Introduction
Bladder cancer, prevalent in men over 65 years old,
is a globally common urinary tract malignancy (1). Generally, bladder cancer refers to
bladder urothelial carcinoma (BLCA), and BLCA accounts for 90% of
bladder cancers. According to studies by the American Cancer
Society, 96% of bladder cancer in situ patients survive 5
years or more. Hence, the prognosis of patients with bladder cancer
is better than that of patients with other malignant tumours such
as pancreatic carcinoma, lung cancer, hepatocellular carcinoma or
oesophageal cancer. However, a high recurrence rate and tumour
progression makes the prognosis of bladder cancer patients worse.
Thus, the five-year survival rate of bladder cancer decreases to
5–70% (2). Currently, tumour node
metastasis (TNM) classification and pathological grade are widely
used clinically to plan treatment and predict prognosis. However,
they are indicators at a population level, incapable of precisely
predicting individual clinical outcomes (3). In clinical practice, the individual
clinical outcome is considered more meaningful to guide clinical
therapy and evaluate prognosis, if we know this personal
information in advance, including disease recurrence risk,
therapeutic response and overall survival (OS). Therefore, new
prognostic and predictive biomarkers are urgently needed for
bladder cancer patients.
Due to The Cancer Genome Atlas (TCGA) public
database collecting a broad range of bladder cancer gene expression
data, TCGA data mining has become an important tool for molecular
analysis, such as tumour molecular subtyping and diagnostic or
prognostic biomarker screening. By analysing gene expression
profiles, a large number of new molecular biomarkers at the
transcriptome level have been identified. Accordingly, studies on
prognosis biomarkers of bladder cancer in recent years are
primarily based on RNA expression. Long non-coding RNAs (a group of
non-protein-coding transcripts longer than 200 nucleotides) and
microRNAs (a class of small non-coding RNAs that have potential in
RNA silencing) have been demonstrated to play essential roles in
regulating gene expression at the transcriptional or
post-transcriptional level and have prognostic value in multiple
tumours (4–8). Concerning bladder cancer, accumulative
studies have revealed the potential of lncRNAs or miRNAs as
prognostic markers. The lncRNA HOTAIR has been demonstrated by
Shang et al as an independent prognostic biomarker of
overall survival in bladder transitional cell carcinoma patients
(9). It was identified by Wu et
al (10) that high tissue
metastasis associated with lung adenocarcinoma transcript 1
(MALAT1) level was associated with an inferior clinical outcome in
various cancers, including bladder cancer. Furthermore, Ratert
et al (11) screened out and
validated 15 bladder cancer-specific miRNAs, among which miR-141
and miR-205 were related to the overall survival time. miR-31,
miR-100 and miR-145 were reported to be independent prognostic
factors in patients with bladder cancer (12–14).
Since the prognostic factors of bladder cancer are complex and
collectively profiled lncRNAs or microRNAs have demonstrated
considerable prognostic power, current studies are focused on
prognostic signatures or combinations of multi-lncRNAs or
multi-microRNAs. Dong et al performed a comprehensive
analysis of 234 BLCA patients and demonstrated that a four-lncRNA
(AC005682.5, CTD-2231H16.1, CTB-92J24.2 and RP11-727F15.13)
signature could be a novel independent biomarker for predicting the
survival of patients with BLCA (15). Zhou et al (16) identified an eight-miRNA signature,
including three upregulated (miR-141, miR-200c and miR-21) and five
downregulated (miR-145, miR-125, miR-199a, let-7c and miR-99a)
miRNAs that could predict overall survival (OS) of bladder cancer.
A four-miRNA (miR-422a-3p, miR-486-3p, miR-103a-3p and miR-27a-3p)
signature was developed, and its considerable predictive potential
was verified for muscle-invasive bladder cancer (17).
Compared with lncRNAs or microRNAs, protein-coding
gene transcripts or mRNA levels directly reflect the gene
expression level, and some studies have confirmed that PCGs can
predict bladder cancer patient survival. For example, Mitra et
al (18) identified a gene
signature comprised of JUN, MAP2K6, STAT3 and ICAM1 that could
predict the clinical outcome of bladder cancer patients. Another
study found that a 24-gene hypoxia signature demonstrated strong
and independent prognostic and predictive value for muscle invasive
bladder cancer patients (19).
In conclusion, lncRNAs, microRNAs and PCGs can be
prognostic markers of bladder cancer. However, there are no studies
that explore the role of a multidimensional transcriptome signature
combining lncRNAs and microRNAs with PCGs in bladder cancer. In the
present study, we investigated the clinical value of a
PCG-lncRNA-microRNA signature by mining the expression profiles of
239 bladder cancer patients.
Materials and methods
lncRNA, microRNA and mRNA expression
data of BLCA patients
We downloaded microRNA (Illumina HiSeq microRNA Seq)
and mRNA (Illumina HiSeq RNA Seq V2) level 3 expression data and
corresponding clinical information of BLCA patients from the TCGA
portal (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/).
lncRNA expression data were obtained from the TANRIC database
(http://ibl.mdanderson.org/tanric/_design/basic/index.html).
Genes with missing expression values in >30% of samples or
patients were removed, and the remaining missing values were
entered by the k-nearest neighbour method. Genes whose RPKM
expression values were 0 in all samples were excluded (20). Following these steps, we obtained a
total of 239 bladder cancer patients or samples for the present
study. Since the data were obtained from TCGA, further approval by
an Ethics Committee was not required. The present study met the
publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines).
Construction of a prognostic
PCG-lncRNA-microRNA signature in the training dataset
We used univariate Cox proportional hazards
regression analysis to assess the relationship between the gene
expression of lncRNAs, microRNAs and PCGs and patient OS in the
training dataset. Considering that a smaller number of lncRNAs,
microRNAs and PCGs in the model render the model more practical, we
used the random survival forests-variable hunting (RSFVH) algorithm
to filter out genes until three lncRNAs, three microRNAs and three
PCGs were screened out (21).
Subsequently, in order to further screen prognostic
genes with better predictive power, we performed a multivariable
Cox regression analysis and developed a model to estimate prognosis
risk as follows:
Risk Score(RS)=∑i=1N(Expi*Coei)
where N is the number of prognostic lncRNAs,
microRNAs or PCGs; Expi is the expression value
of lncRNAs, microRNAs or PCGs; and Coei is the
estimated regression coefficient of lncRNAs, microRNAs or PCGs in
the multivariable Cox regression analysis. Each patient then
obtained 511 risk scores since three lncRNAs, three microRNAs and
three PCGs could form 29−1=511 combinations or
signatures. The time-dependent receiver operating characteristic
(ROC) curve was used to compare the sensitivity and specificity of
the survival prediction risk score of the 511 signatures in the
training dataset. AUC were calculated from the ROC curve. By
comparing the AUC values, we constructed the prognostic
PCG-lncRNA-microRNA signature in the training group.
Statistical analysis
We used the median risk score in the training
dataset as a cut-off value (22),
and the BLCA patients were divided into high- and low-risk groups.
The estimation of survival time and comparison of survival curves
of the high- and low-risk groups were obtained with the
Kaplan-Meier survival analyses, and statistical significance was
assessed using the two-sided log-rank test. We then validated the
prognostic performance of the PCG-lncRNA-microRNA signature in the
test dataset by the Kaplan-Meier survival analysis and ROC values.
We compared the survival prediction power of the
PCG-lncRNA-microRNA signature with TNM stage by the ROC values.
Furthermore, multivariable Cox regression analysis and data
stratification analysis were performed to assess whether the
PCG-lncRNA-microRNA signature was an independent prognostic factor
within the available data. P<0.05 was considered to indicate a
statistically significant difference. All analyses were performed
using the R programme (www.r-project.org), including packages named pROC,
survival and randomForestSRC downloaded from Bio-conductor.
Function prediction of the selected
prognostic lncRNAs, microRNAs and PCGs
The co-expressed relationships between the selected
prognostic genes (2 lncRNAs, 2 microRNAs and 2 PCGs) and
protein-coding genes in BLCA patients were computed using Pearson
correlation coefficient visualized by Cytoscape. Gene Ontology (GO)
and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment
analyses of the co-expressed protein-coding genes were then
performed to predict their biological function. GO analysis was
performed in the ClueGo of the Cytoscape plugin (version 3.2.3)
(23,24), which is a commonly used functional
annotation tool that can assess over-representation of functional
categories among a gene set of interest. Enrichment analysis was
performed using the functional annotation chart and functional
annotation clustering options and was limited to GO terms and KEGG
pathways in the ‘Biological Process’ categories. Functional
annotation with P<0.05 was considered significant.
Results
Patient characteristics and expression
profiles
Expression profiles and corresponding clinical data
of 239 (out of 411) patients diagnosed with BLCA were downloaded
from TCGA database, and 172 (out of 411) patients with missing
clinical data were not included in the present study. The median
age of the enrolled patients was 69 years (60–76 years). Of the
BLCA patients, 235 were stage I, II, III and IV disease, and the
stage of 4 patients was unknown. Other clinical information of
patient characteristics is summarized in Table I. Simultaneously, the expression
values of 6,603 lncRNAs, 429 microRNAs and 14,644 PCGs for the BLCA
patients were obtained after eliminating low-expression genes, and
missing expression values were removed as described in the
Materials and methods. All of these gene expression values were
log2 transformed. Subsequently, we divided the dataset randomly
into two groups (training dataset, n=119; test dataset, n=120) to
explore and validate the prognostic PCG-lncRNA-microRNA signature
of the BLCA patients. The selection process of the prognostic
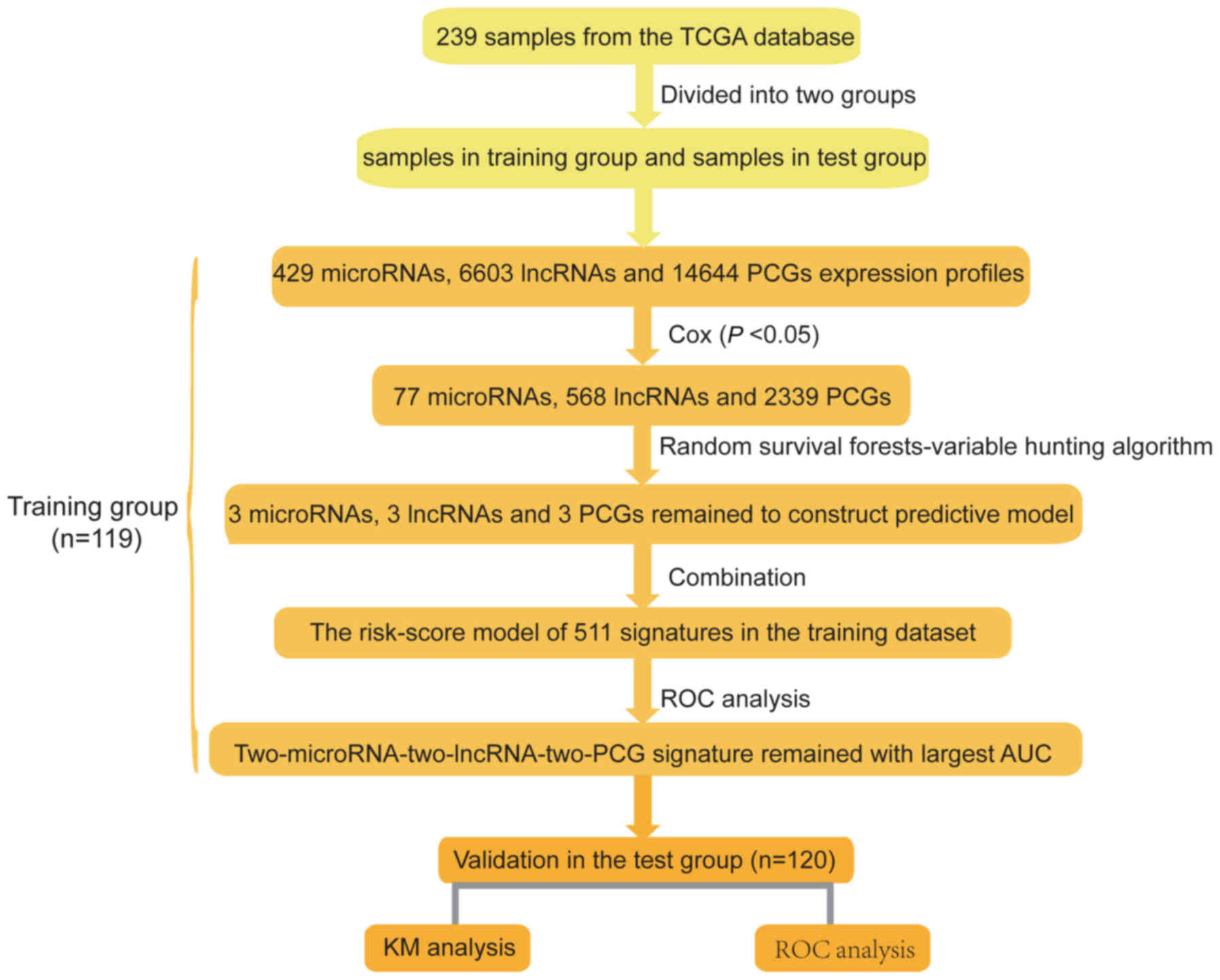
PCG-lncRNA-microRNA signatures is displayed in Fig. 1.
 | Table I.Patient demographics and clinical
characteristics. |
Table I.
Patient demographics and clinical
characteristics.
|
Characteristics | Training set | Testing set | Total |
|---|
| pTNM stage |
|
Unknown | 3 | 1 | 4 |
| stage
I | 1 | 1 | 2 |
| stage
II | 28 | 48 | 76 |
| stage
III | 34 | 46 | 80 |
| stage
IV | 53 | 24 | 77 |
| Vital status |
|
Living | 76 | 82 | 158 |
|
Dead | 43 | 38 | 81 |
| Sex |
|
Female | 32 | 25 | 57 |
|
Male | 87 | 95 | 182 |
| Age (years) |
|
>69 | 47 | 52 | 99 |
|
≤69 | 72 | 68 | 140 |
Identification of three prognostic
lncRNAs, microRNAs and mRNAs from the training dataset
We used the training dataset to select the
prognostic genes. Firstly, using survival time and clinical outcome
as the dependent variable, we performed univariate Cox proportional
hazards regression analysis of all of the lncRNAs, microRNAs and
PCGs expressed in the training dataset and identified a 2,974-gene
set including 568 lncRNAs, 77 microRNAs and 2,339 PCGs, which were
significantly correlated with patient OS (P-value <0.05, data
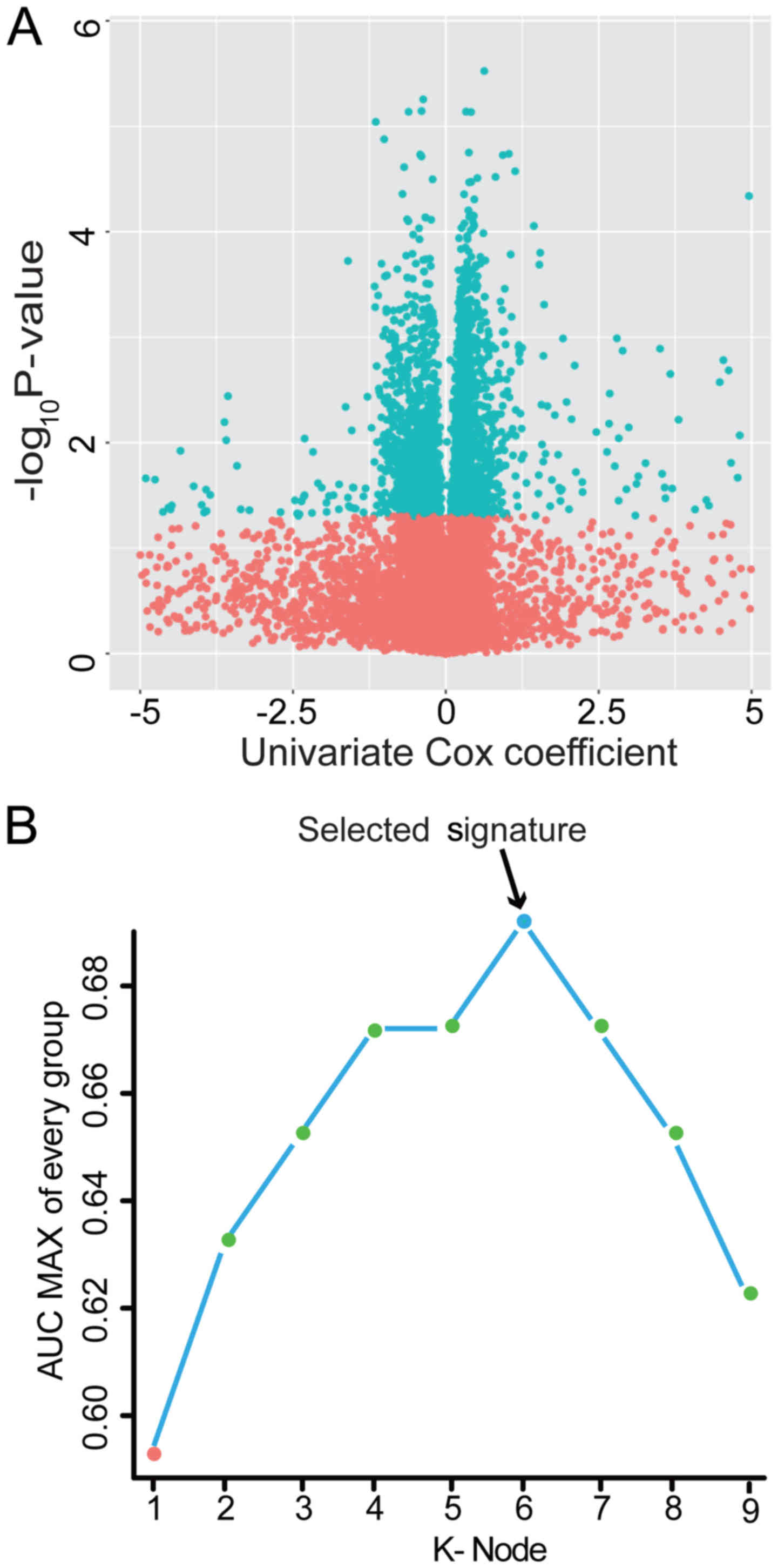
not shown). To visually display these selected genes, we drew a
volcano plot using the univariate Cox coefficient as the x-axis and
-log10 (P-value) as the y-axis. As displayed in Fig. 2A, blue dots in the volcano plot
represent the 2,974 genes with significant differences (P<0.05),
and red dots represent the remaining genes with no significant
differences. Secondly, we further screened the prognostic genes
from the above 2,974 genes using a random forest supervised
classification algorithm, and three lncRNAs (RP11-60L3.1,
CTD-3195I5.3 and TMC4), three microRNAs (hsa-miR-3913-1,
hsa-miR-891a and hsa-mir-1976) and three PCGs (ACADS, C1QTNF9B and
HP) strongly related to patient survival were screened out
according to the permutation important score by the random survival
forests-variable hunting (RSFVH) algorithm.
Construction of the prognostic
PCG-lncRNA-microRNA signature in the training dataset
Three lncRNAs, three microRNAs and three PCGs formed
511 combinations or signatures, and each combination had a risk
score according to the risk score model constructed in the
Materials and methods. To select a signature with the biggest
prediction power from these 511 combinations in the training
dataset, we performed 511 time-dependent ROC analyses using the
patient survival statuses and signature risk scores as variables
and compared their areas under the respective ROC curves (AUC)
(data not shown). Among them, the PCG-lncRNA-microRNA combination
comprised of ACADS, C1QTNF9B, RP11-60L3.1, CTD-3195I5.3,
has-miR-3913-1 and has-miR-891a with the max AUC was screened out
(Fig. 2B and Table II). The risk score model comprised
of ACADS, C1QTNF9B, RP11-60L3.1, CTD-3195I5.3, has-miR-3913-1 and
has-miR-891a was as follows: Risk score = (−0.59 × expression value
of ACADS) + (0.47 × expression value of C1QTNF9B) + (0.11 ×
expression value of RP11-60L3.1) + (−1.54 × expression value of
CTD-3195I5.3) + (−0.46 × expression value of hsa-miR-3913-1) +
(0.38 × expression value of hsa-miR-891a). The AUC of the selected
PCG-lncRNA-microRNA signature in the training group was 0.79,
demonstrating its good survival prediction performance.
 | Table II.Identities of PCGs, lncRNAs and
microRNAs in the prognostic expression signature and their
univariable Cox association with prognosis. |
Table II.
Identities of PCGs, lncRNAs and
microRNAs in the prognostic expression signature and their
univariable Cox association with prognosis.
| Database ID | Gene symbol | Gene name |
Coefficienta |
P-valuea | Gene expression
level association with poor prognosis | Chromosome location
(GRCh38/hg38) |
|---|
|
ENSG00000122971b | ACADS | Acyl-CoA
dehydrogenase | −0.59 | 0.01 | Low |
chr12:120725735-120740008:1 |
|
ENSG00000205863b | C1QTNF9B | C1q and tumor
necrosis factor related protein 9B | 0.47 | 0.00 | High |
chr13:23891099-23902655:-1 |
|
ENSG00000259264b | RP11-60L3.1 |
| 0.11 | 0.05 | High |
chr15:74,202,705-74,221,557:1 |
|
ENSG00000262692b | CTD-3195I5.3 |
| −1.54 | 0.01 | Low | chr
17:3721628-3722488:1 |
|
MI0016417c | hsa-miR-3913-1 |
| −0.46 | 0.00 | Low |
chr12:69584722-69584823: −1 |
|
MI0005524c | hsa-miR-891a |
| 0.38 | 0.03 | High |
chrX:146027794-146027872:-1 |
Validation of the selected
PCG-lncRNA-microRNA signature for survival prediction in the
training and the test dataset
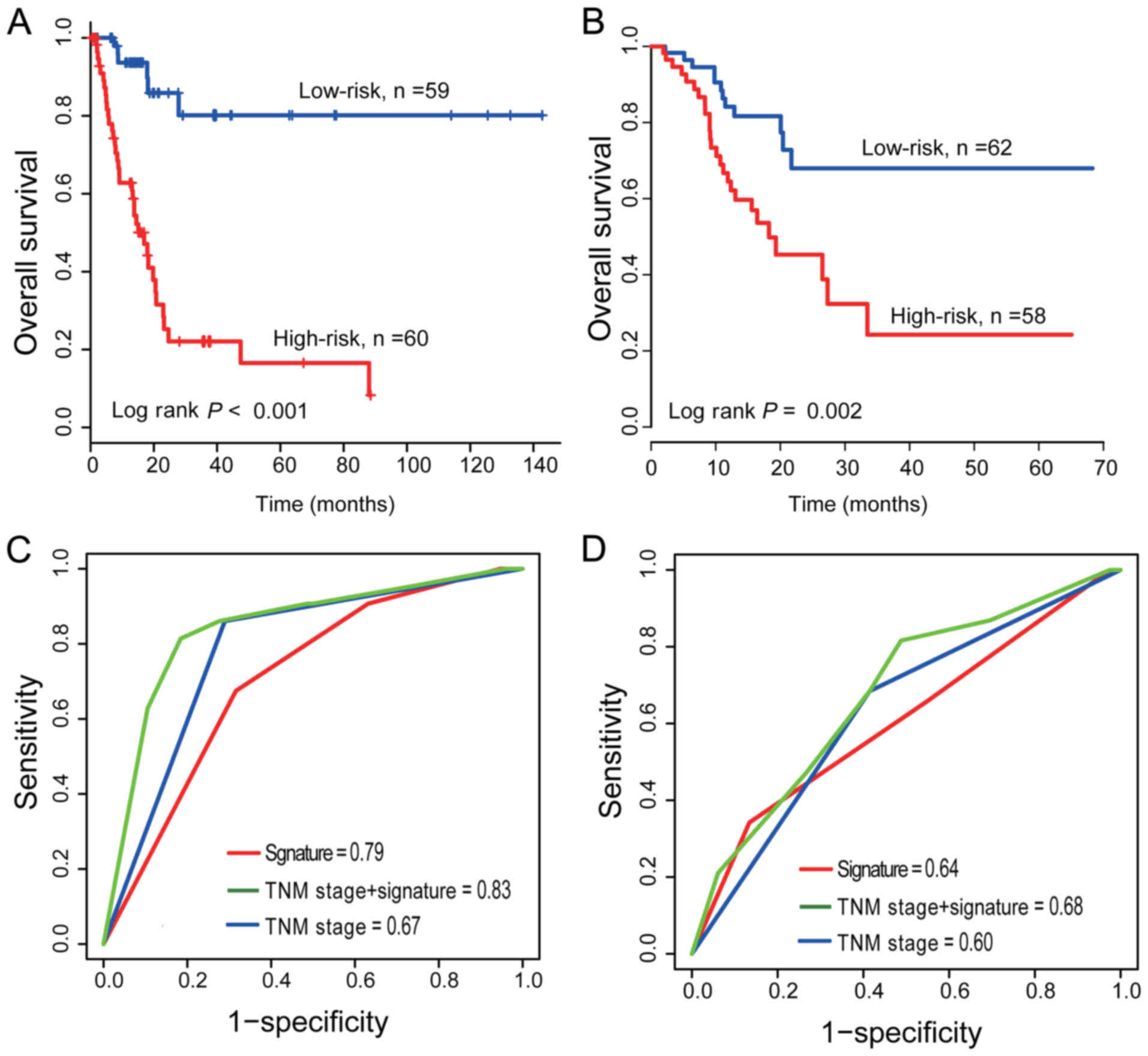
In the training dataset, the selected
PCG-lncRNA-microRNA signature gave each patient a risk score with
the prognostic model and divided patients into a high-risk group
(n=60) and low-risk group (n=59). The median risk score was used as
the cut-off point. Then, Kaplan-Meier survival analysis was
performed to compare the overall survival rates of the two groups
of patients. Patients in the high-risk group had significantly
shorter OS than those in the low-risk group (median survival: 16.9
months vs. 54.9 months, log-rank test P<0.001; Fig. 3A). The 5-year OS rate of the
patients in the high-risk group was <11%, while that of the
patients in the low-risk group was >60%.
To validate the prognostic prediction power of the
signature, the same prognostic risk score model obtained from the
training dataset was used to calculate the PCG-lncRNA-microRNA
signature-based risk scores of 120 patients in the test dataset.
Similarly, the test dataset was divided into two groups using the
same median cut-off point obtained from the training dataset: one
group with a high risk and the other group with a low risk.
Kaplan-Meier curves for the high- and low-risk groups in the test
dataset are displayed in Fig. 3B.
Similar to the results of the training dataset, the OS rate of the
high-risk group in the test dataset was significantly lower than
that of the low-risk group (median survival: 18.2 months vs. 58.9
months, log-rank test P=0.003). The OS rate of the patients in the
high-risk group was ~19% at 5 years in contrast to 45% in the
low-risk group.
The selected PCG-lncRNA-microRNA
signature is an independent prognostic factor
To obtain a better understanding of the clinical
significance of the PCG-lncRNA-microRNA signature in BLCA patients,
we correlated the signature to a series of clinicopathological
parameters in the training groups (n=119). As displayed in Table III, there was an association
between the PCG-lncRNA-microRNA signature and clinicopathological
variables, including regional lymph nodes and TNM stage (Chi-square
test, P<0.05).
 | Table III.Association of the
PCG-lncRNA-microRNA signature with clinicopathological
characteristics in BLCA patients (training group, n=119). |
Table III.
Association of the
PCG-lncRNA-microRNA signature with clinicopathological
characteristics in BLCA patients (training group, n=119).
|
| PCG-lncRNA
signature |
|
|---|
|
|
|
|
|---|
| Variables | Low
riska | High
riska | P-value |
|---|
| Age |
|
| 0.76 |
|
>69 | 28 | 19 |
|
|
≤69 | 32 | 40 |
|
| Sex |
|
| 1.00 |
|
Female | 16 | 16 |
|
|
Male | 44 | 43 |
|
| Pathologic M |
|
| 0.84 |
| M0 | 32 | 31 |
|
| M1 | 1 | 2 |
|
| MX | 27 | 26 |
|
| Regional lymph
nodes |
|
| 0.01 |
| N0 | 32 | 21 |
|
| N1 | 5 | 12 |
|
| N2 | 12 | 21 |
|
| N3 | 1 | 3 |
|
| NX | 10 | 2 |
|
| Primary tumor |
|
| 0.11 |
| T1 | 1 | 0 |
|
| T2 | 15 | 14 |
|
| T3 | 26 | 37 |
|
| T4 | 10 | 6 |
|
| TX | 8 | 2 |
|
| pTNM stage |
|
| 0.02 |
| I | 1 | 0 |
|
| II | 19 | 9 |
|
|
III | 20 | 14 |
|
| IV | 18 | 35 |
|
| stage
X | 2 | 1 |
|
To assess whether the PCG-lncRNA-microRNA signature
was an independent risk factor for survival prediction,
multivariable Cox regression analysis was performed using the
PCG-lncRNA-microRNA signature-based risk score and other clinical
features as covariates. The P-values of the prognostic signature in
multivariable Cox regression analysis from the training datasets
was <0.05, which indicated that the PCG-lncRNA-microRNA
signature risk score was an independent prognostic factor,
following adjustment for other clinical features including sex, age
and pTNM (high-risk group vs. low-risk group, HR=7.42, 95% CI
3.08–17.88, P<0.001, n=119; Table
IV). The same result appeared in the test dataset (HR=2.84, 95%
CI 1.43–5.64, P<0.001, n=120; Table
IV).
 | Table IV.Univariable and multivariable Cox
regression analysis of the PCG-lncRNA-microRNA signature and
survival of BLCA patients in the training, test and entire
group. |
Table IV.
Univariable and multivariable Cox
regression analysis of the PCG-lncRNA-microRNA signature and
survival of BLCA patients in the training, test and entire
group.
|
|
| Training set
(n=119) | Test set
(n=120) | Entire dataset
(n=239) |
|---|
|
|
|
|
|
|
|---|
|
|
|
| 95% CI of HR |
|
| 95% CI of HR |
|
| 95% CI of HR |
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Variables |
| HR | Lower | Upper | P-value | HR | Lower | Upper | P-value | HR | Lower | Upper | P-value |
|---|
| Univariable
analysis |
|
Age | >69 vs. ≤69 | 1.19 | 0.82 | 1.73 | 0.36 | 1.16 | 0.51 | 2.63 | 0.28 | 0.91 | 0.55 | 1.50 | 0.70 |
|
Sex | Male vs.
female | 0.79 | 0.42 | 1.50 | 0.48 | 1.23 | 1.07 | 1.86 | 0.32 | 1.11 | 1.11 | 1.63 | 0.40 |
| pTNM
stage | IV vs.
I+II+III | 2.39 | 1.26 | 4.53 | 0.01 | 2.31 | 1.18 | 4.51 | 0.01 | 2.18 | 1.41 | 3.38 | 0.00 |
| PCG-lncRNA-microRNA
signature | High risk vs. low
risk | 8.29 | 3.49 | 19.70 | 0.00 | 2.84 | 1.43 | 5.64 | 0.00 | 4.68 | 2.77 | 7.91 | 0.00 |
| Multivariable
analysis |
|
Age | >69 vs. ≤69 | 1.23 | 1.06 | 1.98 | 0.43 | 0.91 | 0.83 | 1.76 | 0.36 | 1.19 | 1.00 | 1.79 | 0.37 |
|
Sex | Male vs.
female | 0.68 | 0.35 | 1.30 | 0.24 | 1.24 | 0.53 | 2.89 | 0.61 | 0.92 | 0.56 | 1.53 | 0.76 |
| pTNM
stage | IV vs.
I+II+III | 1.41 | 0.92 | 2.17 | 0.11 | 2.43 | 1.22 | 4.81 | 0.01 | 1.83 | 1.17 | 2.84 | 0.00 |
| PCG-lncRNA-microRNA
signature | High risk vs. low
risk | 7.42 | 3.08 | 17.88 | 0.00 | 3.09 | 1.54 | 6.20 | 0.00 | 4.30 | 2.54 | 7.31 | 0.00 |
Comparison of the survival prediction
power of the PCG-lncRNA-microRNA signature with TNM stage
To compare the survival prediction power of TNM
stage and the PCG-lncRNA-microRNA signature, we performed ROC
analysis considering that a larger AUC usually represented a better
model for prediction (25,26). In the training dataset (n=119), the
AUC of the PCG-lncRNA-microRNA signature was greater than that of
the TNM stage (AUCSignature=0.79 vs.
AUCTNM=0.67, n=119; Fig.
3C), which demonstrated that the signature for survival
prediction in our study had high sensitivity and specificity and
had important clinical significance. The same result was indicated
in the test group (AUCSignature=0.64 vs.
AUCTNM=0.60, n=120; Fig.
3D), while the model combining TNM with the PCG-lncRNA-microRNA
signature had a larger AUC than the TNM stage or signature alone
(AUCTNM+ Signature=0.83/0.68 in the training/test group;
Fig. 3C and D).
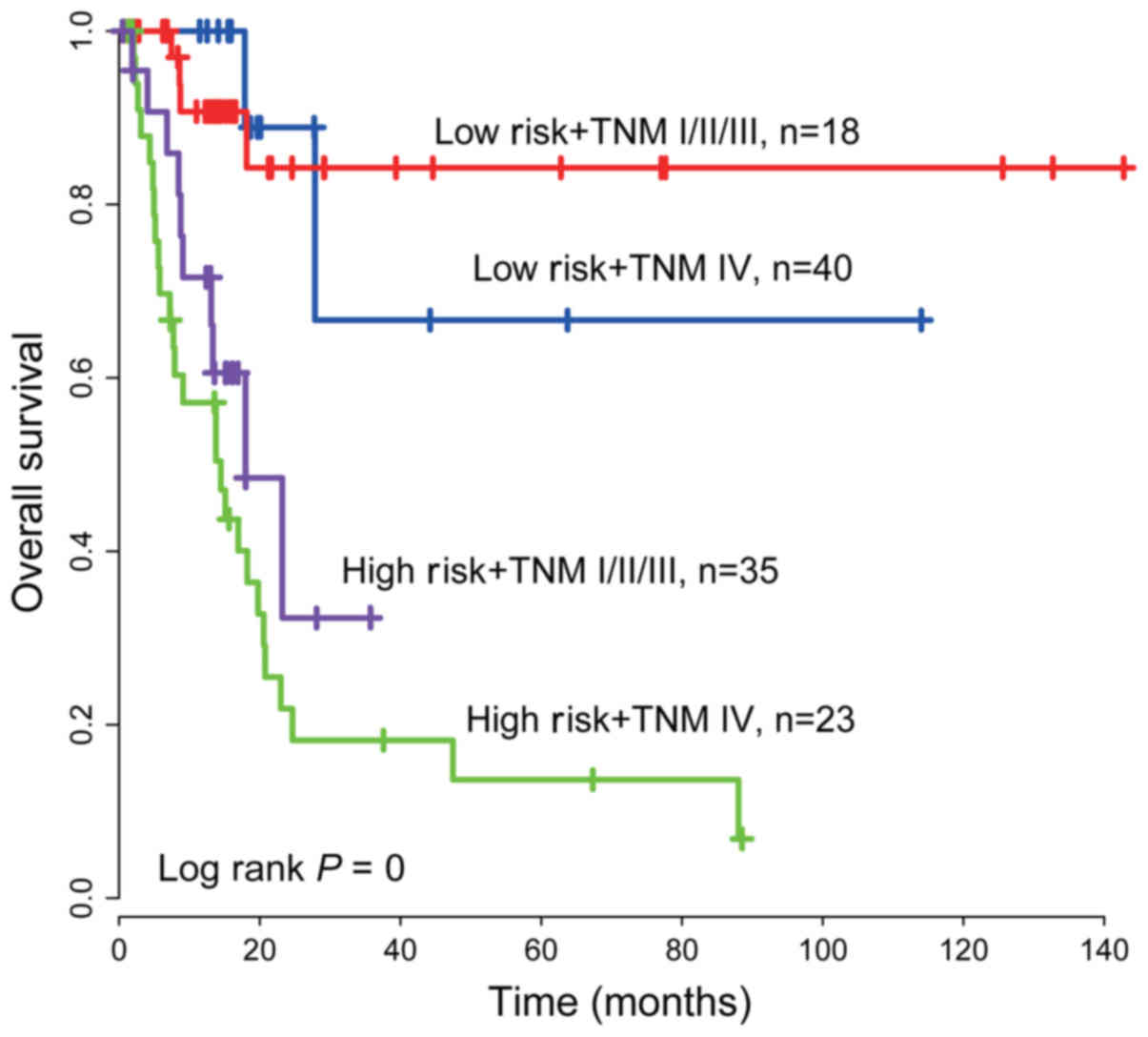
Stratification analysis
Due to the correlation between the TNM stage and the
PCG-lncRNA-microRNA signature (P<0.05, Table III) and the combination of TNM
with the PCG-lncRNA-microRNA signature having a relatively large
AUC, we stratified the TNM stage by the PCG-lncRNA-microRNA
signature risk score, and thus, patients from the training dataset
(n=119) were grouped into four groups: high-risk and TNM IV stage,
high-risk and TNM I+II+III stage, low-risk and TNM IV stage, and
low-risk and TNM I+II+III stage. The Kaplan-Meier test was
performed, and Kaplan-Meier curves revealed that the combination
model could more precisely subdivide patients. The log-rank test
revealed that the signature could further subdivide TNM I+II+III
stage patients into either a high-risk group with shorter survival
or a low-risk group with longer survival (log-rank test
P<0.001). Similarly, the TNM stage IV patients were also divided
into a high-risk group with lower OS and a low-risk group with
higher OS (log-rank test P<0.001, Fig. 4).
Functional characterization of the
selected prognostic lncRNAs, microRNAs and PCGs
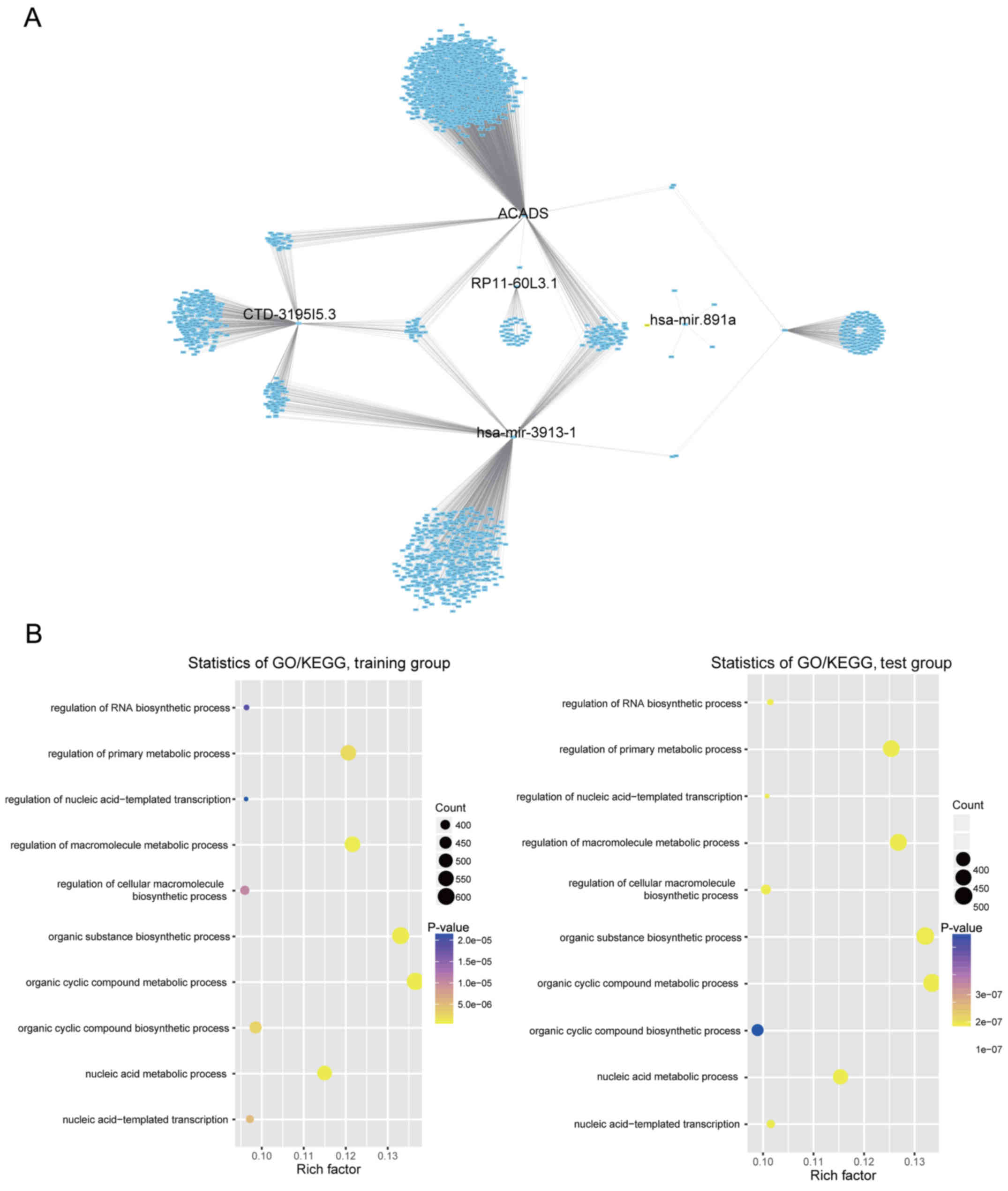
Co-expression relationships of these selected
lncRNAs, microRNAs and PCGs with their corresponding protein-coding
genes were computed using Pearson's correlation coefficient in the
training/test dataset. A total of 1,593/1,643 protein-coding genes
in the training/test dataset were highly correlated with at least
one of the selected lncRNAs, microRNAs and PCGs, and the
interaction network was visualized by Cytoscape (27) (Pearson's correlation coefficient
>0.40, P<0.05; Fig. 5A and
data not shown). GO and KEGG of these co-expressed protein-coding
genes was performed, indicating that in the two datasets
co-expressed protein-coding genes were significantly enriched in 31
different GO terms and KEGG pathways (P<0.05). The six genes may
be involved in tumourigenesis by interacting with those
protein-coding genes that affect the important biological processes
such as cellular nitrogen compound metabolic process,
nucleobase-containing compound metabolic process, heterocycle
metabolic process and cellular aromatic compound metabolic process
(top 5 ranking by P-value, Fig.
5B).
Discussion
In a broad sense, bladder cancer includes not only
bladder urothelial carcinoma (BLCA) but also non-epithelial
cancers, such as lymphoma or sarcoma. However, BLCA is ordinarily
considered as bladder cancer since it accounts for 90% of bladder
cancers. In the present study, we used BLCA samples to explore
bladder cancer prognosis markers. In terms of prognosis, bladder
cancer patients with similar TNM staging have different survival
time. A portion of them may encounter disease recurrence and
progression, which decreases the OS rate to a great extent. Thus,
the 5-year survival rate of BLCA patients varies significantly from
5 to 70%. Thereby, TNM staging, as the dominant prognostic
assessment tool, is not adequate for prediction of individual
outcomes. In the era of precision medicine, gene signature has been
identified as being able to serve as a novel biomarker for
predicting the survival of various carcinomas. In the past decades,
with the development of transcriptomic and bioinformatics studies,
mRNAs as well as non-coding RNAs including lncRNAs and microRNAs,
have become the focus of tumour prognosis-related research.
lncRNAs have been found to be involved in regulating
gene expression at transcriptional, post-transcriptional and
epigenetic levels (4), and
microRNAs could regulate hundreds of downstream genes by targeting
the 3′untranslated region of specific messenger RNAs for
degradation or translational repression (4). Furthermore, similar to protein coding
genes or mRNAs, accumulating evidence has indicated that both
lncRNAs and microRNAs are involved in oncogenic and tumour
suppressive pathways and perform various functions in a wide
variety of important biological processes, such as transcriptional
regulation, cell growth and tumourigenesis (29–33).
Different from protein coding genes, lncRNAs and microRNAs are
tissue-specific and their expression is more closely associated
with biological function and tumour status as their non-coding
feature (30,34–37).
All these characteristics open a door for lncRNAs, microRNAs and
PCGs to be used as cancer diagnostic or prognostic biomarkers.
Their prognostic value in various types of cancer has been
confirmed by numerous studies. As aforementioned, lncRNAs,
microRNAs and PCGs have revealed their prognostic potential in
bladder cancer. However, current studies are limited to one type of
RNA (lncRNA, microRNA or mRNA) or RNA signature, and there is
little research combining three types of RNA and exploring more
powerful markers from a comprehensive and multidimensional
transcriptome level. In the present study, by analysing the BLCA
transcriptome data, we aimed to find a multidimensional
transcriptome gene signature that could predict the survival of
bladder cancer patients. Based on the results of TCGA expression
data mining, we identified a signature involving two lncRNAs, two
microRNAs and two PCGs that was significantly associated with the
OS of BLCA patients in the training dataset. The selected signature
separated BLCA patients into a low-risk group and a high-risk group
with significantly different survival times in the training or test
dataset, demonstrating its good prognostic performance.
Multivariable Cox regression analysis was performed to assess the
independence of the selected PCG-lncRNA-microRNA signature in
predicting OS in the training dataset and test dataset. With sex
and pTNM stage as covariables in the regression analysis, risk
score of the PCG-lncRNA-microRNA signature maintained an
independent correlation with OS. Collectively, these results
indicated that the prognostic power of the PCG-lncRNA-microRNA
signature was independent of other clinical features. In addition,
comparison of the prediction ability of TNM staging and the
multidimensional transcriptome signature confirmed that the
signature was superior to TNM staging. The stratification analysis
found that the PCG-lncRNA-microRNA signature combined with TNM
stage demonstrated a more robust prognostic power, implying that
the signature could be an auxiliary biomarker of TNM stage to more
precisely subdivide BLCA patients.
As for the characteristics of the PCGs, lncRNAs and
microRNAs in the signature, we found that high expression of
C1QTNF9B, RP11-60L3.1 and hsa-miR-891a was associated with a short
survival time (univariable Cox coefficient >0), and high
expression of ACADS, CTD-3195I5.3 and hsa-miR-3913-1 was associated
with a long survival time (univariable Cox coefficient <0) (the
KM analysis of the six genes in the training group are data not
shown). C1QTNF9B is a novel protein that hetero-oligomerizes with
C1q/TNF family members. C1q/TNF family members have been revealed
to play diverse roles in various physiological processes in
different tissue compartments, ranging from development to the
immune, endocrine, skeletal, neuronal, reproductive, sensory and
vascular systems (38). ACADS is
involved in free fatty acid β-oxidation and regulates energy
homeostasis (39). hsa-miR-891a can
target bladder cancer-associated protein (BLCAP) potentially in the
3′UTR (40), while there is a lack
of research for the remaining genes. Although functions of these
lncRNAs, microRNAs and PCGs have been inferred by bioinformatics
analysis, the biological roles of the selected genes in
tumourigenesis are still not clear and should be investigated in
further experimental studies. With the rapidly increasing related
studies, more multidimensional signatures will become available,
such as combining PCGs, lncRNAs and microRNAs with circular RNAs or
other non-coding RNAs.
Limitations in the present study need to be
acknowledged. First, only a fraction of human mRNAs (14,644 out of
30,000+), lncRNAs (6,603 out of 15,000+) and microRNAs (429 out of
2,000+) were included in the present study. Therefore, we may have
missed candidates that are potentially correlated with BLCA overall
survival. Second, we failed to further seek the mechanisms behind
the prognostic values of these PCGs, lncRNAs and microRNAs in BLCA,
and experimental studies on these PCGs, lncRNAs and microRNAs may
provide important information to deepen our understanding of their
functional roles. Finally, although we demonstrated the selected
PCG-lncRNA-microRNA signature may replace or complement the TNM
staging, applying it for clinical use still warrants more confirmed
studies. Despite these drawbacks, however, the PCG-lncRNA-microRNA
signature has been demonstrated to serve as a marker for BLCA
patient prognosis prediction.
In conclusion, the multidimensional transcriptome
signature comprised of ACADS, C1QTNF9B, RP11-60L3.1, CTD-3195I5.3,
hsa-miR-3913-1 and hsa-miR-891a can predict the survival of bladder
cancer patients with more accuracy and further subdivide BLCA
patients combined with TNM stages, revealing that the model has
promising clinical significance.
Acknowledgements
Not applicable.
Funding
The present study was supported by the
Bioinformatics Research Group of the Chinese Academy of
Science.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CPL, JHZ, JL and SCZ collected, analysed,
interpreted and drafted the data. JCG designed and supervised the
study and provided the final approval of the manuscript. JHZ, JL
and JCG provided technical support. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under the ROC curve
|
|
CI
|
confidence interval
|
|
GO
|
Gene Ontology
|
|
HR
|
hazard ratio
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
lncRNAs
|
long non-coding RNAs
|
|
OS
|
overall survival
|
|
BLCA
|
bladder urothelial carcinoma
|
|
SD
|
standard deviation
|
|
TCGA
|
The Cancer Genome Atlas
|
References
|
1
|
Clark PE, Spiess PE, Agarwal N, Bangs R,
Boorjian SA, Buyyounouski MK, Efstathiou JA, Flaig TW, Friedlander
T, Greenberg RE, et al: NCCN Guidelines Insights: Bladder cancer,
version 2.2016. J Natl Compr Canc Netw. 14:1213–1224. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Solomon JP and Hansel DE: Prognostic
factors in urothelial carcinoma of the bladder: Histologic and
molecular correlates. Adv Anat Pathol. 22:102–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dykes IM and Emanueli C: Transcriptional
and post-transcriptional gene regulation by long non-coding RNA.
Genomics Proteomics Bioinformatics. 15:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petrovic N, Ergun S and Isenovic ER:
Levels of microRNA heterogeneity in cancer biology. Mol Diagn Ther.
21:511–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo JC, Li CQ, Wang QY, Zhao JM, Ding JY,
Li EM and Xu LY: Protein-coding genes combined with long non-coding
RNAs predict prognosis in esophageal squamous cell carcinoma
patients as a novel clinical multi-dimensional signature. Mol
Biosyst. 12:3467–3477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo JC, Wu Y, Chen Y, Pan F, Wu ZY, Zhang
JS, Wu JY, Xu XE, Zhao JM, Li EM, et al: Protein-coding genes
combined with long noncoding RNA as a novel transcriptome molecular
staging model to predict the survival of patients with esophageal
squamous cell carcinoma. Cancer Commun. 38:42018. View Article : Google Scholar
|
|
9
|
Shang C, Guo Y, Zhang H and Xue YX: Long
noncoding RNA HOTAIR is a prognostic biomarker and inhibits
chemosensitivity to doxorubicin in bladder transitional cell
carcinoma. Cancer Chemother Pharmacol. 77:507–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Y, Lu W, Xu J, Shi Y, Zhang H and Xia
D: Prognostic value of long non-coding RNA MALAT1 in cancer
patients. Tumour Biol. 37:897–903. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ratert N, Meyer HA, Jung M, Lioudmer P,
Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert
S, et al: miRNA profiling identifies candidate mirnas for bladder
cancer diagnosis and clinical outcome. J Mol Diagn. 15:695–705.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang S, Li Q, Wang K, Dai Y, Yang J, Xue
S, Han F, Zhang Q, Liu J and Wu W: Decreased expression of
microRNA-31 associates with aggressive tumor progression and poor
prognosis in patients with bladder cancer. Clin Transl Oncol.
15:849–854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S, Xue S, Dai Y, Yang J, Chen Z, Fang
X, Zhou W, Wu W and Li Q: Reduced expression of microRNA-100
confers unfavorable prognosis in patients with bladder cancer.
Diagn Pathol. 7:1592012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villadsen SB, Bramsen JB, Ostenfeld MS,
Wiklund ED, Fristrup N, Gao S, Hansen TB, Jensen TI, Borre M,
Ørntoft TF, et al: The miR-143/-145 cluster regulates plasminogen
activator inhibitor-1 in bladder cancer. Br J Cancer. 106:366–374.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bao Z, Zhang W and Dong D: A potential
prognostic lncRNA signature for predicting survival in patients
with bladder urothelial carcinoma. Oncotarget. 8:10485–10497. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou H, Tang K, Xiao H, Zeng J, Guan W,
Guo X, Xu H and Ye Z: A panel of eight-miRNA signature as a
potential biomarker for predicting survival in bladder cancer. J
Exp Clin Cancer Res. 34:532015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang X, Du L, Duan W, Wang R, Yan K, Wang
L, Li J, Zheng G, Zhang X, Yang Y, et al: Serum microRNA expression
signatures as novel noninvasive biomarkers for prediction and
prognosis of muscle-invasive bladder cancer. Oncotarget.
7:36733–36742. 2016.PubMed/NCBI
|
|
18
|
Mitra AP, Pagliarulo V, Yang D, Waldman
FM, Datar RH, Skinner DG, Groshen S and Cote RJ: Generation of a
concise gene panel for outcome prediction in urinary bladder
cancer. J Clin Oncol. 27:3929–3937. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, Taylor J, Eustace A, Irlam JJ,
Denley H, Hoskin PJ, Alsner J, Buffa FM, Harris AL, Choudhury A, et
al: A gene signature for selecting benefit from hypoxia
modification of radiotherapy for high-risk bladder cancer patients.
Clin Cancer Res. 23:4761–4768. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z,
Shao T, Zhang J, Wang L and Li X: The mRNA related ceRNA-ceRNA
landscape and significance across 20 major cancer types. Nucleic
Acids Res. 43:8169–8182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li J, Chen Z, Tian L, Zhou C, He MY, Gao
Y, Wang S, Zhou F, Shi S, Feng X, et al: LncRNA profile study
reveals a three-lncRNA signature associated with the survival of
patients with oesophageal squamous cell carcinoma. Gut.
63:1700–1710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo JC, Xie YM, Ran LQ, Cao HH, Sun C, Wu
JY, Wu ZY, Liao LD, Zhao WJ, Fang WK, et al: L1CAM drives
oncogenicity in esophageal squamous cell carcinoma by stimulation
of ezrin transcription. J Mol Med. 95:1355–1368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tse LA, Dai J, Chen M, Liu Y, Zhang H,
Wong TW, Leung CC, Kromhout H, Meijer E, Liu S, et al: Prediction
models and risk assessment for silicosis using a retrospective
cohort study among workers exposed to silica in China. Sci Rep.
5:110592015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fatica A and Bozzoni I: Long non-coding
RNAs: new players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang XQ, Sun S, Lam KF, Kiang KM, Pu JK,
Ho AS, Lui WM, Fung CF, Wong TS and Leung GK: A long non-coding RNA
signature in glioblastoma multiforme predicts survival. Neurobiol
Dis. 58:123–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wan J, Wu W, Che Y, Kang N and Zhang R:
Insights into the potential use of microRNAs as a novel class of
biomarkers in esophageal cancer. Dis Esophagus. 29:412–420. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Geng Q, Fan T, Zhang B, Wang W, Xu Y and
Hu H: Five microRNAs in plasma as novel biomarkers for screening of
early-stage non-small cell lung cancer. Respir Res. 15:1492014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hauptman N and Glavac D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Du Z, Fei T, Verhaak RG, Su Z, Zhang Y,
Brown M, Chen Y and Liu XS: Integrative genomic analyses reveal
clinically relevant long noncoding RNAs in human cancer. Nat Struct
Mol Biol. 20:908–913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Peterson JM, Wei Z and Wong GW: CTRP8 and
CTRP9B are novel proteins that hetero-oligomerize with C1q/TNF
family members. Biochem Biophys Res Commun. 388:360–365. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen Y and Su Z: Reveal genes functionally
associated with ACADS by a network study. Gene. 569:294–302. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|