Introduction
Breast cancer is the second most commonly diagnosed
invasive cancer in women with 1.15 million cases diagnosed annually
worldwide, and is the main cause of cancer-related mortality
worldwide (1). In Western
countries, studies have shown that one in eight women is likely to
develop breast cancer (2). However,
it remains difficult to elucidate the mechanisms of breast
carcinogenesis. The diverse biochemistry and clinical background of
breast cancer have led researchers to discover more efficacious
markers and evaluation strategies for diagnosis and therapy.
A proteasome is an ATP-dependent multi-catalytic
proteinase complex (3), which is
crucial to the degradation of intracellular targeted proteins and
the maintenance of significant biological functions (4,5).
Emerging evidence indicates that the proteasome plays a novel role
in regulating anti-apoptotic and proliferative signaling pathways
in a variety of malignancies, including breast cancer (6–9).
Indeed, proteasome inhibition provides a unique approach to cancer
therapy by depressing the function of proteasomes. Thus, proteasome
inhibitors, such as carfilzomib and bortezomib, have been proven to
be effective against cancers in human trials by target multifarious
subunits (10,11). PSMB4, a β4 subunit of the 20S
proteasome, is a member of the ubiquitin-proteasome family, and is
involved in the cytoplasmic protein catabolic process (12). More and more studies have revealed
the correlation between PSMB4 expression and tumor tissues
(13–18). The association of PSMB4
overexpression with increased NF-κB activity led to an increase in
the proliferation ability of multiple myeloma and epithelial
ovarian cancer (13,14). In addition, PSMB4 expression is
related to increased viability and the malignant progression of
gliomas (15). Therefore, PSMB4 may
be recognized as an efficacious marker and potential therapeutic
target for breast cancer. However, its expression and extensional
regulatory mechanism in breast cancer remains virtually
unclear.
In the present study, the expression of PSMB4 was
detected in breast cancer cell lines and tissues by
immunohistochemistry and western blot analysis. In addition, the
association of PSMB4 with the clinicopathological characteristics
obtained from breast cancer patients was discovered. Next, the
functional significance of PSMB4 was explored in cultured breast
cancer cells by siRNA gene silencing. These results show that PSMB4
is a novel regulator of breast cancer cell proliferation and a
potential therapeutic target in breast cancer.
Materials and methods
Patients and tissue samples
Breast specimens were obtained from 92 patients who
underwent excision of breast cancer tumors at the Department of
General Surgery, Affiliated Hospital of Nantong University from
2006 to 2012. The present study was approved by the Ethics
Committee of the Affiliated Cancer Hospital of Nantong University
(Nantong, China). Fresh breast cancer tissues were collected and
immediately stored in liquid nitrogen. Lobular carcinoma or other
histological types were confirmed at the Department of Pathology,
the Affiliated Hospital of Nantong University. The detailed
clinical information of patients was obtained by telephone or
interview with the patients. All patients provided a written
informed consent. For histological analysis, the specimens were
fixed in formalin and embedded in paraffin. The protein of 8
tumorous and adjacent non-tumorous tissue samples was analyzed by
western blot analysis. After receiving the authorization of the
patients, the detailed information of the patients, including age,
tumor grade, estrogen receptor (ER) status, progesterone-receptor
(PR) status, Her2 status, tumor size, lymph node status, histologic
grade and Ki-67, were obtained. The data are presented in Table I.
 | Table I.Correlation between PSMB4 expression
and the clinicopathological factors of the breast cancer cases. |
Table I.
Correlation between PSMB4 expression
and the clinicopathological factors of the breast cancer cases.
|
|
| PSMB4
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Patient
characteristics | Total | Low | High |
P-valuea | χ2 |
|---|
| Age (years) |
|
≤50 | 48 | 19 | 29 | 0.297 | 0.586 |
|
>50 | 44 | 15 | 29 |
|
|
| Tumor grade |
| I | 8 | 7 | 1 | 0.005b | 10.578 |
| II | 43 | 16 | 27 |
|
|
|
III | 41 | 11 | 30 |
|
|
| ER status |
|
Negative | 40 | 14 | 26 | 0.733 | 0.116 |
|
Positive | 52 | 20 | 32 |
|
|
| PR status |
|
Negative | 69 | 25 | 44 | 0.803 | 0.062 |
|
Positive | 23 | 9 | 14 |
|
|
| Her2 status |
|
Negative | 68 | 27 | 41 | 0.358 | 0.846 |
|
Positive | 24 | 7 | 17 |
|
|
| Tumor size
(cm) |
|
≤2.5 | 34 | 17 | 17 | 0.047b | 3.938 |
|
>2.5 | 58 | 17 | 41 |
|
|
| Lymph node
status |
|
Negative | 34 | 15 | 19 | 0.276 | 1.187 |
|
Positive | 58 | 19 | 39 |
|
|
| Histology |
|
Ductal | 76 | 28 | 48 | 0.960 | 0.002 |
|
Others | 16 | 6 | 10 |
|
|
| Ki-67 |
|
Low | 44 | 21 | 23 | 0.040b | 4.199 |
|
High | 48 | 13 | 35 |
|
|
Immunohistochemistry (IHC)
The specimens (4-µm thick) were mounted on glass
slides embedded in paraffin and coated with 10% formalin. Next, the
tissue sections were deparaffinized with xylene and rehydrated with
graded alcohol washes. Then, the antigen was retrieved using
citrate buffer (pH 6.0) by heating to 120°C for 3 min. After
douching in phosphate-buffered saline (PBS) (pH 7.2), the sections
were incubated with rabbit anti-PSMB4 antibody (1:1,000; cat. no.
11029-1-AP; ProteinTech Group, Inc., Chicago, IL, USA) and rabbit
anti-Ki-67 antibody (1:1,000; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) at 4°C overnight. The slides were washed three
times with PBS, and were incubated using a 2-step Plus Poly-HRP
anti-mouse/rabbit IgG detection system (ZSGB-Bio; OriGene
Technologies, Inc., Beijing, China) at room temperature for 30 min.
Next, each section was washed with PBS and the sections were
incubated with 3,3′-diaminobenzidine (DAB). After washing in water,
the sections were counterstained with hematoxylin to visualize
normal nuclei, dehydrated in a series of graded alcohol, and
cover-slipped on microslide glass. The observers who evaluated the
sections were blinded to the characteristics of the patients.
Cell culture and siRNA
transfection
MDA-MB-231 and MCF-7 cells were the human breast
cancer cell lines used (Department of Oncology, the Affiliated
Cancer Hospital of Fudan University). MCF-7 was obtained and
cultured in RPMI-1640 medium, while MDA-MB-231 cells were cultured
in L15 medium (both from Gibco-BRL, Grand Island, NY, USA) at 37°C
in an incubator with 5% CO2. Both types of mediums were
supplemented with 10% heat inactivated fetal bovine serum (FBS).
The PSMB4-specific siRNAs were synthesized by Shanghai GeneChem
Co., Ltd., (Shanghai, China). The siRNA target sequence of PSMB4
included the following: PSMB4-siRNA#1, 5-GCUAUAGUCCUAGAGCUAU-3 and
PSMB4-siRNA#2, 5-GCUAUUCAUU CAUGGCUGA-3. According to the
manufacturer's instructions, when 80% confluency was reached,
MDA-MB-231 and MCF-7 cells were transfected with the PSMB4-siRNA or
control siRNA, and were harvested for follow-up experiments after
transfection for 48 h. Cell transfection was performed with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's
instructions.
Protein extraction and western blot
analysis
Cells were washed with ice-cold PBS for three times
and resuspended in lysis buffer (50 mM of Tris-HCl, 120 mM of NaCl,
0.5% of Nonidet P-40, 100 mM of NaF, 200 µM of
Na3VO4, and a protease inhibitor mixture).
Next, the sample was centrifuged at 12,000 rpm for 30 min at 4°C to
obtain the supernatant. Then, the protein concentration was
detected using the BioRad protein assay (Bio-Rad Laboratories,
Hercules, CA, USA). Next, the protein samples were denatured in
water at 100°C for 3 min. Then, the proteins were separated using
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto polyvinylidene difluoride filter
(PVDF) membranes (EMD Millipore Corp., Billerica, MA, USA). After
blocking the membranes with 5% dried skim milk in Tris-buffered
saline Tween-20 (TBST) for 2 h, these were incubated with
antibodies overnight at 4°C and washed with TBST for three times
for 5 min each time. Then, the membranes were incubated with
horseradish peroxidase-linked IgG for 2 h. Finally, the bands were
detected using the ECL detection system (Pierce, Rockford, IL,
USA). The densities of bands were compared using ImageJ (NIH,
Bethesda, MD, USA).
Flow cytometric analyses of cell cycle
distribution and cell apoptosis
After serum deprivation for 72 h, the cells were
incubated with complete medium for 4, 8, 12 and 24 h. Then, the
cells were strictly collected at every time-point. The MDA-MB-231
and MCF-7 cells were transfected with PSMB4-siRNA, according to
manufacturer's instructions. The MDA-MB-231 and MCF-7 cells were
harvested and fixed with 70% ice-cold ethanol for at least 24 h.
Then, DNA-staining solution [25 µg/ml of propidium iodide (PI), 100
µg/ml of RNase A, and 0.5% Nonidet P-40 in PBS] was added to stain
the cells at room temperature for 20 min. Next, the DNA content was
analyzed from 1×104 cells using the Muse™ cell analyzer
(EMD Millipore Corp.). Each experiment was performed in
triplicate.
In order to analyze apoptosis, the cells were
double-stained using an Annexin V-FITC/PI apoptosis detection kit
(BBI), and analyzed using the FACSCalibur flow cytometer.
Plate colony formation assay
The MDA-MB-231 and MCF-7 cells transfected with
PSMB4-siRNA#1 were seeded in 2 ml of L15 or RPMI-1640 supplemented
with 10% FBS in 6-well plates at 5×103 per well. After 2
weeks, each well was washed with PBS. Next, the colonies were fixed
with 4% paraformaldehyde for 20 min. Finally, the colonies were
stained with 0.4% crystal violet for 30 min at room temperature.
The efficiency of the assay was the following: Clone forming
efficiency = number of colonies/number of inoculated cells
×100%.
Immunohistochemical evaluation
Two independent investigators randomly examined
every section in a blinded manner using a Leica fluorescence
microscope (Leica Microsystems GmbH, Wetzlar, Germany). Five fields
were chosen from each slide, counting >1,000 cells on each view
at high magnification. In order to objectively make an assessment
of the immunoreaction of PSMB4, the staining intensity was divided
into scores 0–3 (0, negative staining; 1, weak staining; 2,
moderate staining; 3, strong staining). Based on the proportion of
PSMB4 expression and positive tumor cells, patients were
categorized into four groups: Negative (<10%, 1), low (10–35%,
2), moderate (36–50%, 3), and high (>50%, 4). The immunostaining
score was calculated as the staining intensity score × the positive
cell percentage score. As a result, if the total value was >6,
the sample was assigned a high grade, and if the value was <6,
the sample was assigned a low grade.
Antibodies
The antibodies applied for the immunohistochemistry
and western blot analysis in the present study were as follows:
Rabbit anti-PSMB4 monoclonal antibody (1:1,000; cat. no.
11029-1-AP; ProteinTech Group, Inc., Chicago, IL, USA), mouse
anti-human Ki-67 monoclonal antibody (1:1,000; cat. no. sc-23900),
mouse anti-human cyclin D1 polyclonal antibody (1:1,000; cat. no.
sc-8396) (both from Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA), rabbit anti-P-P65 monoclonal antibody (1:800; cat. no. 3033),
rabbit anti-Iκβα monoclonal antibody (1:1,000; cat. no. 76041)
(both from Cell Signaling Technology, Danvers, MA, USA), rabbit
anti-Bax monoclonal antibody (1:1,000; cat. no. sc-493), mouse
anti-human Bcl-2 monoclonal antibody (1:1,000; cat. no. sc-509),
mouse anti-human PCNA monoclonal antibody (1:1,000; cat. no.
sc-25280) (all from Santa Cruz Biotechnology, Inc.), mouse
anti-human PARP monoclonal antibody (1:800; cat. no. sc-136208;
Cell Signaling Technology) and rabbit anti-human GAPDH polyclonal
antibody (1:5,000; cat. no. 10494-1-AP; ProteinTech Group,
Inc.).
Statistical analysis
The expression of PSMB4 and its correlation with
clinicopathological features of breast cancer were investigated
using the Chi-square (χ2) test. In order to investigate
the survival data, Kaplan-Meier survival plots and a log rank test
were used. In addition, the association between PSMB4 mRNA
expression and relapse-free survival (RFS) was evaluated with the
online database Kaplan-Meier Plotter (www.kmplot.com). Briefly, ‘202244_at’ (PSMB4 probe
number) was entered into the breast cancer-specific area of the
database to obtain the Kaplan-Meier survival curves. Hazard ratios
(HR) with 95% confidence intervals (CI) and P-values were displayed
on the webpage. In addition, the multivariate analysis was
performed with the Cox's proportional hazards model. Statistical
significance was set at P<0.05. All statistical analyses were
performed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) and
SigmaPlot 12 statistical (Systat Software, Inc., San Jose, CA, USA)
software programs. The data are presented as mean ± standard
deviation (SD). All experiments were conducted for at least three
replicates.
Results
PSMB4 expression in human breast
cancer
In order to further elaborate the function of PSMB4
in the development of breast cancer, PSMB4 protein expression was
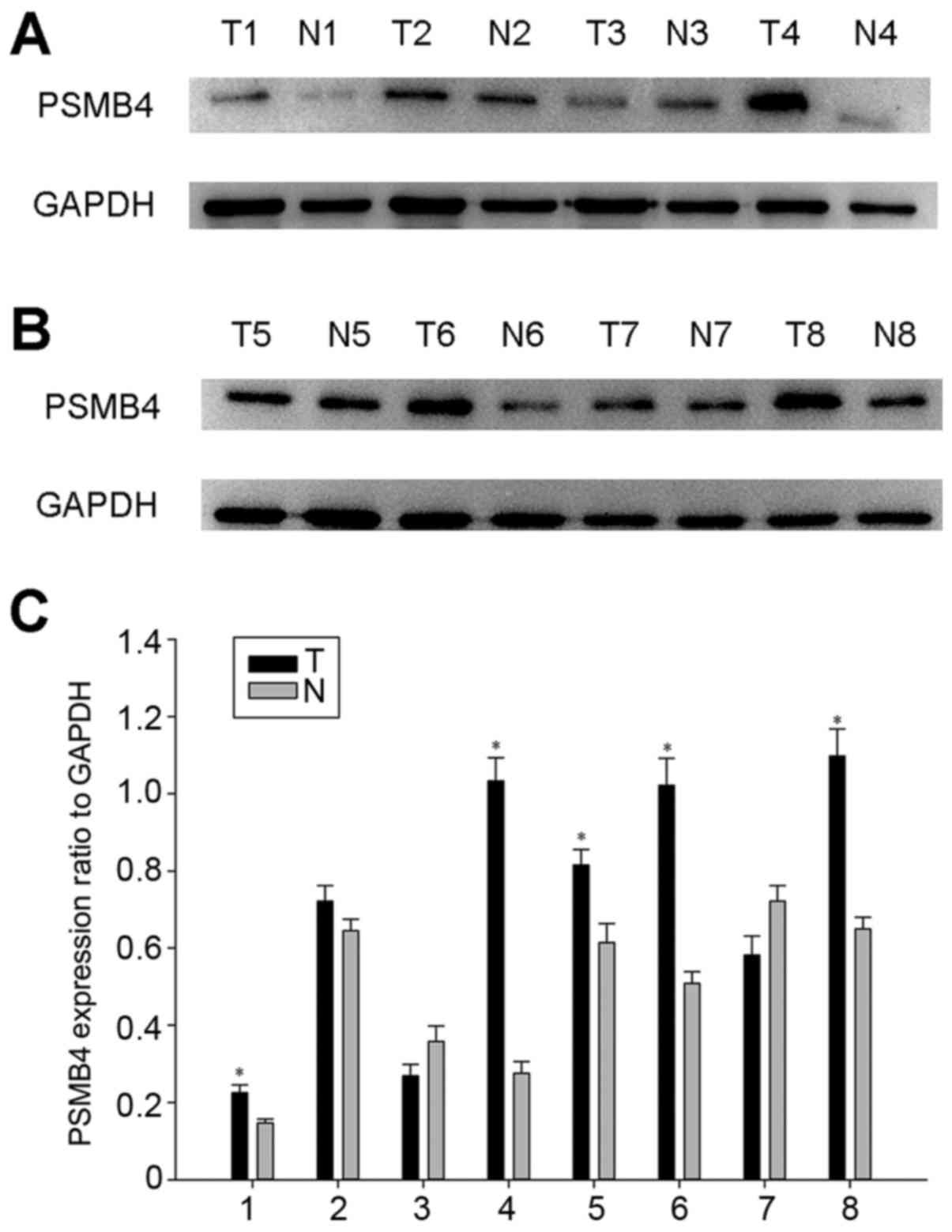
detected by western blot analysis. As shown in Fig. 1, PSMB4 was highly expressed in the 8
tumors, compared with that noted in the adjacent normal tissues.
Taken together, PSMB4 may function as a cancerogenic factor in
breast cancer.
PSMB4 expression and patient
survival
The distribution and expression level of PSMB4 and
Ki-67 in 92 breast cancer tissue sections were examined using
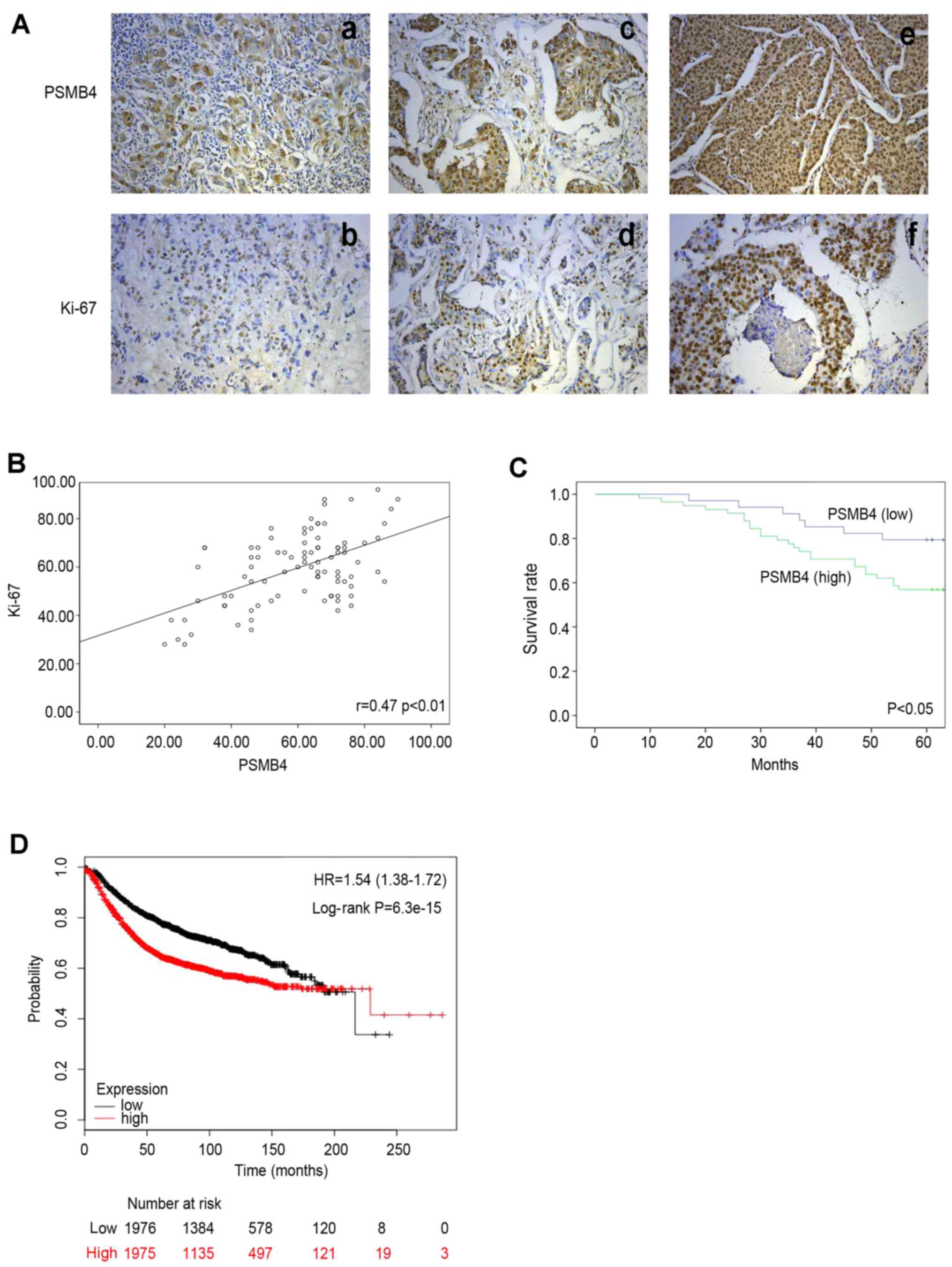
immunohistochemistry. As shown in Fig.
2A, compared with the adjacent normal tissues, PSMB4 was highly
stained mainly in the cytoplasm and nuclei in poorly differentiated
specimens. We next investigated the association between PSMB4
protein level and common clinical parameters of the breast cancer
cases. As presented in Table I,
positive correlations were found between the expression level of
PSMB4 and Ki-67 (P=0.040), tumor grade (P=0.005), and tumor size
(P=0.047). This was compared with the clinicopathological features
listed in Table II. Furthermore,
the univariate survival analysis revealed that tumor grade
(P=0.036), tumor size (P=0.019), PSMB4 expression (P=0.029), and
Ki-67 expression (P=0.040) were prognostic factors for overall
survival (Table II). The
multivariate analysis using Cox's proportional hazards model
revealed that grade (P=0.010), tumor size (P=0.005), PSMB4
expression (P=0.001), and Ki-67 expression (P=0.008) were
prognostic indicators for overall survival (Table III). The present results revealed
that there was a positive correlation between PSMB4 and Ki-67
(Pearson's correlation coefficient: r=0.47, P<0.01) (Fig. 2B). Subsequently, the relationship
between PSMB4 protein level and the survival condition of the 92
patients were examined. As shown in the Kaplan-Meier survival
curves, patients with high expression of PSMB4 exhibited poor
prognosis (Fig. 2C). Then, the
prognostic value of PSMB4 expression was examined by using
Kaplan-Meier Plotter database (Fig.
2D). These results were consistent with the two data
methods.
 | Table II.Univariate analysis of the overall
survival of the breast cancer patients. |
Table II.
Univariate analysis of the overall
survival of the breast cancer patients.
|
|
| Survival
status |
|
|---|
|
|
|
|
|
|---|
| Patient
characteristics | Total | Deceased | Alive |
P-valuea |
|---|
| Age (years) |
|
≤50 | 48 | 17 | 31 | 0.894 |
|
>50 | 44 | 15 | 29 |
|
| Tumor grade |
| I | 8 | 5 | 3 | 0.036b |
| II | 43 | 18 | 25 |
|
|
III | 41 | 9 | 32 |
|
| ER status |
|
Negative | 40 | 13 | 27 | 0.687 |
|
Positive | 52 | 19 | 33 |
|
| PR status |
|
Negative | 69 | 27 | 42 | 0.129 |
|
Positive | 23 | 5 | 18 |
|
| Her2 status |
|
Negative | 68 | 25 | 43 | 0.502 |
|
Positive | 24 | 7 | 17 |
|
| Tumor size
(cm) |
|
≤2.5 | 34 | 17 | 17 | 0.019b |
|
>2.5 | 58 | 15 | 43 |
|
| Lymph node
status |
|
Negative | 34 | 9 | 25 | 0.200 |
|
Positive | 58 | 23 | 35 |
|
| Histology |
|
Ductal | 16 | 5 | 11 | 0.744 |
|
Others | 76 | 27 | 49 |
|
| Ki-67 |
|
Low | 44 | 20 | 24 | 0.040b |
|
High | 48 | 12 | 36 |
|
| PSMB4 |
|
Low | 34 | 7 | 27 | 0.029b |
|
High | 58 | 25 | 33 |
|
 | Table III.Multivariate analysis of overall
survival. |
Table III.
Multivariate analysis of overall
survival.
| Variables | HR | 95.0% CI | P-value |
|---|
| Age | 0.600 | 0.275–1.310 | 0.200 |
| Tumor grade | 0.318 | 0.158–0.640 | 0.001a |
| ER | 1.426 | 0.628–3.239 | 0.397 |
| PR | 0.472 | 0.170–1.308 | 0.149 |
| Her2 | 0.480 | 0.202–1.139 | 0.096 |
| Tumor size | 0.310 | 0.136–0.704 | 0.005a |
| Axillary lymph
node | 1.855 | 0.760–4.528 | 0.175 |
| Histology | 1.154 | 0.410–3.247 | 0.787 |
| Ki-67 | 0.330 | 0.145–0.749 | 0.008a |
| PSMB4 | 9.670 | 3.291–28.410 | 0.001a |
PSMB4 expression in different phases
of the cell cycle
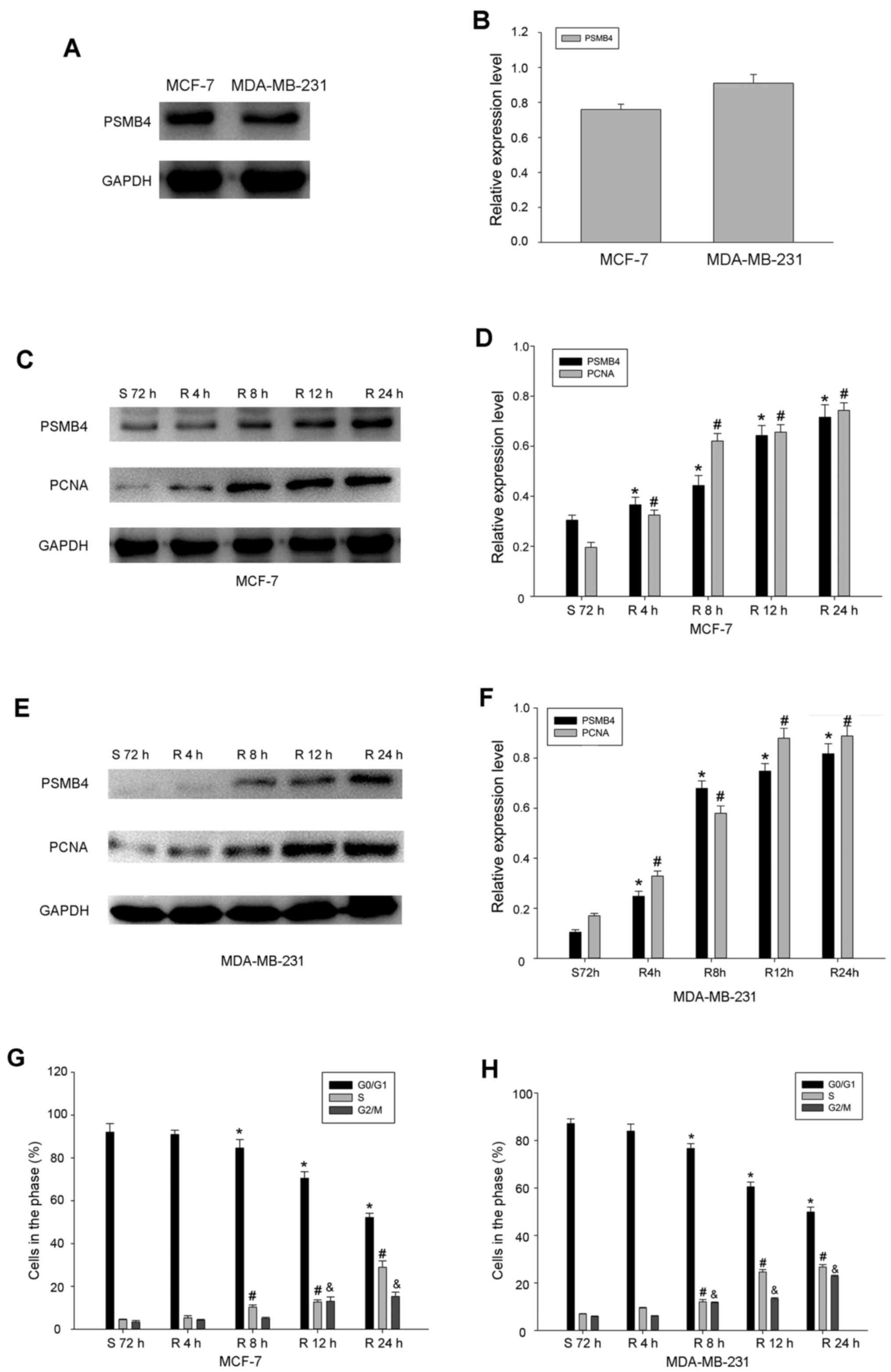
Next, the expression of PSMB4 in several common
breast cancer cell lines was detected by western blot analysis.
Among these cell lines, it was found that PSMB4 was highly
expressed in the MDA-MB-231 and MCF-7 cells (Fig. 3A and B). As shown in the Table I, high levels of PSMB4 expression
were related to Ki-67, which is used to assess cell proliferation.
Therefore, this suggests that the expression of PSMB4 is correlated
with cell proliferation. In order to test this hypothesis, the
change in PSMB4 expression during serum starvation and the
re-feeding process was analyzed. After serum deprivation for 72 h,
PSMB4 expression was lowest in the cells. With the release of
serum, the expression of PSMB4 gradually increased, which was
similar to the cell proliferation marker PCNA (Fig. 3C-F). These results revealed that
PSMB4 plays an integral role in the regulation of cell growth. In
order to further verify the exact function of PSMB4 in the cell
cycle, the level of PSMB4 was detected in the process of serum
starvation and refeeding. Most MDA-MB-231 and MCF-7 cells were
arrested in the G1 phase after the process of serum deprivation for
72 h. Remarkably, within 24 h after the release of serum, the
number of cells blocked in the G1 phase gradually decreased from
~90 to 50%, while the expression of PSMB4 gradually increased,
which was similar to PCNA (Fig. 3G and
H).
PSMB4 knockdown promotes cell cycle
arrest and inhibits cell proliferation through the NF-κB
pathway
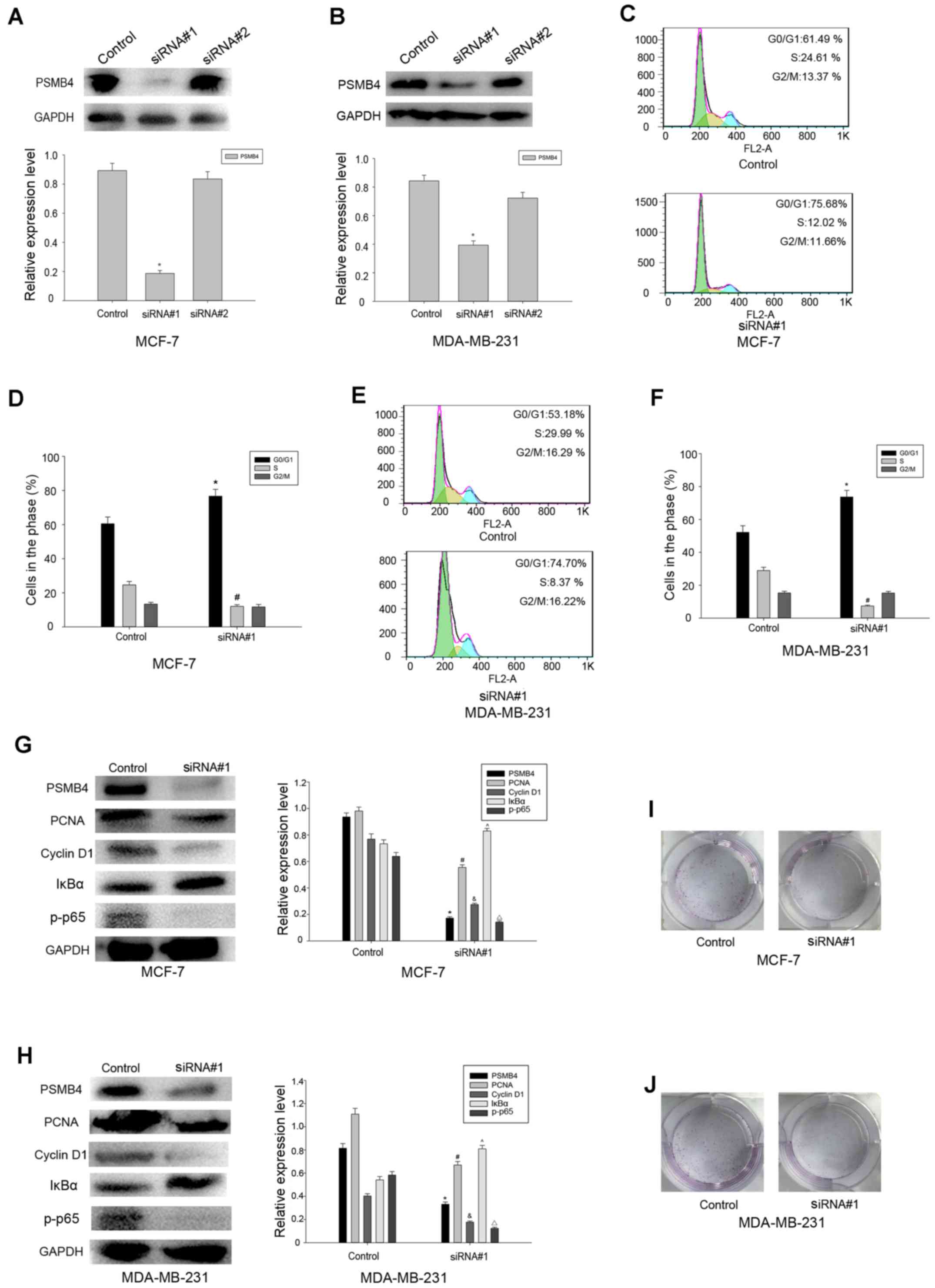
MDA-MB-231 and MCF-7 cells were transfected with
PSMB4 siRNA#1 and control, and the expression of PSMB4 was detected
in the transfected cells by western blot analysis. Consequently, we
determined the effect of PSMB4 knockdown on cell proliferation
(Fig. 4A and B). After the
transfection of these cells, it was found that the percentage of
cells in the G1 phase was significantly higher than that in the
control group (Fig. 4C-F). PSMB4
has been previously reported to modulate the proliferation of
ovarian cancer and multiple myeloma cells via the NF-κB pathway.
Therefore, we investigated whether the NF-κB pathway is involved in
the process of tumor proliferation mediated by PSMB4. Then, a
number of molecules related to the cell cycle was detected, and it
was found that the expression of PCNA, cyclin D1 and p-p65 were
significantly reduced, compared with the control group, while Iκβα
was increased (Fig. 4G and H). The
colony formation analysis also validated our conjecture (Fig. 4I and J). Taken together, it was
found that PSMB4 can promote the proliferation of breast cancer
cells through the NF-κB pathway.
PSMB4 silencing results in the
suppression of cell viability
It has been reported that as a survival gene, PSMB4
knockdown can cause a marked decrease in glioma cell viability. It
was hypothesized that PSMB4 could also maintain the viability of
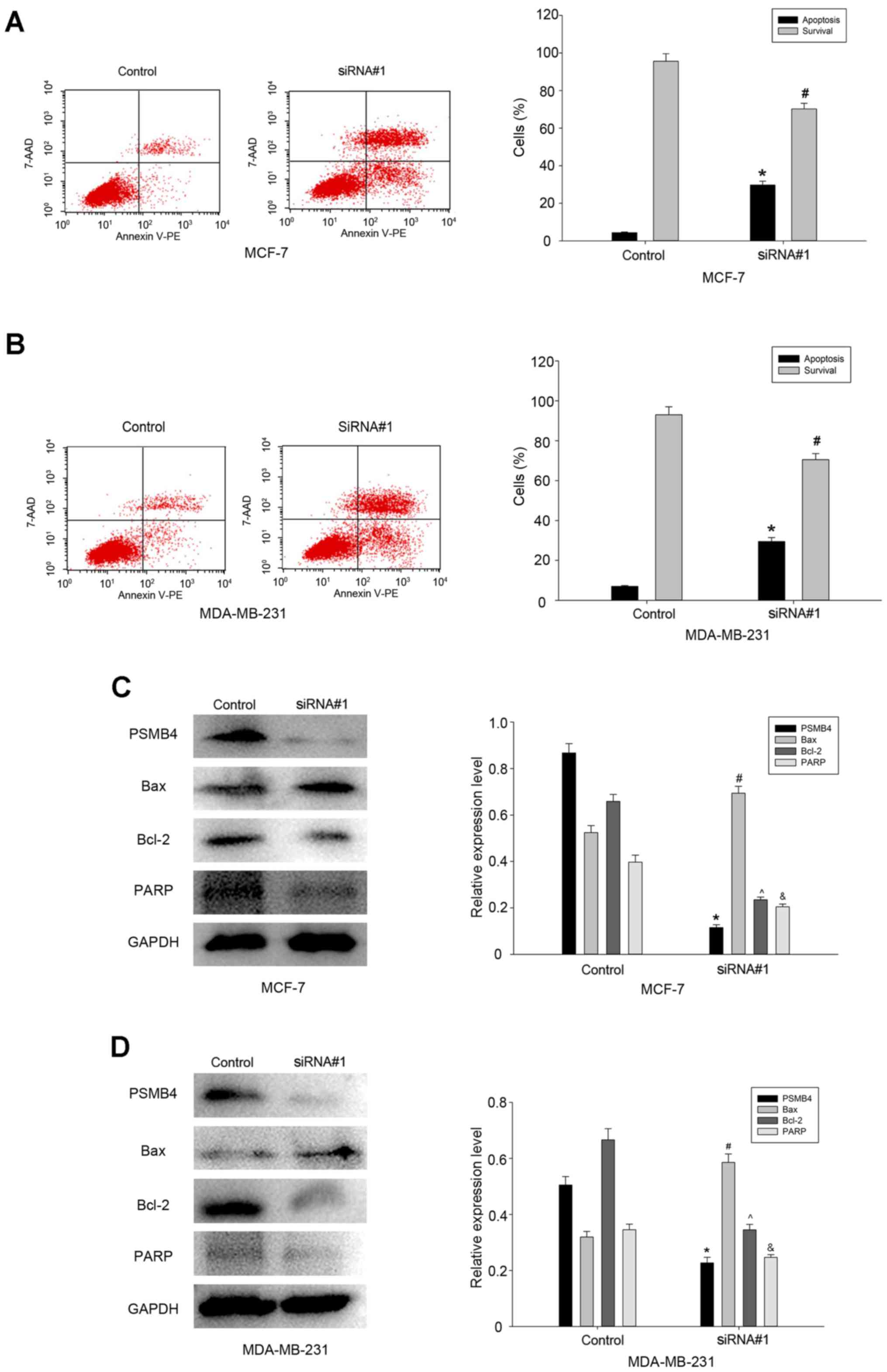
breast cancer MDA-MB-231 and MCF-7 cells. Therefore, the apoptosis
of breast cancer cells transfected with PSMB4-siRNA#1 or controls
was analyzed by flow cytometry (Fig. 5A
and B). The results revealed that compared with the control
group, the percentage of apoptotic cells was higher in the
PSMB4-siRNA#1 group. Next, a series of molecular markers for
cellular apoptosis were analyzed, and it was found that the
expression levels of Bcl-2 and PARP were significantly decreased,
while the level of the apoptotic marker Bax was increased in the
cells transfected with PSMB4-siRNA#1 (Fig. 5C and D). Thus, it is clear that as a
survival gene, PSMB4 can maintain the viability of breast cancer
cells.
Discussion
Breast cancer is the second leading cause of
cancer-related mortality worldwide. Notwithstanding the utilization
of comprehensive diagnostic and therapeutic approaches, the
recurrence and metastasis of breast cancer are commonly observed
(19,20). Although factors including ER, PR and
HER-2 are clinical predictive markers for breast cancer, patients
with diverse molecular subtypes have distinct efficacies.
Therefore, they cannot fully reflect the molecular mechanisms of
breast cancer therapy (21,22). Hence, it is important to identify a
molecule that can reveal the mechanism of breast cancer
progression.
The proteasome is a family of the multi-catalytic
proteinase complex (3), which is
crucial to the degradation of preternatural proteins, and involves
almost all cellular processes (4,5).
Increasing studies have found that the proteasome plays a key role
in regulating cell proliferation and migration signaling pathways
in various malignant tumors. In the future, proteasome inhibitors
are expected to be effective drug targets for the treatment of
breast cancer. PSMB4, a β4 subunit of the 20S proteasome, is a
member of the ubiquitin-proteasome family, and is involved in the
cytoplasmic protein catabolic process (12,13).
More and more studies elaborate the correlation between PSMB4
expression and tumor tissues, including ovarian cancer and
hepatocellular carcinoma. PSMB4 silencing was found to suppress the
viability of cell lines, including glioma and non-glioma cell lines
(MCF7 and A549). PSMB4 siRNA led to decreased protein levels of
other members of the proteasome subunit (PSMB1, PSMB2 and PSMB5)
(15). Furthermore, it has been
reported that the subunit of proteasomes stabilizes other subunits
during assembly, and decreases the protein level of other members
due to the destabilization of the β-ring assembly pathway, leading
to decreased viability (23).
Moreover, the cytotoxic effect of proteasome inhibitors on various
cell types has been reported, and the PARP decrease was obvious in
glioma cells transfected with PSMB4 siRNA (24–26).
Therefore, it was demonstrated that PSMB4 knockdown also caused a
decrease in PARP. During tumor development, proliferation is an
important process that involves many classic pathways, such as the
JAK-STAT, NF-κB, PI3K and mTOR signaling pathways (27–29).
As a transcriptional factor (30,31),
NF-κB can regulate cell proliferation and cell death. Generally
speaking, the inhibitor of NF-κB can be regulated by the
proteasome, and proteasome inhibition can induce depressed NF-κB
activation and increase cell viability (32–36).
Previous studies (13,14) have suggested that PSMB4 can promote
the proliferation of ovarian cancer and multiple myeloma through
the NF-κB signaling pathway. The present study found that PSMB4
knockdown inhibited the activation of the NF-κB signaling pathway
in breast cancer. Consistent with our hypothesis, it was also
verified that there is a PSMB4/NF-κB signaling pathway in breast
cancer. Indeed, these results suggest that PSMB4 can promote
proliferation and inhibit apoptosis through the NF-κB pathway,
thereby enhancing breast cancer patient survival.
In summary, it was found that PSMB4 is overexpressed
in breast cancer, and its interference can inhibit the
proliferation and viability of breast cancer cells. Therefore, it
is expected to be an effective target for the treatment of breast
cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant
from the Jiangsu Province Maternal and Child Health Research
Project (no. F201682).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HW and ZH conceived and designed the study. LX, WZ,
LX and XY performed the experiments. LX and WZ wrote the
manuscript. LX, YX and XR reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Cancer Hospital of Nantong University
(Nantong, China). All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Secginli S: Mammography self-efficacy
scale and breast cancer fear scale: Psychometric testing of the
Turkish versions. Cancer Nurs. 35:365–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peters JM, Franke WW and Kleinschmidt JA:
Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and
their distribution in the nucleus and the cytoplasm. J Biol Chem.
269:7709–7718. 1994.PubMed/NCBI
|
|
4
|
Tsakiri EN and Trougakos IP: The amazing
ubiquitin-proteasome system: Structural components and implication
in aging. Int Rev Cell Mol Biol. 314:171–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ling Q and Jarvis P: Functions of plastid
protein import and the ubiquitin-proteasome system in plastid
development. Biochim Biophys Acta. 1847:939–948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Csizmar CM, Kim DH and Sachs Z: The role
of the proteasome in AML. Blood Cancer J. 6:e5032016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee GY, Haverty PM, Li L, Kljavin NM,
Bourgon R, Lee J, Stern H, Modrusan Z, Seshagiri S, Zhang Z, et al:
Comparative oncogenomics identifies PSMB4 and SHMT2 as potential
cancer driver genes. Cancer Res. 74:3114–3126. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhong J, Shaik S, Wan L, Tron AE, Wang Z,
Sun L, Inuzuka H and Wei W: SCF β-TRCP targets MTSS1 for
ubiquitination-mediated destruction to regulate cancer cell
proliferation and migration. Oncotarget. 4:2339–2353. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim YH, Kim JH, Choi YW, Lim SK, Yim H,
Kang SY, Chung YS, Lee GY and Park TJ: Gankyrin is frequently
overexpressed in breast cancer and is associated with ErbB2
expression. Exp Mol Pathol. 94:360–365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manasanch EE and Orlowski RZ: Proteasome
inhibitors in cancer therapy. Nat Rev Clin Oncol. 14:417–433. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pellom ST Jr and Shanker A: Development of
proteasome inhibitors as therapeutic drugs. J Clin Cell Immunol.
S5:52012.PubMed/NCBI
|
|
12
|
Nandi D, Woodward E, Ginsburg DB and
Monaco JJ: Intermediates in the formation of mouse 20S proteasomes:
Implications for the assembly of precursor beta subunits. EMBO J.
16:5363–5375. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu R, Lu S, Deng Y, Yang S, He S, Cai J,
Qiang F, Chen C, Zhang W, Zhao S, et al: PSMB4 expression
associates with epithelial ovarian cancer growth and poor
prognosis. Arch Gynecol Obstet. 293:1297–1307. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Liu H, Cui M, Liu J, Yi R, Niu Y,
Chen T and Zhao Y: Effect of the HBV whole-X gene on the expression
of hepatocellular carcinoma associated proteins. J Microbiol
Immunol Infect. 49:335–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valdagni R, Rancati T, Ghilotti M,
Cozzarini C, Vavassori V, Fellin G, Fiorino C, Girelli G, Barra S,
Zaffaroni N, et al: To bleed or not to bleed. A prediction based on
individual gene profiling combined with dose-volume histogram
shapes in prostate cancer patients undergoing three-dimensional
conformal radiation therapy. Int J Radiat Oncol Biol Phys.
74:1431–1440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng-Hao D, Ji-Fang W, De-Sheng X and
Jian-Hua Z: Galectin-1 is up-regulated by RASSF1A gene in human
gastric carcinoma cell line SGC7901. APMIS. 120:582–590. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thaker NG, Zhang F, McDonald PR, Shun TY,
Lewen MD, Pollack IF and Lazo JS: Identification of survival genes
in human glioblastoma cells by small interfering RNA screening. Mol
Pharmacol. 76:1246–1255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng P, Guo H, Li G, Han S, Luo F and Liu
Y: PSMB4 promotes multiple myeloma cell growth by activating
NF-κB-miR-21 signaling. Biochem Biophys Res Commun. 458:328–333.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Runowicz CD, Leach CR, Henry NL, Henry KS,
Mackey HT, Cowens-Alvarado RL, Cannady RS, Pratt-Chapman ML, Edge
SB, Jacobs LA, et al: American Cancer Society/American Society of
Clinical Oncology Breast Cancer Survivorship Care Guideline. J Clin
Oncol. 34:611–635. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adamczyk A, Niemiec JA, Ambicka A,
Mucha-Małecka A, Mituś J and Ryś J: CD44/CD24 as potential
prognostic markers in node-positive invasive ductal breast cancer
patients treated with adjuvant chemotherapy. J Mol Histol.
45:35–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen XS, Wu JY, Huang O, Chen CM, Wu J, Lu
JS, Shao ZM, Shen ZZ and Shen KW: Molecular subtype can predict the
response and outcome of Chinese locally advanced breast cancer
patients treated with preoperative therapy. Oncol Rep.
23:1213–1220. 2010.PubMed/NCBI
|
|
22
|
Yao N, Song Z, Wang X, Yang S and Song H:
Prognostic Impact of Progesterone Receptor Status in Chinese
Estrogen Receptor Positive Invasive Breast Cancer Patients. J
Breast Cancer. 20:160–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hirano Y, Kaneko T, Okamoto K, Bai M,
Yashiroda H, Furuyama K, Kato K, Tanaka K and Murata S: Dissecting
beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J.
27:2204–2213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Poulaki V, Mitsiades CS, Kotoula V, Negri
J, McMillin D, Miller JW and Mitsiades N: The proteasome inhibitor
bortezomib induces apoptosis in human retinoblastoma cell lines in
vitro. Invest Ophthalmol Vis Sci. 48:4706–4719. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yin D, Zhou H, Kumagai T, Liu G, Ong JM,
Black KL and Koeffler HP: Proteasome inhibitor PS-341 causes cell
growth arrest and apoptosis in human glioblastoma multiforme (GBM).
Oncogene. 24:344–354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fribley A, Zeng Q and Wang CY: Proteasome
inhibitor PS-341 induces apoptosis through induction of endoplasmic
reticulum stress-reactive oxygen species in head and neck squamous
cell carcinoma cells. Mol Cell Biol. 24:9695–9704. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou B, Damrauer JS, Bailey ST, Hadzic T,
Jeong Y, Clark K, Fan C, Murphy L, Lee CY, Troester MA, et al:
Erythropoietin promotes breast tumorigenesis through
tumor-initiating cell self-renewal. J Clin Invest. 124:553–563.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Templin J, Atanackovic D, Hasche D,
Radhakrishnan SV and Luetkens T: Oscillating expression of
interleukin-16 in multiple myeloma is associated with
proliferation, clonogenic growth, and PI3K/NFKB/MAPK activation.
Oncotarget. 8:49253–49263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eroles P, Bosch A, Pérez-Fidalgo JA and
Lluch A: Molecular biology in breast cancer: Intrinsic subtypes and
signaling pathways. Cancer Treat Rev. 38:698–707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Abdel-Mageed AB and Agrawal KC: Activation
of nuclear factor kappaB: Potential role in
metallothionein-mediated mitogenic response. Cancer Res.
58:2335–2338. 1998.PubMed/NCBI
|
|
31
|
Biswas DK, Cruz AP, Gansberger E and
Pardee AB: Epidermal growth factor-induced nuclear factor kappa B
activation: A major pathway of cell-cycle progression in
estrogen-receptor negative breast cancer cells. Proc Natl Acad Sci
USA. 97:8542–8547. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Grundler K, Rotter R, Tilley S, Pircher J,
Czermak T, Yakac M, Gaitzsch E, Massberg S, Krötz F, Sohn HY, et
al: The proteasome regulates collagen-induced platelet aggregation
via nuclear-factor-kappa-B (NFĸB) activation. Thromb Res.
148:15–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Di Filippo C, Petronella P, Freda F,
Scorzelli M, Ferretti M, Canonico S, Rossi F and D'Amico M:
Involvement of the ubiquitin-proteasome system in the formation of
experimental postsurgical peritoneal adhesions. Mediators Inflamm.
2012:1947232012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pajonk F and McBride WH: The proteasome in
cancer biology and treatment. Radiat Res. 156:447–459. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ehrlich ES, Chmura JC, Smith JC, Kalu NN
and Hayward GS: KSHV RTA abolishes NFκB responsive gene expression
during lytic reactivation by targeting vFLIP for degradation via
the proteasome. PLoS One. 9:e913592014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kiliccioglu I, Konac E, Varol N, Gurocak S
and Bilen Yucel C: Apoptotic effects of proteasome and histone
deacetylase inhibitors in prostate cancer cell lines. Genet Mol
Res. 13:3721–3731. 2014. View Article : Google Scholar : PubMed/NCBI
|