Introduction
Lung cancer is a debilitating neoplasm, and accounts
for significant morbidity and mortality worldwide (1). Despite significant progress seen in
the last decade in regards to treatment regimens including surgery,
radiotherapy and chemotherapy, and ongoing research development,
the survival rate of patients with lung cancer is still less than
satisfactory. A substantial number of patients succumb to the
disease within the weeks following diagnosis, and the 5-year
survival rate does not exceed 15% (2–5). The
poor prognosis of lung cancer mainly results from its high degree
of malignancy (malignant proliferation, invasion and migration).
Most patients with lung cancer do not die of primary cancer, but
rather die of metastatic cancer (6,7).
Therefore, the key molecules that mediate lung cancer metastasis
have become a focus of scientific research.
The histone deacetylases (HDACs) form a family of
enzymes, which have fundamental roles in the epigenetic regulation
of gene expression and contribute to proliferation,
differentiation, apoptosis and cell cycle progression (8–10).
HDACs are frequently dysregulated in human malignancies and have
therefore become therapeutic targets in cancer therapy (11). As a member of the class IIa family
of HDACs, histone deacetylase 5 (HDAC5) is known to undergo
nuclear-cytoplasmic shuttling and to be a prominent regulator of
cellular and epigenetic processes that underlie the progression of
human disease, including cardiac diseases and tumorigenesis
(12–14). A growing body of literature suggests
that HDAC5 is extensively expressed in many cancers. Li et
al demonstrated that HDAC5 was extensively expressed in human
breast cancer tissues, and high HDAC5 expression was associated
with poor patient prognosis. Downregulation of HDAC5 was found to
suppress breast cancer cell proliferation, invasion and migration,
and promote breast cancer cell apoptosis (15). He et al showed that HDAC5 was
upregulated in human colorectal cancer. Overexpression of HDAC5
significantly improved the proliferation of colorectal cancer
cells. On the contrary, HDAC5 knockdown was found to suppress
colorectal tumor cell growth (8).
Feng et al showed that HDAC5 was increased in human
hepatocellular carcinoma. Overexpression of HDAC5 promoted liver
cancer cell proliferation, and inhibition of HDAC5 significantly
inhibited liver cancer cell proliferation (16). Chen et al found that HDAC5
was upregulated in osteosarcoma, and overexpression of HDAC5
promoted the proliferation of osteosarcoma cells. In contrast,
HDAC5 knockdown inhibited the proliferation of osteosarcoma cells
(9). Liu et al demonstrated
that HDAC5 displayed high expression in melanoma cells compared
with normal skin cells. HDAC5 knockdown was found to suppress the
proliferation and metastasis of melanoma cells (17). Milde et al found that HDAC5
displayed a significant upregulation in high-risk medulloblastoma
compared with low-risk medulloblastoma, and the upregulation of
HDAC5 was associated with poor patient survival (18). In summary, HDAC5 has been found to
play an important role in tumorigenesis, metastasis and invasion
(16–18). However, little is known regarding
the specific role of HDAC5 in lung cancer.
In the present study, lung cancer cell lines (A549,
HCC827 and 95-D) and human bronchial epithelial cells (HBE) were
used to detect the expression of HDAC5 by western blotting and
RT-qPCR. The effects of HDAC5 on A549 cell proliferation, apoptosis
and invasion were assayed. In addition, we analyzed the effects of
HDAC5 on the expression of proteins, DLL4 (Delta-like 4), Six1,
Notch 1 and Twist 1 in A549 cells. These data may provide
information for the prediction of lung cancer prognosis and the
establishment of targeted therapies.
Materials and methods
Specimens
The present study was reviewed and approved by the
Ethics Committee of the Affiliated Hospital of Nantong University.
All patients volunteered to participate in the study and signed a
written informed consent. Fresh lung cancer tissues and matched
adjacent non-tumor tissues were collected from 18 non-small cell
lung cancer (NSCLC) patients that underwent surgical resection at
the Affiliated Hospital of Nantong University from July 2015 to
January 2017. The median patient age was 61 years (range, 48–72
years) and 13 patients (72.2%) were male. Before surgery, all the
patients received no radiotherapy and chemotherapy, and had no
other treatment history, nor presented with inflammatory diseases.
All tissue specimens collected from patients with NSCLC were
immediately frozen in liquid nitrogen upon surgery, and were
transported to the laboratory and stored at −80°C for further
tissue preparation.
Materials
All cell culture reagents were obtained from
Gibco/Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Human lung
cancer cell lines (A549, HCC827 and 95-D) and human bronchial
epithelial (HBE) cells (https://www.atcc.org/Products/All/PCS-300-010.aspx)
were purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). Protein extraction buffer, MTT reagent, BCA
protein concentration assay kit, Annexin V-FITC and propidium
iodide (PI) were purchased from Beyotime Institute of Biotechnology
(Haimen, China). Polyvinylidene difluoride (PVDF) membranes were
supplied by Millipore (Bedford, MA, USA). Pierce ECL
chemiluminescence detection kit was obtained from Thermo Fisher
Scientific, Inc. Transwell invasion chamber was supplied by Costar
Corp. (Cambridge, MA, USA). Matrigel was purchased from
Collaborative Biomedical Products (Bedford, MA, USA). The
antibodies used in this study included rabbit anti-HDAC5 polyclonal
antibody (Abcam, Cambridge, UK; cat. no. ab55403), rabbit
anti-delta-like 4 (DLL4) antibody (Cell Signaling Technology, Inc.,
Danvers, MA, USA; cat. no. 2589T), rabbit anti-SIX homeobox 1
(SIX1) antibody (LifeSpan BioSciences, Seattle, WA, USA; cat. no.
LS-C490560-100), mouse anti-Notch 1 monoclonal antibody (Invitrogen
Antibodies/Thermo Fisher Scientific, Inc.; cat. no. MA1-81888),
rabbit anti-Twist 1 polyclonal antibody (Cell Signaling Technology,
Inc.; cat. no. 46702S), mouse anti-β-actin monoclonal antibody
(R&D Systems, Minneapolis, MN, USA; cat. no. MAB8929),
horseradish peroxidase-conjugated goat anti-rabbit (cat. no. 31239)
and goat anti-mouse (cat. no. 31185) IgG polyclonal antibodies
(Invitrogen Antibodies/Thermo Fisher Scientific, Inc.).
Cell culture and treatment
HBE cells and A549, HCC827 and 95-D cells were all
cultured in RPMI-1640 medium containing 10% fetal bovine serum
(FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 1.5 g/l
sodium bicarbonate, 4.5 g/l glucose, 50 U/ml penicillin and 50
µg/ml streptomycin at 37°C in a humidified atmosphere with 5%
CO2. When cells reached 70–80% confluence, trypsin
digestion was performed for passage. Cells in logarithmic growth
phase were digested with 0.25% trypsin and collected for further
experiments.
A549 cells were chosen to perform further
experiments. A549 cells (2×105) were seeded in a 6-well
tissue culture plate with 2 ml antibiotic-free RPMI-1640 medium
supplemented with 10% FBS. When cells reached 60–80% confluence,
the cells were transfected with HDAC5 siRNA, pcDNA3.1-HDAC5 and
control vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China)
using Invitrogen™ Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's specification.
Then, A549 cells were incubated with the compound at 37°C in a
CO2 incubator for 5 h. Following, the transfection
mixture was replaced with fresh medium to culture for 48 h.
Finally, the A549 cells were assayed using the appropriate
protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was selected to determine the expression of
HDAC5 mRNA. The primers for HDAC5 (GenBank: BC051824.1) were: Left
primer gtgacaccgtgtggaatgag and right primer agtccacgatgaggaccttg.
The primers for β-actin (GenBank: M10277.1) were: Left primer
ctcttccagccttccttcct and right primer agcactgtgttggcgtacag. Total
RNA was extracted using Trizol reagent obtained from Beyotime
Institute of Biotechnology (cat. no. R0016). Then mRNAs were
reverse transcribed into cDNA using BeyoRT™ cDNA Synthesis Kit
(Beyotime Institute of Biotechnology; cat. no. D7166). qPCR
analyses were performed using the BeyoFast™ SYBR Green qPCR Mix
(Beyotime Institute of Biotechnology; cat. no. D7260) on Applied
Biosystems® 7500 Real-Time PCR Systems (Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were used
for qPCR. Firstly 95°C for 5 min, followed by 36 cycles of 95°C for
10 sec, 60°C for 30 sec, and 72°C 30 sec. The genes β-actin was
used as internal control for RT-qPCR. All RT-PCRs were performed in
triplicate and the relative fold differences in gene expression
were calculated according to the 2−ΔΔCt method.
Western blot analysis
Total proteins were extracted from lung cancer
tissues, lung cancer-adjacent normal tissues, lung cancer cell
lines (A549, HCC827 and 95-D) and HBE cells using a protein
extraction kit, and were quantified using a BCA protein
concentration assay kit. Proteins were separated by 12%
SDS-polyacrylamide gel electrophoresis and transferred to a PVDF
membrane using a wet-type transblotting apparatus (Bio-Rad
Laboratories, Richmond, CA, USA). Then the PVDF membrane was
blocked with 5% skimmed milk diluted in TBS (10 mM Tris-HCl, pH
7.5, 150 mM NaCl) solution for 1 h, and incubated with the primary
antibody at 4°C overnight. All primary antibodies were diluted
1:2,000 in TBST buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl and
0.1% Tween-20) supplemented with 5% non-fat milk. Next morning, the
PVDF membrane was washed 3×5 min in TBST and incubated with the
corresponding secondary antibody at room temperature for 2 h. All
secondary antibodies were diluted 1:5,000 in TBST buffer.
Following, the PVDF membrane was washed 3×5 min in TBST and bands
were visualized using the ECL chemiluminescence reagent. The
relative expression of the target protein was valuated with the
gray value ratio of target protein content to β-actin (target
protein/β-actin) content by Quantity One software (Bio-Rad
Laboratories, Hercules, CA, USA).
MTT assay
MTT assay was used to examine the effects of HDAC5
on the viability of A549 cells. Briefly, A549 cells were seeded in
96-well plates and allowed to adhere overnight. Then, A549 cells
were incubated with 10 µl of MTT (5 mg/ml) for 4 h. The mixture
culture medium was replaced by 150 µl of dimethyl sulfoxide (DMSO)
to dissolve the crystals. The optical density (OD) values at 570 nm
(test wavelength) and 630 nm (reference wavelength) were examined
on a 96-well micro test spectrophotometer (BioTek Instruments,
Inc., Winooski, VT, USA). The relative cell viability was
calculated by the equation as described in a previous study
(19) and the experiments were run
in triplicate.
Flow cytometric analysis
The effects of HDAC5 on the A549 cell cycle and
apoptosis were determined using flow cytometry. First, A549 cells
were seeded in serum-free RPMI-1640 medium for 24 h to synchronize
and then incubated with complete RPMI-1640 medium for 24 h.
Following, A549 cells were trypsinized, washed, harvested, fixed
with 70% ice-cold ethanol and stored at −20°C. On the next day,
A549 cells were washed with citrate phosphate buffer and PBS in
turn, treated with PBS containing 100 µg/ml of RNase A at 37°C for
30 min, and then cultured in PBS containing 100 µg/ml of propidium
iodide (PI) at room temperature for 30 min. Finally, cell cycle
distribution was determined using flow cytometry (BD Biosciences,
Franklin Lakes, NJ, USA). The experiments were performed in
triplicate.
A549 cell apoptosis was quantitated by staining with
Annexin V-FITC. Briefly, A549 cells were washed, collected, and
resuspended in 195 µl of Annexin V-FITC binding buffer. Following,
5 µl of Annexin V-FITC was added into the Annexin V-FITC binding
buffer and incubated in the dark at room temperature for 10 min.
Then, A549 cells were collected by centrifugation for 5 min at
1,500 × g, and gently resuspended in 190 µl of Annexin V-FITC
binding buffer. Finally, 10 µl of PI staining solution was added
into the Annexin V-FITC binding buffer and kept on ice in the dark
until flow cytometric analysis. CellQuest software (BD Biosciences)
was used to analyze the datum, and the analysis was run in
triplicate.
Transwell invasion analysis
Transwell invasion chamber was used to evaluate the
invasive ability of the A549 cells. Briefly, the chamber filter was
washed with serum-free RPMI-1640 medium, and then the upper side of
the filter was evenly covered with 20 µl of Matrigel (1:2 dilution
with RPMI-1640). The chamber was divided by two compartments
including the supper chamber and the lower chamber. For invasion
assays, 200 µl of serum-free RPMI-1640 medium containing
1×105 A549 cells were added in the upper chamber of the
Transwell invasion system, while 500 µl of RPMI-1640 medium
containing 10% FBS were added into the lower chamber. Then the
Transwell invasion system was incubated for 48 h in an incubator.
Following, the cells on the upper surface of the filter were
removed with a sterile cotton swab. Those cells that invaded to the
lower surface of the filter and invaded in the lower chamber were
collected and assessed by MTT assay. The results are presented as
the mean ± SD, and the experiment was performed in triplicate.
Statistical analysis
All data are expressed as mean ± SD from at least
three independent experiments. SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA) was used to analyze the experimental data with
Student's t-test or one-way ANOVA followed by Tukey's post hoc
test. The results were considered statistically significant at
P<0.05. GraphPad Prism software version 5.0 (GraphPad Software,
Inc., San Diego, CA, USA) was applied to draw the graphs.
Results
Overexpression of HDAC5 in lung cancer
tissues and cell lines
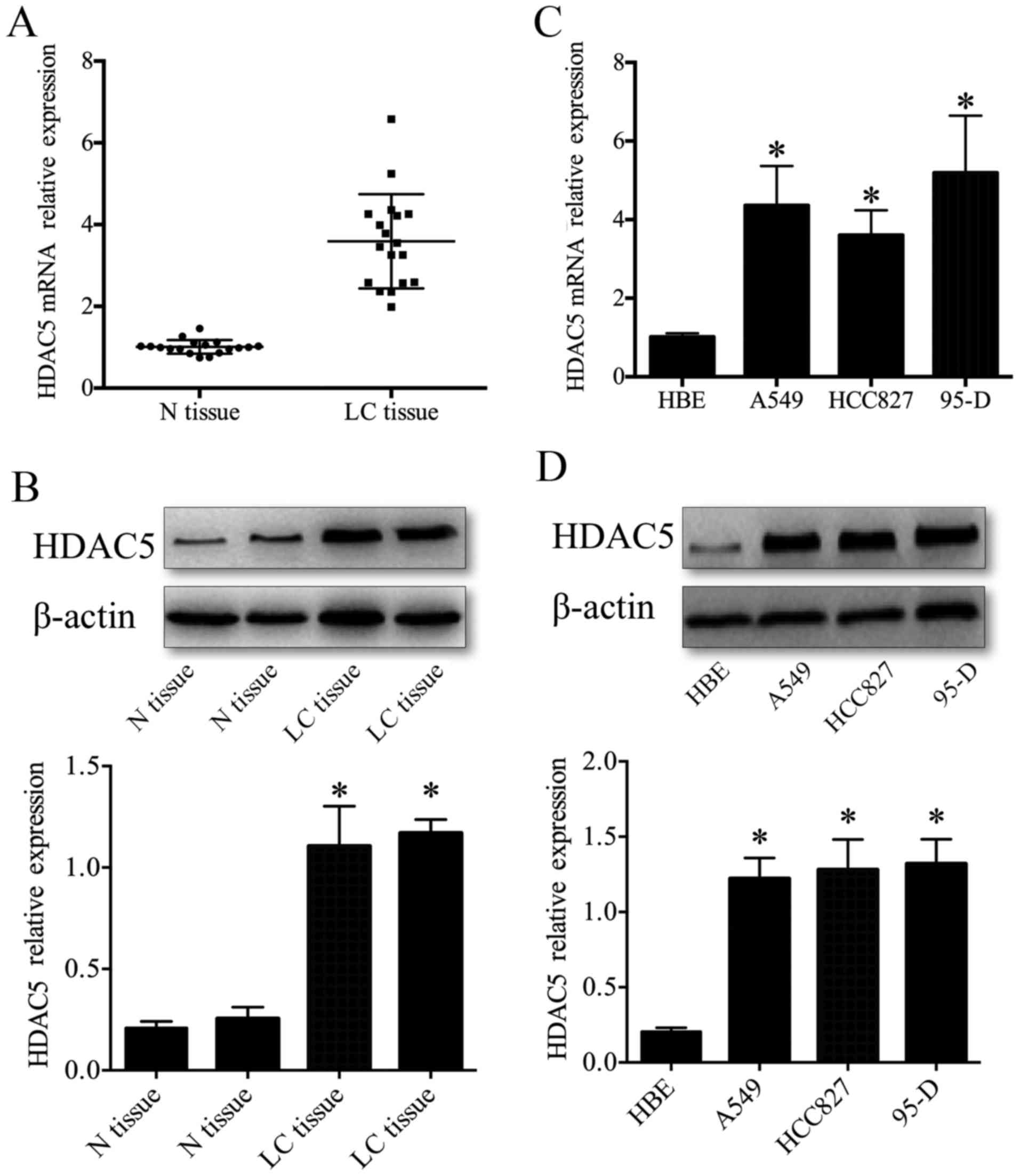
We first examined HDAC5 protein and mRNA levels in
human lung cancer tissues by western blotting and RT-qPCR. The
results showed that the expression of HDAC5 protein and mRNA was
significantly upregulated in lung cancer tissues compared to that
observed in the lung cancer-adjacent normal tissues (P<0.05)
(Fig. 1A and B). The expression
profile of HDAC5 protein and mRNA in lung cancer tissues were
similar to those in lung cancer cell lines, which showed that HDAC5
protein and mRNA displayed significant upregulation in human lung
cancer cell lines (A549, HCC827 and 95-D) compared to that observed
in the human bronchial epithelial (HBE) cells (P<0.05) (Fig. 1C and D). These data demonstrated
that the elevated HDAC5 may be important in the tumorigenesis and
progression of lung cancer.
In order to address the function of HDAC5 in the
tumorigenesis and progression of lung cancer, A549 cells were
chosen for further investigation unless specified otherwise. We
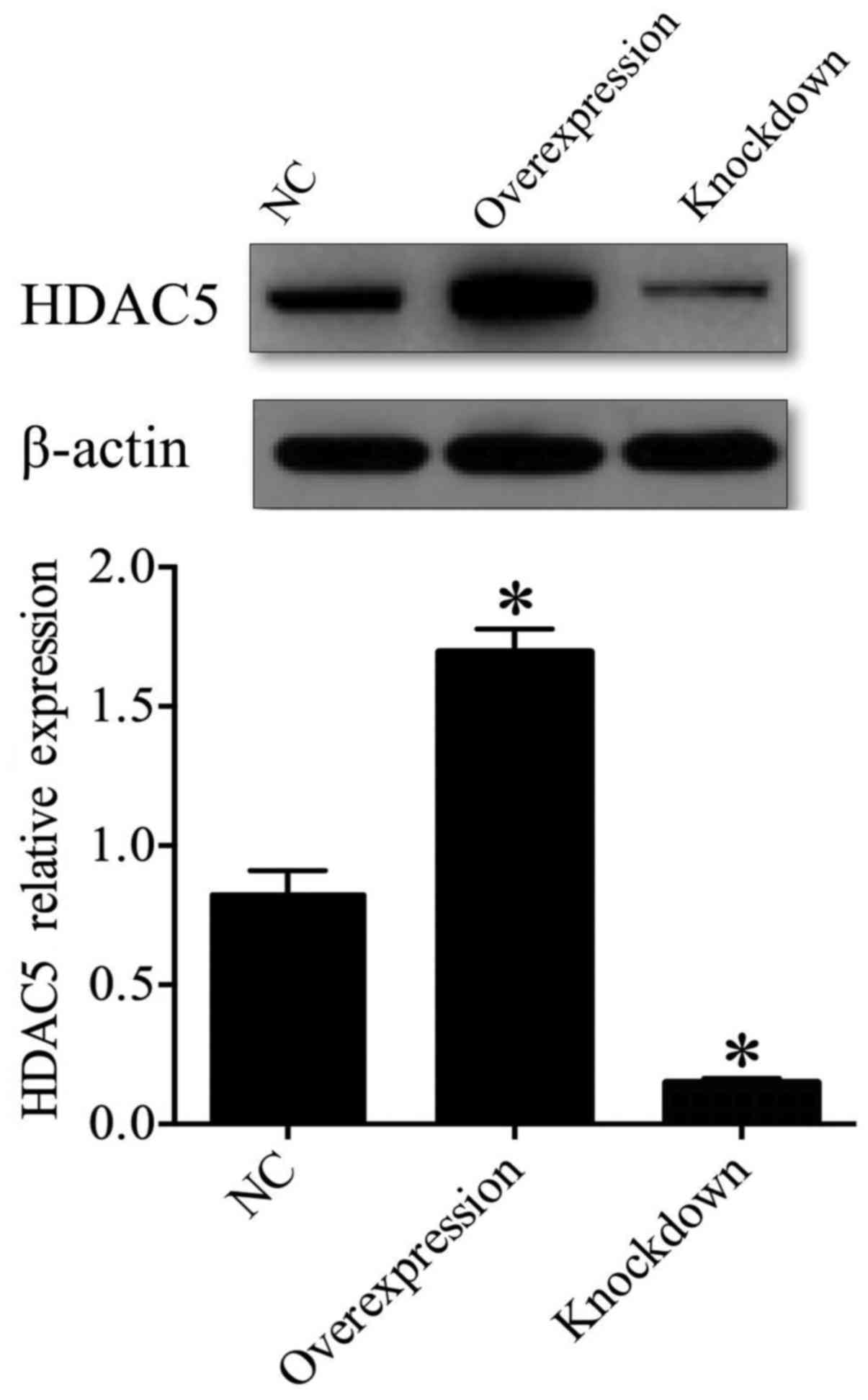
generated A549 cells in which HDAC5 was either overexpressed or
depleted. Western blot analysis indicated that the expression of
HDAC5 protein was significantly upregulated in the HDAC5
overexpression group (transfected with pcDNA3.1-HDAC5) (P<0.05),
and was obviously downregulated in the HDAC5-knockdown group
(transfected with HDAC5 siRNA) compared to the control group (NC,
transfected with vector only) (P<0.05) (Fig. 2), which suggested that A549 cell
models, in which HDAC5 was either overexpressed or depleted, were
successfully established.
HDAC5 enhances A549 cell
viability
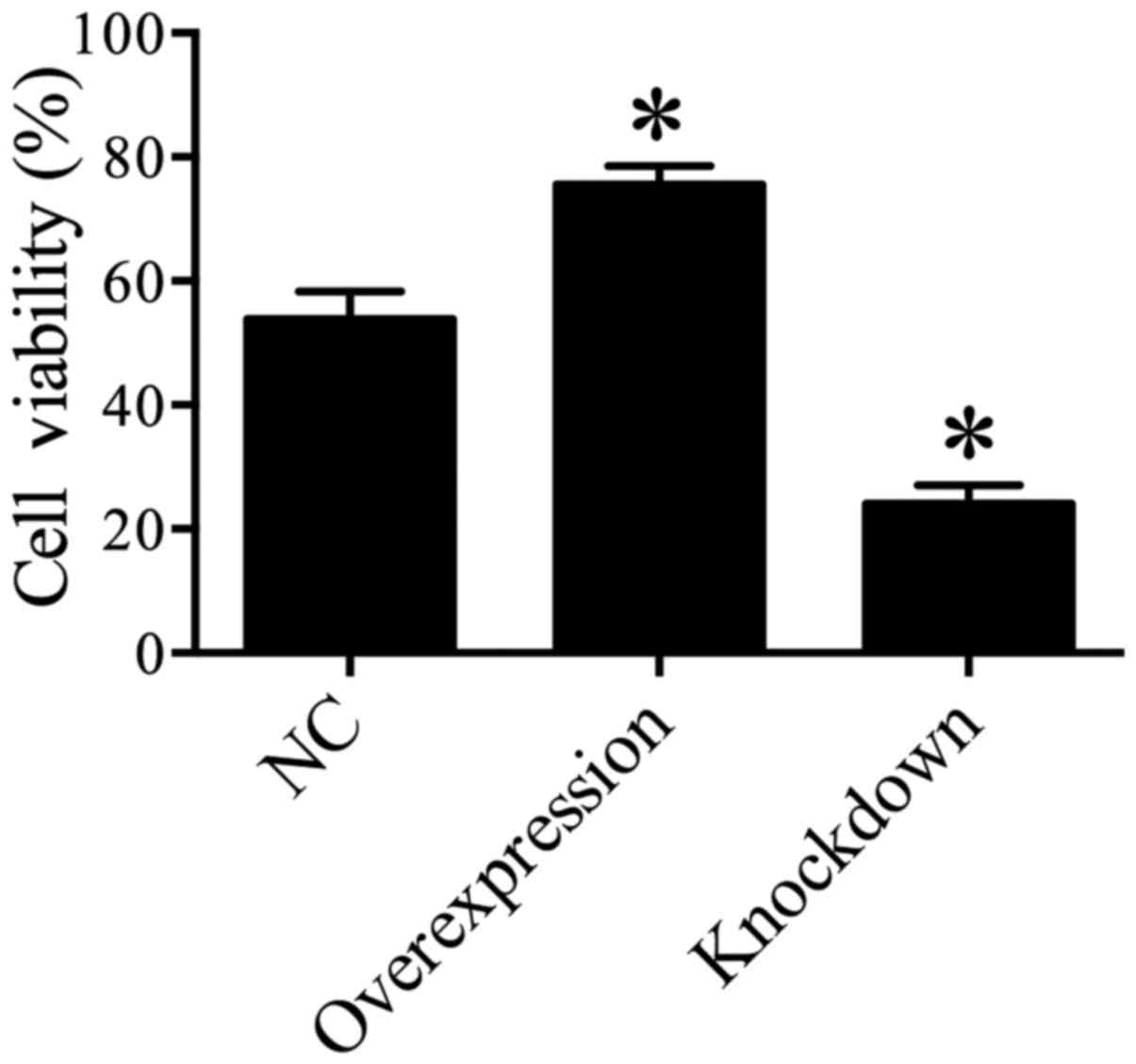
In order to explore the effect of HDAC5 on the cell
viability of lung cancer cells, MTT assay was performed and the
results suggested that A549 cell viability was significantly
enhanced in the HDAC5 overexpression group compared with the
control group (P<0.05), while the cell viability of A549 cells
was markedly inhibited in the HDAC5 knockdown group compared with
the control group (P<0.05) (Fig.
3). These data indicated that elevated HDAC5 may play a crucial
role in the increase in A549 cell viability.
HDAC5 promotes A549 cell cycle
progression
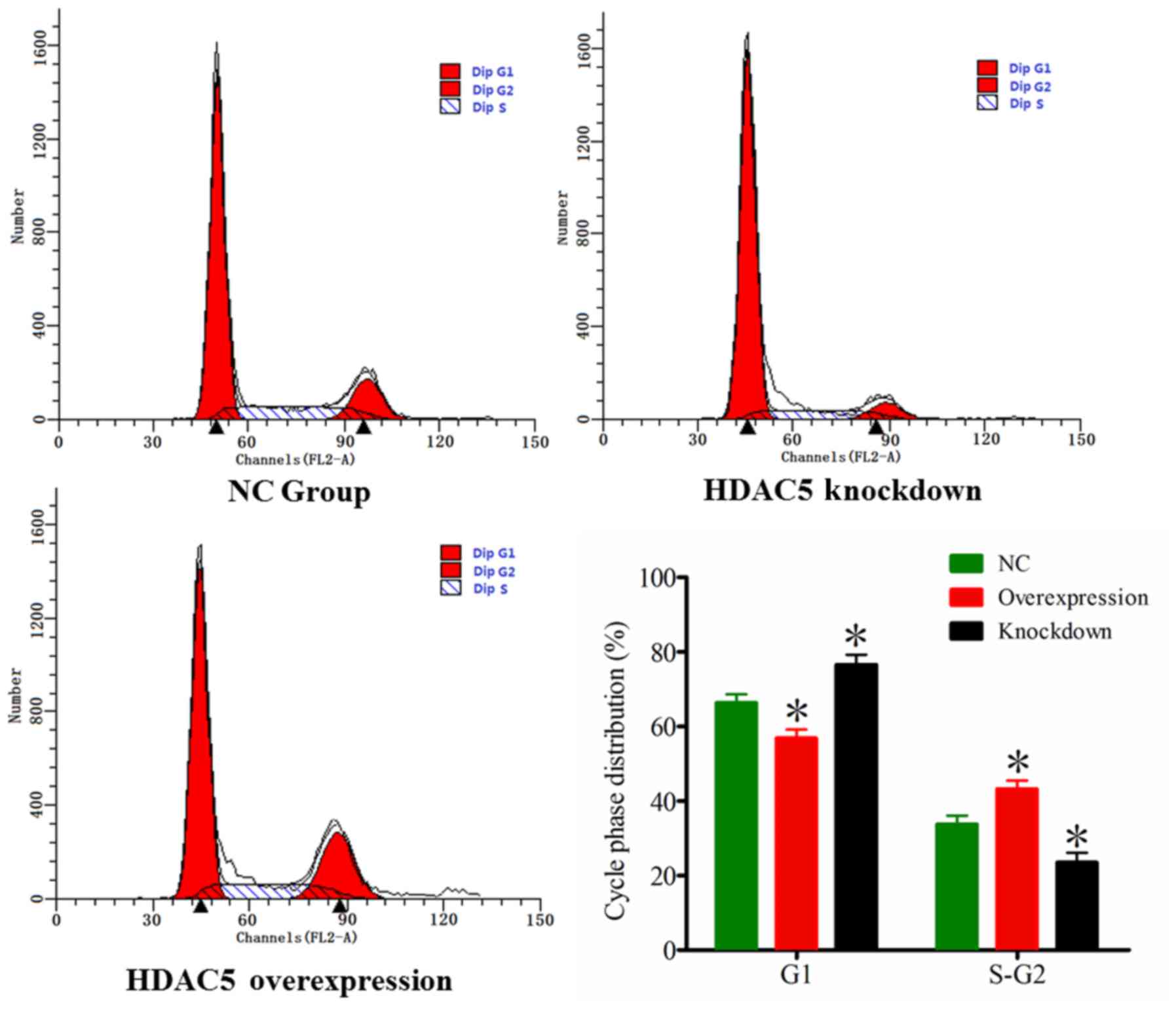
In order to address whether HDAC5 is associated with
A549 cell cycle progression, we evaluated the cell cycle
distribution of A549 cells by FCM. The results indicated that there
were more A549 cells in the S and G2 phases, and less
A549 cells in the G1 phase in the HDAC5 overexpression
group compared to these populations in the control group
(P<0.05). On the contrary, there were less A549 cells in the S
and G2 phases, and more A549 cells in the G1
phase in the HDAC5 knockdown group compared to these populations in
the control group (P<0.05) (Fig.
4). These data demonstrated that HDAC5 may promote A549 cell
cycle progression.
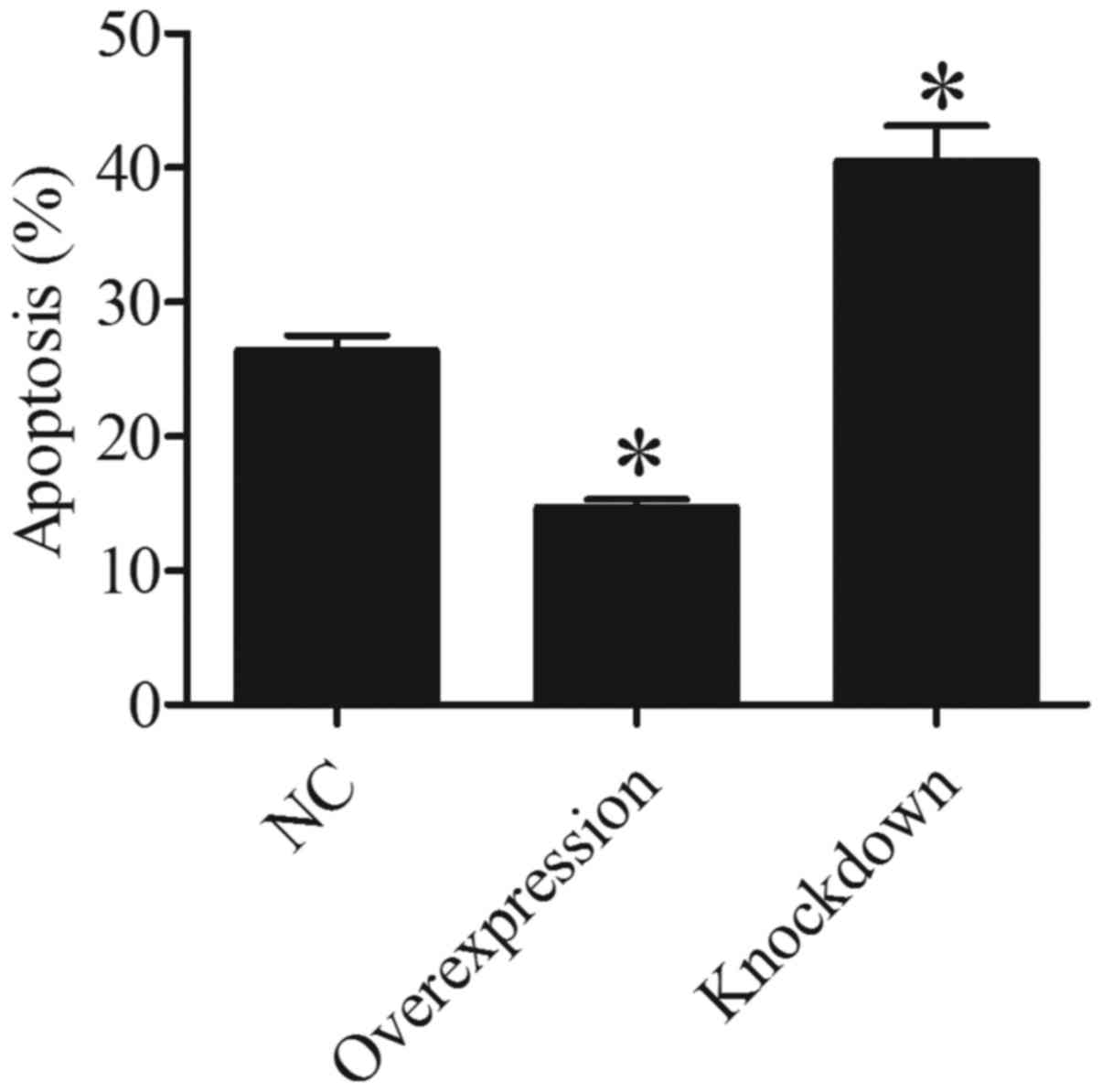
HDAC5 inhibits A549 cell
apoptosis
In order to ascertain whether HDAC5 is associated
with A549 cell apoptosis, we evaluated the cell apoptosis of A549
cells by FCM. The results showed that there were less apoptotic
A549 cells in the HDAC5 overexpression group than that in the
control group (P<0.05). On the contrary, more apoptotic A549
cells were found in the HDAC5 knockdown group compared to that in
the control group (P<0.05) (Fig.
5). These data suggested that HDAC5 plays a crucial role in the
inhibition of A549 cell apoptosis.
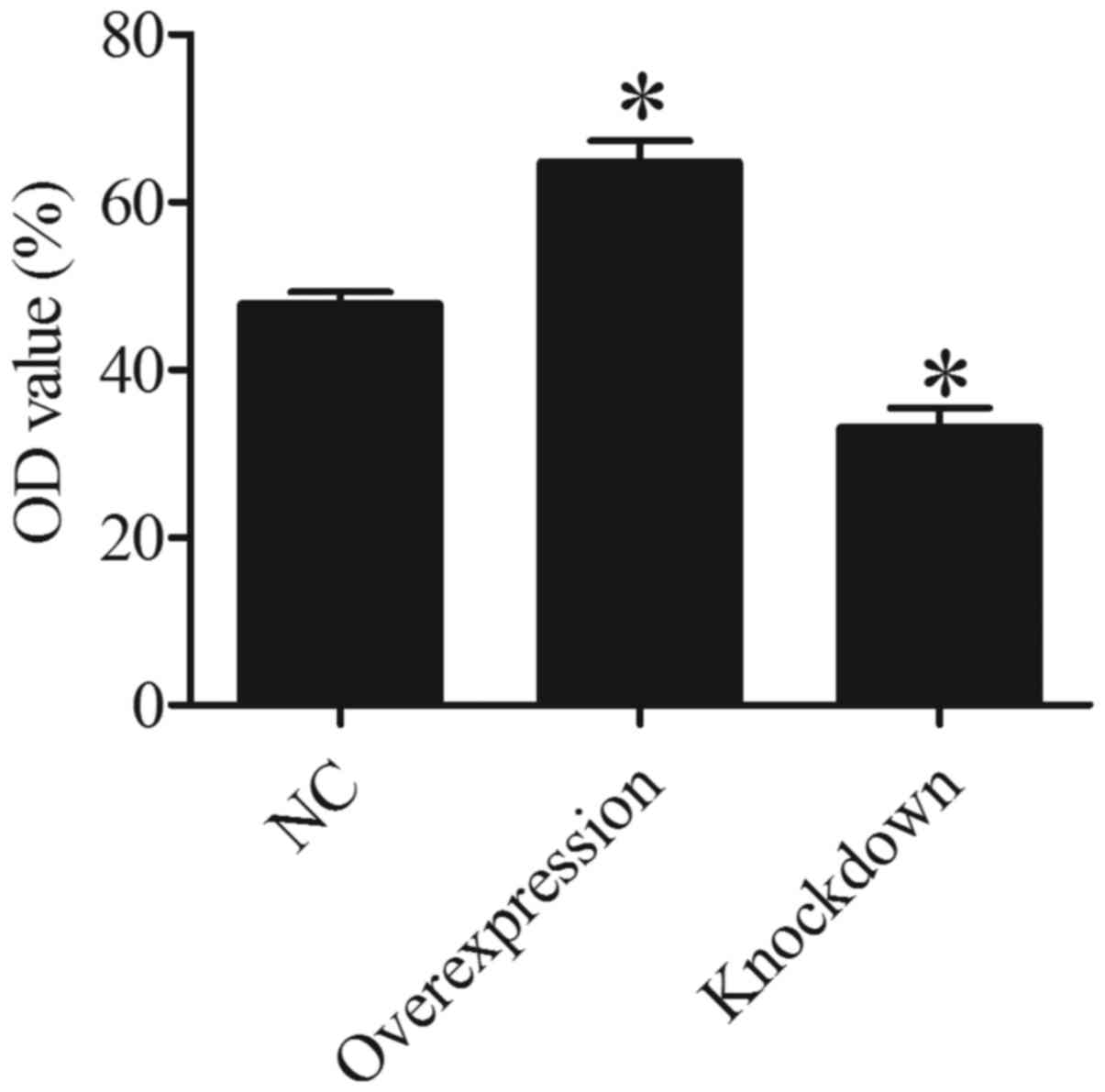
HDAC5 enhances the invasive ability of
A549 cells
To determine whether HDAC5 is associated with A549
cell invasion, Transwell invasion assay was used to evaluate the
effect of HDAC5 on A549 cells. The results suggested that the OD
value of the invaded A549 cells in the HDAC5 overexpression group
was higher than that in the control group (P<0.05). On the
contrary, a lower OD value was found in the HDAC5 knockdown group
compared to that noted in the control group (P<0.05) (Fig. 6). These data demonstrated that more
A549 cells invaded through the polycarbonate membrane in HDAC5
overexpression group, and less A549 cells invaded through the
polycarbonate membrane in HDAC5 knockdown group compared to control
group, which indicated that HDAC5 strengthen the invasive ability
of A549 cells.
HDAC5 increases the expression of
DLL4, Six1, Notch 1 and Twist 1
HDAC5 has fundamental roles in the epigenetic
regulation of gene expression and contributes to proliferation,
differentiation, apoptosis, cell cycle and invasion (8–10).
Previous studies suggest that HDAC5 is an important regulatory
factor for the expression of DLL4, Six1, Notch 1 and Twist 1
(8,9,16,20).
Therefore, the expression of DLL4, Six1, Notch 1 and Twist 1 was
detected in this study, and the results indicated that the
expression of DLL4, Six1, Notch 1 and Twist 1 was significantly
enhanced in the HDAC5 overexpression group, and obviously
suppressed in the HDAC5 knockdown group compared with the control
group (P<0.05) (Fig. 7). These
results suggested that HDAC5 increased the expression of DLL4,
Six1, Notch 1 and Twist 1 in the A549 cells.
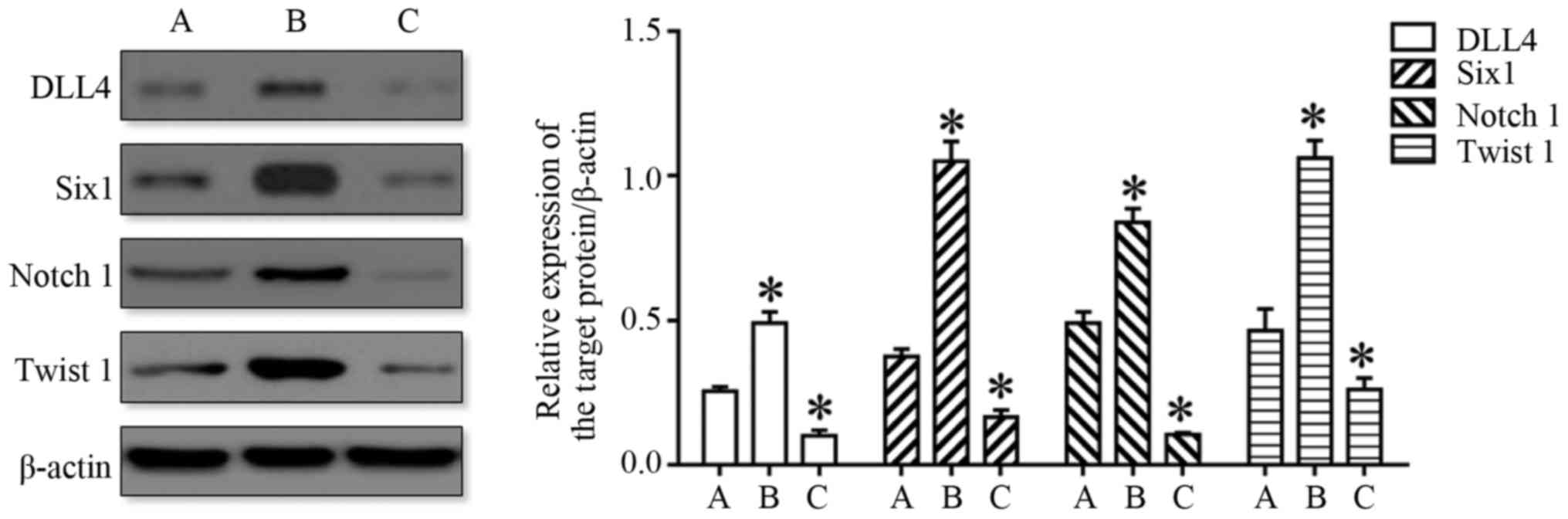 | Figure 7.HDAC5 increases the expression of
DLL4, Six1, Notch 1 and Twist 1 in A549 cells. The image on the
left displays the representative western blot image. The histogram
shown compares the relative expression levels of DLL4, Six1, Notch
1 and Twist 1 in the A549 cells transfected with pcDNA3.1-HDAC5
(overexpression) and HDAC5 siRNA (knockdown) compared with the A549
cells transfected with the vector only (NC). *P<0.05 vs. A. (A,
NC, B, Overexpression, C, Knockdown). HDAC5, histone deacetylase
5. |
Discussion
Lung cancer is one of the most common malignant
neoplasms, as well as the most common cause of cancer-related
mortality. Most lung cancers are squamous cell carcinomas, small
cell carcinomas or adenocarcinomas (21). The prognosis of patients with lung
cancer is still less than satisfactory, and the 5-year survival
rate does not exceed 15% (2–5). This
is mainly due to the fact that many key factors regulating the
malignant phenotype of lung cancer have not been studied clearly.
HDAC5 is frequently dysregulated in human malignancies and has
therefore become a therapeutic target in cancer therapy (8,9,11,16,20).
However, whether HDAC5 is involved in lung cancer incidence,
migration and invasion remains elusive.
In the present study, our data showed that HDAC5
displayed significantly high expression in lung cancer tissues and
cell lines. The expression profile of HDAC5 in lung cancer was
consistent with that in breast cancer, colorectal cancer and glioma
(8,15,22).
Elevated HDAC5 was found to promote the proliferation of colorectal
cancer cells through upregulation of DLL4 (8). HDAC5 was found to be increased in
human glioma tissues and to promote the proliferation of glioma
cells by the upregulation of Notch 1 (20). These data indicate that elevated
HDAC5 may play a central role in the tumorigenesis of lung
cancer.
In order to elucidate the role of HDAC5 in lung
cancer cells, A549 cell models, in which HDAC5 was either
overexpressed or depleted, were generated. HDAC5 displayed a higher
expression in the HDAC5 overexpression group and a lower expression
in the HDAC5 knockdown group compared to the control group, which
indicated that the A549 cell models were successfully established.
In view of the established lung cancer A549 cell models, the
effects of HDAC5 on cell viability, cell cycle distribution,
apoptosis and invasion of A549 cells were determined, and the
results showed that HDAC5 overexpression enhanced the cell
viability and proliferation of A549 cells. On the contrary, HDAC5
inhibition suppressed the cell viability and proliferation of A549
cells, leading to cell growth inhibition and cell cycle
G1 phase arrest in A549 cells. These data indicated that
HDAC5 improved the cell growth and proliferation of A549 cells. The
effect on the proliferation of A549 cells was consistent with that
of other cancers reported by previous studies which demonstrated
that elevated HDAC5 promoted the proliferation of colorectal cancer
cells, hepatocellular carcinoma cells and glioma cells, while
downregulation of HDAC5 caused a significant inhibition of
colorectal cancer cell, hepatocellular carcinoma cell and glioma
cell proliferation (8,16,20).
The results also showed that overexpression of HDAC5 displayed an
obvious inhibition of A549 cell apoptosis, and the inhibition of
HDAC5 facilitated A549 cell apoptosis, which indicated that HDAC5
inhibited A549 cell apoptosis. The results in this study were also
confirmed by previous studies concerning breast cancer and
hepatocellular carcinoma, which reported that knockdown of HDAC5
reduced tumorigenesis and enhanced apoptosis (15,22–24).
HDAC5 was extensively expressed in many human cancers. HDAC5,
overexpressed in neuroblastoma, was found to trigger neuroblastoma
cell invasion and metastasis (25),
and knockdown of HDAC5 restrained breast cancer cell proliferation,
invasion and metastasis (15).
Furthermore, the effect of HDAC5 on A549 cell invasion was
evaluated by Transwell invasion assay. The results revealed that
overexpression of HDAC5 was associated with the increased invasive
capacity of A549 cells, and the inhibition of HDAC5 was associated
with the decreased invasive capacity of A549 cells.
In summary, these results demonstrated that HDAC5
was associated with increased A549 cell growth, proliferation, and
invasion and decreased A549 cell apoptosis. Nevertheless, the
detailed mechanism or the downstream HDAC5 targets in human lung
cancer cells remains unclear. HDAC5 promoted colorectal cancer cell
proliferation by upregulating DLL4 expression (8), glioma cell proliferation by
upregulation of Notch 1 (20),
human hepatocellular carcinoma cell proliferation by upregulating
Six1 expression (16), and
osteosarcoma progression by upregulation of Twist 1 expression
(9). Therefore, the expression
levels of DLL4, Six1, Notch 1 and Twist 1 were also determined in
this study, and the results indicated that HDAC5 increased the
expression of DLL4, Six1, Notch 1 and Twist 1 in the A549 cells.
Our results were in line with the results from previous studies
that found that overexpression of DLL4 was associated with poor
outcomes of patients with pancreatic adenocarcinoma, and elevated
DLL4 promoted renal carcinoma cell metastasis (26,27).
Thus, we may speculate that elevated HDAC5 promotes the expression
of DLL4 in lung cancer, and contributes to poor patient outcomes
and metastasis. Increased Six1 was found to be associated with the
poor prognosis of prostate cancer patients and enhanced pancreatic
cancer cell proliferation through upregulation of cyclin D1
(28,29). Conversely, downregulation of Six1
suppressed colorectal cancer cell growth and invasion (30). From our results, we could infer that
elevated HDAC5 could promote the expression of Six1 in lung cancer,
and contribute to lung cancer cell proliferation and poor
prognosis. Notch 1 signaling, activated in many cancers, promoted
the malignant features including epithelial to mesenchymal
transition of cancers through NF-κB activation (31,32).
Notably, DLL4-Notch signaling was found to participate in the
formation of large vessels in tumors, leading to distant metastasis
of tumors (33). Inhibition of
Notch 1 signaling pathway was found to inhibit breast cancer cell
proliferation and invasion (34).
Twist 1, a key factor in the promotion of metastasis of cancer
cells, promoted cell growth and metastasis in acute myeloid
leukemia (35,36). In view of these data, we may
conjecture that elevated HDAC5 in lung cancer could promote the
expression of Notch 1 and Twist 1, and then promote EMT and distant
metastasis. HDAC5 could repress the expression of miR-125a-5p in
human breast cancer (22). Our
previous study showed that miR-125a-5p was downregulated and acted
as a tumor suppressor in lung carcinoma by directly targeting STAT3
(37). These data may be
responsible for the explanation that HDAC5 promoted A549 cell
growth, proliferation, invasion and inhibited A549 cell
apoptosis.
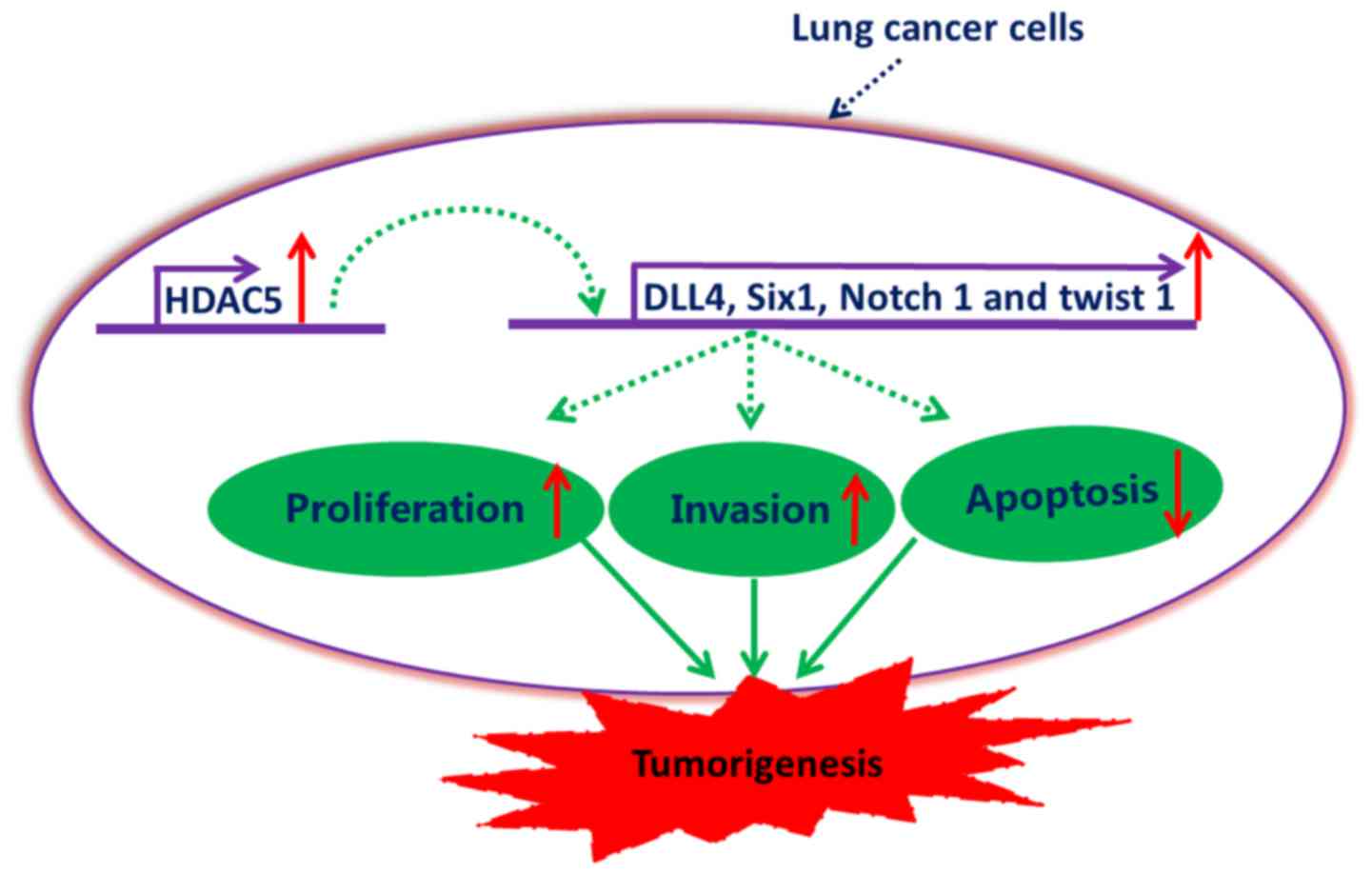
In summary, our data demonstrated that HDAC5 was
significantly upregulated in lung cancer, and elevated HDAC5 may be
involved in the potentiation of proliferation and invasion of lung
cancer cells, as well as the inhibition of lung cancer cell
apoptosis, at least partially, by the upregulation of DLL4, Six1,
Notch 1 and Twist 1 (Fig. 8). This
study provides evidence for the potential application of HDAC5
inhibitors in the therapy of lung cancer.
Acknowledgements
We thank Dr Hualin Sun for his technical
support.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81501967).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
LZ and JHS conceived and designed the study. LZ,
SYS, SMY, MMG and XH performed the experiments. LZ wrote the paper.
LZ, SYS, SMY, MMM, XH and JHS reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was reviewed and approved by the
Ethics Committee of the Affiliated Hospital of Nantong University.
All patients volunteered to participate in the study and signed a
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Denton EJ, Hart D, Wainer Z, Wright G,
Russell PA and Conron M: Changing trends in diagnosis, staging,
treatment and survival in lung cancer: Comparison of three
consecutive cohorts in an Australian lung cancer centre. Intern Med
J. 46:946–954. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Osińska I and Domagała-Kulawik J:
Bronchoalveolar lavage in lung cancer-diagnostic value and
assessment of the anti-cancer immune response. Postepy Hig Med
Dosw. 67:1119–1127. 2013.(In Polish). View Article : Google Scholar
|
|
3
|
Giangreco A, Groot KR and Janes SM: Lung
cancer and lung stem cells: Strange bedfellows? Am J Respir Crit
Care Med. 175:547–553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grivaux M, Debieuvre D, Herman D,
Lemonnier C, Marcos JM, Crequit J, Vuillermoz-Blas S, Barre P,
Saillour M and Martin F: Early mortality in lung cancer: French
prospective multicentre observational study. BMC Pulm Med.
16:452016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whiteside TL: Stimulatory role of exosomes
in the context of therapeutic anti-cancer vaccines. Biotarget.
1:52017. View Article : Google Scholar
|
|
6
|
Al-Mulla F, Bitar MS, Al-Maghrebi M,
Behbehani AI, Al-Ali W, Rath O, Doyle B, Tan KY, Pitt A and Kolch
W: Raf kinase inhibitor protein RKIP enhances signaling by glycogen
synthase kinase-3β. Cancer Res. 71:1334–1343. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noma D, Inamura K, Matsuura Y, Ninomiya H,
Ichinose J, Nakao M, Mun M, Ishikawa Y and Okumura S:
ALK-rearranged lung adenocarcinoma showing intra-bronchial
protrusion: A case of actually peripheral origin with a rare
spreading pattern. Biotarget. 1:152017. View Article : Google Scholar
|
|
8
|
He P, Liang J, Shao T, Guo Y, Hou Y and Li
Y: HDAC5 promotes colorectal cancer cell proliferation by
up-regulating DLL4 expression. Int J Clin Exp Med. 8:6510–6516.
2015.PubMed/NCBI
|
|
9
|
Chen J, Xia J, Yu YL, Wang SQ, Wei YB,
Chen FY, Huang GY and Shi JS: HDAC5 promotes osteosarcoma
progression by upregulation of Twist 1 expression. Tumour Biol.
35:1383–1387. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fang Q, Xu T, Wu C, Zhou S and Sun H:
Biotargets in neural regeneration. Biotarget. 1:62017. View Article : Google Scholar
|
|
11
|
Xiao H, Jiao J, Wang L, O'Brien S, Newick
K, Wang LC, Falkensammer E, Liu Y, Han R, Kapoor V, et al: HDAC5
controls the functions of Foxp3+ T-regulatory and
CD8+ T cells. Int J Cancer. 138:2477–2486. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guise AJ and Cristea IM: Approaches for
studying the subcellular localization, interactions, and regulation
of histone deacetylase 5 (HDAC5). Methods Mol Biol. 1436:47–84.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Greco TM, Yu F, Guise AJ and Cristea IM:
Nuclear import of histone deacetylase 5 by requisite nuclear
localization signal phosphorylation. Mol Cell Proteomics. 10(M110):
0043172011.PubMed/NCBI
|
|
14
|
Lin M, Zhu Q, Wang J, Yang W, Fan H, Yi J
and Jiang M: Molecules involved in acrosomal exocytosis and
cortical granule exocytosis. Biotarget. 1:112017. View Article : Google Scholar
|
|
15
|
Li A, Liu Z, Li M, Zhou S, Xu Y, Xiao Y
and Yang W: HDAC5, a potential therapeutic target and prognostic
biomarker, promotes proliferation, invasion and migration in human
breast cancer. Oncotarget. 7:37966–37978. 2016.PubMed/NCBI
|
|
16
|
Feng GW, Dong LD, Shang WJ, Pang XL, Li
JF, Liu L and Wang Y: HDAC5 promotes cell proliferation in human
hepatocellular carcinoma by up-regulating Six1 expression. Eur Rev
Med Pharmacol Sci. 18:811–816. 2014.PubMed/NCBI
|
|
17
|
Liu J, Gu J, Feng Z, Yang Y, Zhu N, Lu W
and Qi F: Both HDAC5 and HDAC6 are required for the proliferation
and metastasis of melanoma cells. J Transl Med. 14:72016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Milde T, Oehme I, Korshunov A,
Kopp-Schneider A, Remke M, Northcott P, Deubzer HE, Lodrini M,
Taylor MD, von Deimling A, et al: HDAC5 and HDAC9 in
medulloblastoma: Novel markers for risk stratification and role in
tumor cell growth. Clin Cancer Res. 16:3240–3252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT,
Xia YJ, Ye ZY and Tao HQ: MicroRNA-101 is down-regulated in gastric
cancer and involved in cell migration and invasion. Eur J Cancer.
46:2295–2303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Q, Zheng JM, Chen JK, Yan XL, Chen HM,
Nong WX and Huang HQ: Histone deacetylase 5 promotes the
proliferation of glioma cells by upregulation of Notch 1. Mol Med
Rep. 10:2045–2050. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Medenica M, Medenica M, Bojović O,
Soldatović I and Durutović I: Changing trends in incidence of lung
cancer by histological type in Montenegro. Srp Arh Celok Lek.
142:23–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH,
Wu DC, Lee JN, Chang WC and Tsai EM: HDAC inhibitors target HDAC5,
upregulate microRNA-125a-5p, and induce apoptosis in breast cancer
cells. Mol Ther. 23:656–666. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang M, Pan Y, Dorfman RG, Czen Z, Liu F,
Zhou Q, Huang S, Zhang J, Yang D and Liu J: AR-42 induces apoptosis
in human hepatocellular carcinoma cells via HDAC5 inhibition.
Oncotarget. 7:22285–22294. 2016.PubMed/NCBI
|
|
24
|
Fan J, Lou B, Chen W, Zhang J, Lin S, Lv
FF and Chen Y: Down-regulation of HDAC5 inhibits growth of human
hepatocellular carcinoma by induction of apoptosis and cell cycle
arrest. Tumour Biol. 35:11523–11532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fabian J, Opitz D, Althoff K, Lodrini M,
Hero B, Volland R, Beckers A, de Preter K, Decock A, Patil N, et
al: MYCN and HDAC5 transcriptionally repress CD9 to trigger
invasion and metastasis in neuroblastoma. Oncotarget.
7:66344–66359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou L, Yu L, Ding G, Chen W, Zheng S and
Cao L: Overexpressions of DLL4 and CD105 are associated with poor
prognosis of patients with pancreatic ductal adenocarcinoma. Pathol
Oncol Res. 21:1141–1147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang QB, Ma X, Li HZ, Ai Q, Liu SW, Zhang
Y, Gao Y, Fan Y, Ni D, Wang BJ and Zhang X: Endothelial Delta-like
4 (DLL4) promotes renal cell carcinoma hematogenous metastasis.
Oncotarget. 5:3066–3075. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeng J, Shi R, Cai CX, Liu XR, Song YB,
Wei M and Ma WL: Increased expression of Six1 correlates with
progression and prognosis of prostate cancer. Cancer Cell Int.
15:632015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang
L, Li X, Li L, Ma W, Wu J and Zhang M: Six1 promotes proliferation
of pancreatic cancer cells via upregulation of cyclin D1
expression. PLoS One. 8:e592032013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Tian T, Hu X, Zhang X, Li L, Nan F,
Chang Y, Wang X, Sun Z, Lv F, et al: Targeting Six1 by
lentivirus-mediated RNA interference inhibits colorectal cancer
cell growth and invasion. Int J Clin Exp Pathol. 7:631–639.
2014.PubMed/NCBI
|
|
31
|
Fender AW, Nutter JM, Fitzgerald TL,
Bertrand FE and Sigounas G: Notch-1 promotes stemness and
epithelial to mesenchymal transition in colorectal cancer. J Cell
Biochem. 116:2517–2527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Zhao F, Lu J, Li T, Yang H, Wu C and
Liu Y: Notch-1 signaling promotes the malignant features of human
breast cancer through NF-κB activation. PLoS One. 9:e959122014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li JL, Sainson RC, Oon CE, Turley H, Leek
R, Sheldon H, Bridges E, Shi W, Snell C, Bowden ET, et al:
DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy
in vivo. Cancer Res. 71:6073–6083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Q, Yuan Y, Cui J, Xiao T and Jiang
D: Paeoniflorin inhibits proliferation and invasion of breast
cancer cells through suppressing Notch-1 signaling pathway. Biomed
Pharmacother. 78:197–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wushou A, Hou J, Zhao YJ and Shao ZM:
Twist-1 up-regulation in carcinoma correlates to poor survival. Int
J Mol Sci. 15:21621–21630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang N, Guo D, Zhao YY, Dong CY, Liu XY,
Yang BX, Wang SW, Wang L, Liu QG, Ren Q, et al: TWIST-1 promotes
cell growth, drug resistance and progenitor clonogenic capacities
in myeloid leukemia and is a novel poor prognostic factor in acute
myeloid leukemia. Oncotarget. 6:20977–20992. 2015.PubMed/NCBI
|
|
37
|
Zhong L, Sun S, Shi J, Cao F, Han X and
Chen Z: MicroRNA-125a-5p plays a role as a tumor suppressor in lung
carcinoma cells by directly targeting STAT3. Tumour Biol.
39:10104283176975792017.doi: 10.1177/1010428317697579. View Article : Google Scholar : PubMed/NCBI
|