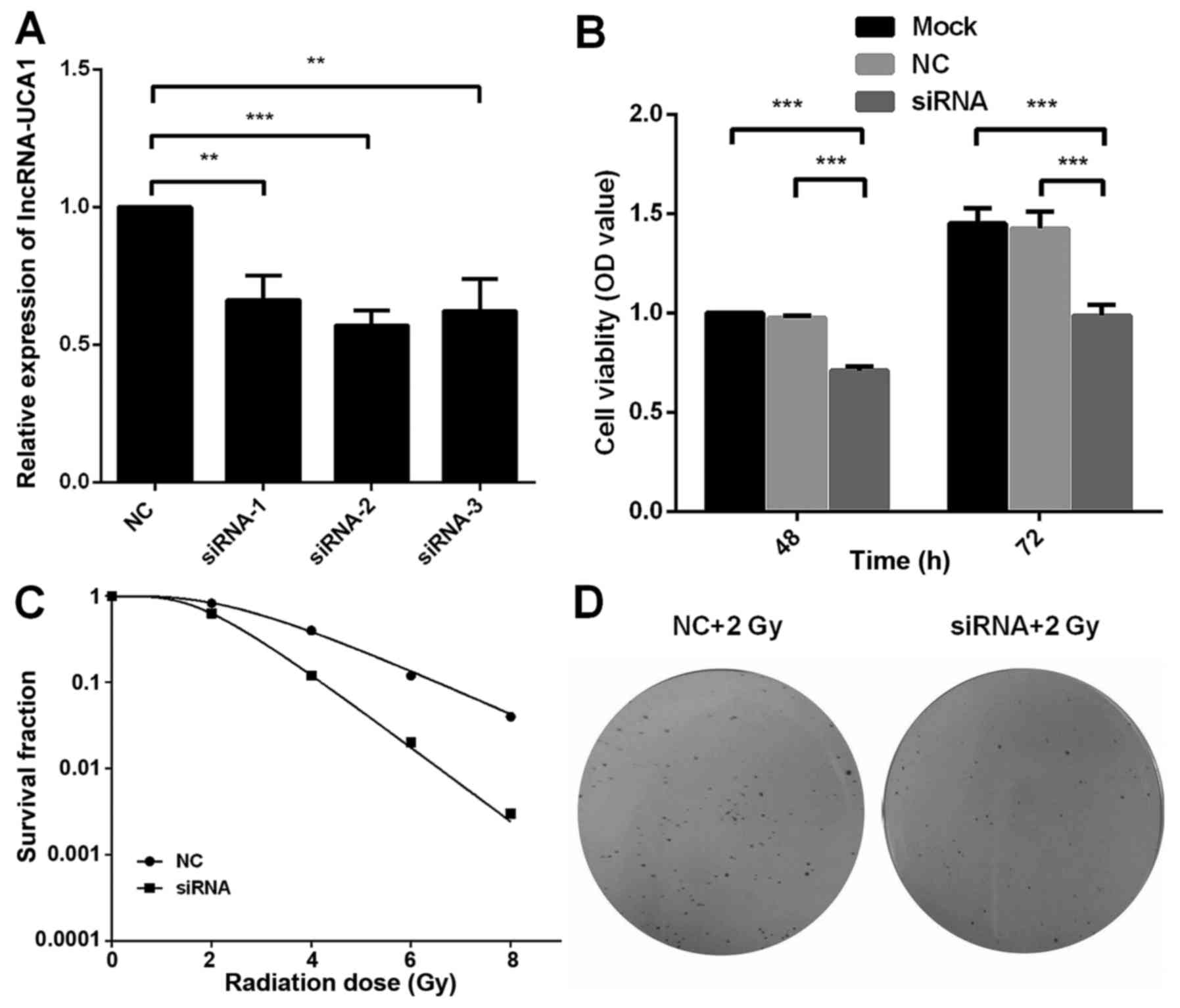
UCA1 regulates the radiosensitivity of
CCL-244 cells
The results of Fig.
2 demonstrated that UCA1 was upregulated in CCL244 cells
following X-ray irradiation. Thus, we conjectured that UCA1 may be
involved in the regulation of radiosensitivity of CRC cells. Three
different siRNA vectors were transfected into CCL244 cells and the
efficiency of transfection was verified by real-time PCR. siRNA-2
exhibited the highest efficiency (57%) of transfection compared to
the other two siRNAs (66 and 62%) in Fig. 3A. Thus, siRNA-2 was then used for
further experiments. siRNA and NC-siRNA were transfected into
CCL244 cells and then the cells were exposed to 8-Gy X-ray
irradiation after transfection for 24 h. After 48 and 72 h, MTT and
colony-formation assays were performed. The MTT assay demonstrated
that the cell viability was reduced after UCA1 downregulation,
compared to the mock-transfected cells (P<0.001) and negative
control UCA1 (P<0.001) (Fig.
3B). CCL244 cells were transfected with siRNA or NC-siRNA 24 h
prior to irradiation at 0, 2, 4, 6 and 8 Gy. The multi-target
single hit model was used to obtain the dose-survival curves. A
dose-dependent radiosensitization on UCA1-silenced cells was also
observed with a sensitizing enhancement ratio (SER) of 1.41
(Fig. 3C). Representative colony
images of UCA1-silenced CCL244 cells compared to the negative
control exposed to 2 Gy of X-ray irradiation are shown in Fig. 3D. These results demonstrated that
downregulation of UCA1 significantly increased the radiosensitivity
of CCL244 cells.
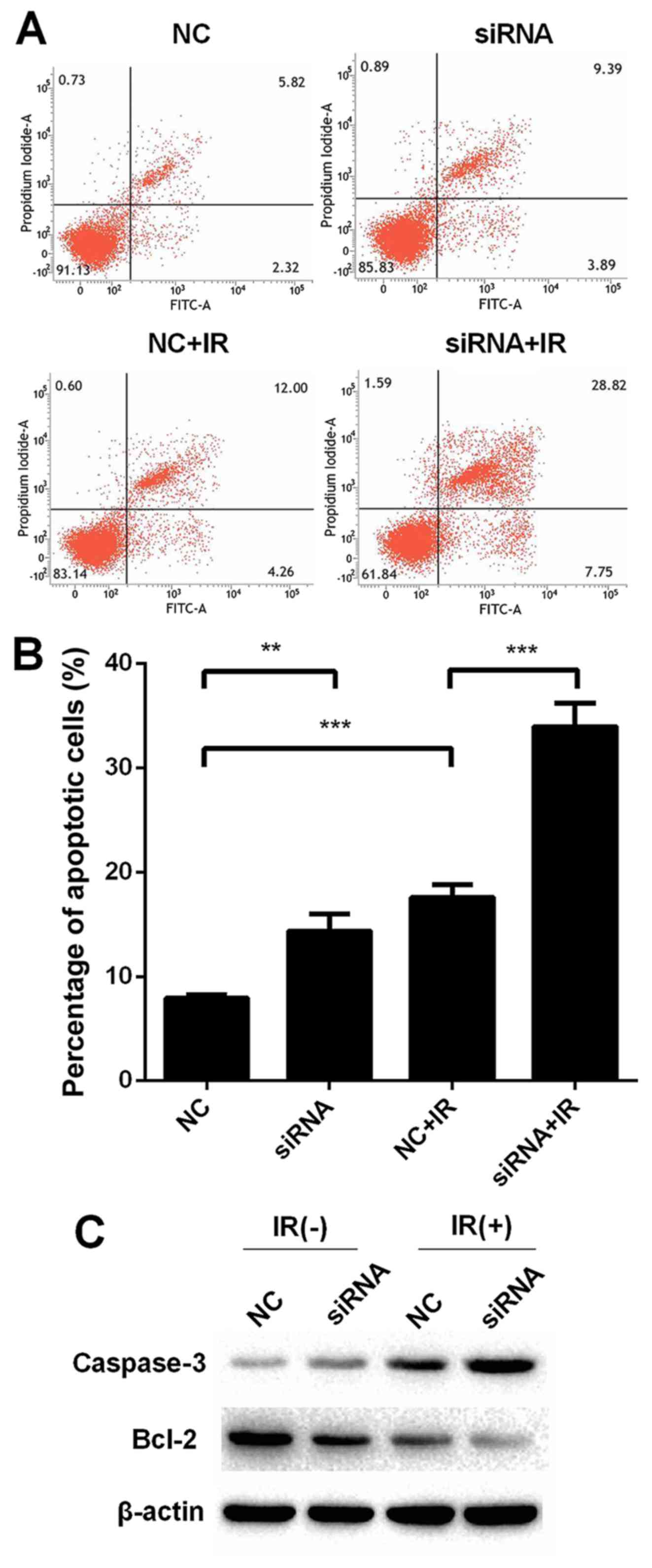
Knockdown of UCA1 promotes
radiation-induced apoptosis in CCL244 cells and regulates the
expression of apoptosis-related proteins
Apoptosis was detected using Annexin V/PI staining
with flow cytometry to investigate the role of UCA1 in
radiation-induced apoptosis in CCL244 cells. Apoptosis was promoted
in the CCL244 cells in the NC+IR group when compared to the NC
group [NC (7.93±0.20%) vs. NC+IR (17.60±0.69%), P<0.001], as
shown in Fig. 4A and B. The
percentage of apoptosis in the irradiation plus the UCA1-knockdown
group was significantly enhanced in contrast with the NC+IR group
[siRNA+IR (33.97±1.03%) vs. NC+IR (17.60±0.69%), P<0.001].
Furthermore, the expression of apoptosis-related molecules such as
Bcl-2 and caspase-3 was evaluated by western blot analysis. As
shown in Fig. 4C, the expression of
Bcl-2 was downregulated after transfection with siRNA compared to
the negative control regardless of irradiation. In contrast with
Bcl-2, the expression of caspase-3 was upregulated. All these
results demonstrated that knockdown of UCA1 promoted
radiation-induced apoptosis in CCL244 cells.
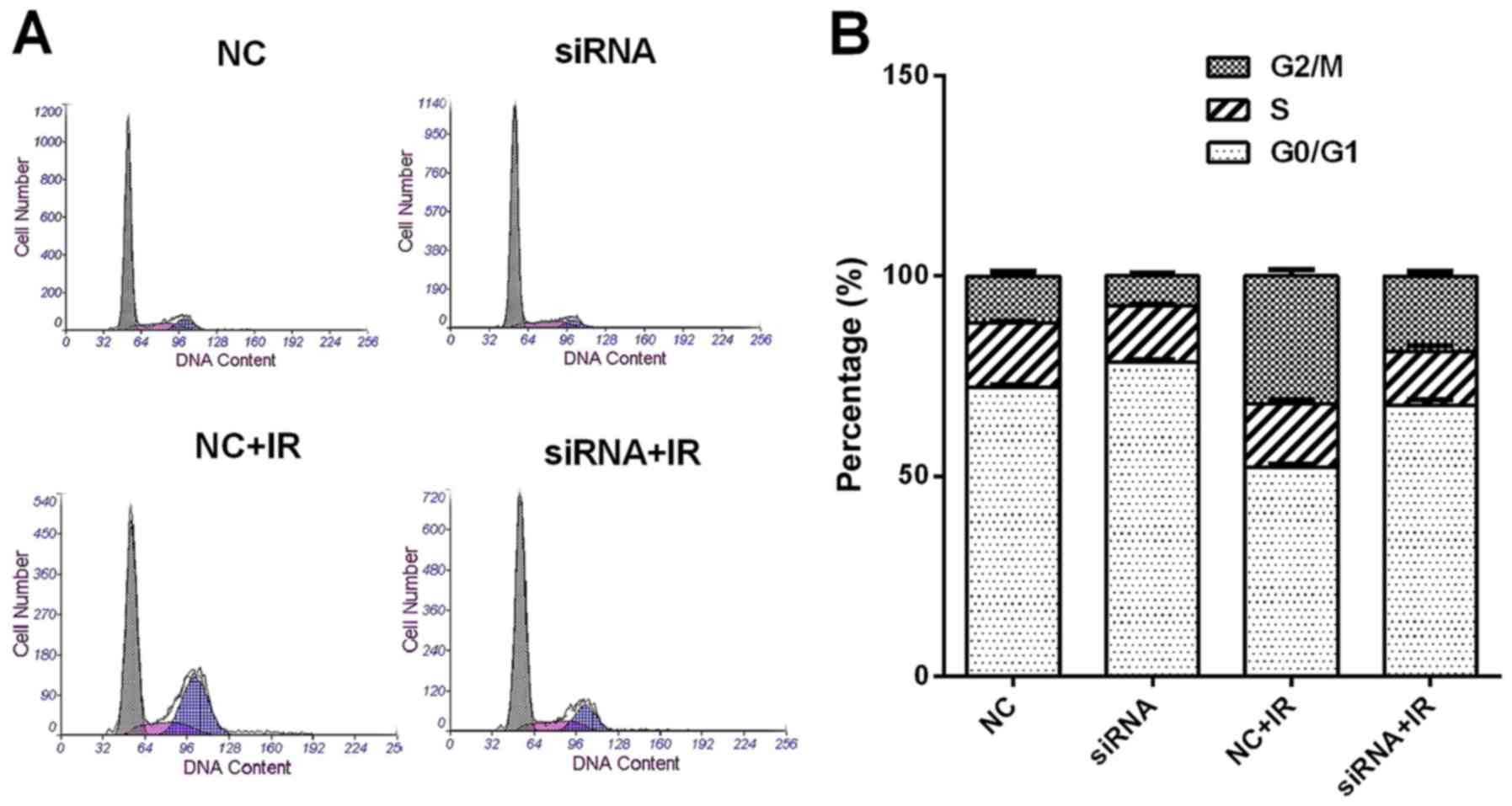
UCA1 silencing attenuates
radiation-induced G2/M arrest
We then studied whether UCA1 had any effects on
radiation-induced cell cycle changes. Transfected CCL244 cells were
subjected to 6-Gy irradiation. Twenty-four hours later, the cells
were harvested and detected by flow cytometry. As shown in Fig. 5, the results revealed that IR caused
cell cycle arrest in the G2/M phase in the NC+IR group when
compared to the NC group [NC (11.77±1.01%) vs. NC+IR (31.81±1.59%),
P<0.001]. However, the percentage of cells in the G2/M phase was
reduced when UCA1 was knocked down after irradiation compared to
the NC+IR group [siRNA+IR (18.87±1.15%) vs. NC+IR (31.81±1.59%)
P<0.01]. The percentage of radiation-induced G2/M arrest in the
siRNA+IR group was reduced by 12.94% compared to the NC+IR group.
These results indicated that knockdown of UCA1 can reduce the
radiation-induced G2/M arrest.
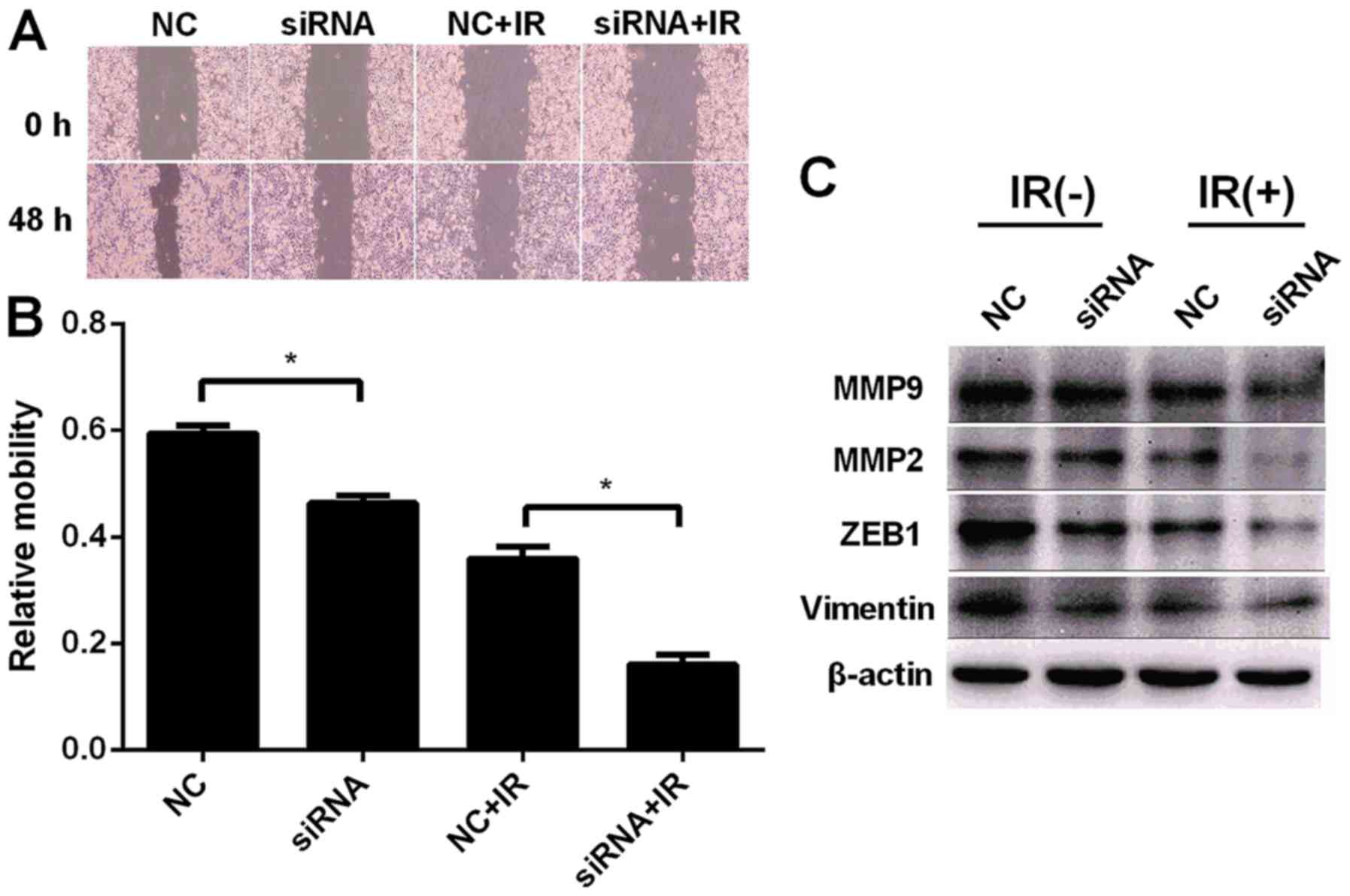
Knockdown of UCA1 inhibits the
migration and EMT of CCL244 cells after irradiation
Metastasis is an important factor of poor prognosis
in CRC. A wound healing assay was carried out to investigate the
effect of UCA1 on the metastasis of CCL244 cells. Fig. 6A and B revealed that downregulation
of UCA1 inhibited the migration of CCL244 cells compared to the
negative control CCL244 cells (46.38 vs. 59.42%, P<0.05). After
transfection and exposure to 6 Gy X-ray irradiation, knockdown of
UCA1 (siRNA+IR) significantly inhibited the migration of CCL244
cells compared to cells with X-ray irradiation alone (NC+IR) (16.06
vs. 35.92%, P<0.05). As widely known, matrix metalloproteinases
(MMPs) are of vital importance in tumor metastasis. ZEB1 promotes
EMT by binding to DNA through E-boxes of target gene promoters such
as E-cadherin (26). Vimentin is
also a biomarker of EMT (27).
Western blot analysis revealed that downregulation of UCA1 plus
irradiation reduced the expression level of MMP2 and MMP9. Similar
results were obtained in the expression levels of ZEB1 and vimentin
(Fig. 6C). These results
demonstrated that knockdown of UCA1 inhibited the migration and EMT
of CCL244 cells after irradiation.
Discussion
Radiotherapy is a primary treatment modality for
early and locally advanced CRC cancer (28). However, radiation resistance has a
greater impact on the treatment effect, which suggests a poor
prognosis and high recurrence rate. Therefore, research has begun
to focus on how to increase the radiosensitivity of tumor cells.
Downregulation of c-myc and MG132
(carbobenzoxyl-leucinyl-leucinyl-leucinal-H) enhanced the
radiosensitivity of A549 cells in vivo and in vitro
(29). In addition, anti-apoptotic
proteins, such as Bcl-2 and Bcl-xL, play an important role in the
radioresistance of cancer cells. When acquired radioresistant
breast cancer cell line MDA-MB-231R was treated with ABT-737, Bcl-2
and Bcl-xL were downregulated, radiation-induced apoptosis was
increased, restoring the radiosensitivity of the MDA-MB-231R cells
(30). The PI3K/Akt/mTOR pathway is
an important intracellular signaling pathway involved in the
regulation of cell radioresistance (31,32).
The PI3K/Akt/mTOR pathway inhibitors BEZ235 and PI103 increase the
radiation sensitivity of prostate cancer cells via induction of
apoptosis and reduction of autophagy (33). In our previous study, CCL244 was
identified as a radioresistant cell line and HCT116 as a
radiosensitive cell line as determined through clone formation
assay (25). lncRNA UCA1 was
identified as a vital lncRNA which was distinctly upregulated in
CCL244 cells after irradiation. UCA1 has been established to be
involved in many of the biological behaviors of tumor. However,
there are no studies concerning UCA1 affecting the radiosensitivity
of tumor cells.
In recent years, as long non-coding RNAs have become
a hot topic, numerous lncRNAs have been demonstrated to be related
with tumor cell radiosensitivity, although UCA1 was not among them.
Downregulation of HOTAIR increased the radiosensitivity of cervical
cancer when p21 was simultaneously upregulated (34). lncRNA-p21 enhanced the sensitivity
of radiotherapy for CRC by targeting the β-catenin signaling
pathway and it may provide a potential target for CRC radiotherapy
(35). lncRNA-LIRR1 was able to
modulate the radiosensitivity of BEAS-2B cells through the DDR
signaling pathways in a p53-dependent manner (36). Downregulation of MALAT1 and
upregulation of miR-145 increased the radio-sensitivity in
HR-HPV+ cervical cancer (37). In the present study, we identified
that UCA1 was upregulated in CCL244 cells after irradiation which
indicated that UCA1 may be involved in radioresistance of CRC
cells. Since UCA1 was highly expressed in tissues and CCL244 cells,
we used small interfering RNA to downregulate UCA1 and investigate
the relevance between lncRNA-UCA1 and the radiosensitivity of CRC
cells. An MTT assay indicated that silencing of UCA1 significantly
decreased the proliferation rate compared to the negative control
siRNA when cells were exposed to X-ray irradiation.
Colony-formation assay revealed that UCA1-silencing was observed
with a sensitizing enhancement ratio (SER) of 1.41 and the colony
survival fraction was significantly reduced. All these results
indicated that downregulation of UCA1 enhanced the radiosensitivity
of CCL244 cells. In addition, UCA1 may be involved in the
modulation of chemosensitivity of CRC cells.
Radiation-induced apoptosis was one of modes of
radiation-induced cell death (38).
lncRNAs were identified to be involved in the modulation of
radiation-induced apoptosis. Downregulation of MALAT1 significantly
enhanced radiation-induced apoptosis, as the expression of Bcl-2
was decreased and that of caspase-3 was increased (39). It was demonstrated that silencing of
HOTAIR increased the radiation-induced apoptosis in CRC cells
(40). In the present study, we
examined the apoptosis of CCL244 cells after transfection and
exposure to X-ray irradiation in order to confirm that UCA1 was
involved in regulating the radiosensitivity of CCL244 cells. It was
revealed that the downregulation of UCA1 resulted in a higher
percentage of radiation-induced apoptosis than the negative
control. Moreover, downregulation of UCA1 reduced the expression of
Bcl-2 and improved the expression level of caspase-3. All of the
aforementioned results demonstrated that downregulation of UCA1
enhanced the radiosensitivity via the promotion of
radiation-induced apoptosis.
Cell cycle regulation may be the most important
determinant of ionizing radiation sensitivity (41). It has been universally acknowledged
that irradiation induces DNA double-strand breaks (DSBs) resulting
in G2 arrest and then DNA DSB repair occurrs. The G2 arrest in
tumor cells provides time for the repair processes to operate which
are critical for ensuring cell survival after DNA damage (41). It was suggested that the combination
of BI-69A11 and celecoxib impaired IR-induced DSB repair,
attenuated the G2/M cell cycle and then enhanced the
radiosensitivity of CRC cancer cells (42). In the present study, CCL244 cells
with irradiation revealed prolonged G2/M phase arrest. However,
downregulation of UCA1 attenuated the radiation-induced arrest of
G2/M cell cycle. These findings indicated that silencing of UCA1
enhanced the radiosensitivity of CRC by impairing G2/M arrest.
In the last decade, EMT has been widely recognized
to play a vital role in the promotion of carcinoma metastasis.
lncRNAs have been reported to be involved in the process of EMT.
Silencing of HOTAIR prevented the EMT process stimulated by TGF-β1
in CRC and breast cancer (43).
EMT-associated proteins (ZEB1, ZEB2, Snail, E-cadherin, MMP2, MMP9
and vimentin) have been studied in the EMT process (27). In the present study, we found that
downregulation of UCA1 plus irradiation reduced the expression
levels of MMP2, MMP9, ZEB1 and vimentin. These results indicated
that silencing of UCA1 significantly inhibited EMT in CCL244
cells.
In summary, the present study demonstrated that UCA1
was highly expressed in CRC cells and that downregulation of UCA1
enhanced the radiosensitivity and suppressed EMT of CCL244 cells.
All these results demonstrated that lncRNA-UCA1 may be a novel
therapeutic target for CRC in radiotherapy.
Acknowledgements
The authors thank Shuyu Zhang, Ming Li and Jianping
Cao for their technical support.
Funding
The present study was partially supported by the
National Natural Science Foundation of China (grant nos. 81672970,
81301933, 81472917 and 81572345), the Natural Science Foundation of
Jiangsu Province (no. BK20160338), the projects of Suzhou
Technology Bureau (nos. SZS201618, SYS201552 and SYSD2015034), the
Focus of Clinical Disease Treatment Technology Special Funds of
Suzhou City (LCZX201505) and the Second Affiliated Hospital of
Soochow University Preponderant Clinic Discipline Group Project
Funding.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CX and ZY conceived and designed the study. WL and
XY performed the experiments and wrote the paper. WL, XY, XX, JZ,
YongW, KZ and SH were involved in the conception of the study. SZ,
ML, YongyouW and JC provided technical support. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integ-rity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Institutional Review Board of the Second Affiliated Hospital of
Soochow University, Suzhou, Jiangsu 215004.
Consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
El-Shami K, Oeffinger KC, Erb NL, Willis
A, Bretsch JK, Pratt-Chapman ML, Cannady RS, Wong SL, Rose J,
Barbour AL, et al: American cancer society colorectal cancer
survivorship care guidelines. CA Cancer J Clin. 65:428–455. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jonker FH, Tanis PJ, Coene PP and van der
Harst E; Dutch Surgical Colorectal Audit Group, : Impact of
neoadjuvant radiotherapy on complications after hartmann procedure
for rectal cancer. Dis Colon Rectum. 58:931–937. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Leersum NJ, Snijders HS, Wouters MW,
Henneman D, Marijnen CA, Rutten HR, Tollenaar RA and Tanis PJ;
Dutch Surgical Colorectal Cancer Audit Group, : Evaluating national
practice of preoperative radiotherapy for rectal cancer based on
clinical auditing. Eur J Surg Oncol. 39:1000–1006. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elferink MA, van Steenbergen LN, Krijnen
P, Lemmens VE, Rutten HJ, Marijnen CA, Nagtegaal ID, Karim-Kos HE,
de Vries E and Siesling S; Working Group Output of the Netherlands
Cancer Registry, : Marked improvements in survival of patients with
rectal cancer in the Netherlands following changes in therapy,
1989–2006. Eur J Cancer. 46:1421–1429. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun QL, Zhao CP, Wang TY, Hao XB, Wang XY,
Zhang X and Li YC: Expression profile analysis of long non-coding
RNA associated with vincristine resistance in colon cancer cells by
next-generation sequencing. Gene. 572:79–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guan D, Zhang W, Zhang W, Liu GH and
Belmonte JC: Switching cell fate, ncRNAs coming to play. Cell Death
Dis. 4:e4642013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han D, Wang M, Ma N, Xu Y, Jiang Y and Gao
X: Long noncoding RNAs: Novel players in colorectal cancer. Cancer
Lett. 361:13–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rupaimoole R, Lee J, Haemmerle M, Ling H,
Previs RA, Pradeep S, Wu SY, Ivan C, Ferracin M, Dennison JB, et
al: Long noncoding RNA ceruloplasmin promotes cancer growth by
altering glycolysis. Cell Rep. 13:2395–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:11480–11484. 2015.PubMed/NCBI
|
|
12
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han Y, Yang YN, Yuan HH, Zhang TT, Sui H,
Wei XL, Liu L, Huang P, Zhang WJ and Bai YX: UCA1, a long
non-coding RNA up-regulated in colorectal cancer influences cell
proliferation, apoptosis and cell cycle distribution. Pathology.
46:396–401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang H: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu W, Zhang S, Li X, Xue M, Cao S and Chen
W: Ets-2 regulates cell apoptosis via the Akt pathway, through the
regulation of urothelial cancer associated 1, a long non-coding
RNA, in bladder cancer cells. PLoS One. 8:e739202013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang C, Li X, Wang Y, Zhao L and Chen W:
Long non-coding RNA UCA1 regulated cell cycle distribution via CREB
through PI3-K dependent pathway in bladder carcinoma cells. Gene.
496:8–16. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
20
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ,
Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, et al: Overexpression
of EIF5A2 promotes colorectal carcinoma cell aggressiveness by
upregulating MTA1 through c-myc to induce
epithelial-mesenchymaltransition. Gut. 61:562–575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cao H, Xu E, Liu H, Wan L and Lai M:
Epithelial-mesenchymal transition in colorectal cancer metastasis:
A system review. Pathol Res Pract. 211:557–569. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Lv Z and Guo E: Knockdown of long
noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation,
metastasis and epithelial-mesenchymal transition. Int J Clin Exp
Pathol. 8:9140–9146. 2015.PubMed/NCBI
|
|
24
|
Sun R, Qin C, Jiang B, Fang S, Pan X, Peng
L, Liu Z, Li W, Li Y and Li G: Down-regulation of MALAT1 inhibits
cervical cancer cell invasion and metastasis by inhibition of
epithelial-mesenchymal transition. Mol Biosyst. 12:952–962. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang XD, Xu XH, Zhang SY, Wu Y, Xing CG,
Ru G, Xu HT and Cao JP: Role of miR-100 in the radioresistance of
colorectal cancer cells. Am J Cancer Res. 5:545–559.
2015.PubMed/NCBI
|
|
26
|
Hill L, Browne G and Tulchinsky E:
ZEB/miR-200 feedback loop: At the crossroads of signal transduction
in cancer. Int J Cancer. 132:745–754. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee WS, Kim N, Park YR, Oh HH, Myung E,
Kim SH, Yu HM, Kim MY, Oak CY, Chung CY, et al: Myeloid cell
leukemia-1 promotes epithelial-mesenchymal transition of human
gastric cancer cells. Oncol Rep. 34:1011–1016. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Drake TM, Ritchie JE, Kanthou C, Staves
JJ, Narramore R and Wyld L: Targeting the endoplasmic reticulum
mediates radiation sensitivity in colorectal cancer. Exp Mol
Pathol. 98:532–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jung J, Kim EJ, Chung HK, Park HJ, Jeong
SY and Choi EK: c-Myc down-regulation is involved in proteasome
inhibitor-mediated enhancement of radiotherapeutic efficacy in
non-small cell lung cancer. Int J Oncol. 40:385–390.
2012.PubMed/NCBI
|
|
30
|
Li JY, Li YY, Jin W, Yang Q, Shao ZM and
Tian XS: ABT-737 reverses the acquired radioresistance of breast
cancer cells by targeting Bcl-2 and Bcl-xL. J Exp Clin Cancer Res.
31:1022012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang L, Graham PH, Hao J, Ni J, Bucci J,
Cozzi PJ, Kearsley JH and Li Y: Acquisition of
epithelial-mesenchymal transition and cancer stem cell phenotypes
is associated with activation of the PI3K/Akt/mTOR pathway in
prostate cancer radioresistance. Cell Death Dis. 4:e8752013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ni J, Cozzi P, Hao J, Beretov J, Chang L,
Duan W, Shigdar S, Delprado W, Graham P, Bucci J, et al: Epithelial
cell adhesion molecule (EpCAM) is associated with prostate cancer
metastasis and chemo/radioresistance via the PI3K/Akt/mTOR
signaling pathway. Int J Biochem Cell Biol. 45:2736–2748. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang L, Graham PH, Hao J, Ni J, Bucci J,
Cozzi PJ, Kearsley JH and Li Y: PI3K/Akt/mTOR pathway inhibitors
enhance radiosensitivity in radioresistant prostate cancer cells
through inducing apoptosis, reducing autophagy, suppressing NHEJ
and HR repair pathways. Cell Death Dis. 5:e14372014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jing L, Yuan W, Ruofan D, Jinjin Y and
Haifeng Q: HOTAIR enhanced aggressive biological behaviors and
induced radio-resistance via inhibiting p21 in cervical cancer.
Tumour Biol. 36:3611–3619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X,
Ma Z, Li X and Zhang Y: LincRNA-p21 enhances the sensitivity of
radiotherapy for human colorectal cancer by targeting the
Wnt/β-catenin signaling pathway. Oncol Rep. 31:1839–1845. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiao Y, Liu C, Cui FM, Xu JY, Tong J, Qi
XF, Wang LL and Zhu W: Long intergenic non-coding RNA induced by
X-ray irradiation regulates DNA damage response signaling in the
human bronchial epithelial BEAS-2B cell line. Oncol Lett.
9:169–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu H, He Y, Lin L, Qi Z, Ma L, Li L and Su
Y: Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+
cervical cancer via sponging miR-145. Tumour Biol. 37:1683–1691.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shinomiya N: New concepts in
radiation-induced apoptosis: ‘Premitotic apoptosis’ and
‘postmitotic apoptosis’. J Cell Mol Med. 5:240–253. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin C, Yan B, Lu Q, Lin Y and Ma L: The
role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal
carcinoma. Tumor Biol. 37:4025–4033. 2016. View Article : Google Scholar
|
|
40
|
Yang XD, Xu HT, Xu XH, Ru G, Liu W, Zhu
JJ, Wu YY, Zhao K, Wu Y, Xing CG, et al: Knockdown of long
non-coding RNA HOTAIR inhibits proliferation and invasiveness and
improves radiosensitivity in colorectal cancer. Oncol Rep.
35:479–487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pawlik TM and Keyomarsi K: Role of cell
cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol
Biol Phys. 59:1–942. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pal I, Dey KK, Chaurasia M, Parida S, Das
S, Rajesh Y, Sharma K, Chowdhury T and Mandal M: Cooperative effect
of BI-69A11 and celecoxib enhances radiosensitization by modulating
DNA damage repair in colon carcinoma. Tumour Biol. 37:6389–6402.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pádua Alves C, Fonseca AS, Muys BR, de
Barros E, Lima Bueno R, Bürger MC, de Souza JE, Valente V, Zago MA
and Silva WA Jr: Brief report: The lincRNA Hotair is required for
epithelial-to-mesenchymal transition and stemness maintenance of
cancer cell lines. Stem Cells. 31:2827–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|