Introduction
Glioma is one the most common and aggressive types
of brain cancer in humans, which is characterized by rapid
proliferation, high invasion and genetic alterations, and is
associated with a poor prognosis in patients (1,2).
Despite the significant advance in cancer treatments, the mortality
rate remains high in malignant glioma (3). Therefore, it is important to
investigate novel interventions or techniques for the treatment of
malignant glioma.
Eukaryotic initiation factors (eIFs) are well known
as key mediators of protein translation initiation in eukaryotic
cells. Among them, eIF4E has been recognized as being key in the
initiation and development of malignant tumors (4–6). eIF4E
can be combined with the scaffolding protein eIF4G and the
ATP-dependent RNA helicase eIF4A to form the eIF4F complex
(7), which is the rate-limiting
factor in a variety of cancer-related behaviors, including cell
proliferation (4), apoptosis
(8,9) and angiogenesis (10,11).
Importantly, the deregulation of eIF4F activity is associated with
the elevation of Myc in the prelymphomatous stage of Eµ-Myc
lymphoma, whereas the inhibition of eIF4F causes the decrease in
cycling pre-B/B cells and tumor onset delay (12). In addition, the inhibition of eIF4F
complex formation either by inhibiting the eIF4E-eIF4G interaction
or by targeting eIF4A, may lead to the death of cancer cells
(13). These lines of evidence
suggest that the eIF4F complex may be a promising molecular target
for the treatment of malignant glioma.
The eIF4E binding proteins (4E-BPs) are a series of
regulatory molecules involved in controlling eIF4F complex
assembly. Following phosphorylation, 4E-BPs inhibit the activity of
translation factor eIF4E through direct interactions (14–16).
Among them, 4E-BP2 is one of the predominant 4E-BPs expressed in
the brain (17), however, the
associated functions remain to be fully elucidated. A previous
study demonstrated that the interaction between eIF4E and 4E-BP2
inhibited eIF4F complex assembly and cap-dependent translation
initiation (18). Therefore, 4E-BP2
may be a potential target for controlling cancer cell proliferation
via affecting eIF4F complex assembly.
In the present study, based on the evidence that
4E-BPs bind to the dorsal side of eIF4E to prevent formation of the
eIF4F complex, phosphorylation-deficient truncated 4E-BP2 (eIF4FD)
was generated and constructed into the eukaryotic expression vector
pSecTag2, and fused with the protein transduction domain (PTD),
which has the potential to permeate cell membranes and deliver
proteins into living cells (19,20).
This truncation was then transfected into the U251 cell line, and
the alterations of biological characteristics and expression of
cancer-related genes were determined in vitro and in
vivo to fully address the effect and mechanism of inhibiting
eIF4F complex assembly on malignant glioma.
Materials and methods
Cell culture
The human U251 glioma cells were purchased from the
Chinese Academy of Sciences Cell Bank (Shanghai, China). The U87-MG
cells were obtained from the American Type Culture Collection
(ATCC; Manassas, VA, USA) and cultured at 37°C, 5% CO2
in Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum (FBS; HyClone Laboratories, Inc., South Logan, UT,
USA), 100 U/ml of penicillin, 100 µg/ml of streptomycin and 2
mmol/l L-glutamine.
Plasmid constructs
Based on the GenBank sequence (NM_004096), upstream
and downstream primers were synthesized using Primer 5.0 software
(Premier Biosoft International, Palo Alto, CA, USA) (forward,
5′-AAAGGATCCACGTCCACTAGCTGCCCCGATTCCC-3′ and reverse,
5′-AAACTCGAGTTATTGTGCGTCATCGGGTATCTCT-3′. Restriction enzyme
(BamHI and XhoI) sites were added to the 5′ end of
the upstream and downstream primers. Using the cDNA as a template,
a phosphorylation-deficient truncated 4E-BP2 sequence with Flag-tag
and BamHI/XhoI restriction sites on each end was
obtained by polymerase chain reaction (PCR). The thermocycling
conditions were as follows: 95°C for 5 min, then 35 cycles of 95°C
for 15 sec, 60°C for 15 sec, and 72°C for 30 sec followed by 72°C
for 10 min. The PCR product was cloned into the pSecTag2-PTD vector
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). As a result,
the recombinant expression vector pSecTag2-PTD-eIF4FD was
obtained.
Transfection
The day preceding transfection, the U251 or U87-MG
cells in the logarithmic growth phase were trypsinized, counted and
seeded in 6-well plates at the density of 1×106
cells/well. When the cells were 80% confluent, the medium was
replaced with serum-free RPMI-1640 medium (Hyclone Laboratories),
and pSecTag2 and pSecTag2-PTD-eIF4FD were transfected into the
culture overnight according to the manufacturer's protocol
(Lipofectamine™ 2000 for plasmid delivery). To set up the U251 cell
line stably expressing eIF4FD, the 5×105 transfected
cells were seeded in 12-well plates and cultured in a complete
medium containing G418 (200 µg/ml). The positive clones were
selected and confirmed by western blot analysis.
Cell cycle distribution analysis
The cells were collected (detached by 0.25%
trypsinization without EDTA) at 0, 12, 24 and 48 h
post-transfection, and then washed twice with phosphate-buffered
saline (PBS) and centrifuged at 12,000 × g for 5 min at 4°C. The
pellet was fixed with 70% ethanol for 1 h at 4°C. Following washing
with PBS, the cells were resuspended with propidium iodide (PI)
solution (0.05 mg/ml) containing RNase and incubated at room
temperature in the dark for 30 min. DNA content was then analyzed
using the FC 500 flow cytometer (Beckman Coulter, Inc., Brea, CA,
USA).
TUNEL staining
Briefly, the U251 cells were fixed with 4%
formaldehyde solution for 20 min at room temperature. The cells
were then permeabilized with 0.2% Trition X-100 for 5 min. The
cells were labeled with fluorescein TUNEL reagent mixture for 60
min at 37°C according to the manufacturer's protocol (Promega
Corp., Madison, WI, USA), and the nuclei were counterstained with
4′,6-diamidino-2-phenylindole. Subsequently, the numbers of
TUNEL-positive cells were counted under fluorescence
microscopy.
Co-immunoprecipitation
Whole U251 cell lysates were obtained by
resuspending U251 cell pellets in RIPA buffer containing 150 mM
NaCl, 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, 1% NP-40, 1%
nadeoxycholate, 0.1% SDS, 1 mM PMSF, 20 mg/ml leupeptin, 20 mg/ml
aprotinin and 3 mg/ml pepstatin A. The lysates were incubated
overnight with Flag antibody (cat. no. ab49763; Abcam, Cambridge,
UK) at 4°C prior to being absorbed with protein A/G PLUS agarose
beads. The precipitated immunocomplexes were released by boiling
with 2X SDS electrophoresis sample buffer and were prepared for
western blot analysis.
Western blot analysis
For western blot analysis, the cells were washed
twice with ice-cold PBS and lysed with RIPA buffer (Beyotime
Institute of Biotechnology, Haimen, China) on ice. Following
centrifugation at 12,000 × g for 10 min at 4°C, the protein
concentration in the supernatant was analyzed using a bicinchoninic
acid assay kit (Thermo Fisher Scientific, Inc.). The total protein
(20 µg) from each sample was separated by SDS-PAGE and transferred
onto a polyvinylidene difluoride membrane (EMD Millipore, Bedford,
MA, USA). The membrane was then blocked with a TBS solution
containing 5% non-fat dry milk at room temperature for 1 h,
followed by incubation with the following corresponding primary
antibodies: B-cell lymphoma 2 (Bcl-2; dilution 1:1,000; cat. no.
ab32124; Abcam, Cambridge, MA, USA), Bcl-extra large (Bcl-xL:
1:1,000; cat. no. ab32370; Abcam), C-myc (1:1,000; cat. no.
ab32072; Abcam), cyclin D1 (dilution 1:1,000; cat. no. 2922; Cell
Signaling Technology, Inc., Beverly, MA, USA), survivin (dilution
1:1,000; cat. no. ab76424; Abcam), β-actin (dilution 1:500; cat.
no. sc-517582; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), β-catenin (dilution 1:500; cat. no. sc-65480; Santa Cruz
Biotechnology, Inc.) overnight at 4°C. The following day, the
membrane was washed with Tris-buffered saline with Tween-20 (0.05%;
TBST) three times (5 min each), and then incubated with HRP-labeled
secondary antibody at 37°C for 1 h (dilution 1:5,000; cat. nos.
BA1050 or BA1054; Boster Bio, Pleasanton, CA, USA). Subsequently,
the membrane was washed with TBST three times and then developed
using an enhanced chemiluminescence reagent. β-actin was used as a
normalization control.
Glioma xenograft model
Male 8-9-week-old BALB/c (nu/nu) nude mice (20–25 g)
were purchased from the Animal Center of the Fourth Military
Medical University (Xi'an, China), and housed under controlled
laboratory conditions on a 12-h light/12-h dark cycle at room
temperature with access to water and food ad libitum. All
procedures were in accordance with the NIH Guide and were approved
by the Ethics Committee of the Fourth Military Medical University.
The surgical procedure was performed as previously described
(21,22). Briefly, the mice were mounted on a
stereotaxic instrument following anesthesia with sodium
pentobarbital (intraperitoneal injection, 50 mg/kg). Following skin
incision, a hole with a diameter of 3 mm was drilled on the skull
at the site of 1.0 mm anterior to the anterior fontanel and 2.0 mm
lateral to the sagittal suture. A microsyringe needle (Hamilton
Bonaduz AG, Bonaduz, Switzerland) was inserted through the hole to
a depth of 3 mm beneath the dura. Subsequently, 5×105
U251 cells in 2 µl PBS were injected slowly. Images of the mice
were captured and the tumor sizes were measured using the
high-efficiency CCD camera system (Xenogen Corp., Alameda, CA, USA)
at ~5 min post-injection with 100 µg of luciferin substrate
(PerkinElmer, Inc., Waltham, MA, USA) in PBS without calcium or
magnesium (Corning Inc., Manassas, VA, USA) as previously described
(23).
Plasmid injections via the tail
vein
Hydrodynamic tail vein injections were performed as
described. The first plasmid delivery was performed on the second
day following U251 cell injection in nude mice. The pSecTag2 or
pSecTag2-PTD-eIF4FD plasmid (50 µg) was diluted in 1.8 ml of QR
buffer (Mirus Bio, Madison, WI, USA) and injected into the tail
vein in 5 sec via a 27-gauge, 0.45-inch needle. The animals were
placed into a restraining device and injected without anesthesia.
The plasmids were administered twice a week for 4 weeks. The tumor
size was measured with CCD camera system every 5 days. The animal
experiments were terminated at 30 days, and all nude mice were
sacrificed. The tumor grafts were isolated for immunohistochemistry
(IHC), western blot analysis and TUNEL staining assays.
IHC analysis
The IHC was performed using the
avidin-biotin-peroxidase method. Tissues were fixed in 4%
paraformaldehyde for 12 h at room temperature, embedded in paraffin
and cut into 5-µm sections. All sections were deparaffinized in
xylene and dehydrated using a concentration gradient of alcohol,
following which endogenous peroxidase activity was blocked using
0.5% H2O2 in methanol for 10 min. When
non-specific binding was blocked, the slides were incubated with
anti-PCNA antibody (dilution 1:200; cat. no. sc-25280; Boster Bio)
overnight at 4°C. Following rinsing with TBS for 15 min,
biotinylated goat anti-rabbit IgG (dilution 1:400; Sigma; EMD
Millipore) was incubated with the sections for 1 h at room
temperature and detected with a streptavidin-peroxidase complex.
The brown color indicative of peroxidase activity was developed by
incubating with 0.1% 3,3-diaminobenzidine (Sigma; EMD Millipore) in
PBS with 0.05% H2O2 for 5 min at room
temperature. The appropriate positive and negative controls were
included in each IHC run under a phase contrast microscopy (Olympus
IX50; Olympus Corp., Tokyo, Japan).
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS 16.0 statistical software (SPSS, Inc., Chicago, IL,
USA) was used to perform the statistical analysis. Comparisons of
the mean values were performed by one-way analysis of variance
(ANOVA), followed by Duncan's multiple-range test. P<0.05 was
considered to indicate a statistically significant difference.
Results
eIF4FD inhibits cell cycle and
suppresses proliferation in glioma cells
The present study first confirmed the protein
overexpression of eIF4FD in pSecTag2-PTD-eIF4FD-transfected U251
cells (Fig. 1A). To determine the
effect of eIF4FD on cell survival, flow cytometry was performed in
U251 cells following pSecTag2-PTD-eIF4FD transfection. The
expression of eIF4FD led to a marked decrease in the DNA synthesis
phase (S) and an increase in the G0-G1 phase
in the U251 cells (Fig. 1B).
Accordingly, cell cycle-related proteins cyclin D1 and C-myc were
reduced 12 h following pSecTag2-PTD-eIF4FD transfection, and were
significantly decreased at 24 and 48 h (Fig. 1C-E). The reductions of cyclin D1 and
C-myc were also observed in U87-MG cells following expression of
eIF4FD (Fig. 1F). Therefore, the
overexpression of eIF4FD inhibited cell cycle progression and
proliferation in the glioma cells. To determine whether the effect
of eIF4FD on cell survival was through inhibition of the eIF4F
complex, the interaction of eIF4FD with eIF4E following
pSecTag2-PTD-eIF4FD transfection was examined. The
co-immunoprecipitation assay showed that eIF4FD directly interacted
with eIF4E but did not affect the expression of eIF4E (Fig. 1G and H). Therefore, eIF4FD inhibited
the eIF4F complex via direct interaction with eIF4E.
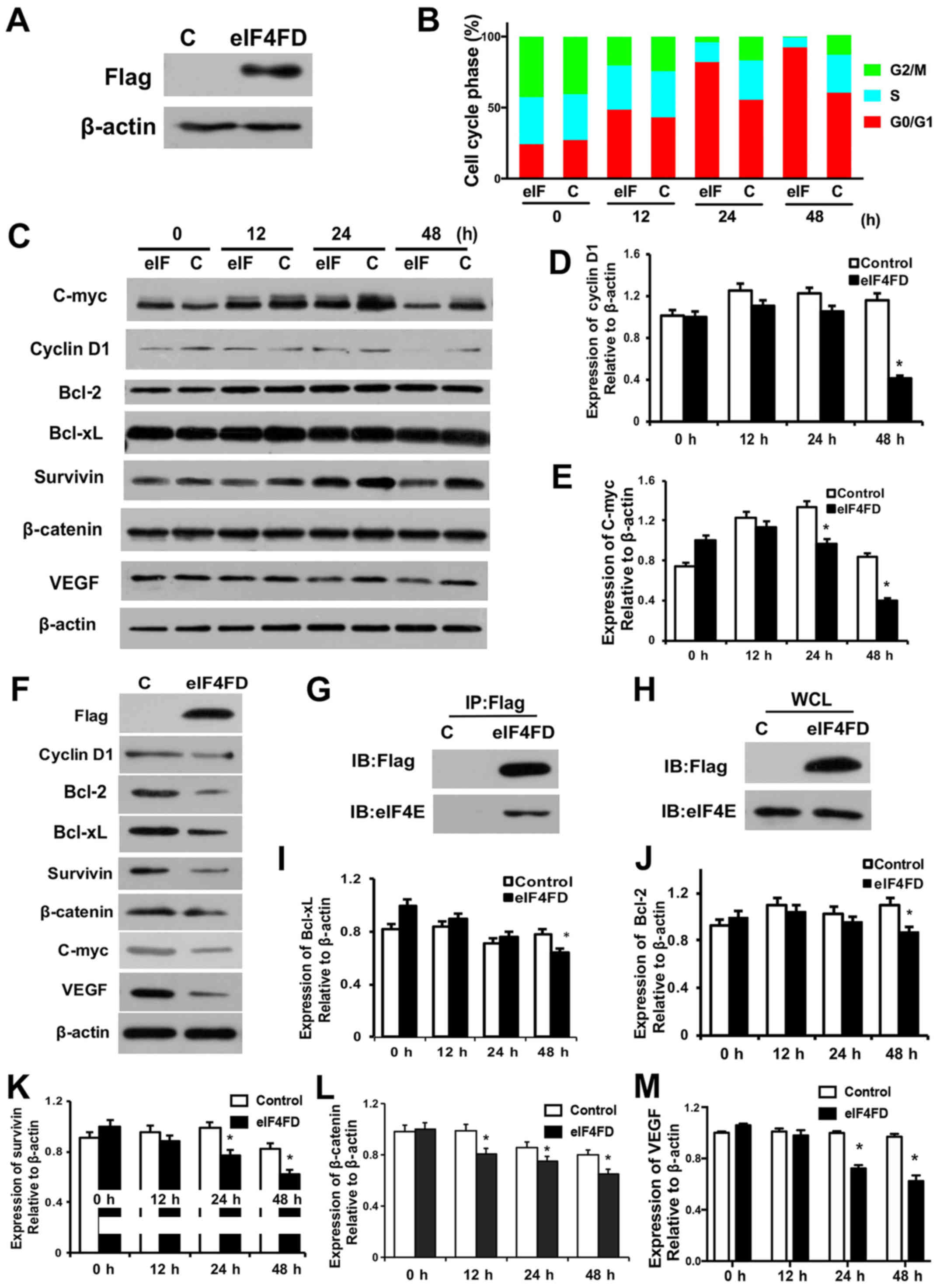 | Figure 1.eIF4FD inhibits cell cycle and
suppresses proliferation in glioma cells. (A) U251 cells were
transfected with pSecTag2 control or pSecTag2-PTD-eIF4FD. The
indicated proteins were determined by western blot analysis 48 h
post-transfection. (B) At 0, 12, 24 and 48 h post-transfection,
cells were collected and subjected to flow cytometric analysis. (C)
Cell lysates were analyzed for the indicated protein levels by
western blot analysis. β-actin was used as a loading control.
Expression levels of (D) cyclin D1 and (E) C-myc were quantified
using ImageJ software and normalized to β-actin. *P<0.05 (F) U87
cells were transfected with pSecTag2 or pSecTag2-PTD-eIF4FD. The
indicated proteins were determined by western blot analysis 48 h
post-transfection. U251 cells with or without pSecTag2-PTD-eIF4FD
transfection were collected for the co-immunoprecipitation assay.
(G) Anti-Flag immunoprecipitates and (H) WCL were resolved by
SDS-PAGE and analyzed by western blot analysis with anti-Flag
antibody or anti-eIF4E antibody. Cell lysates were subjected to
western blot analysis to determine levels of (I) Bcl-xL, (J) BCL2,
(K) survivin, (L) β-catenin and (M) VEGF. WCL whole-cell lysate. C,
control; eIF, eukaryotic initiation factor; PTD, protein
transduction domain; BCL, B-cell lymphoma 2; Bcl-xL, BCL-extra
large; VEGF, vascular endothelial growth factor. |
eIF4FD induces glioma cell apoptosis
and vascularization
To determine whether pSecTag2-PTD-eIF4FD has a tumor
suppressive effect through inducing cell death, the expression
levels of anti-apoptotic genes Bcl-2, Bcl-xL and survivin were
examined. Their expression decreased in the
pSecTag2-PTD-eIF4FD-transfected cells, particularly at 48 h
(Fig. 1C and I-K), indicating that
inhibiting eIF4F complex formation was critical for the induction
of apoptosis through regulating the expression of Bcl-2, Bcl-xL and
survivin. Furthermore, the vascularization associated genes
β-catenin and VEGF were also decreased following the overexpression
of eIF4FD (Fig. 1C, L and M).
Accordingly, the decreases in of Bcl-2, Bcl-xL, survivin, β-catenin
and VEGF were also observed in U87-MG cells following
pSecTag2-PTD-eIF4FD transfection (Fig.
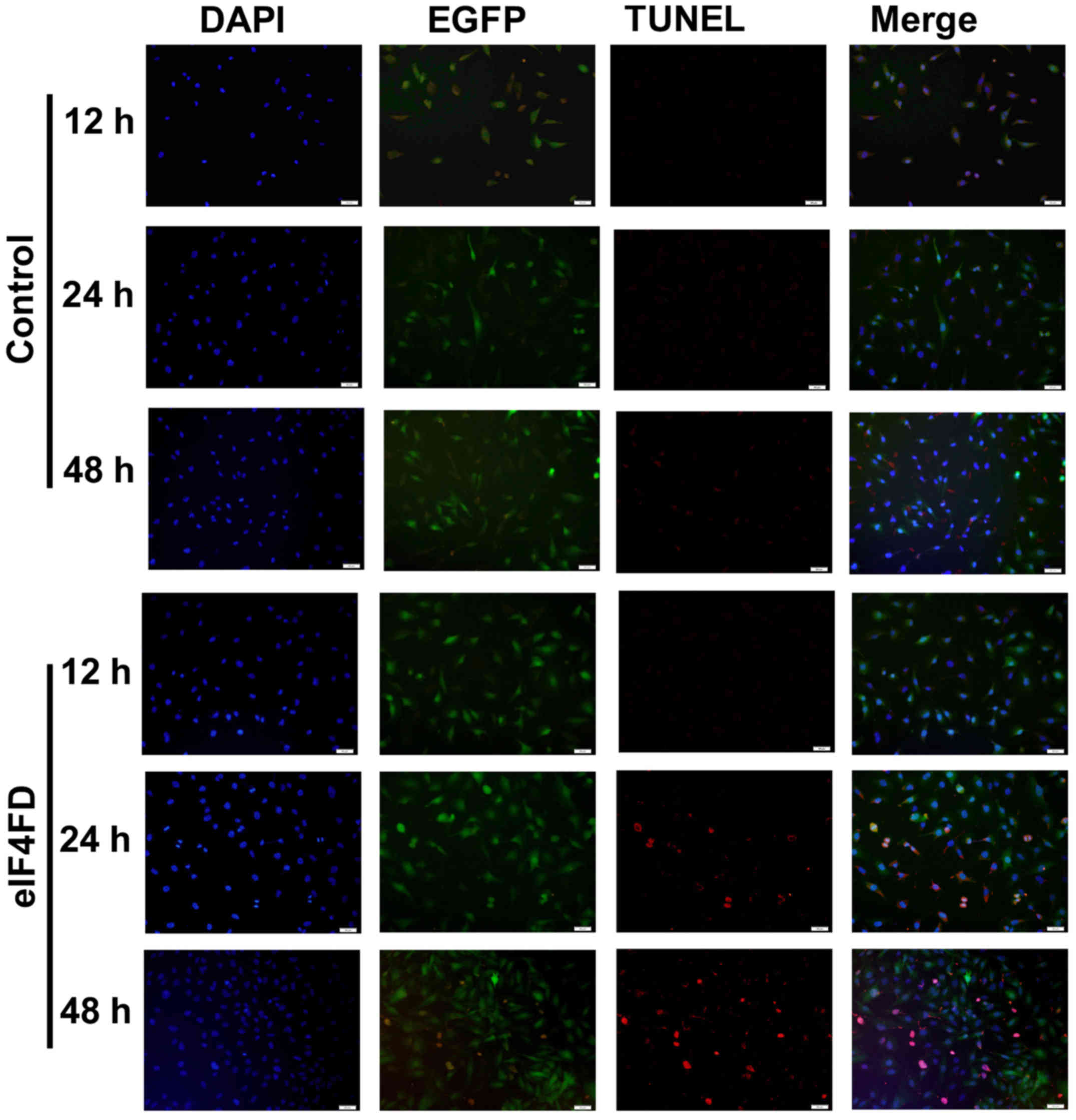
1F). The cell apoptotic rate was also detected through TUNEL
staining of U251 cells (Fig. 2). As
expected, cells transfected with pSecTag2-PTD-eIF4FD resulted in a
significant increase in the number of TUNEL-positive cells.
Therefore, the eIF4FD regulation of cell apoptosis and
vascularization were shown to exist generally in glioma cells.
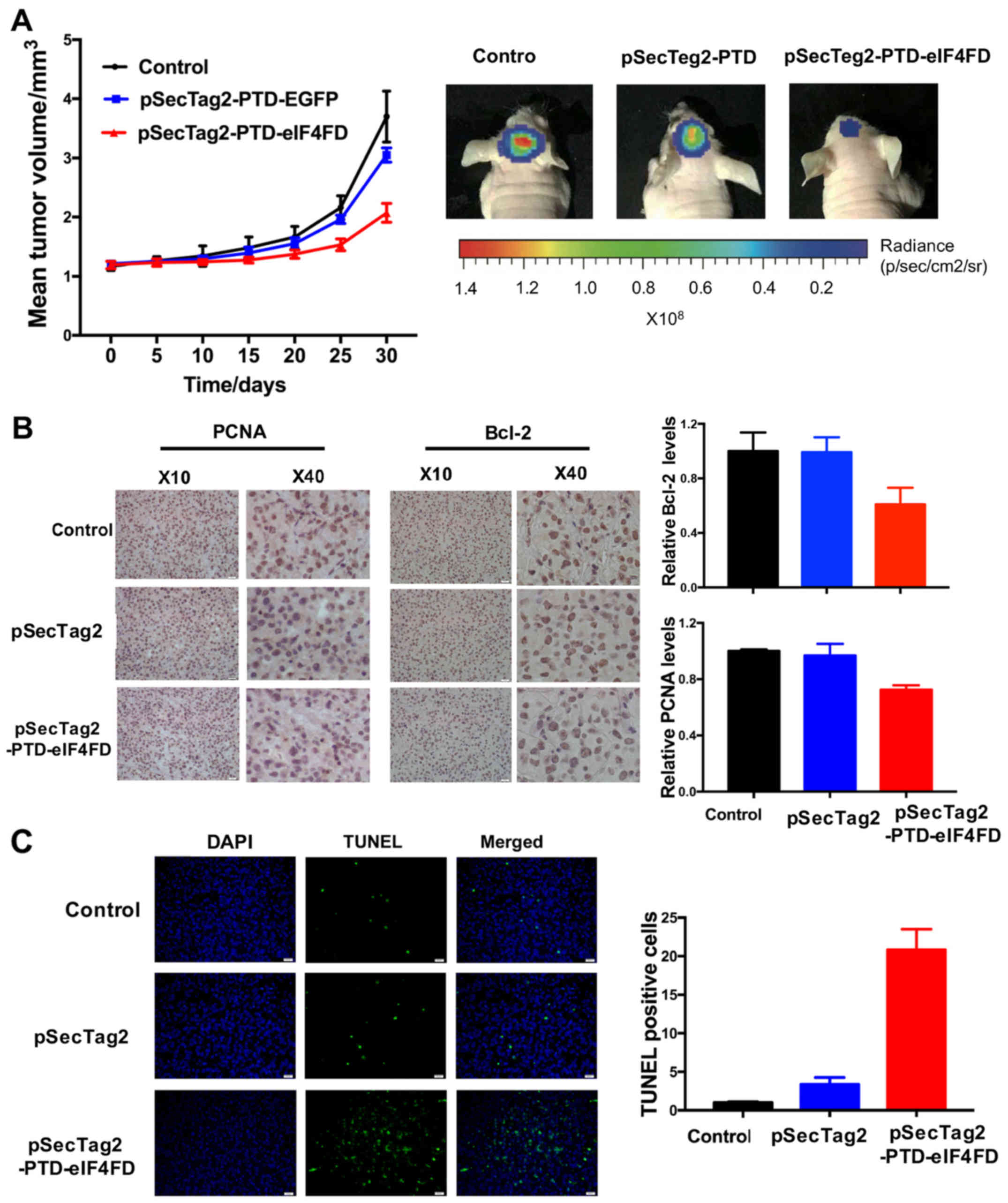
eIF4FD suppresses tumor growth of
glioma cells in nude mice
To further investigate the effect of inhibiting
eIF4F complex assembly via the expression of eIF4FD on cell growth,
U251 cells with stable expression of eIF4FD were established
through transfection with pSecTag2 or pSecTag2-PTD-eIF4FD, and
their effects on tumorigenicity were determined via the
subcutaneous glioma model in nude mice. The mice injected with U251
cells stably expressing eIF4FD developed tumors more slowly than
the other groups (Fig. 3A)
(P<0.01, vs. control; P<0.05, vs. vector). The results of the
IHC assay showed that the positive rates of PCNA and Bcl-2
expression were decreased (Fig.
3B), indicating that the expression of eIF4FD significantly
inhibited tumor growth and induced apoptosis in solid tumors.
Accordingly, TUNEL staining showed that the expression of eIF4FD
promoted the apoptotic cell rates in the tumor tissues (Fig. 3C).
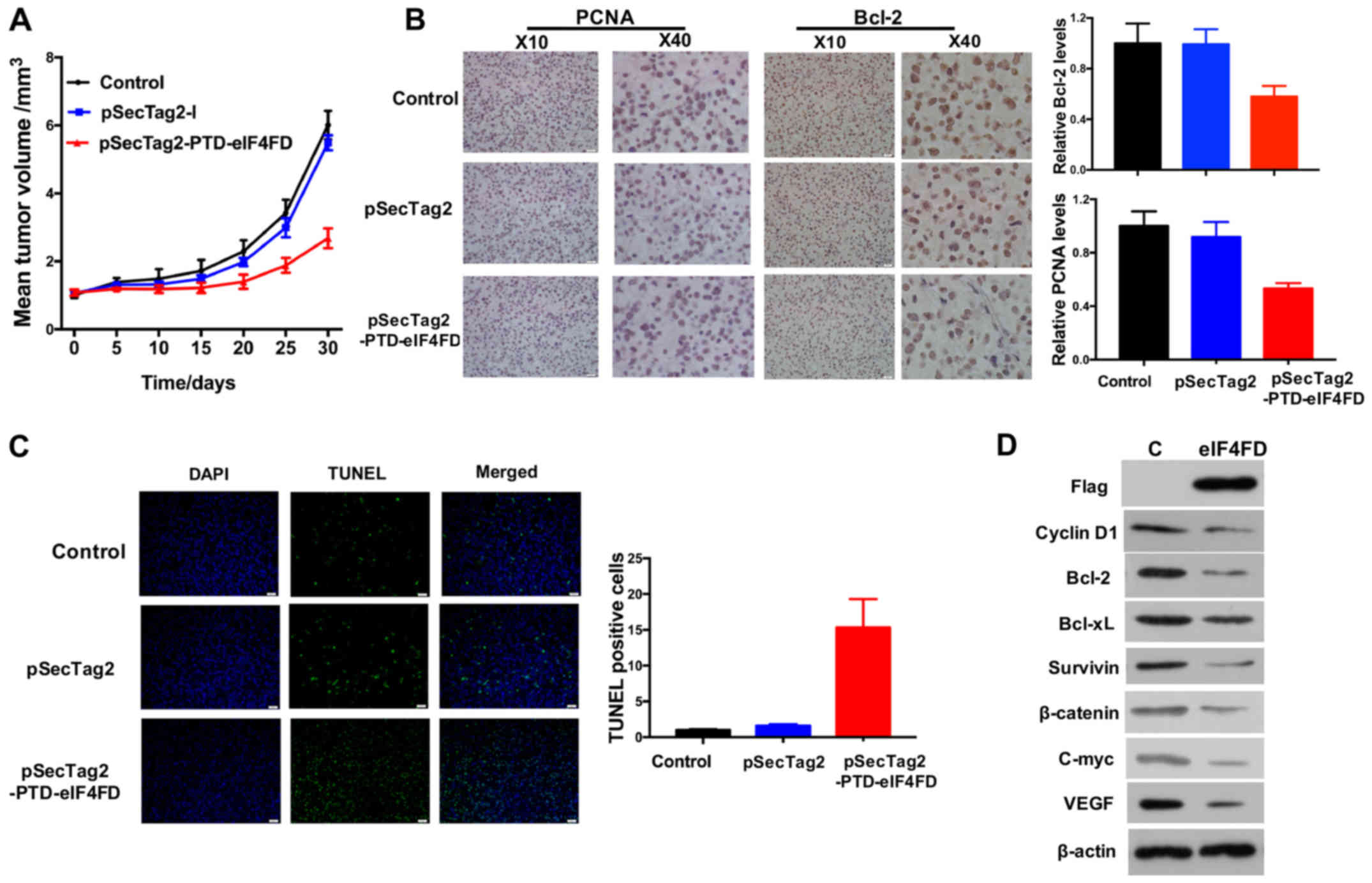
pSecTag2-PTD-eIF4FD injection via the
tail vein represses glioma growth in vivo
To evaluate the therapeutic potential of eIF4FD via
inhibiting eIF4F complex assembly, pSecTag2-PTD-eIF4FD was injected
via the tail vein of nude mice with glioma xenografts. Treatment
with pSecTag2-PTD-eIF4FD had a significant inhibitory effect on
tumor growth compared with the control or empty vector groups
(P<0.05; Fig. 4A). Accordingly,
PCNA and Bcl-2 were reduced and TUNEL staining positive cell
enrichment were observed in glioma tissues (Fig. 4B and C). Genes associated with cell
cycle, apoptosis and vascularization were also decreased in glioma
grafts following pSecTag2-PTD-eIF4FD injection (Fig. 4D). Therefore, inhibiting eIF4F
complex assembly by eIF4FD offers therapeutic potential in the
treatment of gliomas.
Discussion
The eIF4F complex has attracted increasing attention
as a molecular target for malignant tumors. The eIF4F complex is
composed of eIF4E, eIF4G and eIF4A. Progress has been made in
elucidating the association of eIF4E with the occurrence and
development of malignant tumors in previous years. Studies have
shown that eIF4E was expressed at high levels in nasopharyngeal
carcinoma (NPC) and the overexpression of eIF4E promoted NPC growth
and cell cycle progression (24).
eIF4E silencing or inhibition reduced the invasiveness and
metastatic capability of breast cancer cells (25). The inhibition of eIF4E by small
interfering RNA induced cell cycle arrest and suppressed the cell
growth and migration of MDA-MB-231 TN breast cancer cells (26). Furthermore, the overexpression of
eIF4E in renal clear cell carcinoma was correlated with the
presence of phosphorylated (p)4E-BP1. The combined expression of
p4E-BP1 and eIF4E was associated with significantly poorer
disease-free survival rate (27).
Therefore, eIF4E and 4E-BPs may be considered as a potential target
for cancer therapy.
In our previous studies, U251 cells were treated
with the eIF4E inhibitor 4EGI-1, and its suppressive effect on cell
growth was observed (28). It has
been confirmed that the initiation of the translation of cyclin D1
and C-myc are dependent on the eIF4F complex, which directly
promotes the proliferation of cancer cells (4). 4E-BPs act as a post-transcriptional
regulator to inhibit mRNA translation through interacting with
eIF4E (29). The present study
showed that the expression of eIF4FD, phosphorylation-deficient
truncated 4E-BP2, is associated with cell cycle arrest in
G0/G1 phase, which may be due to the
downregulation of growth factors associated with the signal
transduction pathway. The expression of cyclin D1 and C-myc were
determined, which were shown to decrease in glioma cells following
pSecTag2-PTD-eIF4FD transfection. Therefore, cell growth inhibition
by pSecTag2-PTD-eIF4FD-mediated inhibition of the eIF4F complex may
be the result of a reduction in cyclin D1 and C-myc, although the
exact mechanism remains to be elucidated. There is evidence showing
that activation of the eIF4F complex can directly enhance the
expression of Bcl-2, Bcl-xL, survivin and other anti-apoptotic
proteins (8,9). The knockdown or inhibition of survivin
is beneficial in the treatment of glioblastoma (30). In the present study, TUNEL staining
revealed that the number of positive U251 cells was significantly
increased following pSecTag2-PTD-eIF4FD transfection. Accordingly,
the protein levels of anti-apoptotic Bcl-2, Bcl-xL and survivin
were all downregulated in the pSecTag2-PTD-eIF4FD-transfected
cells. These results may explain the higher rates of apoptotic cell
death, which was consistent with the previous studies. The results
showed that the eIF4F complex-dependent initiation of translation
is important in resistance to the apoptosis of cancer cells,
suggesting that preventing eIF4F complex formation may promote
apoptosis via the downregulation of Bcl-2, Bcl-xL and survivin
proteins. Other studies have reported that eIF4F complex activation
can directly promote the 5′-cap structure-dependent mRNA
translation, including survivin and β-catenin (10), and the translation product β-catenin
promotes the transcription of survivin (11). β-catenin can directly enhance the
proliferation and survival of endothelial cells, but also promotes
angiogenesis by inducing the expression of VEGF and promoting
angiogenic progenitor cell mobilization (31). Western blot analysis showed that the
expression levels of β-catenin and VEGF were decreased with the
expression of eIF4FD. The results of the present study indicated
that inhibiting eIF4F complex assembly also led to the inhibition
of tumor angiogenesis by downregulating the expression of β-catenin
and survivin.
To examine the application of the
pSecTag2-PTD-eIF4FD vector as a tool for cancer therapy, the
present study established an orthotopic xenograft model in nude
mice. The data showed that the xenografts overexpressing eIF4FD
showed a significant delay of growth rate and smaller tumor
volumes, accompanied by a high rate of apoptosis. Furthermore, the
injection of pSecTag2-PTD-eIF4FD via the tail vein into glioma
xenograft mice also decreased cell growth rate and induced
apoptosis, indicating a high-efficiency therapeutic potential on
malignant gliomas in vivo. It has been shown that non-small
cell lung cancer cells stably transfected with dominant active
mutant 4E-BP1A37/A46 (HA-TTAA) suppressed growth and
decreased tumorigenicity in xenograft models. Xenograft tumors
expressing HA-TTAA were markedly smaller (32). Breast carcinoma xenografts in
pSecX-t4EBP1 mice also exhibited significantly delayed growth and
smaller tumor volume, with a higher tumor inhibition rate compared
with the control and pSecX groups (33). The results of the present study were
consistent with these findings.
In conclusion, the present study showed that the
eukaryotic expression vector pSecTag2-PTD-eIF4FD, which contains
the phosphorylation defective truncated 4E-BP2, effectively
inhibited eIF4E and prevented eIF4F complex assembly by the
expression of product PTD-eIF4FD. Inhibiting the eIF4F complex
suppressed cell proliferation, induced apoptosis and inhibited
glioma vascularization in vitro, and inhibited tumor growth
and promoted tumor apoptosis in vivo. Therefore, it was
concluded that gene therapy of malignant glioma through targeting
the translation initiation complex eIF4F warrants further
investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Natural Science Foundation of China (grand nos. 81172396, 2011 and
81472358, 2014).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QFD, ZFY and HNZ designed the research; QFD, ZFY,
PQL, XY and JLH performed the research; JLH contributed the new
reagents and analytic tools; QFD, ZFY and HNZ analyzed the data and
wrote the manuscript. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures were in accordance with the NIH Guide
and were approved by the Ethics Committee of the Fourth Military
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bastien JI, McNeill KA and Fine HA:
Molecular characterizations of glioblastoma, targeted therapy, and
clinical results to date. Cancer. 121:502–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jane EP, Premkumar DR, Cavaleri JM, Sutera
PA, Rajasekar T and Pollack IF: Dinaciclib, a Cyclin-Dependent
Kinase inhibitor promotes proteasomal degradation of Mcl-1 and
enhances ABT-737 mediated cell death in malignant human glioma cell
lines. J Pharmacol Exp Ther. 356:354–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rybalkina EY, Pavlova GV and Stavrovskaya
AA: Recent news in the glioblastoma research. Biochem Moscow Suppl
Ser A. 9:1–12. 2015. View Article : Google Scholar
|
|
4
|
Graff JR, Konicek BW, Carter JH and
Marcusson EG: Targeting the eukaryotic translation initiation
factor 4E for cancer therapy. Cancer Res. 68:631–634. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hagner PR, Abraham S and Gartenhaus RB:
Targeting the translational machinery as a novel treatment strategy
for hematologic malignancies. Blood. 115:2127–2135. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsieh AC and Ruggero D: Targeting
eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin
Cancer Res. 16:4914–4920. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rhoads RE: eIF4E: New family members, new
binding partners, new roles. J Biol Chem. 284:16711–16715. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Graff JR, Konicek BW, Vincent TM, Lynch
RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, Dowless MS,
et al: Therapeutic suppression of translation initiation factor
eIF4E expression reduces tumor growth without toxicity. J Clin
Invest. 117:2638–2648. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peponi E, Drakos E, Reyes G, Leventaki V,
Rassidakis GZ and Medeiros LJ: Activation of mammalian target of
rapamycin signaling promotes cell cycle progression and protects
cells from apoptosis in mantle cell lymphoma. Am J Pathol.
169:2171–2180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karni R, Gus Y, Dor Y, Meyuhas O and
Levitzki A: Active Src elevates the expression of beta-catenin by
enhancement of cap-dependent translation. Mol Cell Biol.
25:5031–5039. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Torres VA, Tapia JC, Rodriguez DA, Lladser
A, Arredondo C, Leyton L and Quest AF: E-cadherin is required for
caveolin-1-mediated down-regulation of the inhibitor of apoptosis
protein survivin via reduced beta-catenin-Tcf/Lef-dependent
transcription. Mol Cell Biol. 27:7703–7717. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin CJ, Nasr Z, Premsrirut PK, Porco JA
Jr, Hippo Y, Lowe SW and Pelletier J: Targeting synthetic lethal
interactions between Myc and the eIF4F complex impedes
tumorigenesis. Cell Rep. 1:325–333. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boussemart L, Malka-Mahieu H, Girault I,
Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro
N, Thuaud F, et al: eIF4F is a nexus of resistance to anti-BRAF and
anti-MEK cancer therapies. Nature. 513:105–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou H and Huang S: Role of mTOR signaling
in tumor cell motility, invasion and metastasis. Curr Protein Pept
Sci. 12:30–42. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Robichaud N and Sonenberg N: eIF4E and Its
Binding ProteinsParsyan A: Translat Regulat Cancer Biol Med.
Springer; Dordrecht: pp. 73–113. 2014
|
|
16
|
Satheesha S, Cookson VJ, Coleman LJ,
Ingram N, Madhok B, Hanby AM, Suleman CA, Sabine VS, Macaskill EJ,
Bartlett JM, et al: Response to mTOR inhibition: Activity of eIF4E
predicts sensitivity in cell lines and acquired changes in eIF4E
regulation in breast cancer. Mol Cancer. 10:192011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ayuso MI, Martinez-Alonso E, Salvador N,
Bonova P, Regidor I and Alcázar A: Dissociation of eIF4E-binding
protein 2 (4E-BP2) from eIF4E independent of
Thr37/Thr46 phosphorylation in the ischemic
stress response. PLoS One. 10:e01219582015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lukhele S, Bah A, Lin H, Sonenberg N and
Forman-Kay JD: Interaction of the eukaryotic initiation factor 4E
with 4E-BP2 at a dynamic bipartite interface. Structure.
21:2186–2196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patel LN, Zaro JL and Shen WC: Cell
penetrating peptides: Intracellular pathways and pharmaceutical
perspectives. Pharm Res. 24:1977–1992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wagstaff KM and Jans DA: Protein
transduction: Cell penetrating peptides and their therapeutic
applications. Curr Med Chem. 13:1371–1387. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ozawa T, Wang J, Hu LJ, Bollen AW, Lamborn
KR and Deen DF: Growth of human glioblastomas as xenografts in the
brains of athymic rats. In Vivo. 16:55–60. 2002.PubMed/NCBI
|
|
22
|
Candolfi M, Curtin JF, Nichols WS,
Muhammad AG, King GD, Pluhar GE, McNiel EA, Ohlfest JR, Freese AB,
Moore PF, et al: Intracranial glioblastoma models in preclinical
neuro-oncology: Neuropathological characterization and tumor
progression. J Neurooncol. 85:133–148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Doherty JE, Woodard LE, Bear AS, Foster AE
and Wilson MH: An adaptable system for improving transposon-based
gene expression in vivo via transient transgene repression. FASEB
J. 27:3753–3762. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu M, Liu Y, Di X, Kang H, Zeng H, Zhao Y,
Cai K, Pang T, Wang S, Yao Y and Hu X: EIF4E over-expresses and
enhances cell proliferation and cell cycle progression in
nasopharyngeal carcinoma. Med Oncol. 30:4002013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pettersson F, Del Rincon SV, Emond A, Huor
B, Ngan E, Ng J, Dobocan MC, Siegel PM and Miller WH Jr: Genetic
and pharmacologic inhibition of eIF4E reduces breast cancer cell
migration, invasion, and metastasis. Cancer Res. 75:1102–1112.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou FF, Yan M, Guo GF, Wang F, Qiu HJ,
Zheng FM, Zhang Y, Liu Q, Zhu XF and Xia LP: Knockdown of eIF4E
suppresses cell growth and migration, enhances chemosensitivity and
correlates with increase in Bax/Bcl-2 ratio in triple-negative
breast cancer cells. Med Oncol. 28:1302–1307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Campbell L, Jasani B, Griffiths DF and
Gumbleton M: Phospho-4e-BP1 and eIF4E overexpression
synergistically drives disease progression in clinically confined
clear cell renal cell carcinoma. Am J Cancer Res. 5:2838–2848.
2015.PubMed/NCBI
|
|
28
|
Yang X, Dong QF, Li LW, Huo JL, Li PQ, Fei
Z and Zhen HN: The cap-translation inhibitor 4EGI-1 induces
mitochondrial dysfunction via regulation of mitochondrial dynamic
proteins in human glioma U251 cells. Neurochem Int. 90:98–106.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Di Marino D, D'Annessa I, Tancredi H,
Bagni C and Gallicchio E: A unique binding mode of the eukaryotic
translation initiation factor 4E for guiding the design of novel
peptide inhibitors. Protein Sci. 24:1370–1382. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao A, Shi R, Jiang Y, Tian S, Li P, Song
F, Qu Y, Li J, Yun H and Yang X: SDF-1/CXCR4 axis regulates cell
cycle progression and epithelial-mesenchymal transition via
up-regulation of survivin in glioblastoma. Mol Neurobiol.
53:210–215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim KI, Cho HJ, Hahn JY, Kim TY, Park KW,
Koo BK, Shin CS, Kim CH, Oh BH, Lee MM, et al: Beta-catenin
overexpression augments angiogenesis and skeletal muscle
regeneration through dual mechanism of vascular endothelial growth
factor-mediated endothelial cell proliferation and progenitor cell
mobilization. Arterioscler Thromb Vasc Biol. 26:91–98. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jacobson BA, Alter MD, Kratzke MG,
Frizelle SP, Zhang Y, Peterson MS, Avdulov S, Mohorn RP, Whitson
BA, Bitterman PB, et al: Repression of cap-dependent translation
attenuates the transformed phenotype in non-small cell lung cancer
both in vitro and in vivo. Cancer Res. 66:4256–4262. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang H, Li LW, Shi M, Wang JH, Xiao F,
Zhou B, Diao LQ, Long XL, Liu XL and Xu L: In vivo study of breast
carcinoma radiosensitization by targeting eIF4E. Biochem Biophys
Res Commun. 423:878–883. 2012. View Article : Google Scholar : PubMed/NCBI
|