Introduction
The inhibitor of DNA binding/differentiation (ID)
proteins, as a group of helix-loop-helix transcription factors, are
individually fundamental to development and cell cycle control
(1). Recently, loss- and
gain-of-function studies have indicated the oncogenic effects of ID
proteins in tumors, and have revealed associations between these
proteins and malignant features, including transformative cellular
phenotype, abnormal senility, facilitating proliferation and
distant spreading (2–4).
Different human malignancies have been detected for
ID expression levels (5). For
instance, it has been reported and agreed that increased levels of
IDs were associated with malignant grade and a poor prognosis in
various types of carcinoma (6–9). It
has been revealed that increasing ID1 mRNA expression was observed
in subtypes of human breast tumors and that this acted as a driver
in BC metastasis (10–12). The extracellular micro-environmental
signals can promote a role for ID2 in tumorigenesis (12–14).
Overexpression of ID2 is associated with adverse outcomes in
pancreatic carcinoma, neuroblastoma and pulmonary carcinoma
(15–17). In addition, increased ID2 protein
levels indicated an unfavorable prognosis in patients with
basal-like subtypes of BC (18).
Cellular localization of ID2 has been recognized as an important
factor in determining disease outcome. In BC, cytoplasmic, but not
nuclear, localization of ID2 protein was associated with less
invasive and expansionary phenotypes (19,20).
Kowanetz et al demonstrated that abnormal ID2 or ID3
expression led to proliferation inhibition and
epithelial-mesenchymal transition of BC cells, which was induced by
transforming growth factor β. Furthermore, knockdown of endogenic
ID2 or ID3 enhanced the sensitivity of breast epithelial cells to
bone morphogenetic proteins (BMPs), which resulted in growth
inhibition and poorer differentiation (21). Wen et al (22) reported the amplified ID4 protein in
triple-negative breast carcinoma, while a study undertaken by Thike
et al (23) demonstrated
that ID4 may be downregulated by BRCA1.
Nonetheless, the specific roles of individual ID
family members in BC and the association between IDs and the
clinicopathological features of BC have not yet been elucidated.
Therefore, in-depth investigation and analysis into the potential
roles of ID proteins in BC is required, and may provide an insight
into the molecular mechanisms underpinning the disease.
The present study aimed to systemically investigate
the prognostic values and potential roles of individual IDs in BC
based on a series of large databases. Characterization of the ID
expression status of patients with BC may be valuable, not only for
diagnostic and prognostic assessment, but also for guiding BC
management in the future.
Materials and methods
Oncomine analysis
In order to analyze the expression levels of
specific IDs in a variety of malignancies, an access online
database Oncomine (www.oncomine.org), which is an online cancer
microarray database to facilitate and promote discovery from
genome-wide expression analyses, was used. Paired Student's t-test
was used, and a fold-change of 2 with a P-value of <0.01 was
defined as clinically significant, as previously described
(24).
Breast cancer gene-expression miner
v4.0 analysis
In order to analyze the association among expression
levels of specific IDs and specific clinicopathological features of
BC, including hormonal receptors and lymph nodal status, the open
access database Breast Cancer Gene-Expression Miner v4.0
(bcGenExMiner v4.0), which is comprised of 36 annotated genomic
datasets and three statistical mining functions, was used
(http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php)
(25,26).
GOBO analysis
The mRNA expression levels of IDs were analyzed by
uploading corresponding affymetrix probes to the GOBO
database (http://co.bmc.lu.se/gobo/gsa.pl). GOBO is an
online accessible tool that allows rapid evaluation of gene
expression levels, identification of co-expressed genes and
connection with prognosis for single gene, gene sets or gene
signatures in a BC dataset.
The human protein atlas
The Human Protein Atlas (HPA) is an open access
program that maps all the human proteins in cells, as well as
tissue samples (https://www.proteinatlas.org/) (27). The Pathology Atlas of HPA
demonstrates the association between specific protein levels and
survival of patients with BC (28).
The Kaplan-Meier plotter survival
analysis
To analyze the prognostic values of specific IDs in
BC samples, the Kaplan-Meier plotter (www.kmplot.com) was used to display the relapse-free
survival (RFS). The log-rank P-value was presented on the webpage
(29).
Results
mRNA expression patterns of ID family
members in human BC
Thus far, four ID family members have been
identified in human cancer. The differences in the mRNA expression
of the 4 IDs between tumor and normal tissues in multiple types of
cancer were analyzed using the online Oncomine database. As
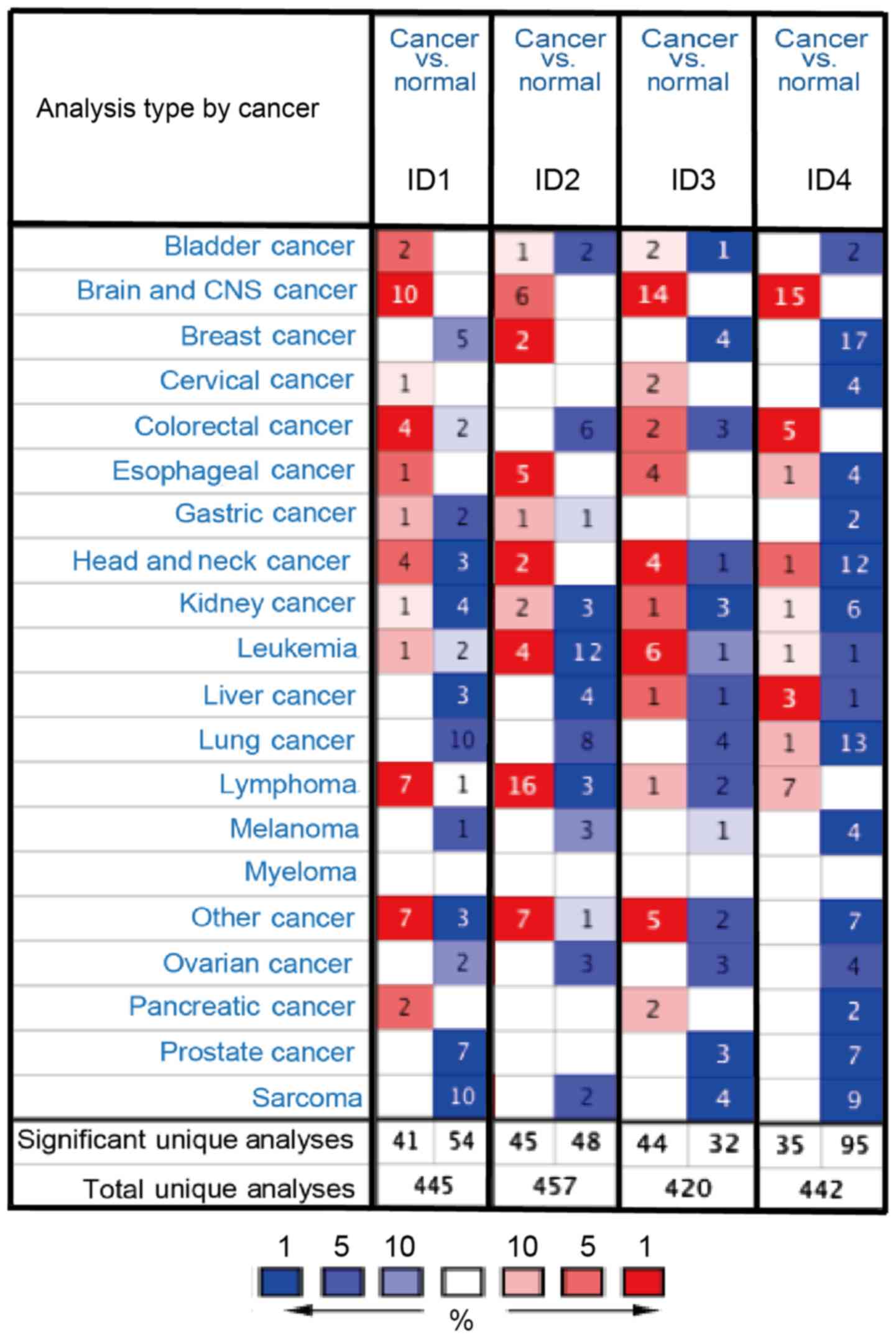
demonstrated in Fig. 1, the
Oncomine database contained a total of 445, 457, 420 and 442 unique
analyses for ID1, ID2, ID3 and ID4, respectively. In BC analysis,
there were two studies revealing a statistically significant
increase in the mRNA expression level of ID2 in BC tissues,
compared with normal tissues. As for ID1, ID3 and ID4, five, four
and 17 unique analyses datasets with statistical significance
revealed higher expression levels in normal tissues than in cancer
tissues. These data indicated that the expression of ID1, ID3 and
ID4 was markedly lower in BC samples than in normal breast tissues
(Fig. 1).
We also analyzed gene alterations using CbioPORTAL.
Based on the analysis of CbioPORTAL, the genetic alteration rate of
ID1-ID4 was 2.2, 2.1, 0.5, and 2.8%, respectively. However, there
was no significant association between gene alteration and without
alteration (data not shown).
Increased ID1 and ID4 expression is
associated with the subgroup with no lymph node metastasis and with
lower tumor grades
To further identify the mRNA expression of ID family
members in different clinicopathological groups of patients,
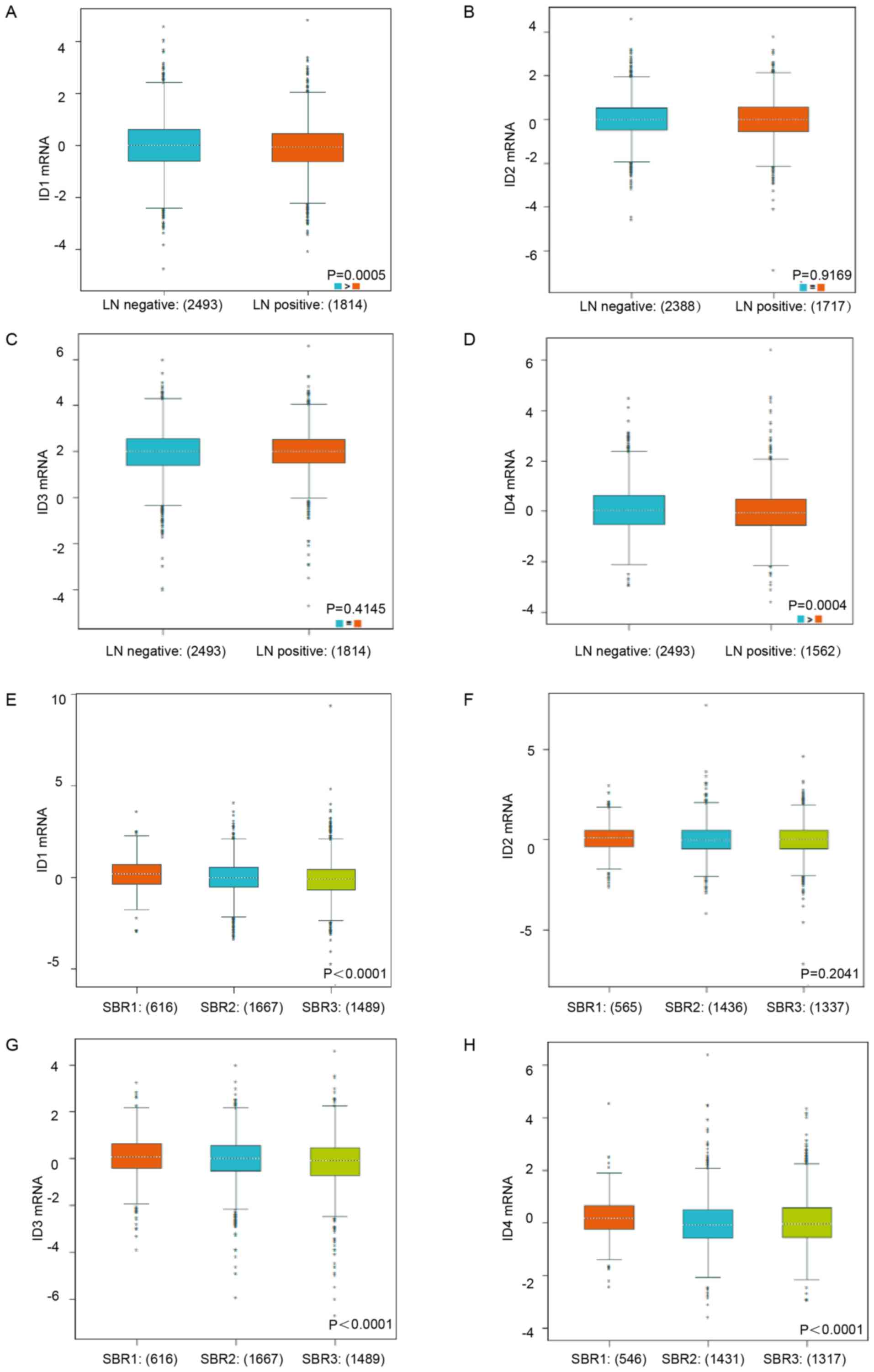
bc-GenExMiner database analysis was used. Increased ID1 mRNA and
ID4 mRNA expression was observed in the negative lymph node
metastasis BC patients (Fig. 2A,
P=0.0005; Fig. 2D, P=0.0004).
Nevertheless, no statistically significant difference in ID2 and
ID3 mRNA expression was observed between patients with LN (+) and
LN (−) BC (Fig. 2B, P=0.9169;
Fig. 2C, P=0.4145). Lower
Scarff-Bloom-Richardson (SBR) status grade (30), was associated with a higher mRNA
expression level of ID1, ID3 and ID4 (Fig. 2E, P<0.0001; Fig. 2G, P<0.0001; Fig. 2H, P<0.0001), but was not
associated with the mRNA expression level of ID2 (Fig. 2F, P=0.2041). All the subgroup
comparisons are presented in Tables
I–III.
 | Table I.Lower SBR grade status is associated
with the enriched mRNA level of ID1. |
Table I.
Lower SBR grade status is associated
with the enriched mRNA level of ID1.
| Group
comparison |
P-valuea |
|---|
| SBR2<SBR1 | P<0.0001 |
| SBR3<SBR1 | P<0.0001 |
| SBR3=SBR2 | P>0.1000 |
 | Table III.Lower SBR grade is associated with
the enriched mRNA level of ID4. |
Table III.
Lower SBR grade is associated with
the enriched mRNA level of ID4.
| Groups
comparison |
P-valuea |
|---|
| SBR2<SBR1 | P<0.0001 |
| SBR3<SBR1 | P<0.0100 |
| SBR3>SBR2 | P<0.1000 |
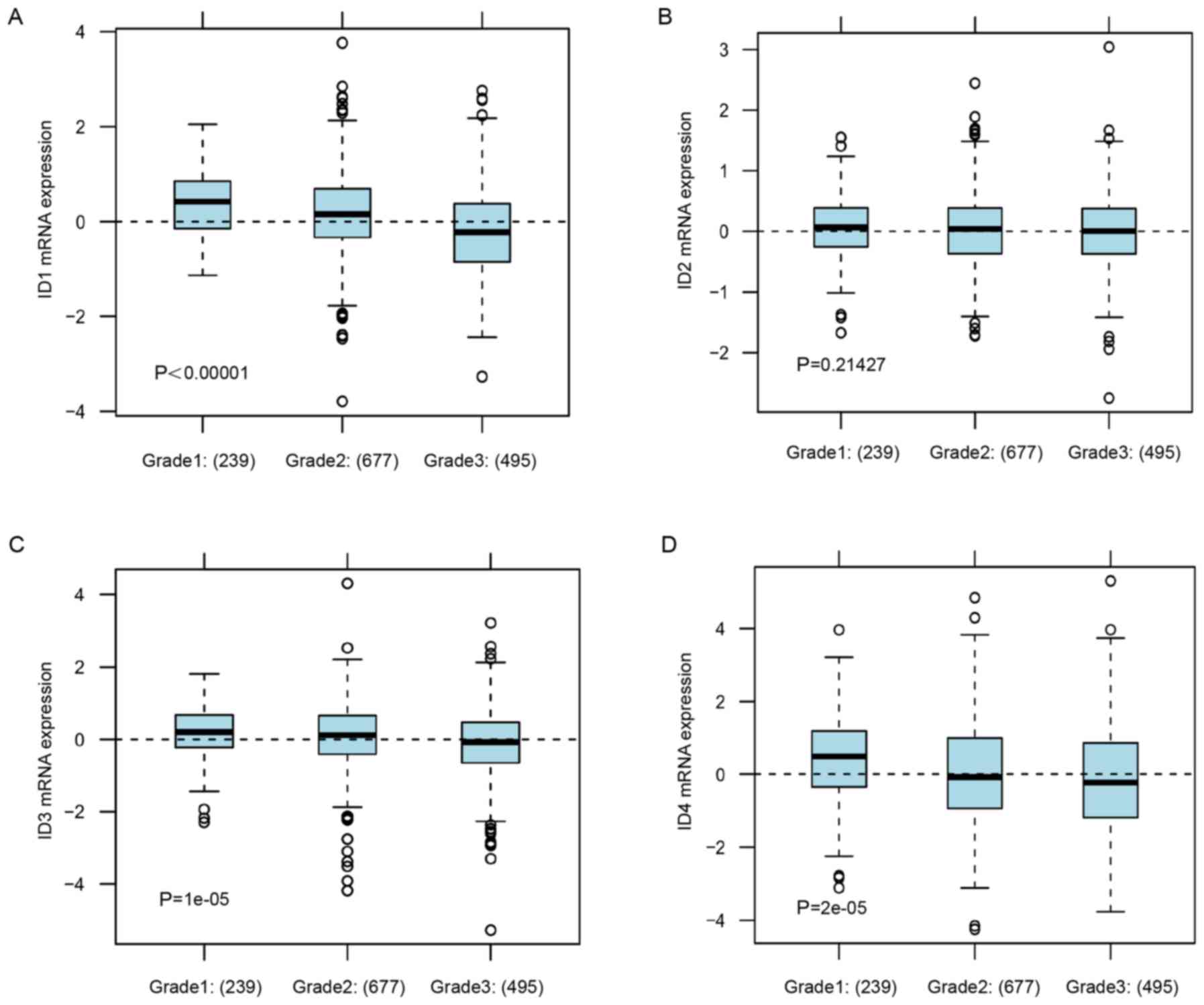
Consistently, in GOBO analysis, the lower
tumor grade subgroup of BC patients was associated with increased
expression levels ID1, ID3 ID4 (Fig.
3A, P<0.00001; Fig. 3C,
P=0.00001; Fig. 3D, P=0.00002), but
was not associated with the mRNA expression level of ID2 (Fig. 3B, P=0.21427). These data indicated
that increased expression of ID1 and ID4 was associated with the
subgroup of BC patients with a low malignant grade and low
metastasis potential.
Increased expression of ID1 and ID4
predicts an improved survival in patients with BC
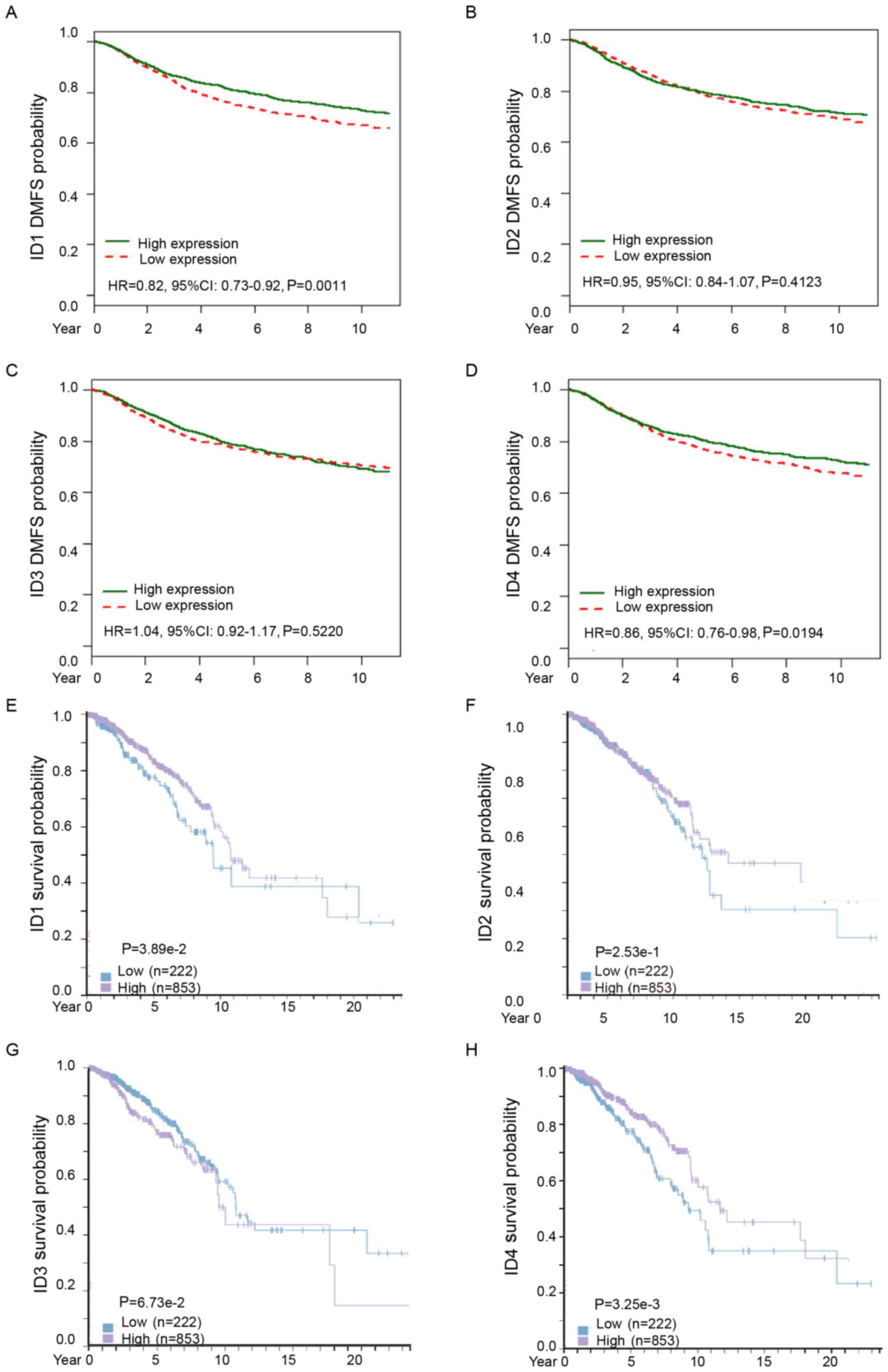
Subsequently, the distant metastasis prognostic
effect of ID family members in BC was analyzed. Analysis from
bc-GenExMiner demonstrated that increased ID1 or ID4 mRNA was
associated with longer distant metastasis-free survival (DMFS)
times in all patients with BC (Fig.
4A, HR=0.82, 95% CI: 0.73–0.92, P=0.0011; Fig. 4D, HR=0.86, 95% CI: 0.76–0.98,
P=0.0194). However, ID2 and ID3 mRNA expression levels were not
significantly associated with DMFS among all BC patients (Fig. 4B, HR=0.95, 95% CI: 0.84–1.07,
P=0.4123; Fig. 4C, HR=1.04, 95% CI:
0.92–1.17, P=0.5220). These results confirmed that higher
expression of ID1 and ID4 predicted a lower potential of metastasis
and improved DMFS in patients with BC.
Consistently, analysis from The Human Protein Atlas
revealed that increased ID1 or ID4 protein expression was
associated with improved survival in patients with BC (Fig. 4E, P=0.0389; Fig. 4H, P=0.00325). However, there was no
significant difference in the protein expression levels of ID2 or
ID3 in patients with BC (Fig. 4F,
P=0.253; Fig. 4G, P=0.0673).
Among the ID family, the expression of ID1 and ID3
have a significant positive correlation in BC (data not shown).
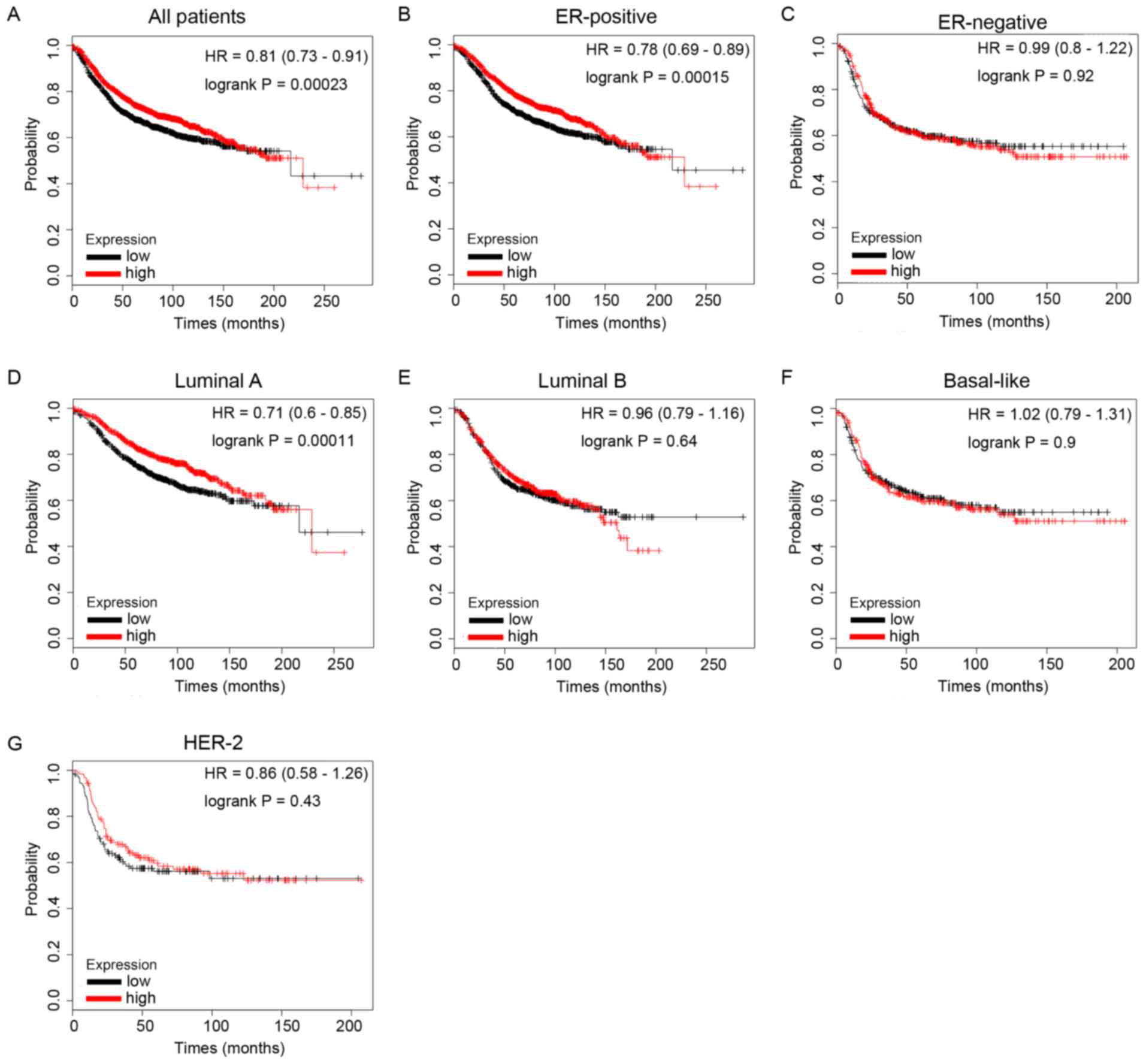
Increased ID1 mRNA expression is
significantly associated with longer RFS times in patients with BC,
particularly in the ER-positive subtypes
The present study then evaluated the prognostic
values of ID family members for BC using the KM Plotter database.
RFS was analyzed for each gene. It was demonstrated that high ID1
mRNA expression predicted longer RFS times in patients with BC
(Fig. 5A, HR=0.81, P=0.00023). In
particular, sub-analysis revealed that elevated ID1 mRNA expression
was associated with longer RFS times in the ER-positive BC subgroup
of patients (Fig. 5B, HR=0.78,
P=0.00015), but not in the ER-negative BC subgroup of patients
(Fig. 5C, HR=0.99, P=0.92).
In addition, increased ID1 mRNA expression was
significantly associated with longer RFS times in BC patients with
special molecular subtype Luminal A tumors (Fig. 5D, HR=0.71, P=0.00011), but not in
other molecular subtypes, including Luminal B (Fig. 5E, HR=0.96, P=0.64), HER-2 type
(Fig. 5G, HR=0.86, P=0.43) and
Basal-like (Fig. 5F, HR=1.02,
P=0.9). These data demonstrated that ID1-overexpression was
associated with longer survival times in patients with BC,
particularly in the ER-positive and Luminal A subtype
subgroups.
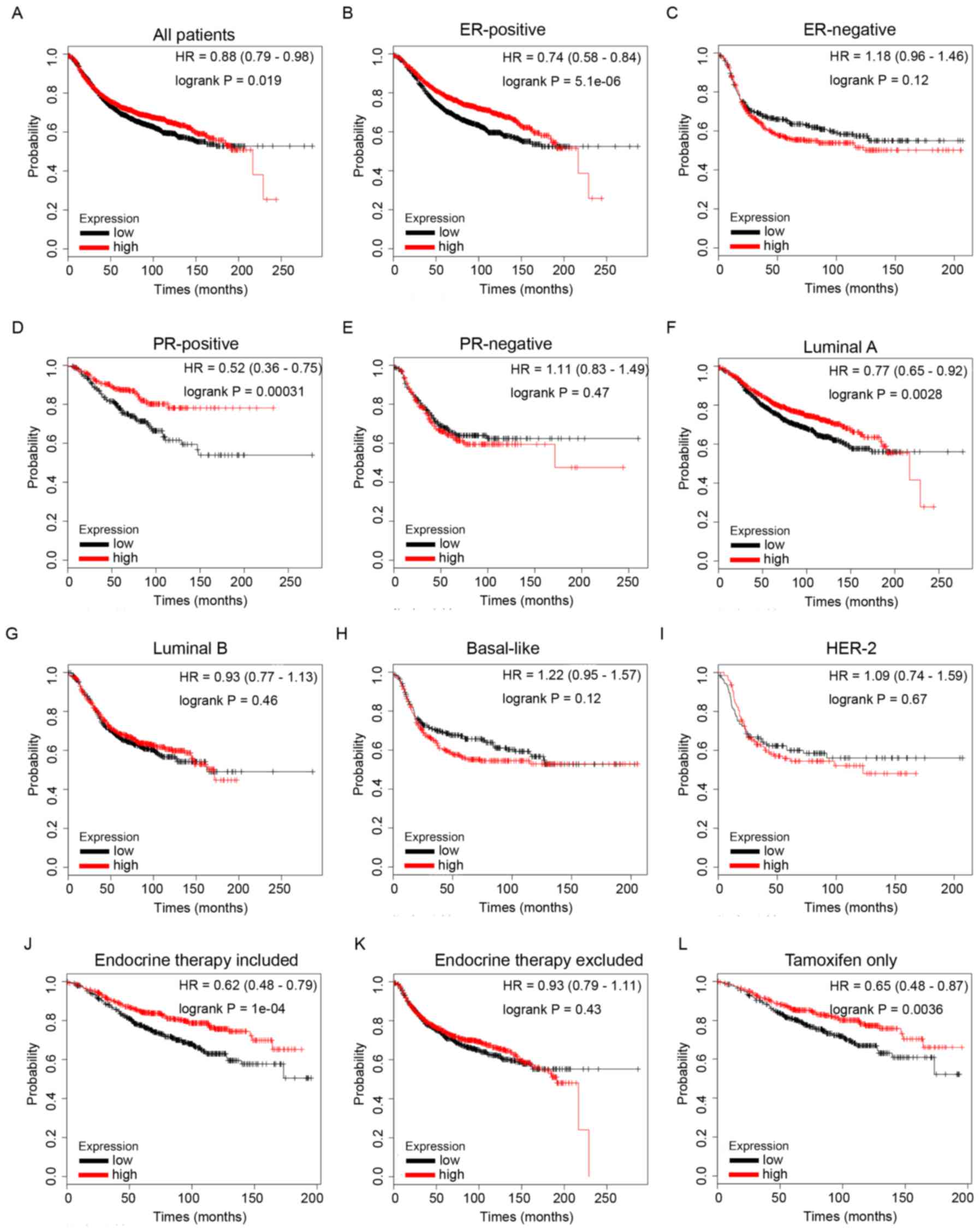
Increased ID4 predicts improved
survival in patients with BC, particularly in the hormone
receptor-positive patients and the subgroup of patients treated
with endocrine therapy
RFS analysis of ID4 from the KM Plotter database
revealed that high expression of ID4 mRNA was associated with
improved RFS in patients with BC (Fig.
6A, HR=0.88, P=1.9e-2). Furthermore, high mRNA expression of
ID4 predicted longer survival times in patients with ER-positive
(Fig. 6B, HR=0.74, P=5.1e-6) and
PR-positive (Fig. 6D, HR=0.52,
P=3.1e-4) disease. However, no statistical significance was
observed in the groups of ER-negative (Fig. 6C, HR=1.18, P=0.12) or PR-negative
(Fig. 6E, HR=1.11, P=0.47)
patients. These data revealed that increased expression of ID4
predicted longer survival times in patients with BC, particularly
in the hormone receptor-positive subgroup of patients.
Furthermore, subgroup analysis revealed that
increased ID4 mRNA expression was significantly associated with
improved RFS in BC patients with Luminal A molecular subtype tumors
(Fig. 6F, HR=0.77, P=2.8e-3), but
not in other molecular subtypes, including Luminal B (Fig. 6G, HR=0.93, P=0.46), Basal-like
(Fig. 6H, HR=1.22, P=0.12) and
HER-2 type (Fig. 6I, HR=1.09,
P=0.67).
Additionally, higher mRNA expression of ID4 was
significantly associated with longer RFS times in patients with BC
who had received endocrine therapy (Fig. 6J, HR=0.62, P=1e-4), but not in the
group that excludes endocrine therapy (Fig. 6K, HR=0.93, P=0.43), indicating a
potential role of ID4 in endocrine therapy sensitivity in BC.
Notably, the results of the present study also revealed that high
mRNA expression of ID4 was associated with longer RFS times in
patients with BC who had received tamoxifen for endocrine therapy
only (Fig. 6L, HR=0.65, P=3.6e-3),
indicating that ID4 may contribute toward tamoxifen therapy
sensitivity in BC.
Discussion
Over recent decades, key molecular signatures
associated with BC, including ER, PR and HER-2, have been
identified and well-characterized (31). Emerging evidence has continued to
identify novel targets and signaling pathways, including PARP,
CDK4/6 and PI3K/Akt/mTOR, which significantly contribute toward the
pathogenesis and development of BC, leading to a paradigm shift in
the treatment of BC (32).
ID family members are pivotal modulatory proteins
that have been recognized to be downstream targets of a number of
oncogenic pathways, making them attractive targets for the
treatment of cancer (4,5,33).
However, there are significant contradictions in the specific roles
of different IDs in BC development (34).
In the present study, analysis of Oncomine, bc-Gen
ExMiner, The Human Protein Atlas and Kaplan-Meier plotter, was used
to systemically depict the expression profiles of individual IDs in
BC, revealing that the IDs exhibited marked differences in mRNA
expression between breast tumor and normal tissues. The results
indicated that the mRNA expression level of ID2 was significantly
higher in BC than in normal tissues, while the mRNA expression
levels of ID1, ID3 and ID4 were significantly lower in BC tissues
than in normal tissues. Wazir et al (35) reported that increased expression of
ID1 in BC was correlated with disease severity and predicted a poor
survival outcome. Nonetheless, the present study revealed
contradictory results in the analysis of ID1 expression in human
BC, which may be primarily attributed toward discrepancies in
detection methods for mRNA and protein among different
investigators.
IDs are capable of promoting the tumorigenesis of BC
by suppressing cell differentiation, activating proliferation and
promoting tumor development (12).
However, individual IDs may serve diverse roles in this process.
Increased ID1 and ID4 expression was associated with the subgroup
without lymph node metastasis and a lower tumor grade. The results
indicated that ID1 and ID4 may serve suppressive roles in the
development of BC.
It has been reported that IDs are important driving
forces involving distant metastasis of a variety of cancer types,
including BC (36,37). Gumireddy et al (38) demonstrated that ID1 facilitated the
spreading of BC via regulation of S100A9 expression. However,
following assessment of the distant metastasis prognostic effect of
ID family members in BC through analysis of the bc-GenExMiner
database, it was revealed that increased ID1 or ID4 mRNA expression
was associated with longer DMFS times in all patients with BC.
The results demonstrated that increased ID1 mRNA
expression was significantly associated with longer RFS times in
patients with BC, particularly in those with ER-positive and
Luminal A subtype tumors. Although the majority of studies have
demonstrated that high ID1 expression was generally an adverse
prognostic indicator (35), the
results have not been entirely consistent. Cheng et al
(39) reported a paradoxical result
that increased ID1 expression predicts a favorable survival outcome
in patients with lung cancer that has been surgically treated,
followed by adjuvant paclitaxel plus cisplatin chemotherapy, and
this distinct role of ID1 possibly stems from the ID1-dependence of
NSCLC cells for survival, thereby rendering the cancer cells to be
more sensitive to a specific chemotherapy regimen. The present
study also demonstrated that increased ID4 expression was a
favorable prognostic factor in patients with BC. Furthermore, it
was revealed that increased ID4 expression predicted improved
survival in the subset of patients with hormone receptor-positive
tumors, as well as the subgroup of patients treated with endocrine
therapy. These results were consistent with those of a study
undertaken by Zhang et al (40), which reported that hypomethylation
of ID4 may act as a critical biomarker for identifying acquired
tamoxifen resistance in BC.
Another study undertaken by Patel et al
(41) suggested that ID4 may serve
a suppressive role in prostate cancer, and that its loss
facilitates CRPC through constitutive activation of the androgen
receptor. The possible mechanism of this is that ID4 acts as a
suppressor of ID-1, −2 and −3, and stimulates transcription
(42). Therefore, we hypothesized
that ID4 may serve an essential role in the sensitivity of
endocrine therapy.
In summary, the IDs exhibited diverse expression
profiles between BC tissues and normal breast tissues. Notably, the
results indicated that increased ID1 and ID4 expression was
associated with low pathological grades of BC and predicted an
improved survival outcome in patients with distinct subtypes of BC.
Future in-depth studies are warranted to elucidate the exact
functions of ID1 and ID4 in oncogenesis and the progression of BC,
which may provide further evidence that ID1 and ID4 could be
critical prognostic indicators and promising therapeutic targets in
BC. However, the in silico analyses are limited; for
example, the data is limited to one dataset (Affymetrix
arrays-based) and RNA-Seq data (TCGA) or other array platforms
(Illumina in the METABRIC) are not taken into consideration.
Nonetheless, the results of the present study indicated that IDs
were potential prognostic indicators in BC and further study on the
values of IDs in the pathogenesis and development of BC is
warranted.
Acknowledgements
We are grateful to Dr Pi Guo for critically reading
the manuscript. We also thank Dr Yin-Sheng Xiao for advice or
technical assistance in the TCGA data analysis.
Funding
The present study was partly supported by The
Natural Science Foundation of Guangdong Province, China (no.
2015A030313429), The Youth Innovative Talent Project of Colleges
and Universities in Guangdong Province, China (nos. 2016KQNCX051
and 2017KQNCX073).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HYL designed the study. YHY, SMS and WQL prepared
the figures and tables. XFL and CFC analyzed and interpreted the
data. XLZ and DZ drafted the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yokota Y and Mori S: Role of Id family
proteins in growth control. J Cell Physiol. 190:21–28. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Patel D, Morton DJ, Carey J, Havrda MC and
Chaudhary J: Inhibitor of differentiation 4 (ID4): From development
to cancer. Biochim Biophys Acta. 1855:92–103. 2015.PubMed/NCBI
|
|
3
|
Kamalian L, Gosney JR, Forootan SS, Foster
CS, Bao ZZ, Beesley C and Ke Y: Increased expression of Id family
proteins in small cell lung cancer and its prognostic significance.
Clin Cancer Res. 14:2318–2325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cruz-Rodriguez N, Combita AL, Enciso LJ,
Quijano SM, Pinzon PL, Lozano OC, Castillo JS, Li L, Bareño J,
Cardozo C, et al: High expression of ID family and IGJ genes
signature as predictor of low induction treatment response and
worst survival in adult Hispanic patients with B-acute
lymphoblastic leukemia. J Exp Clin Cancer Res. 35:642016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perk J, Iavarone A and Benezra R: Id
family of helix-loop-helix proteins in cancer. Nat Rev Cancer.
5:603–614. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
O'Brien CA, Kreso A, Ryan P, Hermans KG,
Gibson L, Wang Y, Tsatsanis A, Gallinger S and Dick JE: ID1 and ID3
regulate the self-renewal capacity of human colon cancer-initiating
cells through p21. Cancer Cell. 21:777–792. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren Y, Cheung HW, von Maltzhan G, Agrawal
A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, et al:
Targeted tumor-penetrating siRNA nanocomplexes for credentialing
the ovarian cancer oncogene ID4. Sci Transl Med. 4:147ra1122012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao TF, Jia HZ, Zhang ZZ, Zhao XS, Zou
YF, Zhang W, Wan J and Chen XF: LncRNA H19 regulates ID2 expression
through competitive binding to hsa-miR-19a/b in acute myelocytic
leukemia. Mol Med Rep. 16:3687–3693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Korang-Yeboah M, Patel D, Morton D, Sharma
P, Gorantla Y, Joshi J, Nagappan P, Pallaniappan R and Chaudhary J:
Intra-tumoral delivery of functional ID4 protein via
PCL/maltodextrin nano-particle inhibits prostate cancer growth.
Oncotarget. 7:68072–68085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massagué J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta GP, Perk J, Acharyya S, de Candia P,
Mittal V, Todorova-Manova K, Gerald WL, Brogi E, Benezra R and
Massagué J: ID genes mediate tumor reinitiation during breast
cancer lung metastasis. Proc Natl Acad Sci USA. 104:19506–19511.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lasorella A, Benezra R and Iavarone A: The
ID proteins: Master regulators of cancer stem cells and tumour
aggressiveness. Nat Rev Cancer. 14:77–91. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sikder HA, Devlin MK, Dunlap S, Ryu B and
Alani RM: Id proteins in cell growth and tumorigenesis. Cancer
Cell. 3:525–530. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun XH, Copeland NG, Jenkins NA and
Baltimore D: Id proteins Id1 and Id2 selectively inhibit DNA
binding by one class of helix-loop-helix proteins. Mol Cell Biol.
11:5603–5611. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rollin J, Bléchet C, Régina S, Tenenhaus
A, Guyétant S and Gidrol X: The intracellular localization of ID2
expression has a predictive value in non small cell lung cancer.
PLoS One. 4:e41582009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen XS, Zhang YH, Cai QY and Yao ZX: ID2:
A negative transcription factor regulating oligodendroglia
differentiation. J Neurosci Res. 90:925–932. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kleeff J, Ishiwata T, Friess H, Buchler
MW, Israel MA and Korc M: The helix-loop-helix protein Id2 is
overexpressed in human pancreatic cancer. Cancer Res. 58:3769–3772.
1998.PubMed/NCBI
|
|
18
|
Li K, Yao L, Chen L, Cao ZG, Yu SJ, Kuang
XY, Hu X and Shao ZM: ID2 predicts poor prognosis in breast cancer,
especially in triple-negative breast cancer, and inhibits
E-cadherin expression. Onco Targets Ther. 7:1083–1094.
2014.PubMed/NCBI
|
|
19
|
Itahana Y, Singh J, Sumida T, Coppe JP,
Parrinello S, Bennington JL and Desprez PY: Role of Id-2 in the
maintenance of a differentiated and noninvasive phenotype in breast
cancer cells. Cancer Res. 63:7098–7105. 2003.PubMed/NCBI
|
|
20
|
Stighall M, Manetopoulos C, Axelson H and
Landberg G: High ID2 protein expression correlates with a
favourable prognosis in patients with primary breast cancer and
reduces cellular invasiveness of breast cancer cells. Int J Cancer.
115:403–411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kowanetz M, Valcourt U, Bergström R,
Heldin CH and Moustakas A: Id2 and Id3 define the potency of cell
proliferation and differentiation responses to transforming growth
factor beta and bone morphogenetic protein. Mol Cell Biol.
24:4241–4254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wen YH, Ho A, Patil S, Akram M, Catalano
J, Eaton A, Norton L, Benezra R and Brogi E: Id4 protein is highly
expressed in triple-negative breast carcinomas: Possible
implications for BRCA1 downregulation. Breast Cancer Res Treat.
135:93–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thike AA, Tan PH, Ikeda M and Iqbal J:
Increased ID4 expression, accompanied by mutant p53 accumulation
and loss of BRCA1/2 proteins in triple-negative breast cancer,
adversely affects survival. Histopathology. 68:702–712. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin HY, Zeng Liang YK, Wei XL and Chen CF:
GATA3 and TRPS1 are distinct biomarkers and prognostic factors in
breast cancer: Database mining for GATA family members in
malignancies. Oncotarget. 8:34750–34761. 2017.PubMed/NCBI
|
|
25
|
Jezequel P, Frenel JS, Campion L,
Guérin-Charbonnel C, Gouraud W, Ricolleau G and Campone M:
bc-GenExMiner 3.0: New mining module computes breast cancer gene
expression correlation analyses. Database. 2013:bas0602013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jezequel P, Campone M, Gouraud W,
Guérin-Charbonnel C, Leux C, Ricolleau G and Campion L:
bc-GenExMiner: An easy-to-use online platform for gene prognostic
analyses in breast cancer. Breast Cancer Res Treat. 131:765–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thul PJ, Akesson L, Wiking M, Mahdessian
D, Geladaki A, Blal Ait H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science. 356:pii:
eaal3321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlen M, Fagerberg L, Hallstrom BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gyorffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Curtit E, Pivot X, Henriques J,
Paget-Bailly S, Fumoleau P, Rios M, Bonnefoi H, Bachelot T, Soulié
P, Jouannaud C, et al: Assessment of the prognostic role of a
94-single nucleotide polymorphisms risk score in early breast
cancer in the SIGNAL/PHARE prospective cohort: No correlation with
clinico-pathological characteristics and outcomes. Breast Cancer
Res. 19:982017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fragomeni SM, Sciallis A and Jeruss JS:
Molecular subtypes and local-regional control of breast cancer.
Surg Oncol Clin N Am. 27:95–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pawlik TM: Innovation in the diagnosis and
management of breast cancer. Surg Oncol Clin N Am. 27:xiii–xiv.
2018. View Article : Google Scholar
|
|
33
|
Murad JM, Place CS, Ran C, Hekmatyar SK,
Watson NP, Kauppinen RA and Israel MA: Inhibitor of DNA binding 4
(ID4) regulation of adipocyte differentiation and adipose tissue
formation in mice. J Biol Chem. 285:24164–24173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang HY, Liu HL, Ke J, Wu H, Zhu H, Liu
JR, Liu LX and Jiang HC: Expression and prognostic value of Id
protein family in human breast carcinoma. Oncol Rep. 23:321–328.
2010.PubMed/NCBI
|
|
35
|
Wazir U, Jiang WG, Sharma AK, Newbold RF
and Mokbel K: The mRNA expression of inhibitors of DNA binding-1
and −2 is associated with advanced tumour stage and adverse
clinical outcome in human breast cancer. Anticancer Res.
33:2179–2183. 2013.PubMed/NCBI
|
|
36
|
Umetani N, Mori T, Koyanagi K, Shinozaki
M, Kim J, Giuliano AE and Hoon DS: Aberrant hypermethylation of ID4
gene promoter region increases risk of lymph node metastasis in T1
breast cancer. Oncogene. 24:4721–4727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li B, Tsao SW, Li YY, Wang X, Ling MT,
Wong YC, He QY and Cheung AL: Id-1 promotes tumorigenicity and
metastasis of human esophageal cancer cells through activation of
PI3K/AKT signaling pathway. Int J Cancer. 125:2576–2585. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gumireddy K, Li A, Kossenkov AV, Cai KQ,
Liu Q, Yan J, Xu H, Showe L, Zhang L and Huang Q: ID1 promotes
breast cancer metastasis by S100A9 regulation. Mol Cancer Res.
12:1334–1343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cheng YJ, Lee YC, Chiu WC, Tsai JW, Su YH,
Hung AC, Chang PC, Huang CJ, Chai CY and Yuan SS: High Id1
expression, a generally negative prognostic factor, paradoxically
predicts a favorable prognosis for adjuvant paclitaxel plus
cisplatin therapy in surgically treated lung cancer patients.
Oncotarget. 5:11564–11575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Zhang B, Fang J and Cao X:
Hypomethylation of DNA-binding inhibitor 4 serves as a potential
biomarker in distinguishing acquired tamoxifen-refractory breast
cancer. Int J Clin Exp Pathol. 8:9500–9505. 2015.PubMed/NCBI
|
|
41
|
Patel D, Knowell AE, Korang-Yeboah M,
Sharma P, Joshi J, Glymph S, Chinaranagari S, Nagappan P,
Palaniappan R, Bowen NJ and Chaudhary J: Inhibitor of
differentiation 4 (ID4) inactivation promotes de novo
steroidogenesis and castration-resistant prostate cancer. Mol
Endocrinol. 28:1239–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sharma P, Chinaranagari S and Chaudhary J:
Inhibitor of differentiation 4 (ID4) acts as an inhibitor of ID-1,
−2 and −3 and promotes basic helix loop helix (bHLH) E47 DNA
binding and transcriptional activity. Biochimie. 112:139–150. 2015.
View Article : Google Scholar : PubMed/NCBI
|