Introduction
Gastric cancer (GC) is the second most frequent
cause of cancer-related deaths worldwide (1). Several studies revealed that a
positive family history of having a first-degree relative with GC
is considered a strong risk factor for the development of GC,
particularly when two or more relatives are affected (2). There is familial aggregation in
~10–20% of GCs and ~1–3% have a clear inherited genetic
conditioning (3). A good
understanding of the genetic mechanism of GC in the family may shed
light on the driving genes and pathways for treatment options and
genetic counselling. However, the genetic events that predispose
individuals to GC have not been clearly understood.
Three hereditary GC syndromes have been described
which are the following: Hereditary diffuse gastric cancer (HDGC),
familial intestinal gastric cancer (FIGC) and the recently proposed
gastric adenocarcinoma and proximal polyposis of the stomach
(GAPPS) (4). Some other hereditary
cancer syndromes such as hereditary non-polyposis colorectal cancer
(HNPCC), Li-Fraumeni syndrome (LFS), familial adenomatous polyposis
(FAP) and Peutz-Jeghers syndrome (PJS) also predispose individuals
to GC (5). Genetic and epigenetic
alterations play key roles in the pathogenesis of familial GC
development (6). Except for HDGC,
the molecular basis for the familial aggregation remains largely
unknown. Identification of new predisposition genes would provide
novel insights regarding the molecular pathogenesis of GC. However,
the pathogenesis and genetic changes of FIGC have not been clearly
elucidated.
Whole-exome sequencing has been widely used to
identify the genomic mutation signatures for uncovering the
predisposing genes in familial cancers (7,8). In
the present study, we explored three genomic variations (ESR1,
IGF2R and EZR) in FIGC by whole-exome sequencing. The oestrogen
receptor α/oestrogen receptor 1 (ERα/ESR1) gene, a well-known
proto-oncogene, is a member of the nuclear hormone receptor family
and plays an important role in hormone binding, DNA binding and
activation of transcription (9).
The mannose-6-phosphate/insulin-like growth factor 2 receptor
(M6P/IGF2R), referred to as IGF2R, is a multifunctional protein
ubiquitously expressed in human tissues and has been recently
identified as a tumour suppressor (10). Ezrin, encoded by the EZR gene, is a
signal transduction component belonging to the ezrin-radixin-moesin
(ERM) protein family; it acts both as a link between the actin
cytoskeleton and plasma membrane proteins and as a substrate for
tyrosine kinase (11).
The detection of these mutations, which appear to
predispose individuals to familial GC, could lead to the
identification of individuals with a risk of familial GC in
affected families and may be useful as biomarkers for confirmatory
diagnosis of FIGC and appropriate treatment, providing new insights
into tumour initiation and the progression of FIGC. Screening for
these genotypes combined with information on the familial
background may help us to identify individuals who are at increased
risk of FIGC.
Materials and methods
Patients
The present study was approved by the Ethics
Committee of the Third Affiliated Hospital of Nanjing University of
Chinese Medicine. All study procedures were performed according to
the Declaration of Helsinki ethical principles. Informed consent
was obtained from all participating patients.
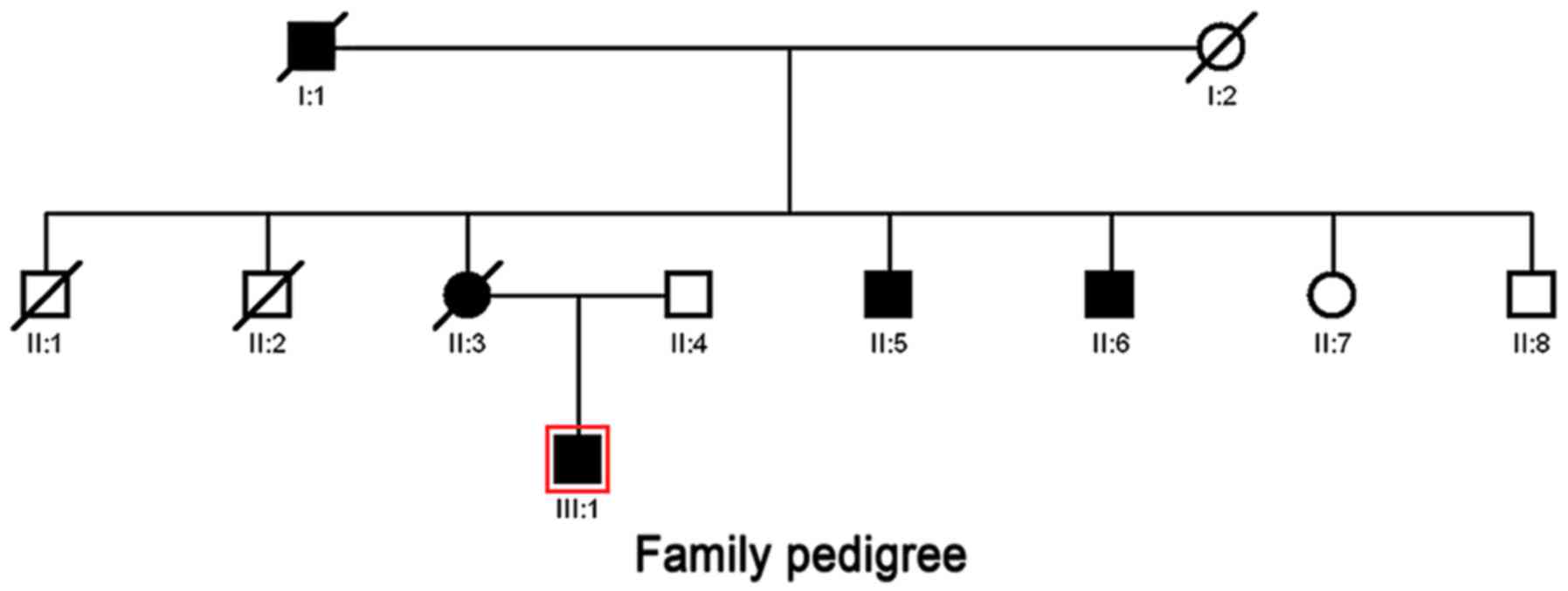
The proband was a 37 year-old man diagnosed with
severe atrophic gastritis in 2014 by gastroscope inspection
(Fig. 1). The probands mother
suffered from chronic atrophic gastritis and died of FIGC at the
age of 63 years in 2014 in our hospital. His maternal grandfather
and grandmother died of GC and lung cancer, respectively. The two
elder brothers of his mother were dead from lung cancer and
oesophagus cancer. The two younger brothers of his mother presented
with chronic atrophic gastritis. His father had no family history
of digestive tract diseases and was considered the normal control.
The relations between individuals are illustrated in the family
pedigree (Fig. 1).
Sample collection and whole-exome
sequencing
A total of 5 ml of peripheral whole blood was
collected from the proband and the family members listed in the
family pedigree (Fig. 1). The blood
samples were collected and stored at −20°C before use. The genomic
DNA was extracted using a DNA extraction kit (Youcheng Biological
Pharmaceutical Technology, Co. Ltd., Jiangsu, China), following the
manufacturers instructions. The library preparation, whole-exome
capture and sequencing were performed at Shanghai GeneChem, Co.,
Ltd. (Shanghai, China). Sequencing was analysed based on the
Illumina PE150 platform (Shanghai Jeayea Biotech Co., Ltd.,
Shanghai, China).
The mean coverage of study samples was ×100. The
variant calling files were created by BCFtools and SAMtools
(http://samtools.sourceforge.net). All
variant annotations were performed using Variant Effect Predictor
(VEP) based on the Ensemble database (http://asia.ensembl.org/info/docs/tools/vep/script/index.html).
Whole-genome analysis
Genes harbouring exonic and/or splice site
variations were filtered and stratified to single-nucleotide
variant (SNV) and insertion-deletion (INDEL) genes in each sample.
Subsequently, the variant genes specific to patient samples were
selected, which had different calls from the normal genotype and
less than two reads in the normal control sample.
The genome variation profiles (accession no.
GSE30833) of the blood and tumour tissues of 2 GC patients were
downloaded from the public Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
The SNVs and INDELs specific to the patients from our dataset and
those from the previous dataset specified above were combined to
identify the overlapped somatic variants.
Function annotation of overlapped
variants
The Database for Annotation, Visualization and
Integration Discovery (DAVID) software allows the functional
annotation of gene sets in terms of biological process (BP),
molecular function (MF), cellular component (CC) and pathway. The
overrepresented Gene Ontology (GO) terms in BPs and the predominant
pathways were visualized by DAVID software (http://david.abcc.ncifcrf.gov/). A P-value <0.05
was set as the cut-off value of significance.
Ingenuity pathway analysis (IPA)
IPA can be used to assign the functional information
and biological relevance of genes in the context of known BPs,
pathways and regulatory networks (12). The canonical pathways involved with
variant genes were analysed by IPA software (Ingenuity Systems,
Redwood City, CA, USA). A score was calculated to identify aberrant
biological functions associated with the gene list.
Protein-protein interaction network
analysis
Osprey served as the biological network visual tool
and provided the direct and indirect protein interaction pairs
(13). The protein-protein
interaction network was established by the Osprey network system
version 1.2.0 (Human GRID; http://osprey.thebiogrid.org/).
PCR amplification and sequencing of
ESR1, ERK and IGF2R
The primers of oestrogen receptor 1 (ESR1), MAPK3/1,
mitogen-activated protein kinase 3/1 (ERK1/2) and insulin-like
growth factor 2 receptor (IGF2R) genes were designed by Primer 5
and synthesized by Shanghai Sangong Pharmaceutical Co., Ltd.
(Shanghai, China). PCR amplification was performed with the KAPA
Taq Extra system (Shanghai Jeayea Biotech Co., Ltd.) in an ABI9700
PCR machine (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The PCR conditions were 94°C for 3 min, 35
cycles at 94°C for 20 sec, 58°C for 15 sec and 72°C for 3 sec,
followed by a final elongation step at 72°C for 3 min. After
amplification, the PCR products were evaluated and sequenced on an
ABI 3730XL automated sequencer (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Three-dimensional protein structure
prediction
The Expert Protein Analysis System (ExPASy) is a web
server for proteomics and protein analysis (14). Based on sequencing the genes of
interest (ESR1, ERK and IGF2R), the nucleotide (DNA) sequences were
translated into protein sequences using the Translate tool of
ExPASy (http://web.expasy.org/translate/). The target amino
acid sequences were submitted to SWISS-MODEL (https://swissmodel.expasy.org/interactive/) to produce
the final 3-dimensional (3D) protein structure.
Immunofluorescence assay
After being embedded in paraffin, the gastric biopsy
specimens of the proband were cut into consecutive 4-µm sections.
The sections were incubated with the primary anti-IGF2R antibody
(1:50; ab32815; Abcam, Cambridge, MA, USA) and anti-ESR1 antibody
(1:40; MA5-13304; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
overnight at 4°C. The sections were washed with phosphate-buffered
saline (PBS) and incubated with fluorochrome-conjugated secondary
antibodies, goat anti-rabbit IgG H&L (DyLight® 594)
(1:200; ab96885; Abcam) and rabbit anti-mouse IgG H&L (Alexa
Fluor® 488) (1:200; ab150125; Abcam), for 1 h at 37°C.
The immunofluorescence staining was observed under a fluorescence
microscope (Olympus Corp., Tokyo, Japan).
Results
Data summary of the exome
sequencing
In total, 571.94 M of raw reads were generated from
the exome sequencing. Following quality control, 565.38 M of
effective reads remained. In each sample, there were >92% of
bases with a Q-value ≥30 and >96.5% of bases with a Q-value ≥20.
Finally, we obtained 2048 INDELs and 15819 SNVs by exome
sequencing.
Overlapped SNVs and INDELs
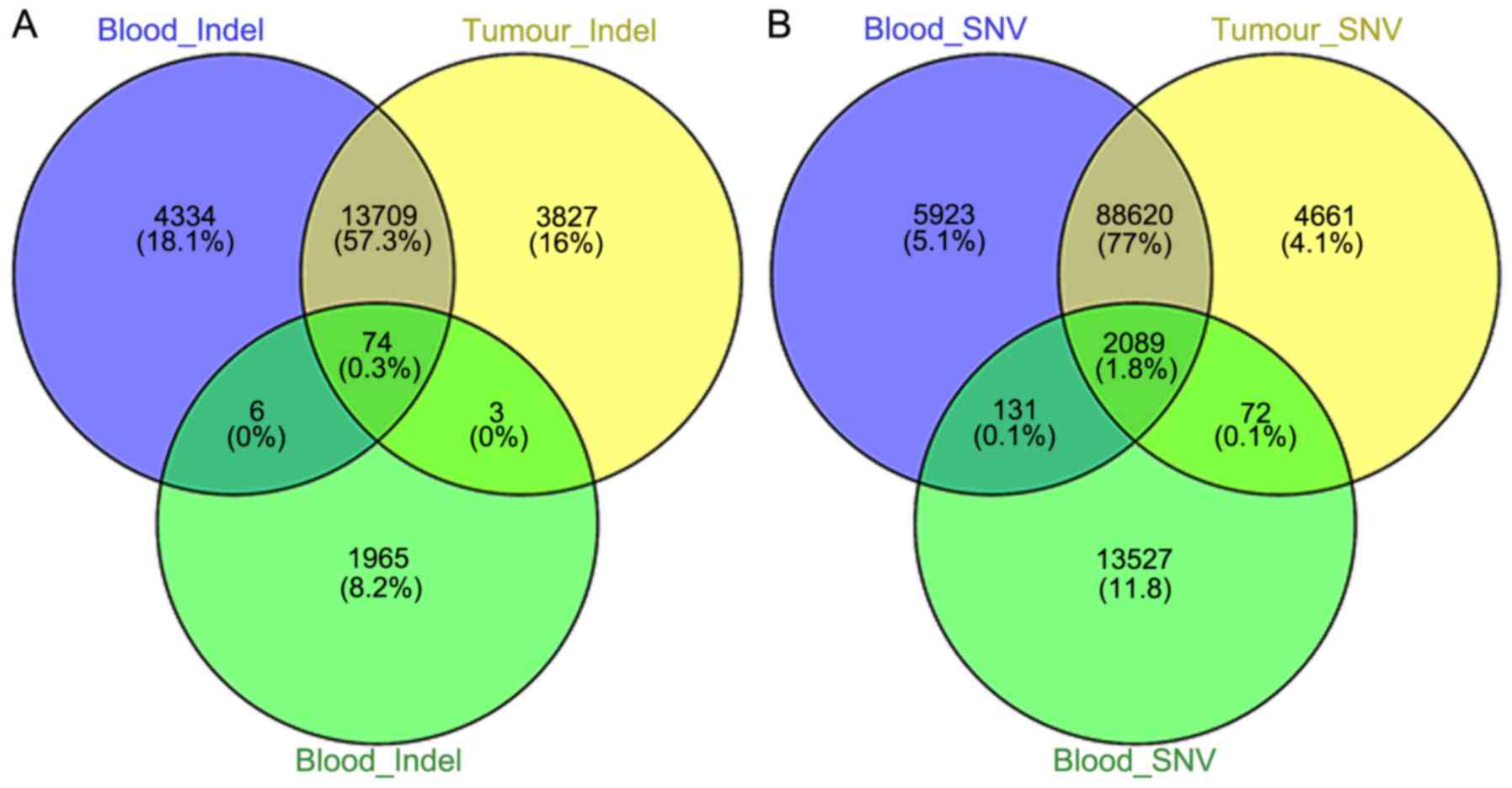
A Venn diagram is a simple and effective procedure
that displays the overlapped gene list from different groups
(15). The overlapped variant
genes, compared with the public exome sequencing data of the blood
and tissue samples of GC patients, are displayed in Fig. 2. The overlapped INDELs and SNVs were
identified to be 74 and 2089, respectively, for further
analysis.
Significant GO terms and pathways
To understand the function of gene variants at the
molecular level, the SNVs and INDELs were subjected to GO and
pathway enrichment analysis, respectively. As displayed in Table I, INDELs were closely associated
with DNA packaging, neurological system processes and BPs related
to nucleosome assembly. The significant pathways for INDELs
included regulation of the actin cytoskeleton, systemic lupus
erythematosus and natural-killer-cell-mediated cytotoxicity. The
cell-function-related BPs were perturbed by SNVs, such as cell
adhesion, motility and motion. The pathways related to cancers such
as bladder, non-small cell lung, thyroid and endometrial cancer
were significantly enriched by SNVs (Table I).
 | Table I.Top 10 significant GO and pathway
terms associated with INDELs and SNVs. |
Table I.
Top 10 significant GO and pathway
terms associated with INDELs and SNVs.
| Gene variants | Term | Count | P-value |
|---|
| Indels | GO:0006323~DNA
packaging | 4 | 0.008699692 |
|
|
GO:0050877~neurological system
process | 10 | 0.027926677 |
|
|
GO:0006334~nucleosome assembly | 3 | 0.037114373 |
|
|
GO:0031497~chromatin assembly | 3 | 0.039558685 |
|
|
GO:0065004~protein-DNA complex
assembly | 3 | 0.042910555 |
|
|
GO:0050890~cognition | 8 | 0.044035891 |
|
|
GO:0034728~nucleosome organization | 3 | 0.044625241 |
|
| GO:0016567~protein
ubiquitination | 3 | 0.06905842 |
|
| GO:0007600~sensory
perception | 7 | 0.070872214 |
|
|
GO:0043087~regulation of GTPase
activity | 3 | 0.073137761 |
| KEGG | hsa04810:
Regulation of actin cytoskeleton | 4 | 0.038043588 |
|
| hsa05322: Systemic
lupus erythematosus | 2 | 0.298474207 |
|
| hsa04650: Natural
killer cell mediated cytotoxicity | 2 | 0.379898987 |
| SNVs | GO:0007155~cell
adhesion | 79 | 3.99E-06 |
|
|
GO:0022610~biological adhesion | 79 | 4.23E-06 |
|
| GO:0048870~cell
motility | 42 | 1.47E-05 |
|
|
GO:0051674~localization of cell | 42 | 1.47E-05 |
|
| GO:0000902~cell
morphogenesis | 44 | 1.03E-04 |
|
| GO:0006928~cell
motion | 54 | 1.40E-04 |
|
| GO:0032989~cellular
component morphogenesis | 47 | 1.61E-04 |
|
| GO:0000904~cell
morphogenesis involved in differentiation | 32 | 3.86E-04 |
|
|
GO:0030855~epithelial cell
differentiation | 21 | 7.22E-04 |
| KEGG | hsa04320:
Dorso-ventral axis formation | 7 | 0.004403386 |
|
| hsa05219: Bladder
cancer | 9 | 0.00487317 |
|
| hsa04810:
Regulation of actin cytoskeleton | 25 | 0.005877286 |
|
| hsa05223: Non-small
cell lung cancer | 10 | 0.0072363 |
|
| hsa04370: VEGF
signalling pathway | 12 | 0.008400522 |
|
| hsa04360: Axon
guidance | 17 | 0.008753884 |
|
| hsa05216: Thyroid
cancer | 7 | 0.009484706 |
|
| hsa05211: Renal
cell carcinoma | 11 | 0.014021769 |
|
| hsa04960:
Aldosterone-regulated sodium reabsorption | 8 | 0.015080678 |
|
| hsa05213:
Endometrial cancer | 9 | 0.017688902 |
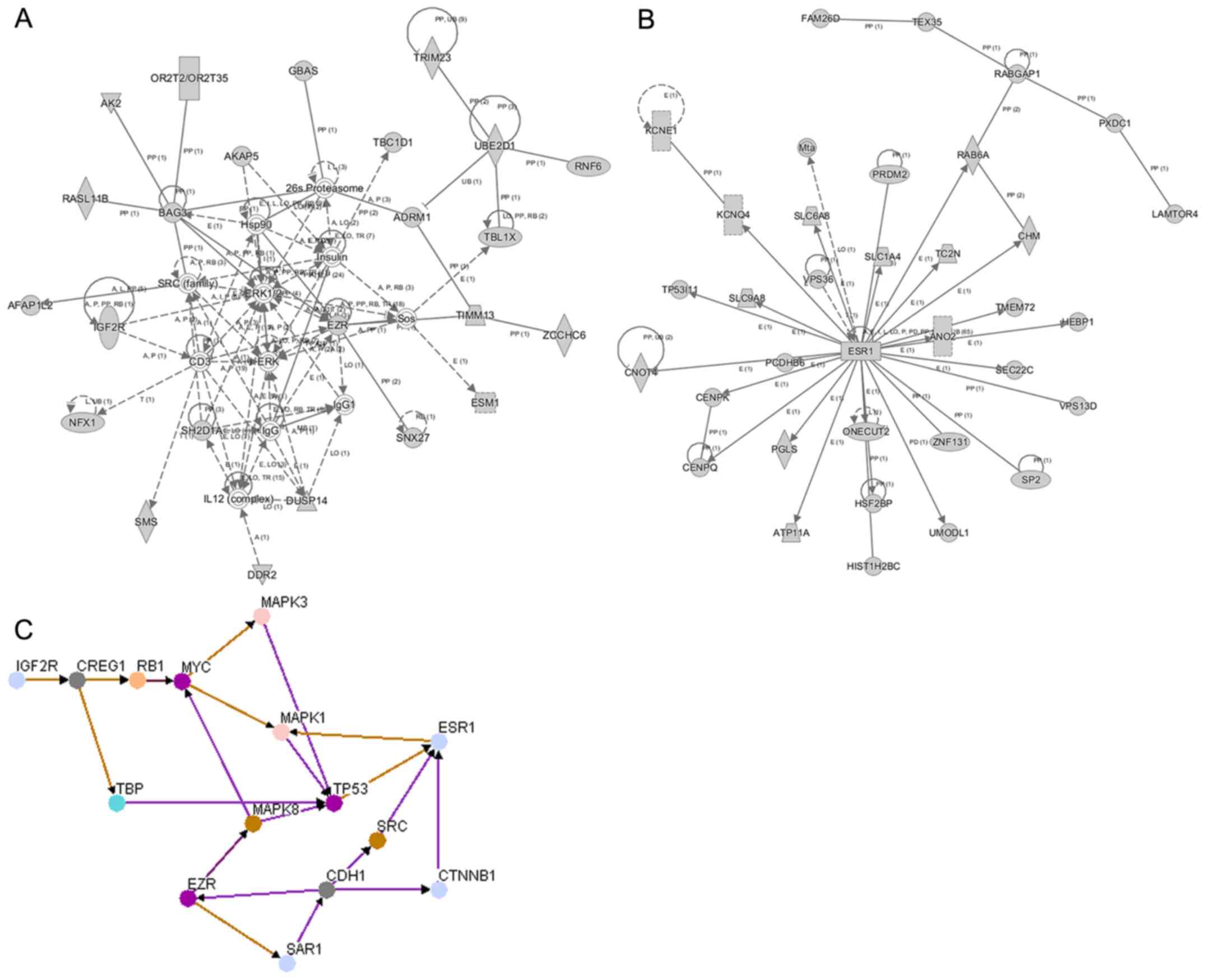
IPA network analysis
To identify the potential molecular function and
pathways perturbed by gene variations, SNVs and INDELs identified
in our study, respectively, were subjected to IPA. As displayed in
Fig. 3A the ERK1/2 (MAPK1/3)
pathway, interacting with EZR and IGF2R (M6P), was the key node in
the INDEL network. The major molecules such as ERK1/2, EZR and
IGF2R were mainly involved in cell-to-cell signalling and closely
related to interaction and connective tissue disorders and
developmental disorders. As displayed in Fig. 3B, a network centred on ESR1 was
constructed for SNVs. ESR1 was mainly associated with auditory
disease, hereditary disorders and neurological disease, with a
highest score of 44.
Protein-protein interaction
The protein interaction network with the major
proteins was constructed by Osprey software. In the present study,
4 proteins (ESR1, ERK, EZK and IGF2R) were selected as the origin
nodes. CDH1 was also included in particular to analyse the
interactions with the four proteins related to GC. As displayed in
Fig. 3C, all proteins were
assembled in one protein interaction network and had direct or
indirect interactions with other proteins. IGF2R demonstrated
regulatory interactions with MAPK3 and MAPK1 by interacting with
cellular suppressor of E1A-stimulated genes (CREG1), retinoblastoma
1 (RB1) and myelocytomatosis oncogene (MYC). ESR1 directly
interacted with MAPK1, and CDH1 demonstrated a direct interaction
with EZR.
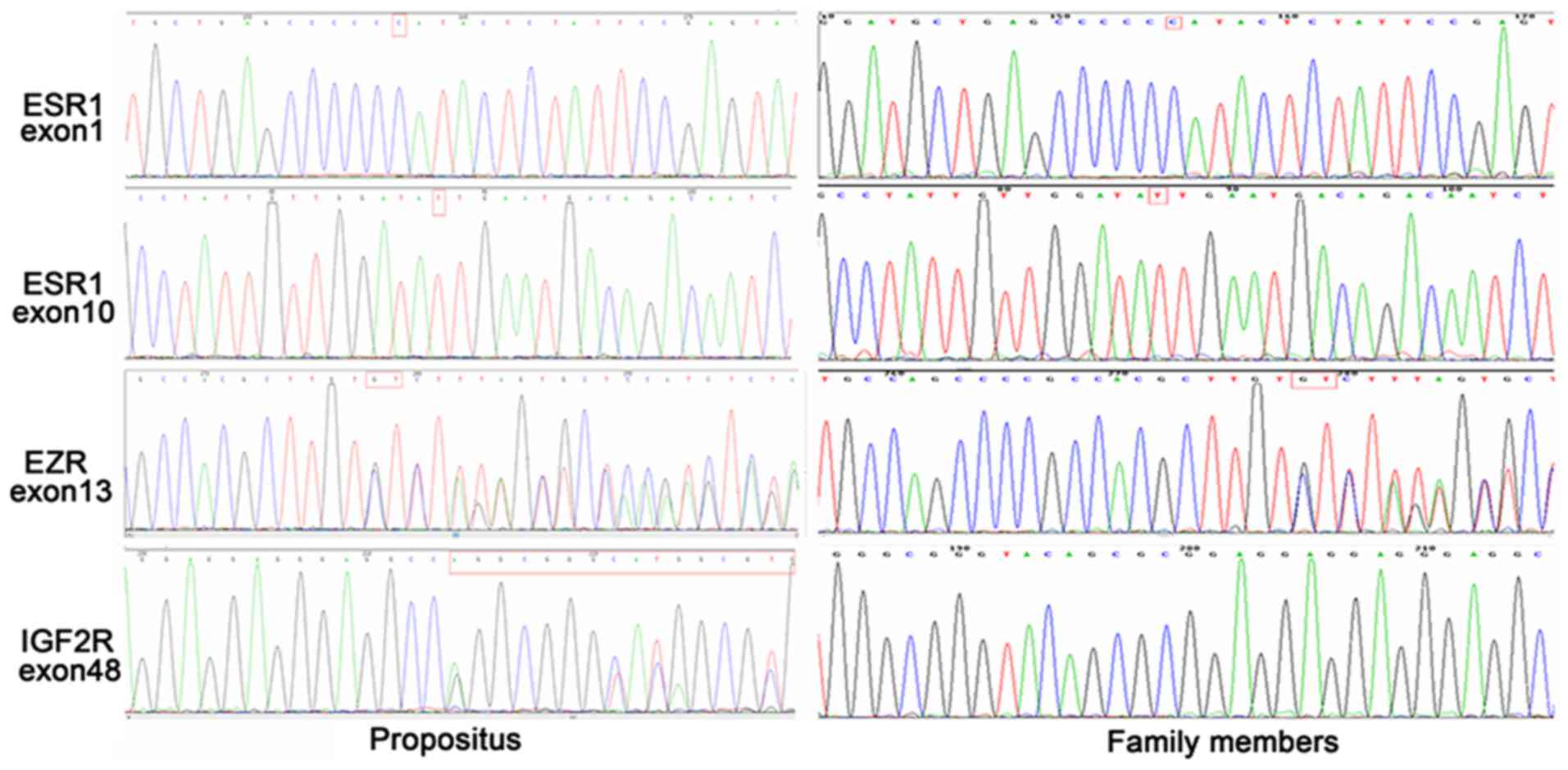
Mutations of IGF2R, EZR and ESR1
The mutations of IGF2R, EZR and ESR1 were determined
by PCR amplification and sequencing. The ESR1 gene showed
homozygous mutations in exon 1 (216G > C) and exon 10 (2234C
> T) in the proband patient. An heterozygous deletion of 68–69
GT nucleotides was detected in exon 13 of the EZR1 gene. In
addition, there was an heterozygous insertion of the 1100GGGCG
GGTACAGCGCGGAGGAGGAGGGAGGCC1131 nucleotide sequence in exon 48 of
the IGF2R gene. ESR1 and EZR carried the same mutations in other
family members, while no consistent mutations were detected for
IGF2R in other family members (Fig.
4).
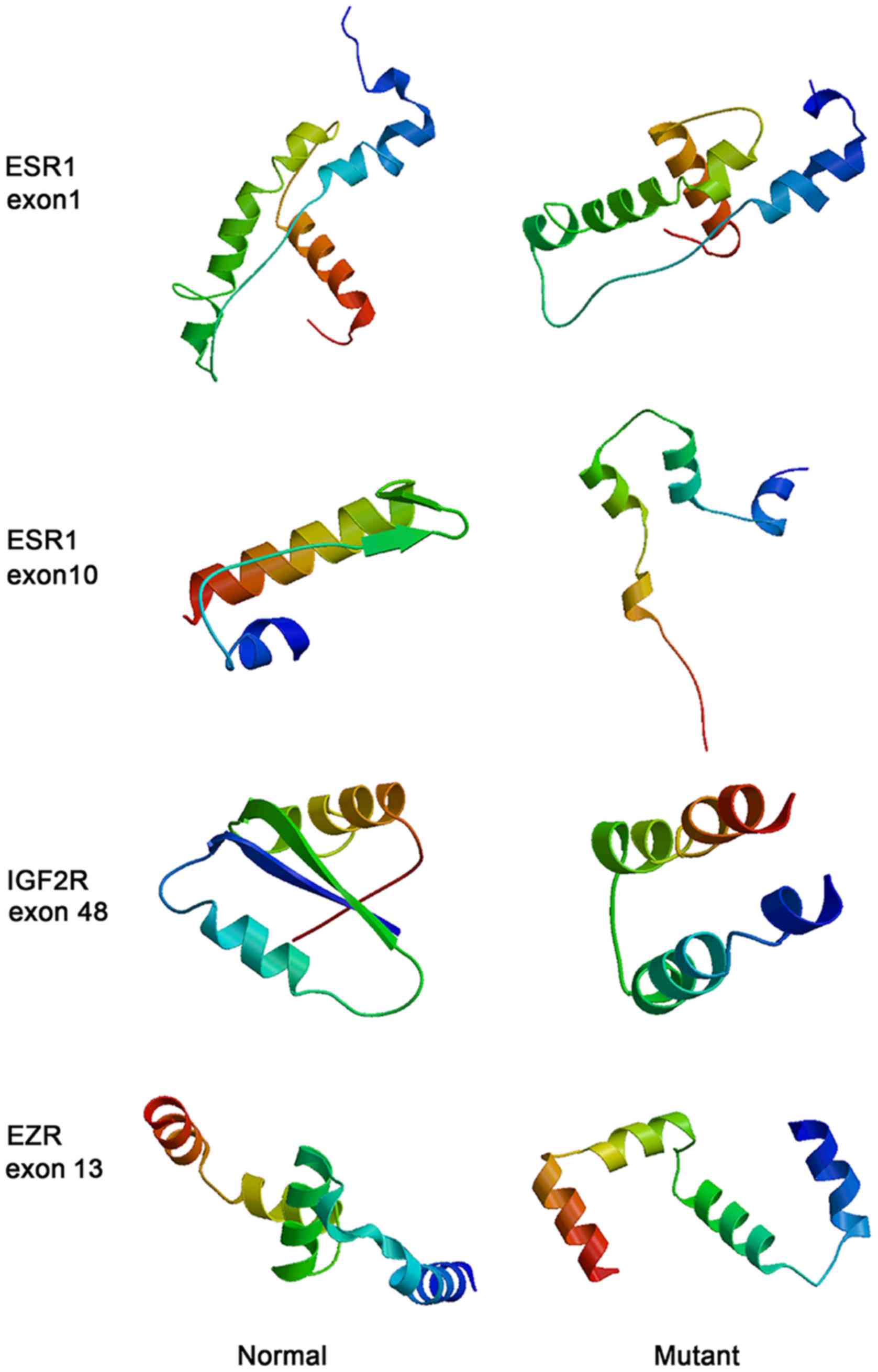
Protein structure modelling
The protein structures before and after mutations
were predicted by SWISS-MODEL. As illustrated in Fig. 5 the protein structures of ESR1,
IGF2R and EZR became loose after mutation, particularly for ESR1
with a mutation in exon 10.
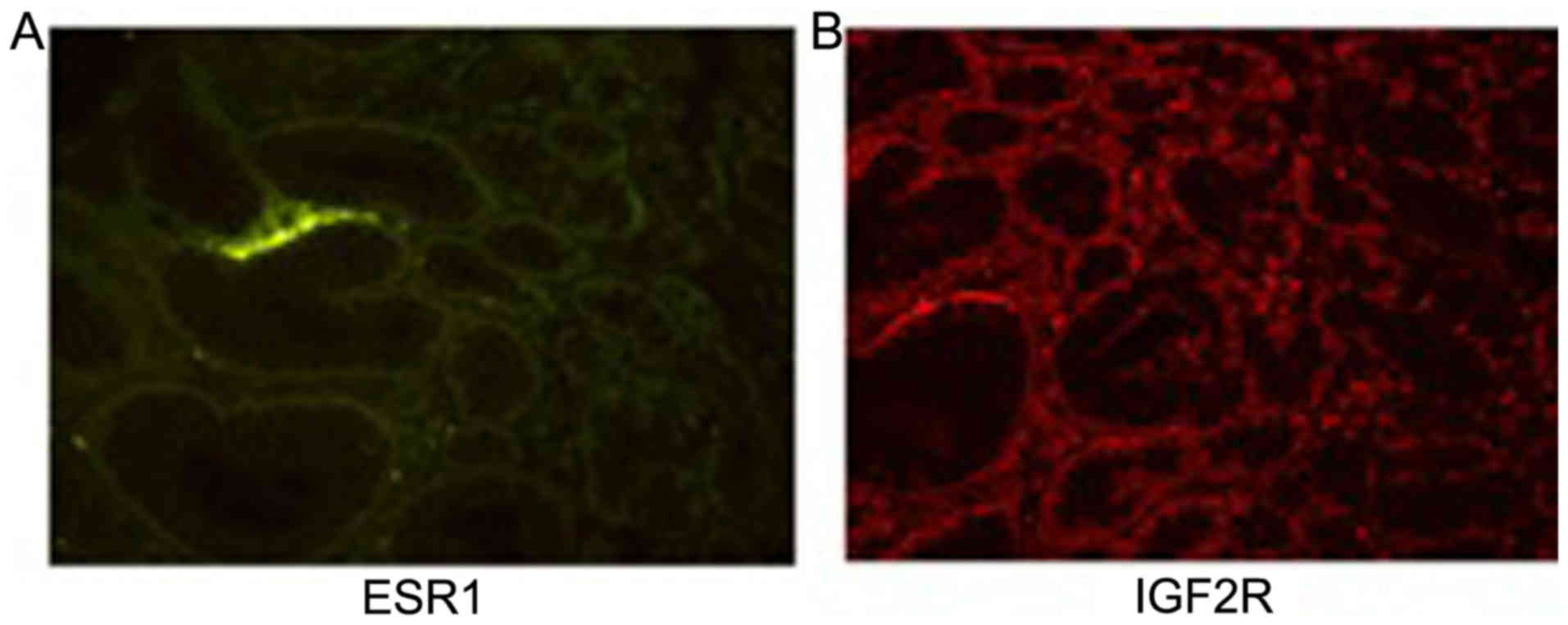
Immunofluorescence staining of IGF2R
and ESR1
The immunofluorescence staining of ESR1 and IGF2R
was observed under the green and red channel, respectively. As
displayed in Fig. 6 the green
staining was less pronounced and not specific for ESR1. Bright red
staining was observed throughout the tissue section for IGF2R.
Discussion
According to Laurens widely used histological
classification, gastric cancer (GC) can be divided into intestinal
and diffuse types of adenocarcinoma. Histopathologically, the
Lauren GC intestinal histotype is more strongly associated with the
GC familial history than the diffuse histotype (16). Incomplete intestinal metaplasia
strongly increases the risk of GC and is regarded as a precursor of
GC (17). Although the role of
intestinal metaplasia in GC has been determined in previous studies
(18) and several studies associate
some genetic factors with GC among individuals with a family
history of cancer (19), to date,
the genetic cause for FIGC has not been well-identified. The
identification of new markers predicting FIGC is important and has
become the focus of intense research. Exome sequencing applied in
the present study has contributed to shaping the complexity of
cancers. The present study was designed to identify the putative
predisposing gene defects underlying FIGC by comparing the mutation
patterns with controls in a family with intestinal-type GC. In the
present study, we reported an intestinal-type GC pedigree,
displaying the features of family members harbouring GC with an
intestinal histotype.
Based on the current dataset of exome sequencing,
2048 INDELs and 15819 SNVs were identified in the blood samples of
subjects with a strong familial history of intestinal-type GC.
Compared with the public exome sequencing data of the blood and
tissue samples of GC patients, 74 and 2,089 overlapped genes
harbouring INDELs and SNVs, respectively, were analysed. According
to the GO functional enrichment analysis, the genes with INDELs
were closely related to DNA packaging and nucleosome assembly,
while SNV genes were enriched in cell-function-related BPs. Genes
harbouring INDELs and SNVs may play different roles in the
development and progression of FIGC. Pathway analysis revealed that
genes harbouring SNVs were closely involved in cancer-related
pathways, which proved that our findings were reliable and that
genes harbouring SNVs may be centred to tumourigenesis from the
intestinal metaplasia.
IPA network analysis revealed several hub genes of
centrality. ERK1/2 had a remarkable centrality in the INDELs
network, interacting with EZR and IGF2R. The ESR1-centred
interaction network was constructed for SNVs. ERK1/2, belonging to
the MAPK family, is expressed in mammalian cells (20). MAPKs, a family of mitogen-activated
protein kinases, play regulatory roles in cell growth,
differentiation and apoptosis (21). Accumulating evidence indicate that
the activation of the MAPK/ERK signalling pathway is a common event
in tumour development and invasion (22,23).
The ESR1 gene, encoding oestrogen receptor α, is a well-known
proto-oncogene. The activation of ESR1 induced ERK phosphorylation
in a mouse spermatocyte-derived cell line, leading to apoptosis
(24). In addition, IGF2R is a
multiple ligand-binding cell surface receptor, the sequence of
which corresponds to the bovine calcium-independent M6-P receptor.
Insulin-like growth factor 2 regulates cell proliferation,
apoptosis, migration and invasive ability and functions as a tumour
suppressor. It has been reported that IGF2R also plays a key role
in activating the downstream ERK/MAPK pathway (25). Ezrin-mediated early metastasis was
reported in osteosarcoma and was partially dependent on the
activation of the ERK/MAPK pathway (26). In the present study, the predicted
protein-protein interaction network indicated that EZR played a
regulatory role in the MAPK signalling pathway. Furthermore,
previous evidence indicated that aberrant regulation of the MAPK
pathway was closely associated with the development of cancer
(27). Thus, we speculated that
genes harbouring INDELs and SNVs perturbed the MAPK/ERK signalling
pathway mediated by IGF2R and EZR, which further affected the
downstream genes involved in cell apoptosis, differentiation and
proliferation underlying FIGC development.
In the present study, the mutations of ESR1, EZR and
IGF2R were identified in a family with intestinal-type GC by
sequencing analysis. Most ESR1 mutations such as p.Leu536Gln,
p.Tyr537Ser, p.Asp538Gly and D538G mutations (28–30)
were identified in the ligand-binding domain (LBD) of oestrogen
receptors. These mutations appear to be driver mutations, leading
to a constitutively active form of ER that becomes
oestrogen-independent (29). ESR1
genetic variations promote the development and progression of
various cancers by altering oestrogen metabolism and play an
important role in hormone binding, DNA binding and the activation
of transcription to stimulate the alteration of the expression of
downstream genes (30).
Hypermethylation of ESR1 is associated with a loss of expression of
oestrogen receptor-α, which may play a critical role in the
carcinogenesis, development and prognosis of GC (31). IGF2R opposes the growth-promoting
effects of IGF-2 and acts as a tumour suppressor gene for several
cancers (10). It has also been
demonstrated to be mutated in multiple human cancers (32,33).
IGF2R was identified to be mutated in exon 27, 28 and 40 for
hepatocellular carcinomas (34,35)
and in exon 31 and 48 for breast cancer (36). When IGF2R is mutated, it loses its
anti-oncogenic activity and neoplastic transformation may be caused
by IGF 2 overaccumulation (33).
IGF2R also inhibits the IGFR signalling pathway. The IGFR
signalling pathway plays an important role in regulating cell
proliferation, differentiation, apoptosis and development (37). As a multifunctional protein
receptor, IGF2R can bind IGF2 at the cell surface and regulates the
IGFR signalling pathway (10).
Ezrin has been reported to have a crucial role in the dissemination
of several tumours (38). However,
EZR mutations in cancers have been rarely reported, particularly
for GC.
To our knowledge, in the present study we reported
for the first time a novel EZR deletion mutation in exon 13, ESR1
gene homozygous mutations in exon 1 (216G > C) and exon 10
(2234C > T) and an IGF2R insertion mutation in exon 48 of the
intestinal-type GC family.
Protein models built by SWISS-MODEL revealed that
the protein structure for ESR1, IGF2R and EZR had significant
changes upon mutations. The genetic mutations may cause alterations
in protein structure and affect the protein function, further
altering the phenotype. Based on our findings, we speculated that
the protein structure changes for ESR1, IGF2R and EZR may affect
signal transduction upstream and downstream of the MAPK/ERK
signalling pathway, which is related to tumourigenesis. We also
speculated that the mutation of ESR1 affected protein structure and
expression, leading to the dysregulation of oestrogen signalling
pathways, which may contribute to tumourigenesis. Further research
is warranted to prove this hypothesis.
Currently, it is recognized that patients with a
familial history of GC and precancerous conditions and lesions of
the stomach may benefit from periodic surveillance (39). Therefore, for individuals with
genetic mutations, we recommend intensive endoscopic surveillance
annually to ensure that there is no evidence of clinically
significant lesions.
Several limitations of the present study should be
mentioned. Firstly, the small sample size and number of specimens
limited the validation of our results. Secondly, the protein
functions affected by gene mutations were not deeply investigated.
Our exploratory study seeking an association between genetic
variation and subjects with a family history of GC indicated that
ESR1, EZR and IGF2R are candidate mutations associated with FIGC.
Further biological and clinical studies should be performed to
ascertain the intricate mechanisms and clinical physiological
relevance of correlation in terms of mutation biology, gastric
tumourigenesis and therapeutic response.
In conclusion, exome sequencing of the members of a
FIGC family outlined the pathogenesis and revealed that the driving
mutations in ESR1, EZR and IGF2R play the hub roles in GC
pathogenesis. These mutations show potential as candidate
biomarkers and as therapeutic targets in the treatment of FIGC. The
variants presented here have not been previously reported. The
present study provided a novel insight into the pathogenesis of GC
as well as guides for the counselling of predisposed
individuals.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant
from the National Natural Science Foundation of China (Key Program
81502401).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors contributions
HC and JW conceived and designed the study. YZ and
HW performed the experiments. JW wrote the paper. HC and JW
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
Research Ethics Committee of The Third Affiliated Hospital of
Nanjing University of Chinese Medicine and with the 1964 Helsinki
declaration and its later amendments or comparable ethical
standards. Informed consent was obtained from all patients or their
relatives for the use of their tissues in the experimental
procedures.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rugge M, Fassan M and Graham DY:
Epidemiology of Gastric Cancer. Springer International Publishing;
pp. 23–34. 2015, PubMed/NCBI
|
|
2
|
Yaghoobi M, Bijarchi R and Narod SA:
Family history and the risk of gastric cancer. Br J Cancer.
102:237–242. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vogelaar IP, van der Post RS, Bisseling
TM, van Krieken JHJ, Ligtenberg MJ and Hoogerbrugge N: Familial
gastric cancer: Detection of a hereditary cause helps to understand
its etiology. Hered Cancer Clin Pract. 10:182012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Caldas C, Carneiro F, Lynch HT, Yokota J,
Wiesner GL, Powell SM, Lewis FR, Huntsman DG, Pharoah PD, Jankowski
JA, et al: Familial gastric cancer: Overview and guidelines for
management. J Med Genet. 36:873–880. 1999.PubMed/NCBI
|
|
5
|
Carneiro F: Hereditary gastric cancer.
Pathologe. 33 Suppl 2:S231–S234. 2012. View Article : Google Scholar
|
|
6
|
Oliveira C, Pinheiro H, Figueiredo J,
Seruca R and Carneiro F: Familial gastric cancer: Genetic
susceptibility, pathology, and implications for management. Lancet
Oncol. 16:e60–e70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagarajan N, Bertrand D, Hillmer AM, Zang
ZJ, Yao F, Jacques PÉ, Teo AS, Cutcutache I, Zhang Z, Lee WH, et
al: Whole-genome reconstruction and mutational signatures in
gastric cancer. Genome Biol. 13:R1152012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suleiman SH, Koko ME, Nasir WH, Elfateh O,
Elgizouli UK, Abdallah MO, Alfarouk KO, Hussain A, Faisal S,
Ibrahim FM, et al: Exome sequencing of a colorectal cancer family
reveals shared mutation pattern and predisposition circuitry along
tumor pathways. Front Genet. 6:2882015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sommer S and Fuqua SA: Estrogen receptor
and breast cancer. Semin Cancer Biol. 11:339–352. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schaffer BS, Lin MF, Byrd JC, Park JH and
MacDonald RG: Opposing roles for the insulin-like growth factor
(IGF)-II and mannose 6-phosphate (Man-6-P) binding activities of
the IGF-II/Man-6-P receptor in the growth of prostate cancer cells.
Endocrinology. 144:955–966. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vaheri A, Carpén O, Heiska L, Helander TS,
Jääskeläinen J, Majander-Nordenswan P, Sainio M, Timonen T and
Turunen O: The ezrin protein family: Membrane-cytoskeleton
interactions and disease associations. Curr Opin Cell Biol.
9:659–666. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Krämer A, Green J, Pollard J Jr and
Tugendreich S: Causal analysis approaches in Ingenuity Pathway
Analysis. Bioinformatics. 30:523–530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Breitkreutz BJ, Stark C and Tyers M:
Osprey: A network visualization system. Genome Biol. 4:R222003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gasteiger E, Gattiker A, Hoogland C,
Ivanyi I, Appel RD and Bairoch A: ExPASy: The proteomics server for
in-depth protein knowledge and analysis. Nucleic Acids Res.
31:3784–3788. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pirooznia M, Nagarajan V and Deng Y:
GeneVenn - A web application for comparing gene lists using Venn
diagrams. Bioinformation. 1:420–422. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bernini M, Barbi S, Roviello F, Scarpa A,
Moore P, Pedrazzani C, Beghelli S, Marrelli D and de Manzoni G:
Family history of gastric cancer: A correlation between
epidemiologic findings and clinical data. Gastric Cancer. 9:9–13.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Filipe MI, Potet F, Bogomoletz WV, Dawson
PA, Fabiani B, Chauveinc P, Fenzy A, Gazzard B, Goldfain D and
Zeegen R: Incomplete sulphomucin-secreting intestinal metaplasia
for gastric cancer. Preliminary data from a prospective study from
three centres. Gut. 26:1319–1326. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cassaro M, Rugge M, Gutierrez O, Leandro
G, Graham DY and Genta RM: Topographic patterns of intestinal
metaplasia and gastric cancer. Am J Gastroenterol. 95:1431–1438.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ghosh A, Hartge P, Purdue MP, Chanock SJ,
Amundadottir L, Wang Z, Wentzensen N, Chatterjee N and Wacholder S:
Assessing disease risk in genome-wide association studies using
family history. Epidemiology. 23:616–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cavalli V, Vilbois F, Corti M, Marcote MJ,
Tamura K, Karin M, Arkinstall S and Gruenberg J: The stress-induced
MAP kinase p38 regulates endocytic trafficking via the GDI: Rab5
complex. Mol Cell. 7:421–432. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Junttila MR, Li SP and Westermarck J:
Phosphatase-mediated crosstalk between MAPK signaling pathways in
the regulation of cell survival. FASEB J. 22:954–965. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reddy KB, Nabha SM and Atanaskova N: Role
of MAP kinase in tumor progression and invasion. Cancer Metastasis
Rev. 22:395–403. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dhillon AS, Hagan S, Rath O and Kolch W:
MAP kinase signalling pathways in cancer. Oncogene. 26:3279–3290.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chimento A, Sirianni R, Casaburi I,
Ruggiero C, Maggiolini M, Andò S and Pezzi V: 17β-Estradiol
activates GPER- and ESR1-dependent pathways inducing apoptosis in
GC-2 cells, a mouse spermatocyte-derived cell line. Mol Cell
Endocrinol. 355:49–59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Buchanan CM, Phillips AR and Cooper GJ: A
novel two-chain IGF-II-derived peptide from purified β-cell
granules. Growth Horm IGF Res. 20:360–366. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khanna C, Wan X, Bose S, Cassaday R, Olomu
O, Mendoza A, Yeung C, Gorlick R, Hewitt SM and Helman LJ: The
membrane-cytoskeleton linker ezrin is necessary for osteosarcoma
metastasis. Nat Med. 10:182–186. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Robinson DR, Wu YM, Vats P, Su F, Lonigro
RJ, Cao X, Kalyana - Sundaram S, Wang R, Ning Y, Hodges L, et al:
Activating ESR1 mutations in hormone-resistant metastatic breast
cancer. Nat Genet. 45:1446–1451. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Toy W, Shen Y, Won H, Green B, Sakr RA,
Will M, Li Z, Gala K, Fanning S, King TA, et al: ESR1
ligand-binding domain mutations in hormone-resistant breast cancer.
Nat Genet. 45:1439–1445. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schiavon G, Hrebien S, Garcia-Murillas I,
Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I,
Lopez-Knowles E, Ribas R, et al: Analysis of ESR1 mutation
in circulating tumor DNA demonstrates evolution during therapy for
metastatic breast cancer. Sci Transl Med. 7:313ra1822015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Woo IS, Park MJ, Choi SW, Kim SJ, Lee MA,
Kang JH, Hong YS and Lee KS: Loss of estrogen receptor-alpha
expression is associated with hypermethylation near its ATG start
codon in gastric cancer cell lines. Oncol Rep. 11:617–622.
2004.PubMed/NCBI
|
|
32
|
Kong FM, Anscher MS, Washington MK,
Killian JK and Jirtle RL: M6P/IGF2R is mutated in squamous
cell carcinoma of the lung. Oncogene. 19:1572–1578. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oka Y, Waterland RA, Killian JK, Nolan CM,
Jang HS, Tohara K, Sakaguchi S, Yao T, Iwashita A, Yata Y, et al:
M6P/IGF2R tumor suppressor gene mutated in hepatocellular
carcinomas in Japan. Hepatology. 35:1153–1163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamada T, De Souza AT, Finkelstein S and
Jirtle RL: Loss of the gene encoding mannose
6-phosphate/insulin-like growth factor II receptor is an early
event in liver carcinogenesis. Proc Natl Acad Sci USA.
94:10351–10355. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
De Souza AT, Hankins GR, Washington MK,
Orton TC and Jirtle RL: M6P/IGF2R gene is mutated in human
hepatocellular carcinomas with loss of heterozygosity. Nat Genet.
11:447–449. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hankins GR, De Souza AT, Bentley RC, Patel
MR, Marks JR, Iglehart JD and Jirtle RL: M6P/IGF2 receptor: A
candidate breast tumor suppressor gene. Oncogene. 12:2003–2009.
1996.PubMed/NCBI
|
|
37
|
Yu H and Rohan T: Role of the insulin-like
growth factor family in cancer development and progression. J Natl
Cancer Inst. 92:1472–1489. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mäkitie T, Carpén O, Vaheri A and Kivelä
T: Ezrin as a prognostic indicator and its relationship to tumor
characteristics in uveal malignant melanoma. Invest Ophthalmol Vis
Sci. 42:2442–2449. 2001.PubMed/NCBI
|
|
39
|
Hirota WK, Zuckerman MJ, Adler DG, Davila
RE, Egan J, Leighton JA, Qureshi WA, Rajan E, Fanelli R,
Wheeler-Harbaugh J, et al: Standards of Practice Committee,
American Society for Gastrointestinal Endoscopy: ASGE guideline:
The role of endoscopy in the surveillance of premalignant
conditions of the upper GI tract. Gastrointest Endosc. 63:570–580.
2006. View Article : Google Scholar : PubMed/NCBI
|