Introduction
Glioma is the most common malignant tumor of the
central nervous system (1,2). Based on the World Health Organization
(WHO) classification, gliomas are divided into four grades: WHO
grade I–IV (3), in which WHO grade
III (anaplastic glioma) and WHO grade IV [glioblastoma (GBM)]
commonly constitute high-grade glioma (HGG), whereas WHO grade II
is generally referred to as low-grade glioma (LGG) (4). HGG is highly lethal and aggressive,
and complete removal is difficult, which ultimately leads to
recurrence post-surgery. Despite advances in standard treatment,
including surgical resection followed by radiotherapy and
chemotherapy, satisfactory outcomes for patients with HGG are often
limited (5). Compared with LGG, the
migratory and invasive capability of HGG is more potent, and the
doubling time of tumor cells is shorter. HGG tumor cells also have
the ability to deeply infiltrate into adjacent normal brain tissue,
which can result in incomplete resection, which is the main cause
of postoperative recurrence. The occurrence and development of
glioma is a multi-gene, multi-step and multi-stage process.
Furthermore, various tumor-specific molecular alterations have been
associated with glioma, in particular, isocitrate dehydrogenase 1
(IDH1) mutations (6), 1p/19q
co-deletion (7), O6-methylguanine
DNA methyltransferase (MGMT) promoter methylation (8) and epidermal growth factor receptor
variant III amplification (9,10) have
been identified as predictive and prognostic indicators, and/or
therapeutic targets for patients with glioma. It is necessary to
identify novel biomarkers to provide more options for the treatment
of patients with glioma.
Cytoskeletal-associated protein 2 (CKAP2), which is
also known as tumor-associated microtubule-associated protein
(TMAP), is localized on microtubule organizing centers and
microtubules, and has an important role in cell mitosis (11). Previous research has revealed that
CKAP2 is required for appropriate chromosome segregation and the
maintenance of genomic stability (12,13).
CKAP2 was recently reported to be a substrate of cyclin-dependent
kinase (CDK)1-cyclin B and was indicated to be essential for
bipolar spindle formation (14).
The oncogenic nature of CKAP2 in the occurrence and development of
gastric (15), ovarian (16) and breast cancer (17), and other malignant tumors (18–21)
has been elaborated. However, limited studies have assessed the
biological characteristics and significance of CKAP2 in HGG.
The present study investigated the expression of
CKAP2, and its association with the clinical features of glioma,
using patient information from the Chinese Glioma Genome Atlas
(CGGA) microarray database. The results demonstrated that the
expression levels of CKAP2 were higher in patients with HGG
compared with in patients with LGG. Subsequently, the patients with
HGG were divided into two subgroups (cut off at 50% of the entire
group) according to their CKAP2 expression levels; the survival
analysis indicated that patients with HGG and lower CKAP2
expression levels exhibited a better prognosis. Subsequent studies
using The Cancer Genome Atlas (TCGA) RNA sequencing database
further validated these findings. Furthermore, univariate and
multivariate Cox regression analyses demonstrated that high CKAP2
expression appeared to be a predictor for poor clinical outcomes in
patients with HGG. Subsequently, CKAP2 positively correlated genes
were analyzed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID) and gene set enrichment analysis
(GSEA). In addition, Gene Ontology (GO) analysis indicated
significant enrichment of the genes in the cell cycle, mitotic and
cell proliferation pathways. In conclusion, the present study
demonstrated that CKAP2 may be associated with glioma malignancy
and could promote malignant progression by stimulating glioma cell
proliferation. These characteristics of CKAP2 suggest that it may
serve as a potential prognostic factor and a candidate gene for
gene therapy of malignant glioma.
Materials and methods
CGGA microarray databases
A total of 301 patients with histologically
confirmed glioma were enrolled in our study and their data was made
available in the GGA database. Microarray data generated using the
Agilent Whole Human Genome Array platform (Agilent Technologies,
Santa Clara, CA, USA) (22) and
follow-up information were obtained from all 301 patients.
Following surgical resection, the histological diagnosis was
performed independently by two experienced neuropathologists
according to the 2016 WHO classification (3). During the study, none of the patients
succumbed to other diseases or accidents. Written informed consent
was obtained from all patients and the present study was approved
by the ethics committee of Beijing Tiantan Hospital (Beijing,
China). Details regarding the establishment and management of our
CGGA database have been specified in our previous publication
(23).
CKAP2 expression analysis in
databases
A total of 301 glioma samples (122 grade II, 51
grade III and 128 grade IV samples) were obtained from the CGGA
microarray database (http://www.cgga.org.cn/index.php?m=Page&a=index&id=42)
as the discovery set. Data from TCGA RNA sequencing database
(http://cancergenome.nih.gov; https://portal.gdc.cancer.gov/projects/TCGA-GBM and
http://portal.gdc.cancer.gov/projects/TCGA-LGG), which
contains 633 glioma samples (223 grade II, 242 grade III and 168
grade IV samples), were obtained as validation sets. TCGA RNA
sequencing data were log2-transformed. In the two databases, only
samples with definite WHO classification were included for
expression analysis. Patients who were not clearly classified were
excluded.
Cell transfection
Human astrocytes (HA; cat. no. 1800; ScienCell
Research Laboratories, Inc., San Diego, CA, USA) cell line and
human glioma cell lines H4, U-87MG ATCC (U87), LN229, U-118MG
(U118) and U-251MG (U251) were purchased from the Institute of
Biochemistry and Cell Biology, Chinese Academy of Sciences
(Beijing, China). The CGGA-N33 cell line is a patient-derived GBM
cell line established by the CGGA. Astrocyte Medium (cat. no. 1801;
ScienCell Research Laboratories, Inc.) and Dulbecco's modified
Eagle's medium (DMEM)/F12 (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) were used to culture HA and
CGGA-N33 cell lines, respectively. Human glioma cell lines were
cultured in DMEM supplemented with 10% FBS. All cell lines were
maintained in an incubator at 37°C in an atmosphere containing 5%
CO2. The following CKAP2 small interfering (si)RNA
sequences were used in the present study: siRNA-1,
5′-GCACTACATCTCAGAACAC-3′; siRNA-2, 5′-GGACTACCATGGCAGAAGA-3′ and
siRNA-3, 5′-GAGACGTTCTCGACGTCTT-3′. CKAP2 siRNA and negative
control (NC; cat. no. siN05815122147) siRNA were synthesized by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Once cell density
reached 30–50%, U87 and CGGA-N33 cell lines were transfected with
siRNA or NC (50 nM) at 37°C using the riboFECT CP Transfection kit
(cat. no. C10511-1; Guangzhou RiboBio Co., Ltd.). After 48 h at
37°C, fresh medium without siRNA was added to the cells.
Western blot analysis
Total proteins were extracted using
radioimmunoprecipitation assay buffer (Cell Signaling Technology,
Inc., Danvers, MA, USA) supplemented with phenylmethylsulfonyl
fluoride (1 mM; Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China). Pierce Bicinchoninic Acid Protein Assay kit
(Thermo Fisher Scientific, Inc.) was used to quantify protein
concentration. The proteins were heated at 95–100°C with SDS-PAGE
loading buffer (Beijing Solarbio Science & Technology Co.,
Ltd.). Equal amounts of total protein (20 µg) were separated by 10%
SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD
Millipore, Billerica, MA, USA). The membrane was then blocked with
5% skim milk (BD Biosciences, Franklin Lakes, NJ, USA) at room
temperature for 1 h. Subsequently, the membrane was incubated with
primary antibodies at 4°C overnight and with secondary antibodies
at room temperature for 1 h. The results were detected using an
Enhanced Chemiluminescence Western Blotting Detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). In the present
study, western blot analysis was performed using rabbit anti-CKAP2
polyclonal antibody (1:800; cat. no. 25486-1-AP; Wuhan Sanying
Biotechnology, Wuhan, China) and β-tubulin (1:5,000; cat. no.
CW0098M; CWBIO, Beijing, China) was used as the loading control.
Goat anti-rabbit and goat anti-mouse immunoglobulin G-horseradish
peroxidase (1:5,000; cat. nos. ZB-2301 and ZB-2305; OriGene
Technologies, Inc., Beijing, China,) were used as secondary
antibodies to detect CKAP2 and β-tubulin, respectively.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Post-transfection with CKAP2 and NC siRNA, total RNA
was extracted from U87 and CGGA-N33 glioma cells using RNAprep Pure
kit for cell/bacteria (Tiangen Biotech Co., Ltd., Beijing, China).
RNA was stored at −80°C and was then reverse transcribed using the
RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc), according to the manufacturer's protocol. To
detect alterations in the relative mRNA expression levels of CKAP2
post-transfection with specific siRNA, RT-qPCR was performed using
the SYBR SuperMix kit (Bio-Rad Laboratories, Inc.) and the 7500
Fast Real-Time PCR system (Applied Biosystems, USA), according to
the manufacturer's protocols. The thermocycling conditions were
conducted according to the manufacturer's protocol, as follows: i)
Uracil DNA glycosylase activation at 50°C for 2 min; ii) 95°C for 2
min; iii) 40 cycles at 95°C for 15 sec and 60°C for 1 min.
Fluorescence measurements were taken at each cycle. The relative
mRNA expression levels of CKAP2 were normalized to GAPDH and were
calculated using the 2−ΔΔCq method (24). The primer sequences of CKAP2 and
GAPDH endogenous control were purchased from GENEWIZ (Beijing,
China). The primer sequences were as follows: CKAP2, forward
5′-GCAAGATGCTAACATGCCCAA-3′, and reverse
5′-TGGCTTTAGGTATAGTGGCTGA-3′; and GAPDH, forward
5′-GGAGCGAGATCCCTCCAAAAT-3′, and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Clonogenic assay
U87 and CGGA-N33 glioma cells in culture plates were
digested with 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific,
Inc.) and dissociated. Subsequently, cells were seeded into 12-well
culture plates (200 cells/well). Following adherence, cells were
transfected with CKAP2 or NC siRNA and the medium was replaced
after 12 h. Plates were incubated at 37°C in an atmosphere
containing 5% CO2 for 12 days before the cell colonies
were stained with 0.2% crystal violet at room temperature for 5
min. An inverted fluorescence microscope (Carl Zeiss AG,
Oberkochen, Germany) was used for in vitro analysis.
Bioinformatics analysis
Pearson's correlation analysis was performed using R
language 3.2.5 (https://cran.r-project.org/bin/windows/base/old/3.2.5/)
to determine the correlation between CKAP2 and other genes in the
CGGA microarray database and TCGA RNA sequencing database.
Positively correlated CKAP2 genes (r>0.4, P<0.01) were
analyzed using DAVID (http://david.abcc.ncifcrf.gov/home.jsp) and GSEA to
detect the association between biological processes and CKAP2
expression in glioma. GO and Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analyses, conducted using DAVID, were
the main methods of bioinformatics analysis used in the present
study. Gene set variation analysis (GSVA) with CKAP2 was analyzed
using a GSVA package of R (25).
The list of CKAP2-related functions and pathways in GSVA were
obtained from the GO database website (http://amigo.geneontology.org/amigo). Protein-protein
interactions of CKAP2 were analyzed using the STRING v10.0 online
tool (https://string-db.org/).
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(SPSS, Inc., Chicago, IL, USA), R language 3.2.5 and GraphPad Prism
7.0 statistical software (GraphPad Software, Inc., La Jolla, CA,
USA). Student's t-test was used to compare the difference in
expression levels between patients with HGG and LGG. One-way
analysis of variance followed by Holm-Sidak test was used to
compare the difference in CKAP2 expression between various
histological grades and molecular subtypes of glioma; the RT-qPCR
results were also analyzed in this manner. Kaplan-Meier (K-M)
survival analysis with long-rank test was used to assess the
predictive value of CKAP2 expression for overall survival (OS) in
the two databases and progression-free survival (PFS) in the CGGA
microarray database between various grades of gliomas. In the K-M
survival analysis, the patients with HGG were divided into two
subgroups (cut off at 50% of the entire group) according to the
expression levels of CKAP2. OS was calculated from the data
obtained from histological diagnosis until mortality, whereas PFS
was calculated until tumor recurrence, which was diagnosed by
magnetic resonance imaging. Univariate and multivariate Cox
regression analyses, including age at diagnosis, gender, IDH1
mutation status and CKAP2 expression were used to assess the
prognostic value of CKAP2 in HGG. A two-sided P<0.05 was
considered to indicate a statistically significant difference.
Results
Baseline clinical characteristics
The clinical and microarray data were obtained from
301 patients with glioma in the CGGA microarray database, including
122 patients with LGG (WHO grade II) and 179 patients with HGG (49
patients with WHO grade III glioma and 130 patients with GBM). The
baseline characteristics of all patients are summarized in Table I. In TCGA RNA sequencing database,
data from a total of 636 patients, including 223 patients with LGG
and 413 patients with HGG were obtained. The expression levels of
CKAP2 and the clinical characteristics of patients were used for
validation analysis.
 | Table I.Clinical characteristics of 301
patients with glioma in the Chinese Glioma Genome Atlas
database. |
Table I.
Clinical characteristics of 301
patients with glioma in the Chinese Glioma Genome Atlas
database.
| Variable | Low-grade
gliomaa | High-grade
glioma |
|---|
| Number | 122 | 179 |
| Gender,
male/female | 71/51 | 109/70 |
| Median age (range),
years | 37.5 (18–61) | 45 (13–70) |
| IDH1 wild-type | 36 | 129 |
| IDH1 mutation | 82 | 50 |
Expression of CKAP2 in gliomas of
different grades and subtypes
The present study screened for the differentially
expressed genes between LGG and HGG in the CGGA microarray database
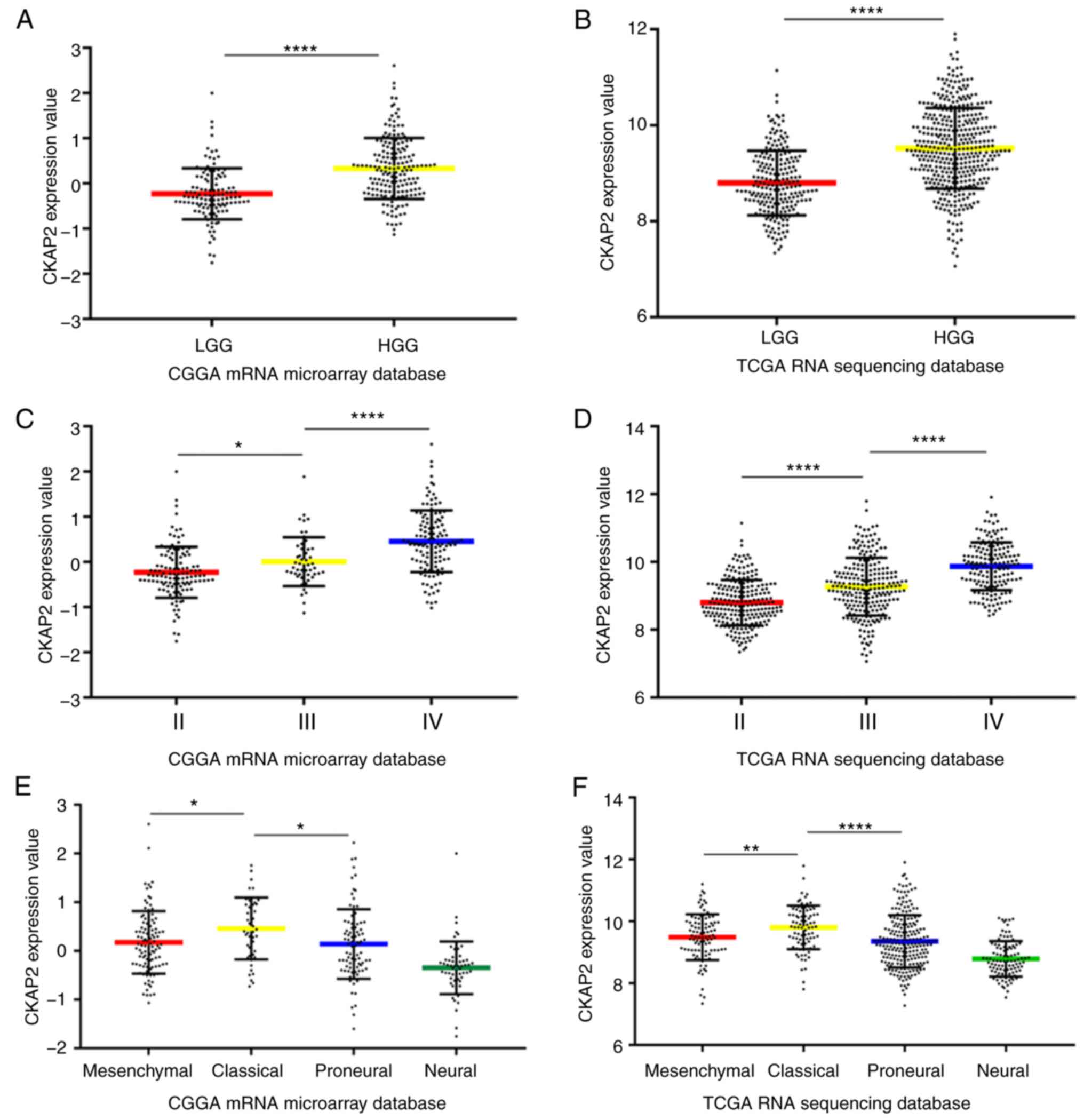
and demonstrated that CKAP2 was significant. Compared with in
patients with LGG, the mRNA expression levels of CKAP2 were
significantly higher in HGG patient samples (Fig. 1A). Subsequently, the mRNA expression
levels of CKAP2 were analyzed in patients with glioma in TCGA RNA
sequencing database. Similarly, CKAP2 mRNA expression was
significantly higher in patients with HGG than in patients with LGG
(Fig. 1B). Furthermore, after the
differential expression of CKAP2 was screened from all patients
from the CGGA microarray database and TCGA RNA sequencing database,
it was demonstrated that CKAP2 expression was positively associated
with tumor grade (Fig. 1C and D).
In the CGGA database, CKAP2 mRNA expression levels were
significantly different in four molecular subtypes of glioma, as
defined by a TCGA network. Compared with the other subtypes, the
classical subtype primarily exhibited the higher CKAP2 expression
(Fig. 1E); as validation, analysis
of TCGA database exhibited similar results (Fig. 1F). These results suggested that
CKAP2 may have the potential to serve as a biomarker for the
classical subtype of glioma.
Association between CKAP2 mRNA
expression and clinical outcomes
The association between CKAP2 mRNA expression and
the clinical outcomes of patients with HGG was investigated by K-M
survival curve analysis, using data from 301 patients with glioma
from the CGGA microarray database and 633 patients with glioma from
TCGA RNA sequencing database. Patients with HGG were divided into
two subgroups (cut off at 50% of the entire group) according to
CKAP2 expression levels. K-M analysis indicated that there was a
significant difference in PFS (P<0.001, log-rank test; Fig. 2A) and OS (P<0.001, log-rank test;
Fig. 2B) between high and low CKAP2
expression groups in the CGGA microarray database. Patients with
HGG and high CKAP2 mRNA expression were observed to have a worse
prognosis compared with those with low CKAP2 expression. In TCGA
RNA sequencing database, the patients with HGG and high CKAP2
expression also exhibited worse OS compared with those with low
CKAP2 expression (P=0.023, log-rank test; Fig. 2C). Conversely, neither PFS nor OS
were significantly altered in patients with LGG according to CKAP2
expression, as determined using the CGGA microarray database
(P=0.647 and 0.991, respectively, log-rank test; Fig. 2D and E) and TCGA RNA sequencing
database (P=0.963, log-rank test; Fig.
2F). Taken together, these results indicated that CKAP2
expression may serve as a prognostic factor for patients with HGG.
In consideration of the heterogeneity across different glioma
grades, the present study further investigated the prognostic value
of CKAP2 expression in patients with GBM from the CGGA database
(P=0.015 and 0.01, OS and PFS, respectively, log-rank test;
Fig. 2G and H) and observed a
similar pattern according to the K-M curves.
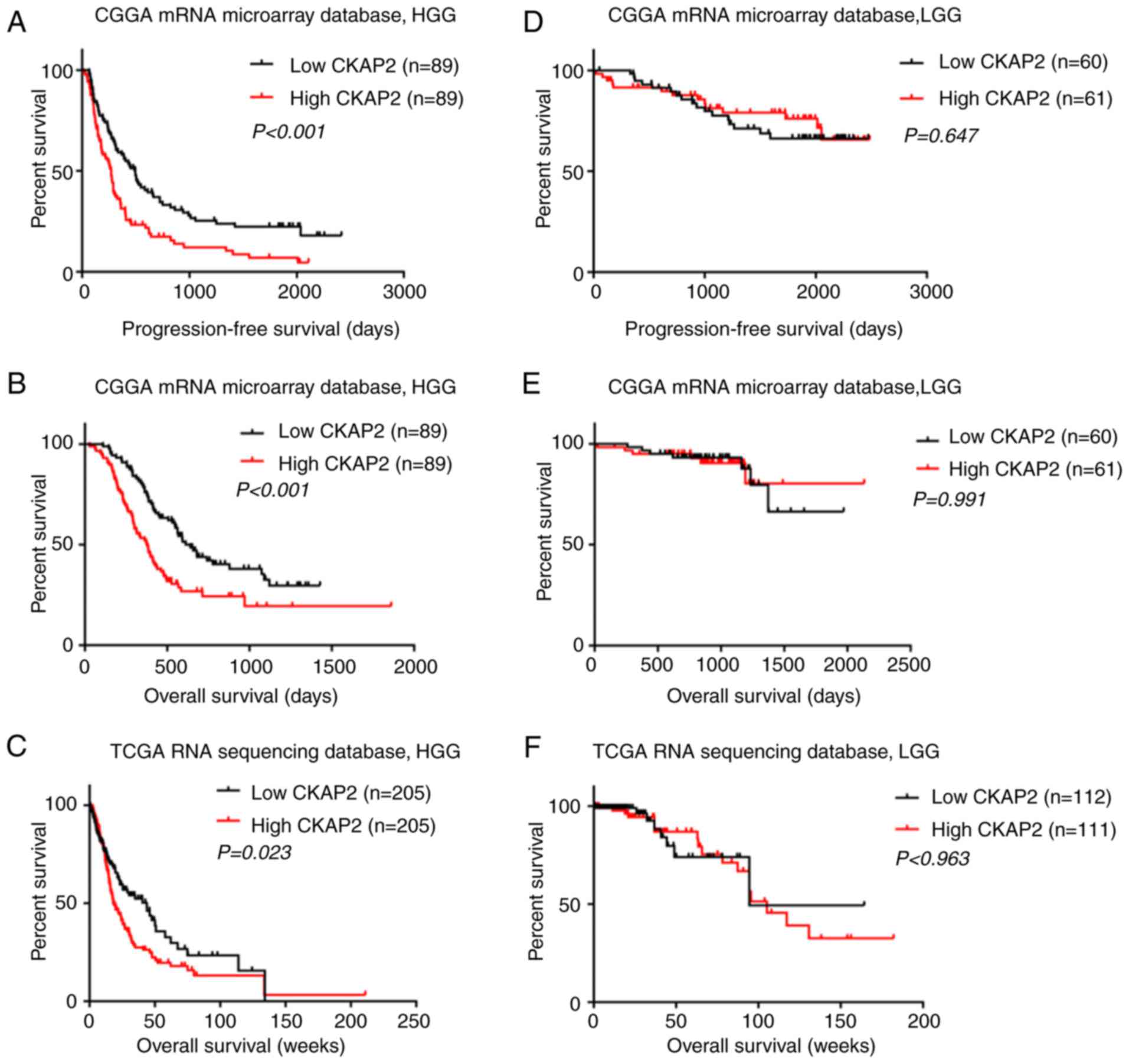 | Figure 2.High CKAP2 expression is associated
with poor prognosis in patients with HGG. (A and B) PFS and OS of
patients with HGG from the CGGA database separated into high and
low CKAP2 expression groups. (C) OS of patients with HGG from TCGA
database separated into high and low CKAP2 expression groups. (D
and E) PFS and OS of patients with LGG from the CGGA database
separated into CKAP2 high and low expression groups. (F) OS of
patients with LGG from TCGA database separated into CKAP2 high and
low expression groups. (G and H) OS and PFS of patients with GBM
from the CGGA database separated into high and low CKAP2 expression
groups. (I) Correlation of CKAP2 expression with well-known genomic
alterations in glioma. Ampli, amplification; CGGA, Chinese Glioma
Genome Atlas; CKAP2, cytoskeletal-associated protein 2; expre,
expression; H, high; HGG, high-grade glioma; L, low; LGG, low-grade
glioma; mut, mutation; OS, overall survival; PFS, progression-free
survival; TCGA, The Cancer Genome Atlas. |
After exploring CKAP2-associated genomic
alterations, an overview of the associations between CKAP2
expression and well-known genomic or transcriptional alterations in
glioma was obtained (Fig. 2I).
According to these results, the incidence of the biomarkers that
indicate a poor prognosis, including high Ki67 expression and
phosphatase and tensin homolog mutations, were higher in patients
with glioma and higher CKAP2 expression. These findings further
confirmed that CKAP2 expression was associated with poor clinical
outcomes.
CKAP2 is a novel independent
prognostic biomarker for patients with HGG
Univariate Cox regression analysis was performed on
data obtained from the CGGA database to further determine the value
of CKAP2 and other variables in predicting OS and PFS of patients
with HGG (Tables II and III). The results demonstrated that high
CKAP2 expression was a risk factor for patients with HGG (P=0.003
for OS and P=0.001 for PFS). Furthermore, other factors, including
age and IDH1 mutation state, were also significantly associated
with the OS and PFS of patients with HGG. A multivariate Cox
regression analysis was performed incorporating CKAP2 expression,
age, WHO grade, Karnofsky Performance Status score, MGMT
methylation state, IDH1 mutation state, chemotherapy and
radiotherapy (Tables II and
III). The results revealed that
CKAP2 was an independent predictive factor for the OS and PFS of
patients with HGG (P=0.039 for OS and P=0.007 for PFS). In
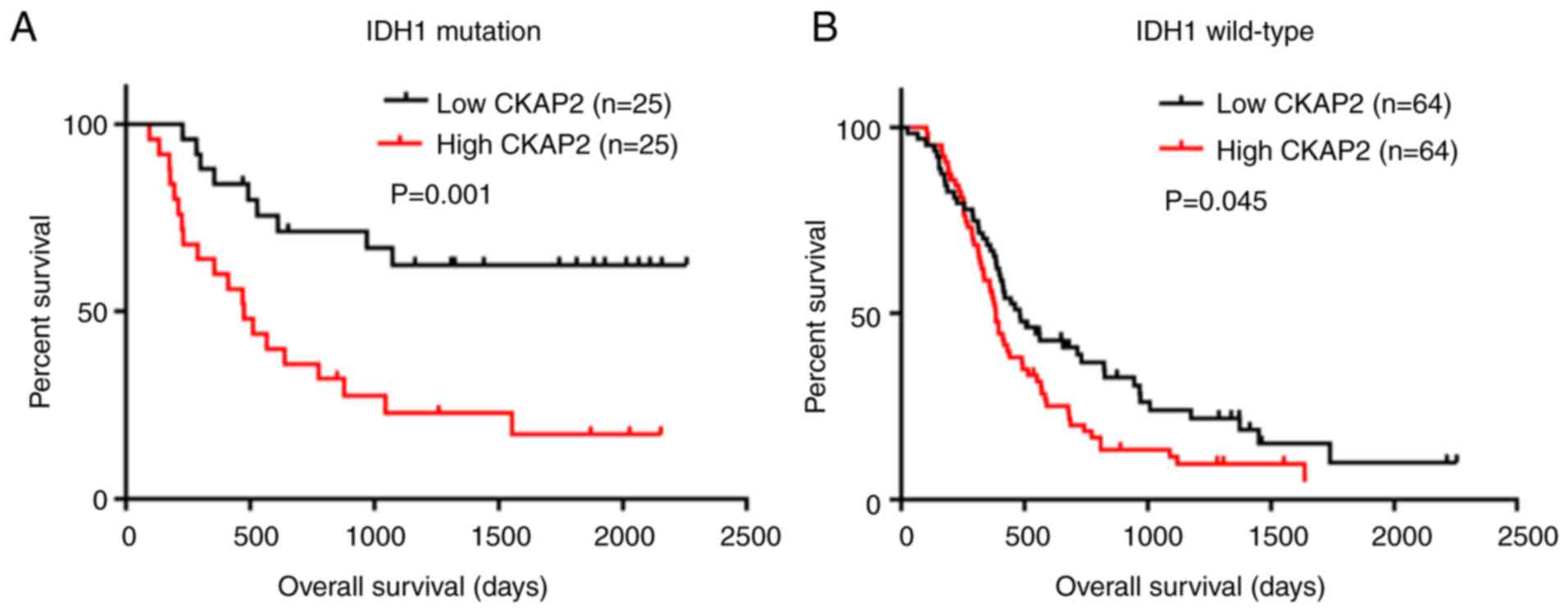
addition, the results of K-M survival curve analysis indicated that
high expression levels of CKAP2 were closely associated with poor
prognosis in patients with HGG with or without IDH1 mutations
(P=0.001 for IDH1 mutation and P=0.045 for IDH1 wild-type, log-rank
test; Fig. 3A and B).
 | Table II.Univariate and multivariate analysis
of overall survival in the Chinese Glioma Genome Atlas microarray
database. |
Table II.
Univariate and multivariate analysis
of overall survival in the Chinese Glioma Genome Atlas microarray
database.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| CKAP2
expression | 1.530
(1.160–2.018) | 0.003 | 1.704
(1.027–2.827) | 0.039 |
| Age | 1.018
(1.002–1.034) | 0.031 | 0.987
(0.961–1.013) | 0.325 |
| Gender | 1.186
(0.813–1.732) | 0.376 |
|
|
| Grade | 2.266
(1.435–3.579) | <0.001 | 0.983
(0.425–2.274) | 0.968 |
| KPS | 0.968
(0.953–0.983) | <0.001 | 0.959
(0.942–0.976) | <0.001 |
| IDH1 mutation | 0.550
(0.353–0.855) | 0.008 | 0.539
(0.239–1.214) | 0.136 |
| MGMT
methylation | 0.572
(0.381–0.858) | 0.007 | 0.499
(0.246–1.012) | 0.054 |
| Chemotherapy | 0.449
(0.307–0.657) | <0.001 | 0.550
(0.292–1.037) | 0.065 |
| Radiotherapy | 0.414
(0.259–0.664) | <0.001 | 0.607
(0.282–1.308) | 0.203 |
 | Table III.Univariate and multivariate analysis
of progression-free survival in the Chinese Glioma Genome Atlas
microarray database. |
Table III.
Univariate and multivariate analysis
of progression-free survival in the Chinese Glioma Genome Atlas
microarray database.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| CKAP2
Expression | 1.486
(1.171–1.884) | 0.001 | 2.003
(1.207–3.324) | 0.007 |
| Age | 1.021
(1.007–1.035) | 0.003 | 0.990
(0.966–1.015) | 0.430 |
| Gender | 1.114
(0.798–1.555) | 0.526 |
|
|
| Grade | 1.927
(1.307–2.842) | 0.001 | 0.662
(0.300–1.461) | 0.307 |
| KPS | 0.978
(0.965–0.992) | 0.002 | 0.971
(0.965–0.987) | <0.001 |
| IDH1 mutation | 0.546
(0.370–0.807) | 0.002 | 0.583
(0.276–1.232) | 0.158 |
| MGMT
methylation | 0.617
(0.430–0.887) | 0.009 | 0.531
(0.270–1.044) | 0.067 |
| Chemotherapy | 0.448
(0.317–0.633) | <0.001 | 0.580
(0.323–1.042) | 0.068 |
| Radiotherapy | 0.600
(0.385–0.935) | 0.024 | 0.909
(0.437–1.891) | 0.799 |
CKAP2 is associated with the cell
cycle, mitosis and cell proliferation
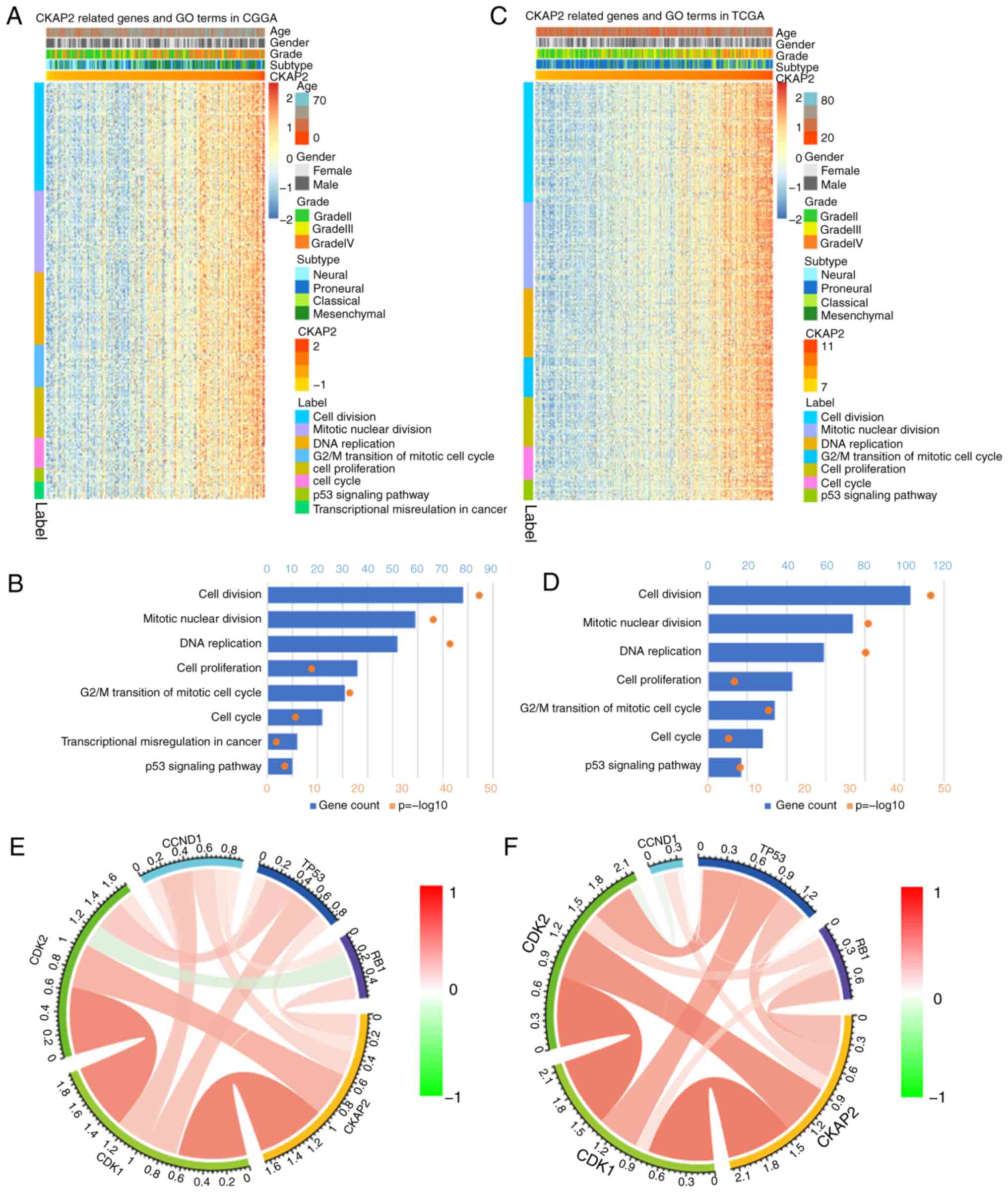
To identify the association between CKAP2 expression
and other genes in glioma, Pearson's correlation analysis was used
on data obtained from the CGGA and TCGA databases. The results
demonstrated that 560 genes in the CGGA database and 805 genes in
TCGA database were positively correlated with CKAP2 (r>0.4,
P<0.01). Subsequently, these genes were subjected to GO
analysis. The top GO terms indicated that CKAP2 was significantly
associated with gene sets associated with the cell cycle, mitotic
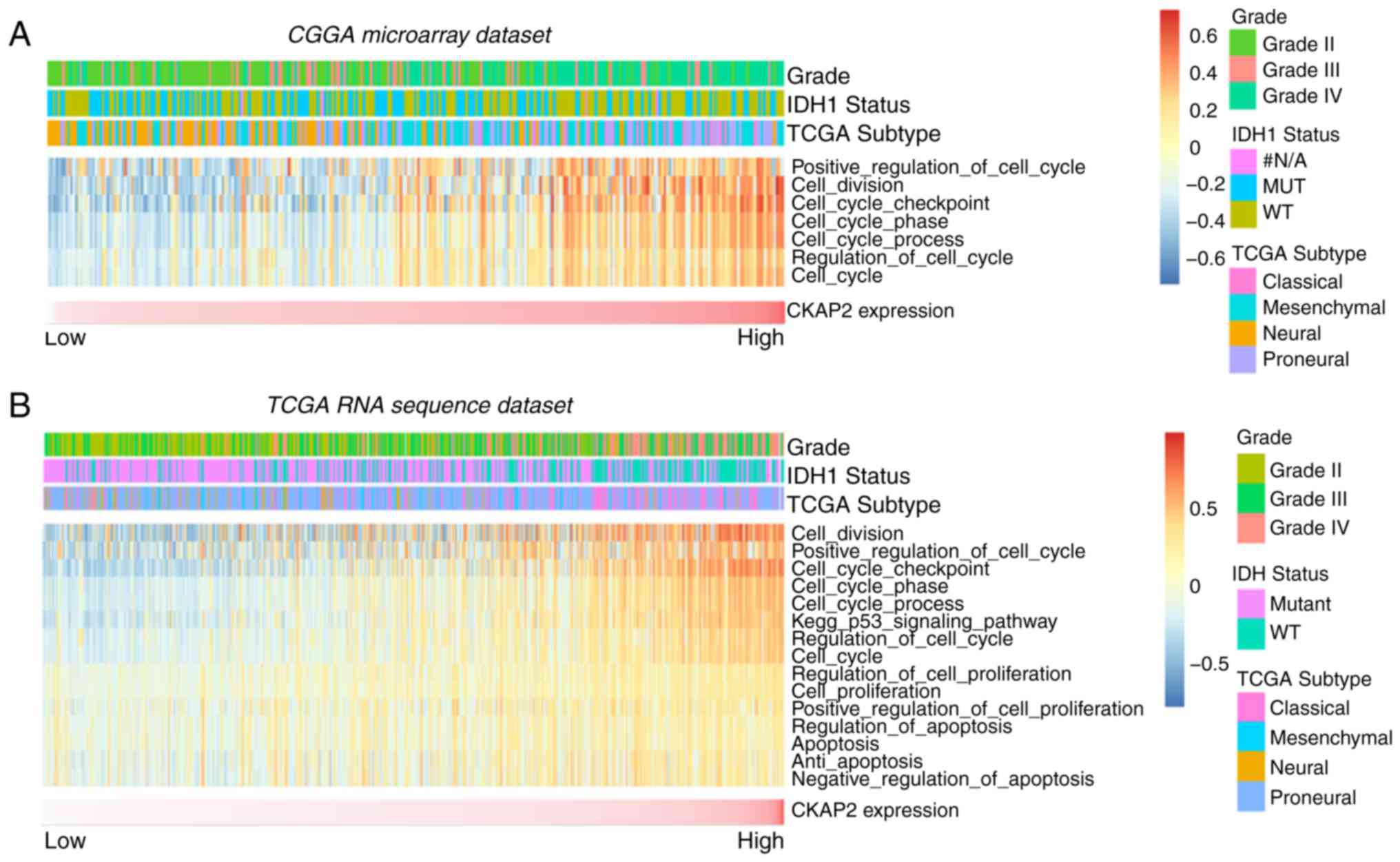
nuclear division and cell proliferation (Fig. 4A-D). Circos plots demonstrated that
CKAP2 expression was closely associated with CDK1 and CDK2
(Fig. 4E and F), both of which
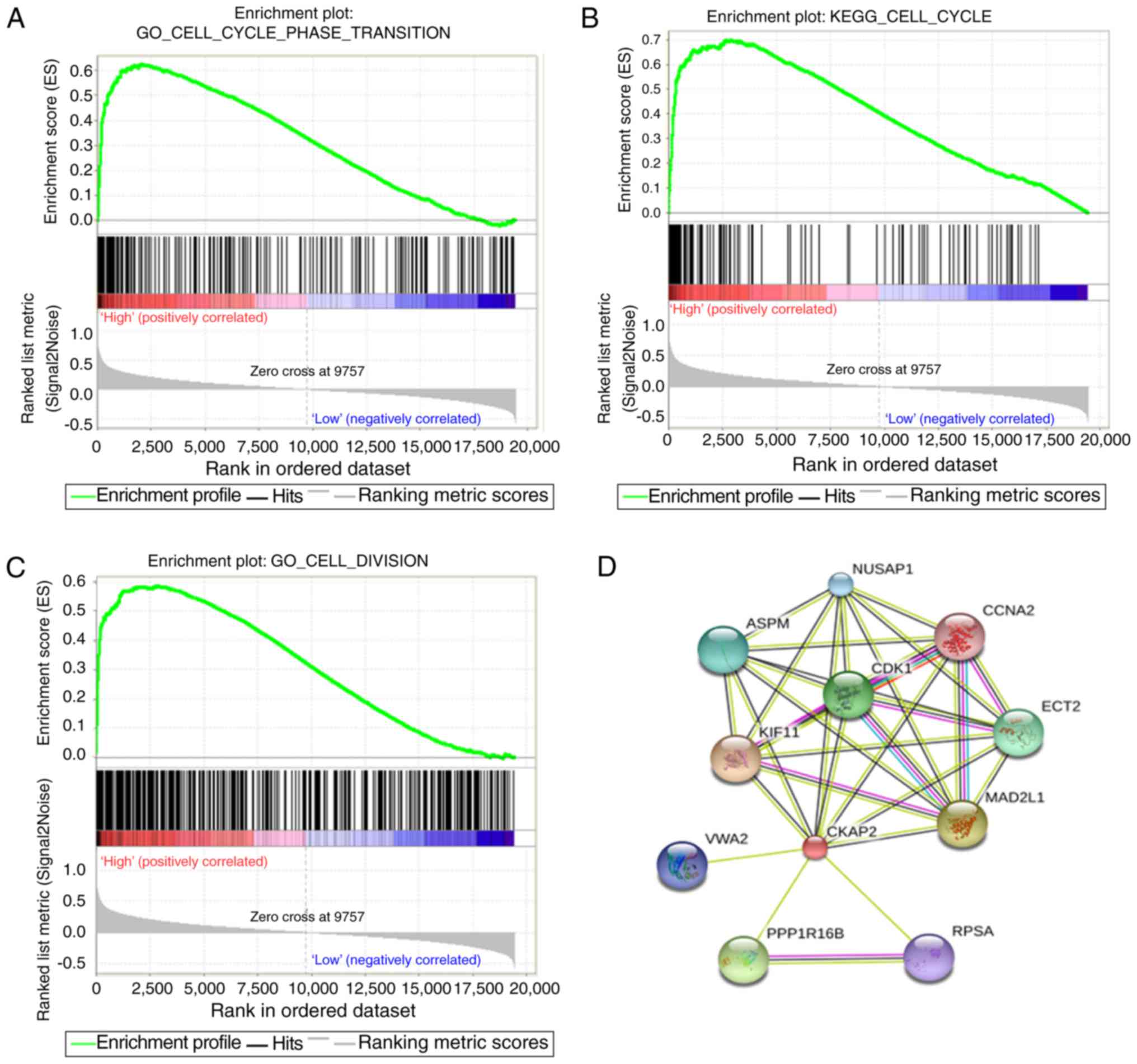
serve vital roles in the cell cycle and cell proliferation. GSEA
results were similar to those obtained from GO analysis (Fig. 5A-C). The protein-protein
interactions of CKAP2 analyzed by STRING indicated that CKAP2 was
closely associated with kinesin family member 11, CDK1 and mitotic
arrest deficient 2 like 1 (Fig.
5D). Notably these three genes have been identified to serve a
vital role in the cell cycle and cell mitosis (26–28).
Furthermore, by applying GSVA on CGGA and TCGA data, the previously
reported cell cycle-, mitotic- and cell proliferation-associated
genes also exhibited a higher enrichment score in the high-risk
group (Fig. 6A and B). These
analyses indicated that CKAP2 may have an essential role in the
cell cycle, mitosis and proliferation of glioma cells.
CKAP2 promotes the proliferation of
glioma cells in vitro
As indicated by GO analysis, the overexpression of
CKAP2 was positively associated with cell division and cell
proliferation. To further determine the functional roles of CKAP2
in glioma, a series of in vitro experiments were performed.
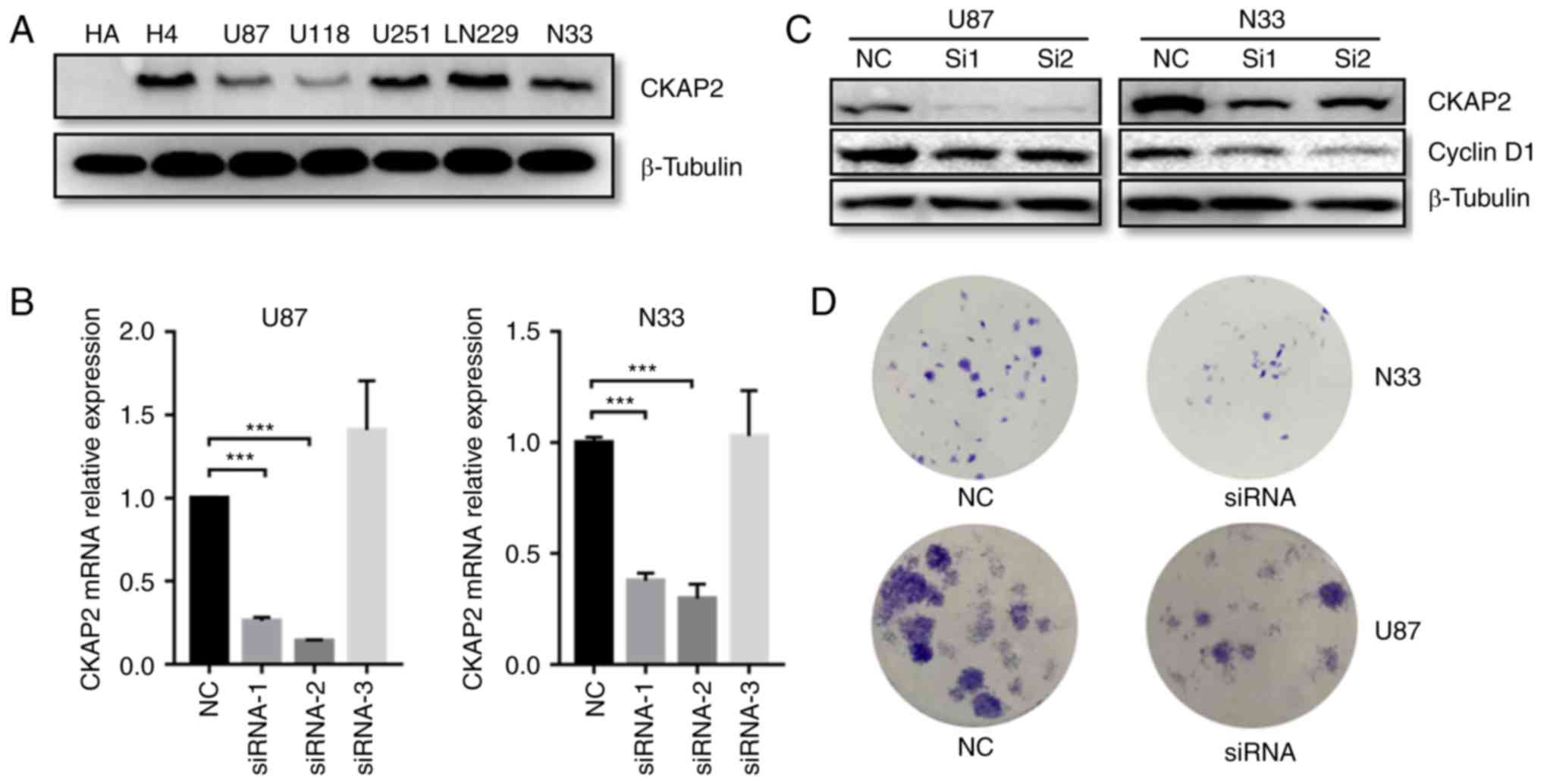
The protein expression levels of CKAP2 were initially detected in
H4, LN229, U87, U251, U118 and CGGA-N33 glioma cell lines, and the
HA cell line. Western blot analysis indicated that all glioma cells
exhibited a higher CKAP2 expression compared with HA (Fig. 7A). In addition, CKAP2 siRNA was
transfected to knockdown CKAP2 expression in U87 and CGGA-N33
cells. RT-qPCR demonstrated that the mRNA expression levels of
CKAP2 were significantly decreased in U87 and CGGA-N33 cells
post-transfection with siRNA-1 and siRNA-2 (Fig. 7B). Furthermore, western blot
analysis indicated that the protein expression levels of the cell
mitotic biomarker cyclin D1 were also significantly decreased in
siRNA-transfected U87 and CGGA-N33 cells (Fig. 7C). These findings indicated that
CKAP2 may have a significant impact on cell division. Additionally,
to evaluate the function of CKAP2 in glioma cell lines, a
clonogenic assay was performed. The results indicated that
silencing CKAP2 expression suppressed the proliferative capacity
and clonogenicity of glioma cells (Fig.
7D).
Discussion
HGG refers to WHO grade III and IV glioma (4). HGG is highly malignant and inevitably
recurs following surgical resection, due to its highly invasive
behavior; therefore, patients with HGG have a typically poor
outcome (29). Therefore, there is
an urgent requirement to understand the mechanism of occurrence and
development of this disease and to identify useful prognostic
biomarkers for the OS of patients with HGG. Tumor growth is a
complex and multistep process; during this process, the cell cycle,
mitosis and cell proliferation all have crucial roles and are
influenced by numerous regulators.
CKAP2, also known as TMAP, serves a vital role in
cell mitosis and cell death in a P53-dependent manner (30). Previous research has been performed
on various human tumors, including gastric cancer, breast cancer,
ovarian cancer and prostate cancer (15–21).
Notably, CKAP2 has been reported to be overexpressed in the
aforementioned cancer types and reduces the OS of patients by
various mechanisms. For example, gastric adenocarcinoma, which is a
poorly differentiated, highly malignant adenocarcinoma, possesses a
relatively high number of cytoplasmic CKAP2-positive cells
(15). In addition, in ovarian
cancer, the OS of patients with high levels of CKAP2 is shorter,
and CKAP2, along with butyrylcholinesterase, claudin 10 and
oviductal glycoprotein 1 mediates resistance to chemotherapy
(16). In a previous study of
early-stage breast cancer, high CKAP2 expression was revealed to
promote the proliferation of tumor cells and significantly shorten
relapse-free survival (17).
Additionally, overexpression of CKAP2 activates the focal adhesion
kinase (FAK)-extracellular signal-regulated kinase 2 signaling
pathway, and induces cell proliferation and migration; however,
this effect can be blocked by FAK inhibitors (18). In addition to CKAP2, other members
of the CKAP family have also demonstrated similar effects on tumor
cells, including CKAP4. Notably, after binding to Dickkopf 1, CKAP4
forms a complex with phosphoinositide 3-kinase (PI3K) and leads to
activation of the PI3K-protein kinase B signaling pathway and tumor
cell proliferation (31). These
previous findings suggested that CKAP2 may potentially have an
important role in the progression of cancer.
The expression levels and functions of CKAP2 in
glioma have not been fully elucidated. To the best of our
knowledge, the present study is the first to investigate CKAP2
expression in a large number of patients with glioma. The
retrospective study was performed to evaluate the expression of
CKAP2 and the clinical data of 934 patients with glioma in the CGGA
microarray database and TCGA RNA sequencing database. The results
demonstrated that the expression levels of CKAP2 were closely
associated with glioma grade. Furthermore, in both databases, the
OS of patients with HGG and high CKAP2 expression was shorter.
Additionally, analysis of the CGGA database indicated that high
CKAP2 expression shortened the PFS of patients with HGG. The
univariate and multivariate Cox regression analyses revealed that
high CKAP2 expression may be an independent predictor for poor
clinical outcomes in patients with HGG. Assessment of the
association between CKAP2 expression and biological processes
indicated that CKAP2 and its associated genes were enriched in the
cell cycle, mitosis and cell proliferation in patients with glioma.
The present study also demonstrated that CKAP2 may act as a
biomarker in glioma progression and has the potential as a novel
target for the treatment of HGG. However, further studies are
required to identify the potential mechanisms by which CKAP2
regulates glioma cell biological functions.
The in vitro experiments further confirmed
the key role of CKAP2 in the cell cycle and cell proliferation of
glioma. Notably, the U-87MG and U-118MG cell lines have been
reported to be contaminated, and are not the originally established
cell lines (32,33). However, U-87MG was confirmed to be
derived from GBM and is still one of the most commonly used cell
lines in glioma research (34–36).
The American Type Culture Collection website declares that U-118MG
and U-138MG are very similar cell lines, whereas they are not the
same (https://www.atcc.org/Products/All/HTB-15.aspx#characteristics).
Furthermore, since both U-118MG and U-138MG glioma cell lines are
derived from malignant gliomas, these misidentifications should not
change the interpretation of the present results.
In conclusion, the present study revealed that CKAP2
was closely associated with tumor grade and could serve as an
independent prognostic marker for OS and PFS in HGG. Furthermore,
the results indicated that CKAP2 may act as an oncogene that
promotes glioma tumor growth by activating the cell cycle, mitosis
and cell proliferation pathways. However, the association between
clinical features and CKAP2 expression was not fully elucidated. It
is therefore necessary to further study the expression pattern and
functional role of CKAP2 in the progression of glioma.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Nature
Science Foundation of China (grant nos. 81502495 and 81702460).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the CGGA repository (http://www.cgga.org.cn/index.php?m=Page&a=index&id=42).
Authors' contributions
TJ and HH designed the experiments. RH, GL and FZ
analyzed the data and contributed analytical tools. KW, ZZ and YL
performed the experiments. RH and KW wrote the paper. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Beijing
Tiantan Hospital institutional review board (Beijing, China) and
informed consent was obtained from all individual participants
included in the study.
Patient consent for publication
Consent was obtained from all individual
participants included in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jiang T, Mao Y, Ma W, Mao Q, You Y, Yang
X, Jiang C, Kang C, Li X, Chen L, et al: CGCG clinical practice
guidelines for the management of adult diffuse gliomas. Cancer
Lett. 375:263–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Brada M, van den Bent MJ, Tonn JC
and Pentheroudakis G: ESMO Guidelines Working Group: High-grade
glioma: ESMO clinical practice guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Demuth T and Berens ME: Molecular
mechanisms of glioma cell migration and invasion. J Neurooncol.
70:217–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Eng J
Med. 360:765–773. 2009. View Article : Google Scholar
|
|
7
|
Mellai M, Annovazzi L, Senetta R,
Dell'Aglio C, Mazzucco M, Cassoni P and Schiffer D: Diagnostic
revision of 206 adult gliomas (including 40 oligoastrocytomas)
based on ATRX, IDH1/2 and 1p/19q status. J Neurooncol. 131:213–222.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brandes AA, Franceschi E, Tosoni A,
Benevento F, Scopece L, Mazzocchi V, Bacci A, Agati R, Calbucci F
and Ermani M: Temozolomide concomitant and adjuvant to radiotherapy
in elderly patients with glioblastoma: Correlation with MGMT
promoter methylation status. Cancer. 115:3512–3518. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Su HK, Zhao HF, Chen ZP and To SS:
Progress in the application of molecular biomarkers in gliomas.
Biochem Biophys Res Commun. 465:1–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huse JT and Aldape KD: The evolving role
of molecular markers in the diagnosis and management of diffuse
glioma. Clin Cancer Res. 20:5601–5611. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hong KU, Choi YB, Lee JH, Kim HJ, Kwon HR,
Seong YS, Kim HT, Park J, Bae CD and Hong KM: Transient
phosphorylation of tumor associated microtubule associated protein
(TMAP)/cytoskeleton associated protein 2 (CKAP2) at Thr-596 during
early phases of mitosis. Exp Mol Med. 40:377–386. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Case CM, Sackett DL, Wangsa D, Karpova T,
McNally JG, Ried T and Camps J: CKAP2 ensures chromosomal stability
by maintaining the integrity of microtubule nucleation sites. PLoS
One. 8:e645752013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hong KU, Kim E, Bae CD and Park J:
TMAP/CKAP2 is essential for proper chromosome segregation. Cell
Cycle. 8:314–324. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoo BH, Kang DS, Park CH, Kang K and Bae
CD: CKAP2 phosphorylation by CDK1/cyclinB1 is crucial for
maintaining centrosome integrity. Exp Mol Med. 49:e3542017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim YW, Eom BW, Kook MC, Kim HS, Kim MK,
Hwang HL, Chandra V, Poojan S, Song Y, Koh JS, et al: Clinical
implications of proliferation activity in T1 or T2 male gastric
cancer patients. Exp Mol Med. 47:e1932015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao Y, Liu X, Li T, Wei L, Yang A, Lu Y,
Zhang J, Li L, Wang S and Yin F: Cross-validation of genes
potentially associated with overall survival and drug resistance in
ovarian cancer. Oncol Rep. 37:3084–3092. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim HS, Koh JS, Choi YB, Ro J, Kim HK, Kim
MK, Nam BH, Kim KT, Chandra V, Seol HS, et al: Chromatin CKAP2, a
new proliferation marker, as independent prognostic indicator in
breast cancer. PLoS One. 9:e981602014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo QS, Song Y, Hua KQ and Gao SJ:
Involvement of FAK-ERK2 signaling pathway in CKAP2-induced
proliferation and motility in cervical carcinoma cell lines. Sci
Rep. 7:21172017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Viticchie G, Lena AM, Latina A, Formosa A,
Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli
LG, et al: MiR-203 controls proliferation, migration and invasive
potential of prostate cancer cell lines. Cell Cycle. 10:1121–1131.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Lee YC, Cheng CJ, Wu CF, Song JH,
Gallick GE, Yu-Lee LY, Kuang J and Lin SH: RSK promotes prostate
cancer progression in bone through ING3, CKAP2, and PTK6-mediated
cell survival. Mol Cancer Res. 13:348–357. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hayashi T, Ohtsuka M, Okamura D, Seki N,
Kimura F, Shimizu H, Yoshidome H, Kato A, Yoshitomi H, Furukawa K,
et al: Cytoskeleton-associated protein 2 is a potential predictive
marker for risk of early and extensive recurrence of hepatocellular
carcinoma after operative resection. Surgery. 155:114–123. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan W, Zhang W, You G, Zhang J, Han L, Bao
Z, Wang Y, Liu Y, Jiang C, Kang C, et al: Molecular classification
of gliomas based on whole genome gene expression: A systematic
report of 225 samples from the Chinese Glioma Cooperative Group.
Neuro Oncol. 14:1432–1440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K,
Ye WL, Hu BQ, Yan W, Zhang W, Akers J, et al: RNA-seq of 272
gliomas revealed a novel, recurrent PTPRZ1-MET fusion
transcript in secondary glioblastomas. Genome Res. 24:1765–1773.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Asbaghi Y, Thompson LL, Lichtensztejn Z
and McManus KJ: KIF11 silencing and inhibition induces
chromosome instability that may contribute to cancer. Genes
Chromosomes Cancer. 56:668–680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parrilla A, Cirillo L, Thomas Y, Gotta M,
Pintard L and Santamaria A: Mitotic entry: The interplay between
Cdk1, Plk1 and Bora. Cell Cycle. 15:3177–3182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Carli E, Wang X and Puget S:
IDH1 and IDH2 mutations in gliomas. N Eng J Med.
360:2248author reply 2249. 2009. View Article : Google Scholar
|
|
29
|
DeAngelis LM: Brain tumors. N Eng J Med.
344:114–123. 2001. View Article : Google Scholar
|
|
30
|
Tsuchihara K, Lapin V, Bakal C, Okada H,
Brown L, Hirota-Tsuchihara M, Zaugg K, Ho A, Itie-Youten A,
Harris-Brandts M, et al: Ckap2 regulates aneuploidy, cell cycling,
and cell death in a p53-dependent manner. Cancer Res. 65:6685–6691.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kikuchi A, Fumoto K and Kimura H: The
Dickkopf1-cytoskeleton-associated protein 4 axis creates a novel
signalling pathway and may represent a molecular target for cancer
therapy. Br J Pharmacol. 174:4651–4665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Capes-Davis A, Theodosopoulos G, Atkin I,
Drexler HG, Kohara A, MacLeod RAF, Masters JR, Nakamura Y, Reid YA,
Reddel RR and Freshney RI: Check your cultures! A list of
cross-contaminated or misidentified cell lines. Int J Cancer.
127:1–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re3532016. View Article : Google Scholar
|
|
34
|
Li H, Chen L, Li JJ, Zhou Q, Huang A, Liu
WW, Wang K, Gao L, Qi S and Lu YT: miR-519a enhances
chemosensitivity and promotes autophagy in glioblastoma by
targeting STAT3/Bcl2 signaling pathway. J Hematol Oncol. 11:702018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jantas D, Grygier B, Golda S, Chwastek J,
Zatorska J and Tertil M: An endogenous and ectopic expression of
metabotropic glutamate receptor 8 (mGluR8) inhibits proliferation
and increases chemosensitivity of human neuroblastoma and glioma
cells. Cancer Lett. 432:1–16. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo B, Sheng Z, Hu D, Li A, Xu S,
Manghnani PN, Liu C, Guo L, Zheng H and Liu B: Molecular
engineering of conjugated polymers for biocompatible organic
nanoparticles with highly efficient photoacoustic and photothermal
performance in cancer theranostics. ACS Nano. 11:10124–10134. 2017.
View Article : Google Scholar : PubMed/NCBI
|