Introduction
According to the World Cancer Report published in
January 2014 by the World Health Organization (WHO) (1), breast cancer is the most commonly
diagnosed cancer in women globally, and has a very high incidence
compared to other cancers; thus, breast cancer is undoubtedly the
world's major women's health issue. Plastics are necessary
materials and widely used in modern society (2), and a variety of environmental
chemicals are classified as endocrine-disrupting
chemicals/substances (EDCs/EDSs) (3). Endocrine disorders are a special form
of intoxication, and natural or man-made chemicals, now known as
EDCs, induce adverse health effects by destroying the endogenous
hormone system (4). According to
the U.S. Environmental Protection Agency (EPA) definition, EDCs
refer to the artificial manufacture of foreign objects called
endocrine disruptors or environmental hormones that imitate or
interfere with endogenous hormones maintaining the body
homeostasis, reproduction, development and behavior (5). When EDCs enter the body, they have
been shown to bind to the hormone receptor, affecting the
synthesis, secretion, transmission and binding activity of the
original endocrine mechanism (6).
EDCs are known to affect reproductive function and sex hormones
primarily in humans because of their estrogenic and antiandrogenic
properties. Phthalates and bisphenol A (BPA) are two well-known
EDCs (7). The American
Endocrinology Society says that EDCs have an impact on
neuroendocrine, thyroid, metabolism, obesity, cardiovascular
endocrinology, both male and female reproduction, prostate cancer,
breast development and breast cancer formation (5).
The a disintegrin and metalloproteinase domain 33
(ADAM33) gene, which is located on chromosome 20p13, is a member of
the ADAM family of genes, consisting of 812 amino acid residues and
22 exons (8). The ADAM proteins are
transmembrane glycoproteins with a variety of different functions,
including cell adhesion and proteolysis, and some members of the
ADAM family are associated with extracellular matrix remodeling and
cellular adhesion modifications that underlie some pathologies and
the development of cancer (9). The
increase of ADAM33 expression may play a critical role in the
pathogenesis of gastric cancer and laryngeal carcinoma (10,11).
Epigenetic regulation involves the methylation of cytosine residues
in human DNA by covalent modification without altering the DNA
sequence (12) and has also been
associated with the regulation of gene expression and the
progression of breast cancer (13,14).
Only 2 studies have explored the relationship between DNA
methylation of ADAM33 and breast cancer. Seniski et al
indicated that selective DNA hypermethylation leads to the
downregulation of ADAM33 expression and is likely to occur in
breast carcinomas, especially in invasive lobular carcinoma (ILC);
therefore, ADAM33 gene promoter methylation can differentiate ILC
and invasive ductal carcinoma (IDC) (9). Furthermore, Manica et al showed
that low ADAM33 expression is associated with shorter overall
survival and metastasis-free survival and ADAM33 may be an
important prognostic marker of triple-negative breast cancer (TNBC)
and basal-like breast cancer (BLBC) (15). It has been demonstrated that
hypermethylation of the ADAM23 promoter (a disintegrin and
metalloproteinase domain 23, another ADAM family member)
downregulates its expression and is associated with tumor
progression and metastasis in breast cancer; consequently,
epigenetic silencing of other members of the ADAM family may be
associated with the development of breast cancer (16). With the exception of Seniski et
al and Manica et al however, few studies have explored
the association between breast cancer and the ADAM33 methylation
profile (9,15).
In the present study, we utilized CpG island
microarray datasets to determine the ADAM33 methylation profile in
subjects. We further examined ADAM33 expression and performed
bisulfite sequencing PCR (BSP), nested PCR and bisulfite sequencing
to evaluate the DNA methylation status of intron 1 in ADAM33 from
peripheral blood mononuclear cells (PBMCs). BPA and phthalates are
epigenetically toxic and affect human health and cause disease
through epigenetic mechanisms (17). The purpose of this study was to test
the hypothesis that exposure to BPA and phthalate metabolites,
estimated from urinary concentrations, would be associated with
ADAM33 expression and methylation profile between breast cancer
patients and healthy controls.
Materials and methods
Study subjects
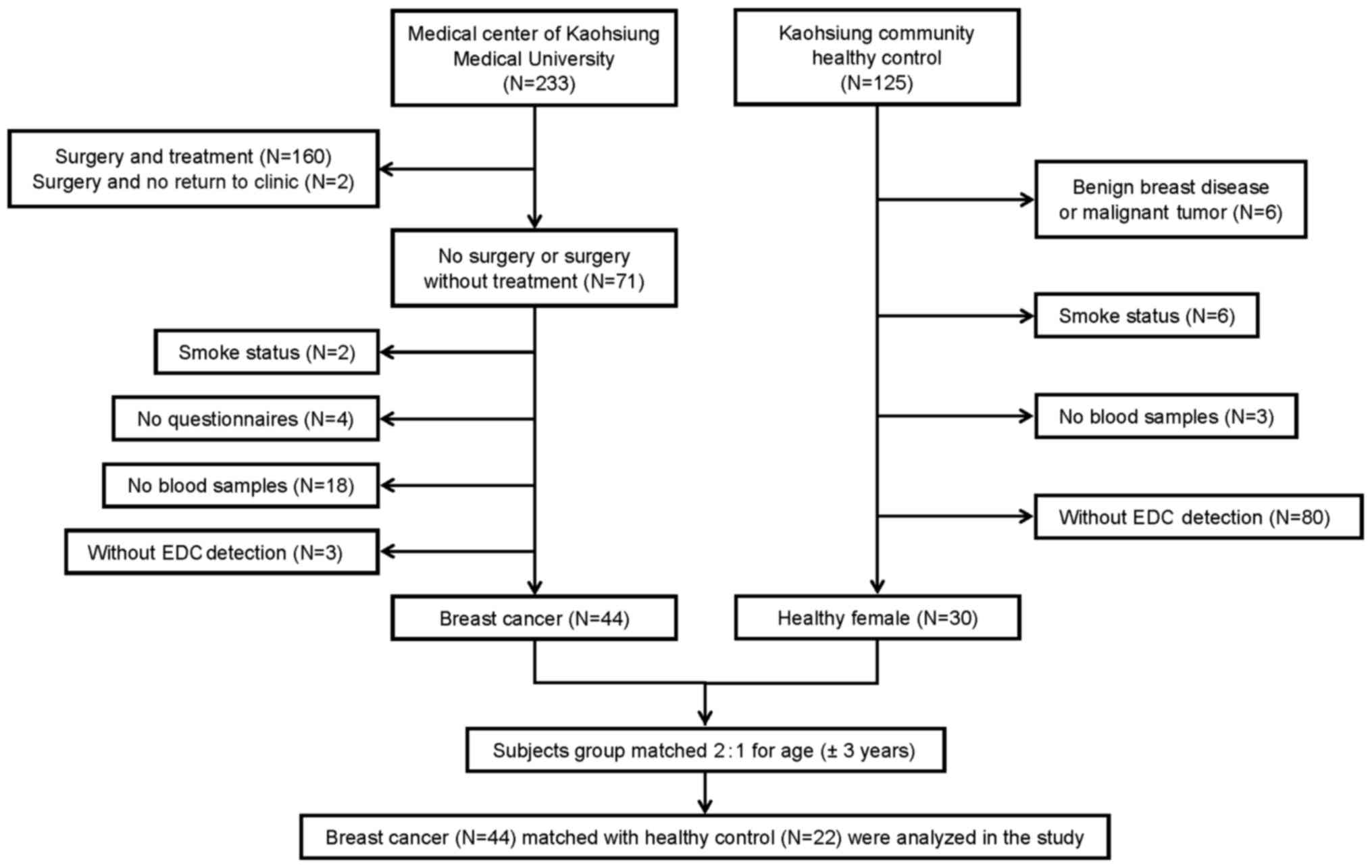
We conducted a case-control study to examine a
hypothesis concerning breast cancer risk and ADAM33 expression and
methylation profile. A total of 233 newly diagnosed breast cancer
patients with histologically verified disease were recruited at the
Medical Center of Kaohsiung Medical University in southern Taiwan
between September 2013 and June 2014. The clinical stages of the
breast cancer specimens were classified according to the American
Joint Commission on Cancer (AJCC) criteria (18). To avoid any effects on gene
methylation associated with treatment, we selected only patients
who had not received any treatment prior to their participation in
this study and 71 patients were eligible eventually. Twenty-seven
breast cancer patients were excluded from the study for the
following reasons: i) 2 patients had a smoking habit; ii) 4 breast
cancer patients did not complete questionnaires; iii) 18 breast
cancer patients refused to provide blood samples; and iv) 3
patients were without detection of EDCs. Ultimately, 44 newly
diagnosed female breast cancer patients were recruited and included
in the analysis. Between September 2013 and June 2014, 125 healthy
women from the same communities in southern Taiwan were recruited,
and 95 subjects were excluded for the following reasons: i) 6
controls had benign breast diseases or a malignant tumor; ii) 6
controls had a smoking habit; iii) 3 controls refused to provide
blood samples; and iv) 80 controls were without detection of EDCs.
Twenty-two community controls were group-matched for age (±3 years)
and paired 1:2 with the 44 breast cancer patients. All cases and
controls were women between the ages of 30 and 70 years and without
presentation of any other cancers (Fig.
1).
Interview questionnaires and
collection of specimens
The participants provided blood and urine samples
and completed questionnaires to collect information regarding
smoking habit, family history of breast cancer, reproductive
factors, and environmental exposure factors. The study protocol was
approved by the Institutional Review Board (IRB) of Kaohsiung
Medical University (IRB no. KMUHIRB-20120104). Written informed
consent was obtained from the study subjects.
Methylation microarray datasets and
analysis
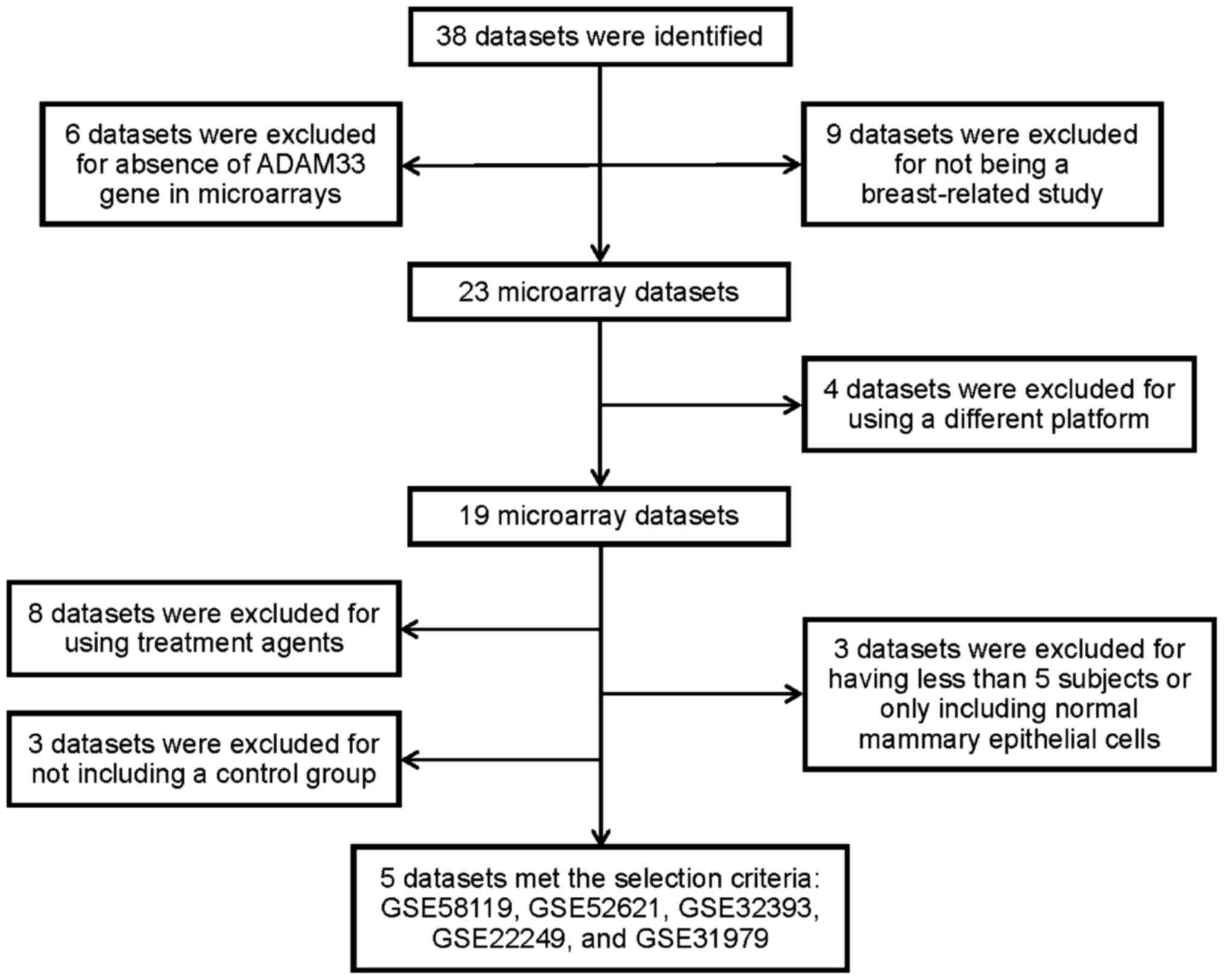
We searched the methylation microarray datasets from
the Gene Expression Omnibus (GEO) on the National Center for
Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/gds) and selected
references published online prior to 15 August 2015. The selection
criteria for breast cancer-related datasets were established using
the following keywords: breast cancer, methylation microarray
and/or Homo sapiens, and pathologies associated with benign
breast disease or recurrence were excluded. Thirty-eight datasets
associated with breast cancer were selected, but 33 datasets were
excluded. Finally, 5 datasets, including GSE58119 (19), GSE52621 (20), GSE32393 (21), GSE22249 (22), and GSE31979 (23), [all using the GPL8490 Illumina
HumanMethylation27 BeadChip (HumanMethylation27_270596_v.1.2)] met
our selection criteria (Fig. 2).
The β value for each CpG locus was used as a measure of methylation
levels (24,25).
Screening of CpG islands and analysis
of methylation levels
The identification of CpG islands in the ADAM33
gene, which was acquired from the NCBI website, was determined
using the CpG Islands Searcher website (http://www.cpgislands.com) (26). Our criteria for screening the CpG
islands were %GC=60%, ObsCpG/ExpCpG=0.7, and a minimum length of
500 bp. The design of the BSP primers was performed using an online
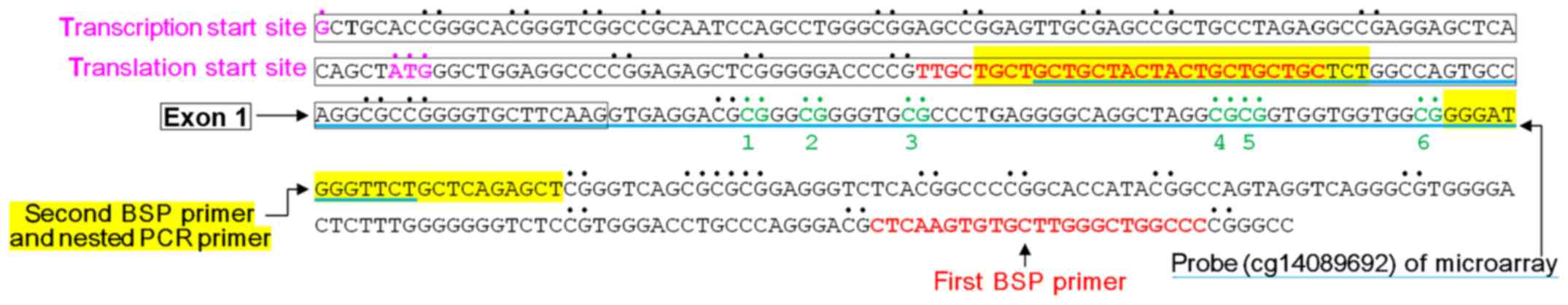
biological information website: MethPrimer (http://urogene.org/methprimer/index1.html) (27). The BSP and nested PCR products
included 9 CpG sites in exon 1 and intron 1 in the ADAM33 gene.
However, the first 3 CpG sites could only be sequenced in less than
90% of subjects in our study, thus we presented information for
only 6 CpG sites (named CpG site 1 to 6) in intron 1 in ADAM33. The
nested PCR product contained the ADAM33 gene probe (cg14089692) in
methylation microarray datasets. The BISMA (Bisulfite Sequencing
DNA Methylation Analysis) website was used to perform the DNA
methylation sequencing analysis (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/
manual_unique.php) (28).
Bisulfite sequencing PCR (BSP) and
bisulfite sequencing
Methylation bisulfite conversion, BSP and nested PCR
were performed using genomic DNA extracted from PBMC samples of
study participants and PBMCs were isolated by Ficoll-Paque Plus
density gradient centrifugation (Amersham Biosciences, Piscataway,
NJ, USA) using the Gentra Puregene Blood Kit according to the
manufacturer's instructions (Gentra Systems Inc., Minneapolis, MN,
USA). Genomic DNA (400 ng) was modified with sodium bisulfite using
the EZ DNA Methylation-Gold™ Kit (Zymo Research
Corporation, Orange, CA, USA) prior to BSP and nested PCR. The
primers of BSP did not include the CpG sites. The first set of
primers for BSP included the forward primer,
5′-TTGTTGTTGTTGTTATTATTGTTGTTGT-3′ and the reverse primer,
5′-AAACCAACCCAAACACACTTAAA-3′. The BSP products (266 bp) were used
as templates for nested PCR amplification. The second set of BSP
primers for nested PCR included the forward primer,
5′-TGTTGTTGTTATTATTGTTGTTGTTTT-3′ and the reverse primer,
5′-AACTCTAAACAAAACCCATCCC-3′, and the final products were 136 bp.
These primers for nested PCR were designed to include the probe
(cg14089692) in the ADAM33 gene from the microarray datasets. The
BSP and nested PCR conditions were as follows: 95°C for 5 min, 38
cycles of 95°C for 30 sec, 62°C for 1 min, 72°C for 30 sec and 72°C
for 7 min. The Universal Methylated Human DNA Standard kit (Zymo
Research Corp.) was used as a positive control DNA for bisulfite
conversion. Positive primers were used for positive controls, and
samples lacking DNA were used as negative controls for the BSP
experiments. The nested PCR products from all case and control
subjects were sequenced. Bisulfite sequencing was conducted with
the ABI Reaction Kit (BigDye® Terminator v3.1 Cycle
Sequencing Kit, Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and analyzed with an ABI Sequencer (Applied
Biosystems ABI 3730×l DNA Analyzer; Thermo Fisher Scientific,
Inc.).
Urinary concentrations of BPA and
phthalate metabolites
The participants provided a spot first morning urine
sample. We measured the urinary concentrations of bisphenol A (BPA)
using high-performance liquid chromatography (HPLC). Seven
phthalate metabolites including monoethyl phthalate (MEP),
mono-n-butyl phthalate (MBP), mono-isobutyl phthalate
(MIBP), mono(2-ethylhexyl) phthalate (MEHP),
mono(2-ethyl-5-hydroxyhexyl) phthalate (MEHHP),
mono(2-ethyl-5-carboxypentyl) phthalate (MECPP), and
mono(2-ethyl-5-oxohexyl) phthalate (MEOHP) were measured with
liquid chromatography-mass spectrometry (LC-MS). The urinary
concentrations of bisphenol A (BPA) and phthalate metabolites were
adjusted by creatinine, and these values of BPA and phthalate
metabolite concentrations less than the limit of detection (LOD)
were assigned a value of half the LOD (LOD/2) for the analysis.
Gene expression by RT-PCR
The ADAM33 gene probe was Hs00905552_m1 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Total RNA from the
PBMCs was extracted using TRIzol (Life Technologies, Inc.; Thermo
Fisher Scientific, Inc.). Then, glycogen (Roche Diagnostics,
Indianapolis, IN, USA) was used to increase nucleic acid recovery
and isopropanol was added to precipitate RNA (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). The quantity and quantify of RNA were
determined at OD260nm/OD280nm using a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc.).
According to the manufacturer's instructions, 1000 ng of total RNA
from each sample was reverse-transcribed in 20 µl reactions with
the High-Capacity cDNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). RT-PCR primers were
designed using the Web-based software ProbeFinder (Roche Applied
Science, Indianapolis, IN, USA). Complementary DNA (20 ng) as a
template with Power SYBR® Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was assayed in a
ViiA™ 7 Real-Time PCR System (Thermo Fisher Scientific
Inc.). Cycling conditions were 50°C for 2 min, 95°C for 10 min, 40
cycles of 95°C for 15 sec and 60°C for 1 min. The expression of
each gene was measured in triplicate for each sample. The relative
changes in gene expression were quantified using the
2−ΔΔCq method relative to the GAPDH expression
(Hs02758991_g1, Applied Biosystems; Thermo Fisher Scientific, Inc.)
(29).
Statistical analysis
The case and control group comparisons were
conducted using a two-tailed t-test for continuous data, including
demographics, clinical characteristics, overall methylation status
of intron 1 in ADAM33, ADAM33 expression and EDC concentrations.
Nonparametric statistics were performed to analyze the difference
in methylation levels (β-values) determined by the CpG island
microarray datasets between cases and controls, while the β-values
was calculated by M/(M+U+100), with M representing the methylated
signal intensity and U representing the unmethylated signal
intensity (24,25). The χ2 test was performed
to determine methylation levels of the 6 CpG sites of intron 1 in
ADAM33 between 2 groups. As ADAM33 expression and the urinary
concentrations of EDCs were not normally distributed, we
transformed the values to logarithmic scales and then used linear
regression to estimate the association of EDC concentrations, the
methylation status of intron 1 in ADAM33, and ADAM33 expression in
the case and control group, respectively. False discovery rate
(FDR) was used to verify multiple comparisons. All P-values were
2-sided with statistical significance set at P<0.05 and were
performed using the Statistical Package for Social Sciences (SPSS)
software (version 20; IBM Corp., Armonk, NY, USA) and SAS 9.4 (SAS
Institute Inc., Cary, NC, USA.
Results
Methylation microarray datasets
In the present study, the five datasets that met our
selection criteria were as follows: GSE58119 (19), GSE52621 (20), GSE32393 (21), GSE22249 (22) and GSE31979 (23). The ADAM33 gene probe (cg14089692) in
the microarray was located in the region between +132 bp and +253
bp, downstream of the transcriptional start site. The β-value
(methylation level) between the case and control group was
significantly different in the GSE32393 dataset (P=0.016) (Table I).
 | Table I.β-values of the ADAM33 gene probe
(cg14089692) in five datasets. |
Table I.
β-values of the ADAM33 gene probe
(cg14089692) in five datasets.
|
|
|
| Cases | Controls |
|
|---|
|
|
|
|
|
|
|
|---|
|
|
|
|
β-valuea |
β-valuea |
|
|---|
|
|
|
|
|
|
|
|---|
| Series | Platforms | ID_REF | Average | Median | IQR | Average | Median | IOR |
P-valueb |
|---|
| GSE58119 | GPL8490 | cg14089692 | 0.130 (N=132) | 0.107 | 0.053 | 0.124 (N=148) | 0.107 | 0.057 | 0.286 |
| GSE52621 | GPL8490 | cg14089692 | 0.137 (N=11) | 0.086 | 0.048 | 0.083 (N=25) | 0.080 | 0.026 | 0.089 |
| GSE32393 | GPL8490 | cg14089692 | 0.098 (N=114) | 0.085 | 0.032 | 0.081 (N=23) | 0.074 | 0.021 | 0.016c |
| GSE22249 | GPL8490 | cg14089692 | 0.080 (N=117) | 0.070 | 0.040 | 0.063 (N=8) | 0.060 | 0.038 | 0.286 |
| GSE31979 | GPL8490 | cg14089692 | 0.085 (N=103) | 0.069 | 0.035 | 0.081 (N=21) | 0.074 | 0.029 | 0.324 |
Participant characteristics
The basic characteristics and reproductive traits,
including age, education, weight, height, BMI, menarche age,
menopause age and the use of oral contraceptives were not
significantly different between the case and control groups
(P>0.05 for all factors) (Table
II). No patient smoked or drank alcohol in either the case or
control group (data not shown).
 | Table II.Demographics and clinical
characteristics of the participants. |
Table II.
Demographics and clinical
characteristics of the participants.
|
Characteristics | Cases | Controls |
P-valueb |
|---|
| N | 44 | 22 |
|
| Age ±
SDa (years) | 52.16±8.58 | 51.18±11.90 | 0.734 |
| Education, n
(%) |
|
| 0.262 |
| No
university | 28 (63.6) | 17 (77.3) |
|
|
University and higher | 16 (36.4) | 5
(22.7) |
|
| Weight ±
SDa (kg) | 59.22±11.25 | 58.26±11.86 | 0.749 |
| Height ±
SDa (cm) | 157.92±5.01 | 157.68±6.31 | 0.868 |
| BMI ± SD
(kg/m2) | 23.78±4.69 | 23.43±4.36 | 0.770 |
| Age at menarche ±
SDa (years) | 13.95±1.59 | 13.68±1.25 | 0.516 |
| Age at menopause ±
SDa (years) | 47.83±4.69 | 50.30±2.95 | 0.064 |
| Oral contraceptive,
n (%) |
|
| 1.000 |
| No | 42 (95.5) | 20 (100.0) |
|
|
Yes | 2 (4.5) | 0 (0.0) |
|
| Grade, n (%) |
|
|
|
| 1 | 6 (14.6) | – |
|
| 2 | 18 (43.9) |
|
|
| 3 | 17 (41.5) |
|
|
| Stage, n (%) |
| Stage
0/I/II | 38 (90.5) | – |
|
| Stage
III/IV | 4 (9.5) |
|
|
| Tumor size (cm), n
(%) |
|
|
|
| ≤2 | 26 (63.4) | – |
|
|
>2 | 15 (36.6) |
|
|
| Invasiveness, n
(%) |
|
Absence | 10 (24.4) | – |
|
|
Presence | 31 (75.6) |
|
|
| ER status, n
(%) |
|
Negative | 9 (20.9) | – |
|
|
Positive | 34 (79.1) |
|
|
| PR status, n
(%) |
|
Negative | 16 (38.1) | – |
|
|
Positive | 26 (61.9) |
|
|
| Her2 status, n
(%) |
|
Negative | 18 (42.9) | – |
|
|
Positive | 24 (57.1) |
|
|
Methylation profiles
We used nested PCR to amplify a 136-bp product in
the region between bp +128 and +263 using bisulfite-treated DNA as
a template (Fig. 3). Bisulfite
sequencing demonstrated that the methylation frequencies at CpG
site 3 were significantly different between the 2 groups (P=0.005);
therefore, the methylation statuses of CpG site 3 in intron 1 of
the ADAM33 gene may be correlated with breast cancer (Table III).
 | Table III.The methylation levels of intron 1 in
the ADAM33 gene of the participants. |
Table III.
The methylation levels of intron 1 in
the ADAM33 gene of the participants.
| Methylation
statusa | Cases (N=44) | Controls
(N=22) |
P-valueb |
|---|
| Overall | 0.70±0.27 | 0.79±0.17 | 0.152 |
| CpG 1, n (%) |
|
Methylated | 38 (86.4) | 22 (100.0) | 0.167c |
|
Unmethylated | 6 (13.6) | 0 (0.0) |
|
| CpG 2, n (%) |
|
Methylated | 39 (88.6) | 20 (90.9) | 1.000c |
|
Unmethylated | 5 (11.4) | 2 (9.1) |
|
| CpG 3, n (%) |
|
Methylated | 20 (45.5) | 18 (81.8) | 0.005d |
|
Unmethylated | 24 (54.5) | 4 (18.2) |
|
| CpG 4, n (%) |
|
Methylated | 28 (63.6) | 17 (77.3) | 0.262 |
|
Unmethylated | 16 (36.4) | 5 (22.7) |
|
| CpG 5, n (%) |
|
Methylated | 28 (63.6) | 17 (77.3) | 0.262 |
|
Unmethylated | 16 (36.4) | 5 (22.7) |
|
| CpG 6, n (%) |
|
Methylated | 31 (70.5) | 10 (45.5) | 0.050 |
|
Unmethylated | 13 (29.5) | 12 (54.5) |
|
ADAM33 expression and urinary
concentrations of EDCs
ADAM33 expression was significantly higher
(3.29±6.56) in the control group than that (0.83±1.04) in the case
group (P=0.007). The concentration of BPA was significantly higher
in the case group (P=0.033). The phthalate metabolites, including
MEP, MBP, MIBP, MEHP, MEHHP, MECPP, MEOHP and Σ4MEHP
(the sum of MEHP, MECPP, MEHHP and MEOHP) were not significantly
different between the case and control group (P>0.05) (Table IV). The adjusted P-values for the
FDR method were not significant.
 | Table IV.ADAM33 gene expression and urinary
concentrations of EDCs in the participants. |
Table IV.
ADAM33 gene expression and urinary
concentrations of EDCs in the participants.
| Gene
expression | Cases (N=44) | Controls
(N=22) |
P-valued |
|
|---|
|
|
|---|
| ADAM33 gene ±
SDa | 0.83±1.04 | 3.29±6.56 | 0.007f |
|
|---|
|
|---|
| EDCsb | Cases (N=44) | Controls
(N=22) |
P-valued | FDR P-value |
|---|
| BPAc | 14.17 (8.75,
22.93) | 5.95 (3.39,
10.46) | 0.033e | 0.297 |
| MEPc | 41.48 (26.53,
64.86) | 40.14 (24.66,
65.35) | 0.925 | 0.948 |
| MBPc | 27.43 (20.21,
37.22) | 28.89 (20.67,
40.37) | 0.831 | 0.948 |
| MIBPc | 20.95 (14.08,
31.18) | 25.02 (16.89,
37.06) | 0.566 | 0.948 |
| MEHPc | 19.07 (14.44,
25.17) | 14.24 (10.46,
19.37) | 0.189 | 0.851 |
| MEHHPc | 10.71 (7.47,
15.36) | 10.39 (7.09,
15.23) | 0.914 | 0.948 |
| MECPPc | 33.61 (23.79,
47.48) | 39.82 (29.46,
53.83) | 0.519 | 0.948 |
| MEOHPc | 12.13 (8.79,
16.75) | 11.93 (8.20,
17.35) | 0.948 | 0.948 |
|
Σ4MEHPc | 83.89 (63.47,
110.89) | 78.76 (58.15,
106.69) | 0.775 | 0.948 |
Association of urinary concentrations
of EDCs, intron 1 methylation profile in the ADAM33 gene, and
ADAM33 gene expression
In the case group, we found a significant positive
association between the methylation level of intron 1 and ADAM33
expression (coefficient = 1.147; 95% CI: 0.268, 2.027; P=0.012) as
well as EDC concentrations, including MEHHP (P=0.008), MECPP
(P=0.013), MEOHP (P=0.028) and Σ4MEHP (P=0.010)
(Table V).
 | Table V.Association of urinary concentrations
of EDCs, intron 1 methylation profile in ADAM33 gene, and ADAM33
gene expression. |
Table V.
Association of urinary concentrations
of EDCs, intron 1 methylation profile in ADAM33 gene, and ADAM33
gene expression.
|
| Cases (N=44) | Cases (N=44) |
|---|
|
|
|
|
|---|
|
| Univariate
model | Univariate
model |
|---|
|
|
|
|
|---|
|
| Methylation status
of intron 1 | ADAM33 gene
expression |
|---|
|
|
|
|
|---|
| EDCsa | Coefficient (95%
CI) | Sth. β |
P-valueb | FDR P-value | Coefficient (95%
CI) | Sth. β |
P-valueb | FDR P-value |
|---|
| BPA | 0.003 (−0.050,
0.055) | 0.017 | 0.912 | 0.912 | −0.034 (−0.198,
0.131) | −0.066 | 0.681 | 0.764 |
| MEP | 0.046 (−0.012,
0.105) | 0.245 | 0.118 | 0.168 | −0.081 (−0.265,
0.102) | −0.146 | 0.375 | 0.680 |
| MBP | 0.048 (−0.036,
0.132) | 0.177 | 0.256 | 0.288 | −0.040 (−0.310,
0.229) | −0.049 | 0.764 | 0.764 |
| MIBP | 0.050 (−0.014,
0.114) | 0.240 | 0.121 | 0.168 | −0.060 (−0.261,
0.140) | −0.099 | 0.545 | 0.701 |
| MEHP | 0.070 (−0.022,
0.161) | 0.234 | 0.131 | 0.168 | 0.169 (−0.116,
0.454) | 0.192 | 0.236 | 0.680 |
| MEHHP | 0.091 (0.025,
0.158) | 0.398 | 0.008d | 0.039c | 0.114 (−0.107,
0.335) | 0.167 | 0.302 | 0.680 |
| MECPP | 0.090 (0.020,
0.161) | 0.377 | 0.013c | 0.039c | 0.081 (−0.155,
0.316) | 0.112 | 0.492 | 0.701 |
| MEOHP | 0.086 (0.010,
0.162) | 0.335 | 0.028c | 0.063 | 0.110 (−0.137,
0.358) | 0.145 | 0.373 | 0.680 |
|
Σ4MEHP | 0.116 (0.030,
0.203) | 0.391 | 0.010c | 0.039c | 0.127 (−0.161,
0.415) | 0.143 | 0.378 | 0.680 |
|
| Methylation status
of intron 1 |
|
|
| 1.147 (0.268,
2.027) | 0.389 | 0.012c |
|
|
|
| Controls
(N=22) | Controls
(N=22) |
|
|
|
|
|
| Univariate
model | Univariate
model |
|
|
|
|
|
| Methylation
status of intron 1 | ADAM33 gene
expression |
|
|
|
|
|
EDCsa | Coefficient (95%
CI) | Std. β |
P-valueb | FDR
P-value | Coefficient (95%
CI) | Std. β |
P-valueb | FDR
P-value |
|
| BPA | −0.032 (−0.104,
0.040) | −0.216 | 0.360 | 0.658 | −0.174 (−0.681,
0.334) | −0.185 | 0.477 | 0.537 |
| MEP | 0.056 (−0.012,
0.124) | 0.359 | 0.101 | 0.658 | −0.701 (−1.171,
−0.231) | −0.620 | 0.006d | 0.054 |
| MBP | −0.011 (−0.117,
0.095) | −0.050 | 0.826 | 0.826 | −0.917 (−1.729,
−0.105) | −0.514 | 0.029c | 0.131 |
| MIBP | 0.025 (−0.065,
0.115) | 0.128 | 0.570 | 0.658 | −0.103 (−0.873,
0.668) | −0.071 | 0.781 | 0.781 |
| MEHP | −0.031 (−0.145,
0.084) | −0.123 | 0.585 | 0.658 | −0.789 (−1.701,
0.122) | −0.417 | 0.085 | 0.255 |
| MEHHP | 0.052 (−0.038,
0.142) | 0.261 | 0.240 | 0.658 | −0.442 (−1.172,
0.288) | −0.305 | 0.218 | 0.366 |
| MECPP | 0.047 (−0.068,
0.163) | 0.188 | 0.403 | 0.658 | −0.357 (−1.327,
0.614) | −0.191 | 0.447 | 0.537 |
| MEOHP | 0.039 (−0.054,
0.133) | 0.193 | 0.389 | 0.658 | −0.434 (−1.182,
0.314) | −0.294 | 0.236 | 0.366 |
|
Σ4MEHP | 0.040 (−0.076,
0.156) | 0.158 | 0.481 | 0.658 | −0.538 (−1.480,
0.404) | −0.290 | 0.244 | 0.366 |
|
| Methylation status
of intron 1 |
|
|
| −0.682 (−4.253,
2.888) | −0.101 | 0.691 |
|
We observed an inverse correlation between ADAM33
expression and EDC concentrations, including MEP (coefficient =
−0.701; 95% CI: −1.171, −0.231; P=0.006), and MBP (coefficient =
−0.917; 95% CI: −1.729, −0.105; P=0.029) in the control group
(Table V). The adjusted P-values
were calculated by FDR, leaving only MEHHP, MECPP and
Σ4MEHP significant.
Discussion
This is the first study to explore the associations
among breast cancer, endocrine-disrupting chemicals (EDCs), and
methylation and expression of the ADAM33 gene. We found that ADAM33
expression was significantly higher in the controls. In cases, we
also found a significant positive association between intron 1
methylation levels and ADAM33 gene expression as well as phthalate
metabolites, including MEHHP, MECPP, MEOHP and Σ4MEHP
(the sum of MEHP, MECPP, MEHHP and MEOHP). We suggest that the
secondary metabolites of DEHP are related to the increase in intron
1 methylation and ADAM33 expression, which is associated with a
reduction in breast cancer risk. The concentration of BPA was
higher in the cases, thus we suggest that BPA exposure could be
associated with breast cancer. MEP and MBP were inversely
correlated with ADAM33 expression in the controls, which may be
correlated with breast cancer.
Cancer development is affected by environmental
factors, lifestyle, and genetic mutations, the interaction between
tumor cells, their surrounding stroma and transmembrane proteins
altered via epigenetics. The tumor stroma primarily comprises the
basement membrane, endothelial cells, extracellular matrix (ECM),
fibroblasts, immune cells, inflammatory cells and vasculature, and
it plays an important role in cancer progression and metastasis
(30). Yang et al predicted
that transcriptional activity of the ADAM33 gene promoter was
associated with the region between bp −550 to +87 (31). Exon 1 in ADAM33 is responsible for
the translation of the signal sequence and inserts proper
localization in endoplasmic reticulum during protein synthesis
(8,32). If ADAM33 pre-mRNA splicing is
incomplete, the mis-localization of ADAM33 may result in the loss
of cell adhesion and the development of disease. Methylation of the
ADAM33 gene promoter may function as a molecular marker for
distinguishing invasive lobular carcinoma (ILC) from invasive
ductal carcinoma (IDC) and this suggests that ADAM33 is a novel
tumor-suppressor gene (9).
Hypermethylation of the promoter in the ADAM23 gene is strongly
associated with decreased mRNA and protein expression (16). Early growth response 2 (EGR2)
functions as a tumor suppressor and its expression in human tumors
and cancer cell lines is often decreased; additionally, a high
level of methylation in intron 1 of EGR2 could upregulated EGR2
gene expression (33). This study
also demonstrated that methylation levels at CpG site 3 in intron 1
were significantly higher in the control group, with CpG site 3 in
intron 1 being located 21 bp downstream of exon 1 end. We also
found that the ADAM33 expression in controls was significantly
higher and positively associated with intron 1 methylation;
therefore, we suggest that the overall decrease in intron 1
methylation was related to the reduction of ADAM33 expression and
may be associated with the development of breast cancer.
Ligand activation of PPARγ is associated with
differentiation of adipocytes, lipid accumulation, and a reduction
in growth in breast cancer (34,35).
One study showed that PPARα and PPARγ were induced by MBzP, MBuP
and MEHP (36). MEHP activated both
human PPARα and PPARγ rather than PPARβ whereas MBP could not
activate any PPAR isoforms (37).
López-Carrillo et al indicated that MBP, MBzP and MCPP were
inversely associated with breast cancer (38). At present, only two studies have
been found to investigate the correlation between ADAM10 and ADAM17
and BPA. ADAM17 is implicated in the shedding of membrane
receptors, and as BPA and nonylphenol (NP) were found to stimulate
the shedding of heparin-binding epidermal growth factor (HB-EGF)
via activation of ADAM17 or ADAM10, this mechanism may be a
potential target for the treatment of disease (39). Another study suggested that BPA and
NP could induce germ cell apoptosis regulated by the activation of
ADAM17 and p38 MAPK (40); however,
no study has been conducted to investigate the relationship between
ADAM33 and EDCs. According to the above-mentioned literature,
another potential underlying mechanism may exist to explain the
positive association between secondary metabolites of DEHP and
intron 1 methylation. We suggest that MEHHP, MECPP, MEOHP and
Σ4MEHP may have a protective effect on reducing the risk
of breast cancer by increasing intron 1 methylation to increase
ADAM33 expression.
Phthalates and BPA may dysregulate tumor-suppressor
gene (TSG) and breast cancer by epigenetics. When the gene
expression of TSG is defective, it can amplify the effect of BPA on
tumor induction (41). Patients
with BRCA1 mutation are more sensitive to BPA exposure and show an
increased number of invasive masses (42). Furthermore, one study suggested that
phthalates could active the aryl hydrocarbon receptor (AhR),
upregulate HDAC6 and c-Myc oncogenes and induce proliferation of
ER- breast cancer (43). Fetal BPA
exposure was found to alter DNA methylation in rat mammary glands
and appeared to change stromal-epithelial interactions in the fetal
mammary gland, associated with development of pre-neoplastic and
neoplastic lesions during adulthood (44). Breast cancer MCF7 cells treated with
BBP resulted in the demethylation of estrogen receptor α (ERα)
promoter, causing ERα gene re-expression (45). A review study about BPA and
phthalates on epigenetic effects showed that BPA and phthalate
caused variant methylation levels in different genes and species
(17); therefore, the region
associated with methylation is a critically important factor in
breast cancer. Regardless of the relative degree of methylation,
hypermethylation or hypomethylation is an abnormal methylation
phenomena in different sequences such as introns or exons that are
likely to cause disease (46);
nevertheless, the mechanism by which these phenomena regulate gene
expression remain unclear, thus the relationship between EDCs and
ADAM33 methylation for breast cancer requires further
exploration.
Oral administration of BPA in the human body will be
quickly metabolized into monoglucuronide and excreted in the urine;
thus assessing the concentration of BPA in the urine is considered
an appropriate method (47,48). As phthalates in the human body are
also rapidly metabolized through hydrolysis and subsequent
oxidation reactions and then finally excreted as glucuronides in
urine, measures of the urinary concentration of phthalate
metabolite could represent the exposure to the respective parent
phthalate within 24 h. Since the beginning of the millennium,
studies on the investigation of phthalate exposure have increased
rapidly by measuring urinary concentrations, thus the
concentrations of phthalate metabolites in the urine could be a
useful biomarker for phthalate exposure (49,50).
The methylation level and gene expression of ADAM33 in this study
were measured in the blood, while BPA and phthalate metabolites
were evaluated in the urine. Although the two test items were from
different samples, BPA is mainly metabolized into
BPA-monoglucuronide in the intestine and liver, and then these
metabolites reach the kidneys via the blood circulation system,
thus PBMC exposure to BPA and its metabolites is unavoidable
(51). As a result, we considered
it appropriate to evaluate the urinary concentration of BPA and
phthalate metabolites to assess the epigenetic effects of EDCs on
ADAM33 expression in PBMCs.
Epigenetic profile of circulating white blood cells
(WBCs) is directly altered by the toxic components of cigarette
smoke entering the bloodstream and this may increase cancer risk
(52); therefore, we excluded
subjects who were smokers in both the case and control groups to
control the potentially confounding effect of smoking. Other
limitations of this study should be noted. Firstly, the smoking
status and reproductive factors of the breast cancer subjects were
evaluated by a self-reported questionnaire, which increases recall
bias. Secondly, this was a case-control study; therefore, the
causal relationship between the observed differences could not be
determined. Thirdly, we excluded subjects with a smoking habit to
control a potentially confounding effect; however, we did not
consider the potential influence of nutrition on epigenetic status.
Fourthly, the microarray datasets had different criteria to recruit
and exclude patients. However, we used an identical gene
methylation expression platform (GPL8490 Illumina
HumanMethylation27 BeadChip) to reduce any bias from inconsistent
relative intensity values for a candidate gene and the
quantification of different initial gene sets. Fifthly, only a
first-in-the-morning urine sample from each woman was collected.
One study also used a single measurement assessment of the exposure
to phthalates and believed the frequency of use of these personal
products containing phthalates was constant. Thus phthalate
metabolites in the urine were considered to be stable
concentrations (38). Two
first-morning samples of phthalates for 2 consecutive days showed
good reproducibility (53). A study
also measured a spot urinary sample of BPA to evaluate the effects
of BPA on gene expression changes (51). Nepomnaschy et al demonstrated
that a correlation between urinary samples of BPA over 2 weeks was
>0.5, which indicates that the daily exposure in a short time is
similar. In addition, the first-morning urine from the same person
showed a daily consistency more than a single-point urine sample,
because the measurement appeared unaffected by diurnal changes
(54). The detection of single-spot
urinary samples of BPA and phthalates is a limitation for long-term
exposure measurements, but based on the above literature, there was
certainly feasibility in detecting the first-morning urinary
concentration of BPA and phthalates in our study. A greater number
of samples and the implementation over a longer period would be
better.
Our study also had several strengths worth noting.
Firstly, we used multiple available methylation microarray datasets
to identify potential biomarkers, and the DNA region (probe
cg14089692) identified in the microarray datasets was verified by
nested PCR and bisulfite sequencing in a case-control study.
Secondly, unlike other studies using breast cancer biopsy tissue,
this study used PBMCs to perform nested PCR to analyze the ADAM33
methylation profile. Early detection of breast cancer can improve
its cure rate, but there are currently significant limitations in
detecting breast cancer in asymptomatic patients. Sharma et
al first demonstrated that the gene expression test using
peripheral blood cell samples had the potential to detect early
stages of breast cancer progression (55). Thus, this non-invasive method, which
is not only used in breast cancer patients, can be also used to
detect the methylation level of genes to predict the chances of
suffering from breast cancer. Finally, all breast cancer subjects
were diagnosed by physicians, and their disease pathology was
histologically verified.
To the best of our knowledge, this study is the
first to explore the epigenetic effects of EDCs on methylation and
expression of ADAM33 gene associated with breast cancer. We
demonstrated that high methylation of intron 1 in ADAM33 may be
related to the elevation in ADAM33 expression and reduced risk of
breast cancer and MEHHP, MECPP, MEOHP and Σ4MEHP may
possess protective effects on reducing the risk of breast cancer.
We also found that high urinary concentration of BPA may be
associated with breast cancer. In addition, MEP and MBP were
negatively associated with ADAM33 expression; this result deserves
further evaluation.
Further investigations into the methylation of this
region are required to validate the prognostic and predictive roles
of the ADAM33 gene in breast cancer. In addition, a larger
population must be evaluated to determine the role of ADAM33
methylation and epigenetic mechanisms by EDCs in breast cancer and
to determine whether the methylation level of intron 1 in ADAM33
has the potential to serve as a diagnostic or prognostic tumor
marker in breast cancer.
Acknowledgements
We thank all breast cancer patients and health women
adults who participated in the study and all staff who have worked
in the study.
Funding
The present study was supported by grants from the
National Science Council of Taiwan (grant no. NSC
102-2632-B-037-001-MY3), and partially from the Kaohsiung Medical
University ‘Aim for the Top Universities Grant (grant no.
KMU-TP103A16, KMU-TP104A01, KMU-TP105A05’). The funding agencies
had no role in the study design, data collection, data analysis,
data interpretation, or writing of the report.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PJY and TNW conceived and designed the study. PJY
drafted the manuscript and performed the experiments. EMT, CYP, SSL
and CCC were involved in the conception of the study. MFH, FOY and
JYK treated the patients and collected the data. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The study protocol was approved by the Institutional
Review Board (IRB) of Kaohsiung Medical University (IRB no.
KMUHIRB-20120104).
Patient consent for publication
Written informed consent was obtained from the study
subjects.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stewart BW and Wild C: World Cancer Report
2014. Lyon, France: International Agency for Research on Cancer.
World Health Organization. 630:2014.
|
|
2
|
Halden RU: Plastics and health risks. Annu
Rev Public Health. 31:179–194. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Erkekoglu P and Kocer-Gumusel B:
Environmental effects of endocrine-disrupting chemicals: A special
focus on phthalates and bisphenol A. Environ Health. 6:1–36.
2016.
|
|
4
|
Solecki R, Kortenkamp A, Bergman Å,
Chahoud I, Degen GH, Dietrich D, Greim H, Håkansson H, Hass U,
Husoy T, et al: Scientific principles for the identification of
endocrine-disrupting chemicals: A consensus statement. Arch
Toxicol. 91:1001–1006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Diamanti-Kandarakis E, Bourguignon JP,
Giudice LC, Hauser R, Prins GS, Soto AM, Zoeller RT and Gore AC:
Endocrine-disrupting chemicals: An Endocrine Society scientific
statement. Endocr Rev. 30:293–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schug TT, Janesick A, Blumberg B and
Heindel JJ: Endocrine disrupting chemicals and disease
susceptibility. J Steroid Biochem Mol Biol. 127:204–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buluş AD, Aşci A, Erkekoglu P, Balci A,
Andiran N and Koçer-Gümüşel B: The evaluation of possible role of
endocrine disruptors in central and peripheral precocious puberty.
Toxicol Mech Methods. 26:493–500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orth P, Reichert P, Wang W, Prosise WW,
Yarosh-Tomaine T, Hammond G, Ingram RN, Xiao L, Mirza UA, Zou J, et
al: Crystal structure of the catalytic domain of human ADAM33. J
Mol Biol. 335:129–137. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seniski GG, Camargo AA, Ierardi DF, Ramos
EA, Grochoski M, Ribeiro ES, Cavalli IJ, Pedrosa FO, de Souza EM,
Zanata SM, et al: ADAM33 gene silencing by promoter
hypermethylation as a molecular marker in breast invasive lobular
carcinoma. BMC Cancer. 9:802009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Topal O, Erinanc H, Ozer C, Canpolat ET,
Celik SB and Erbek SS: Expression of ‘a disintegrin and
metalloproteinase-33’ (ADAM-33) protein in laryngeal squamous cell
carcinoma. J Laryngol Otol. 126:511–515. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim KE, Song H, Hahm C, Yoon SY, Park S,
Lee HR, Hur DY, Kim T, Kim CH, Bang SI, et al: Expression of ADAM33
is a novel regulatory mechanism in IL-18-secreted process in
gastric cancer. J Immunol. 182:3548–3555. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Treviño LS, Wang Q and Walker CL:
Hypothesis: Activation of rapid signaling by environmental
estrogens and epigenetic reprogramming in breast cancer. Reprod
Toxicol. 54:136–140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nephew KP and Huang TH: Epigenetic gene
silencing in cancer initiation and progression. Cancer Lett.
190:125–133. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong C, Fujino K, Monteiro LJ, Gomes AR,
Drost R, Davidson-Smith H, Takeda S, Khoo US, Jonkers J, Sproul D,
et al: FOXA1 repression is associated with loss of BRCA1 and
increased promoter methylation and chromatin silencing in breast
cancer. Oncogene. 34:5012–5024. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Manica GC, Ribeiro CF, Oliveira MA,
Pereira IT, Chequin A, Ramos EA, Klassen LM, Sebastião AP,
Alvarenga LM, Zanata SM, et al: Down regulation of ADAM33 as
a predictive biomarker of aggressive breast cancer. Sci Rep.
7:444142017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Costa FF, Verbisck NV, Salim AC, Ierardi
DF, Pires LC, Sasahara RM, Sogayar MC, Zanata SM, Mackay A, O'Hare
M, et al: Epigenetic silencing of the adhesion molecule ADAM23 is
highly frequent in breast tumors. Oncogene. 23:1481–1488. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh S and Li SS: Epigenetic effects of
environmental chemicals bisphenol A and phthalates. Int J Mol Sci.
13:10143–10153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB; and Cancer AJCo, : AJCC Cancer
Staging Handbook: From The AJCC Cancer Staging Manual. Springer;
New York, NY: 2010
|
|
19
|
Anjum S, Fourkala EO, Zikan M, Wong A,
Gentry-Maharaj A, Jones A, Hardy R, Cibula D, Kuh D, Jacobs IJ, et
al: A BRCA1-mutation associated DNA methylation signature in
blood cells predicts sporadic breast cancer incidence and survival.
Genome Med. 6:472014. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fackler MS, Bujanda ZL, Umbricht C, Teo W,
Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C, Lyman JP, et al:
Detection of hypermethylated circulating serum DNA in metastatic
breast cancer and confirmation by the cMethDNA assay. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52621March
1–2014
|
|
21
|
Zhuang J, Jones A, Lee SH, Ng E, Fiegl H,
Zikan M, Cibula D, Sargent A, Salvesen HB, Jacobs IJ, et al: The
dynamics and prognostic potential of DNA methylation changes at
stem cell gene loci in women's cancer. PLoS Genet. 8:e10025172012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dedeurwaerder S, Desmedt C, Calonne E,
Singhal SK, Haibe-Kains B, Defrance M, Michiels S, Volkmar M,
Deplus R, Luciani J, et al: DNA methylation profiling reveals a
predominant immune component in breast cancers. EMBO Mol Med.
3:726–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fackler MJ, Umbricht CB, Williams D,
Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P,
Visvananthan K, et al: Genome-wide methylation analysis identifies
genes specific to breast cancer hormone receptor status and risk of
recurrence. Cancer Res. 71:6195–6207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bibikova M, Lin Z, Zhou L, Chudin E,
Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen X, Li S, Zhang L, Li H, Hong G, Zhou
X, Zheng T, Zhang W, Hao C, Shi T, et al: An integrated approach to
uncover driver genes in breast cancer methylation genomes. PLoS
One. 8:e612142013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takai D and Jones PA: The CpG island
searcher: A new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
27
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rohde C, Zhang Y, Reinhardt R and Jeltsch
A: BISMA - fast and accurate bisulfite sequencing data analysis of
individual clones from unique and repetitive sequences. BMC
Bioinformatics. 11:2302010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bremnes RM, Dønnem T, Al-Saad S, Al-Shibli
K, Andersen S, Sirera R, Camps C, Marinez I and Busund LT: The role
of tumor stroma in cancer progression and prognosis: Emphasis on
carcinoma-associated fibroblasts and non-small cell lung cancer. J
Thorac Oncol. 6:209–217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang Y, Haitchi HM, Cakebread J, Sammut D,
Harvey A, Powell RM, Holloway JW, Howarth P, Holgate ST and Davies
DE: Epigenetic mechanisms silence a disintegrin and metalloprotease
33 expression in bronchial epithelial cells. J Allergy Clin
Immunol. 121:1391–1399. 2008. View Article : Google Scholar
|
|
32
|
Yoshinaka T, Nishii K, Yamada K, Sawada H,
Nishiwaki E, Smith K, Yoshino K, Ishiguro H and Higashiyama S:
Identification and characterization of novel mouse and human
ADAM33s with potential metalloprotease activity. Gene. 282:227–236.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Unoki M and Nakamura Y: Methylation at CpG
islands in intron 1 of EGR2 confers enhancer-like activity. FEBS
Lett. 554:67–72. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mueller E, Sarraf P, Tontonoz P, Evans RM,
Martin KJ, Zhang M, Fletcher C, Singer S and Spiegelman BM:
Terminal differentiation of human breast cancer through PPAR gamma.
Mol Cell. 1:465–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Elstner E, Müller C, Koshizuka K,
Williamson EA, Park D, Asou H, Shintaku P, Said JW, Heber D and
Koeffler HP: Ligands for peroxisome proliferator-activated
receptorgamma and retinoic acid receptor inhibit growth and induce
apoptosis of human breast cancer cells in vitro and in BNX
mice. Proc Natl Acad Sci USA. 95:8806–8811. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hurst CH and Waxman DJ: Activation of
PPARalpha and PPARgamma by environmental phthalate monoesters.
Toxicol Sci. 74:297–308. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Venkata NG, Robinson JA, Cabot PJ, Davis
B, Monteith GR and Roberts-Thomson SJ: Mono(2-ethylhexyl)phthalate
and mono-n-butyl phthalate activation of peroxisome
proliferator activated-receptors α and γ in breast. Toxicol Lett.
163:224–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
López-Carrillo L, Hernández-Ramírez RU,
Calafat AM, Torres-Sánchez L, Galván-Portillo M, Needham LL,
Ruiz-Ramos R and Cebrián ME: Exposure to phthalates and breast
cancer risk in northern Mexico. Environ Health Perspect.
118:539–544. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Urriola-Muñoz P, Li X, Maretzky T,
McIlwain DR, Mak TW, Reyes JG, Blobel CP and Moreno RD: The
xenoestrogens biphenol-A and nonylphenol differentially regulate
metalloprotease-mediated shedding of EGFR ligands. J Cell Physiol.
233:2247–2256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Urriola-Muñoz P, Lagos-Cabré R and Moreno
RD: A mechanism of male germ cell apoptosis induced by bisphenol-A
and nonylphenol involving ADAM17 and p38 MAPK activation. PLoS One.
9:e1137932014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Romagnolo DF, Daniels KD, Grunwald JT,
Ramos SA, Propper CR and Selmin OI: Epigenetics of breast cancer:
Modifying role of environmental and bioactive food compounds. Mol
Nutr Food Res. 60:1310–1329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fernandez SV, Huang Y, Snider KE, Zhou Y,
Pogash TJ and Russo J: Expression and DNA methylation changes in
human breast epithelial cells after bisphenol A exposure. Int J
Oncol. 41:369–377. 2012.PubMed/NCBI
|
|
43
|
Hsieh TH, Tsai CF, Hsu CY, Kuo PL, Lee JN,
Chai CY, Wang SC and Tsai EM: Phthalates induce proliferation and
invasiveness of estrogen receptor-negative breast cancer through
the AhR/HDAC6/c-Myc signaling pathway. FASEB J. 26:778–787. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dhimolea E, Wadia PR, Murray TJ, Settles
ML, Treitman JD, Sonnenschein C, Shioda T and Soto AM: Prenatal
exposure to BPA alters the epigenome of the rat mammary gland and
increases the propensity to neoplastic development. PLoS One.
9:e998002014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kang SC and Lee BM: DNA methylation of
estrogen receptor alpha gene by phthalates. J Toxicol Environ
Health A. 68:1995–2003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ehrlich M: DNA methylation in cancer: Too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Völkel W, Colnot T, Csanády GA, Filser JG
and Dekant W: Metabolism and kinetics of bisphenol A in humans at
low doses following oral administration. Chem Res Toxicol.
15:1281–1287. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Calafat AM, Kuklenyik Z, Reidy JA, Caudill
SP, Ekong J and Needham LL: Urinary concentrations of bisphenol A
and 4-nonylphenol in a human reference population. Environ Health
Perspect. 113:391–395. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wittassek M, Koch HM, Angerer J and
Brüning T: Assessing exposure to phthalates - the human
biomonitoring approach. Mol Nutr Food Res. 55:7–31. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kay VR, Chambers C and Foster WG:
Reproductive and developmental effects of phthalate diesters in
females. Crit Rev Toxicol. 43:200–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Melzer D, Harries L, Cipelli R, Henley W,
Money C, McCormack P, Young A, Guralnik J, Ferrucci L, Bandinelli
S, et al: Bisphenol A exposure is associated with in vivo
estrogenic gene expression in adults. Environ Health Perspect.
119:1788–1793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shenker NS, Polidoro S, van Veldhoven K,
Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P
and Flanagan JM: Epigenome-wide association study in the European
Prospective Investigation into Cancer and Nutrition (EPIC-Turin)
identifies novel genetic loci associated with smoking. Hum Mol
Genet. 22:843–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hoppin JA, Brock JW, Davis BJ and Baird
DD: Reproducibility of urinary phthalate metabolites in first
morning urine samples. Environ Health Perspect. 110:515–518. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nepomnaschy PA, Baird DD, Weinberg CR,
Hoppin JA, Longnecker MP and Wilcox AJ: Within-person variability
in urinary bisphenol A concentrations: Measurements from specimens
after long-term frozen storage. Environ Res. 109:734–737. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sharma P, Sahni NS, Tibshirani R, Skaane
P, Urdal P, Berghagen H, Jensen M, Kristiansen L, Moen C, Sharma P,
et al: Early detection of breast cancer based on gene-expression
patterns in peripheral blood cells. Breast Cancer Res. 7:R634–R644.
2005. View Article : Google Scholar : PubMed/NCBI
|