Introduction
Liver cancer ranks as one of the most lethal
malignancies worldwide, with 42,220 new cases and 30,200 deaths
estimated for the United States for 2018 (1). Hepatocellular carcinoma (HCC) is the
predominant histological subtype of liver cancer and places a heavy
burden on human health (2,3). HCC is associated with multiple
etiological factors, including hepatitis C and B virus infection,
exposure to toxins and alcohol abuse (4,5).
Therapeutic strategies combining surgical resection and molecular
targeted treatment have provided encouraging results and improved
the outcomes for HCC patients. However, the prognosis for patients
with unresectable advanced-stage HCC remains poor (6,7).
Sorafenib was the only systemic drug available for treating
advanced HCC until the recent approval of regorafenib, another
multi-kinase inhibitor (8,9). However, sorafenib and regorafenib have
a low durable response rate and their benefit for survival is
limited (10). Hence, novel
prognostic biomarkers and a deeper understanding of the exact
molecular mechanisms in HCC are urgently required to improve its
clinical management.
While it was previously assumed that genes encoding
non-coding RNAs have no function, accumulating evidence has proved
that several non-coding RNAs, including long non-coding RNA
(lncRNA) and microRNA (miRNA), have vital regulatory roles in
cellular biology and physiological processes. Of note, small
nucleolar RNAs (snoRNAs), which are non-coding RNAs that are 60–300
nucleotides in length, were proposed to be closely associated with
various human diseases, including cancer (11,12).
Several studies have reported that certain snoRNAs act as
diagnostic or prognostic biomarkers and as therapeutic targets for
HCC (13,14). Several in vitro and in
vivo studies indicated that certain snoRNAs are involved in the
regulation of the genesis and biological behavior of HCC, including
cell proliferation, migration, apoptosis, cell cycle and
metastasis. In spite of this, the function of snoRNAs in HCC
remains limited and requires further elucidation. Systematic
investigation of the expression profiles and clinical significance
of snoRNAs in HCC may provide a deeper understanding of their roles
in HCC and contribute to the development of novel therapeutic
strategies. Gong et al (15)
developed an online database of snoRNAs in cancers (SNORic), which
provides expression profiles in >10,000 samples of different
tumor types using calculations based on The Cancer Genome Atlas
(TCGA) database. The database provides expression profiles of
snoRNAs for analysis.
The present study comprehensively analyzed
differentially expressed snoRNAs in HCC and provided an overview of
their clinical significance. Subsequently, several functional
enrichment analyses were performed to elucidate the functional
roles of key snoRNAs. More importantly, survival-associated snoRNAs
were identified to develop a prognostic index (PI), which may be
utilized as a risk score model for HCC patients. Via these efforts,
the present study aimed to propose a foundation and comprehensive
view of snoRNAs in HCC and identify novel biomarkers to effectively
predict clinical outcomes.
Materials and methods
Data preparation and
pre-processing
The snoRNA gene expression profiles of 1,524 HCC
patients were downloaded from the online database SNORic
(http://bioinfo.life.hust.edu.cn/SNORic/) (15). The expression value of snoRNAs was
normalized and quantified as reads per kilobase per million mapped
reads (RPKM). Only snoRNAs with an average RPKM of >1 across all
samples were used for further analysis. The corresponding clinical
information of the HCC patients was also downloaded from the TCGA
database (https://portal.gdc.cancer.gov/).
Screening of differentially expressed
snoRNAs
Two different strategies, including analysis with
the ‘limma’ package in R (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
and an independent-samples t-test via SPSS 24.0 (IBM Corp., Armonk,
NY, USA), were used to identify snoRNAs which were significantly
differentially expressed between HCC and non-tumor tissues. For the
limma test, the threshold for the significantly differentially
expressed snoRNAs was considered a |fold change|≥2 and a false
discovery rate (FDR) of <0.05. For the t-test, snoRNAs were
considered differentially expressed when P<0.05. Genes that were
identified by the two differential analyses simultaneously were
defined as differentially expressed snoRNAs.
Functional annotation of snoRNAs
To further explore the potential functional roles of
the differentially expressed snoRNAs, the top 10 most significantly
differentially expressed snoRNAs were selected and messenger RNAs
(mRNAs) with expression levels correlated with these snoRNAs were
obtained from SNORic. Next, these mRNAs were subjected to
functional enrichment analysis using the ClusterProfiler package
(16), in order to identify the
enrichment of the snoRNAs in various Gene Ontology (GO) categories
and Kyoto Encyclopedia of Genes and Genomes (KEGG) processes.
ClusterProfiler also calculated corrected P-values to prevent a
high FDR in multiple testing. GO and KEGG terms/pathways with
corrected P-values of <0.05 were considered to be significantly
enriched among the associated genes.
Survival analysis of key driver
genes
snoRNAs that were candidate prognostic biomarkers
were then selected. To obtain more accurate results/avoid
immortal-time bias, patients with <90 days of overall survival
(OS) were removed prior to survival analysis (17). The patients were followed up for a
duration of 91–3,675 days. The association between snoRNA
expression and OS was performed by using univariate Cox regression.
Candidate prognostic snoRNAs were then subjected to multivariate
Cox regression. A survival-predicting algorithm PI, an index
calculated for each patient according to their snoRNA expression
pattern, was built according to the expression values of each
independent snoRNA and weighted by the contribution of each snoRNA
to OS (18). The ‘survivalROC’
package in R (https://CRAN.R-project.org/package=survivalROC) was
used to evaluate the performance of the algorithm in predicting the
prognosis of the HCC patients. The ability of the models (PI) to
predict the outcomes was calculated at 2,000 days, as only few
events occurred after this time-point.
Gene set enrichment analysis
(GSEA)
GSEA was performed to analyze the deregulated
pathways between patients with a high and low risk according to the
predictive model/PI established (19). First, GSEA generated an ordered list
of all genes based on their association with the PI. Subsequently,
the pre-defined KEGG pathways were calculated with an enrichment
score (ES) and nominal P-value. Finally, each pathway was given a
normalized ES (NES) and an FDR calculated for the ES. Pathways with
NES >1 and FDR <0.05 were considered significant. The
different risk groups served as phenotype labels.
Results
Identification of differentially
expressed snoRNAs
A total of 372 HCC and 50 non-tumor tissues were
included in the present analysis. A total of 453 snoRNAs with an
average RPKM of >1 were obtained. Of these, 133 differentially
expressed snoRNAs were assessed using the limma statistical
package, including 119 that were upregulated and 14 that were
downregulated (|fold change|≥2 and FDR <0.05). As indicated in
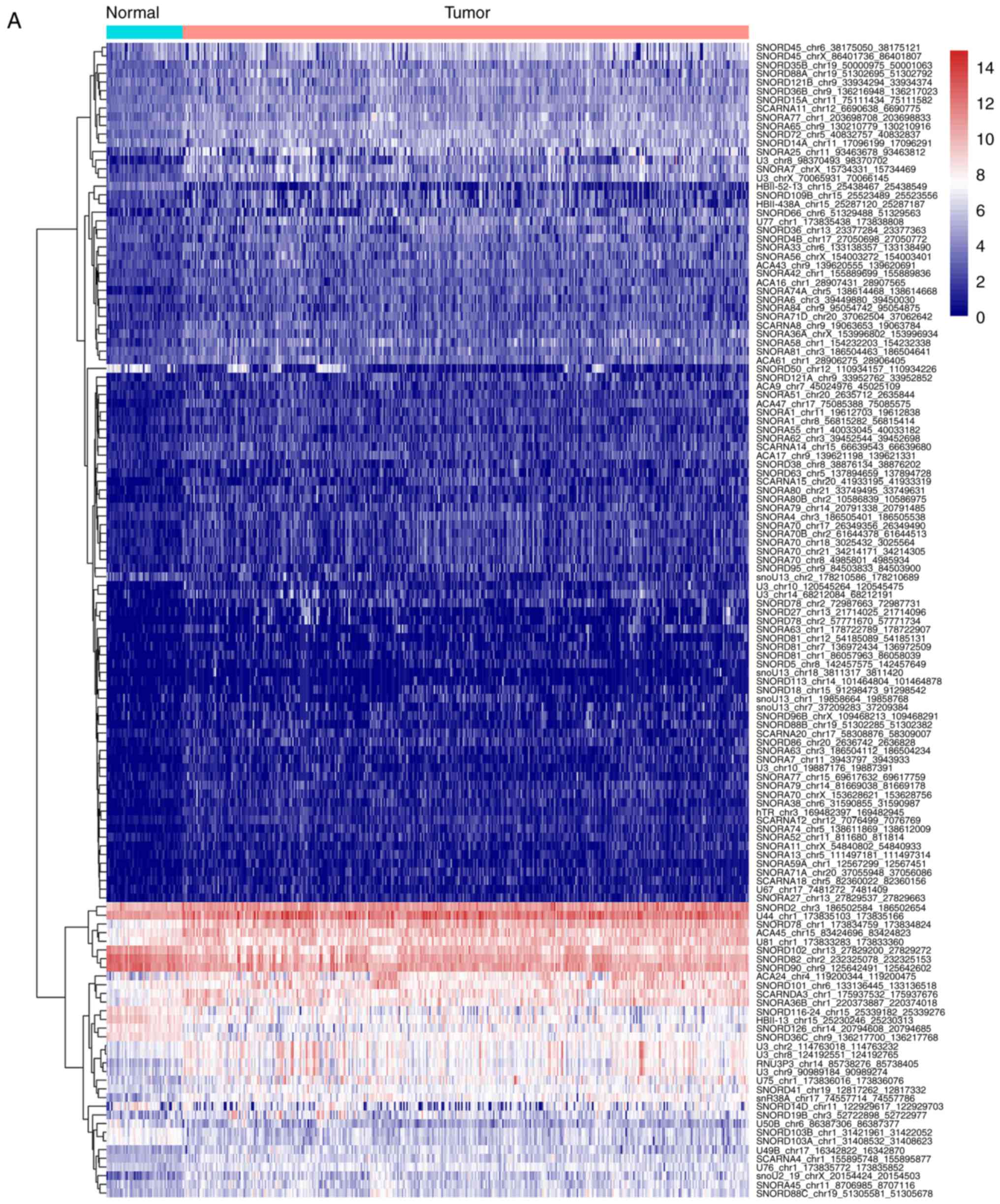
the volcano plot, most of these differentially expressed snoRNAs
were upregulated (Fig. 1).
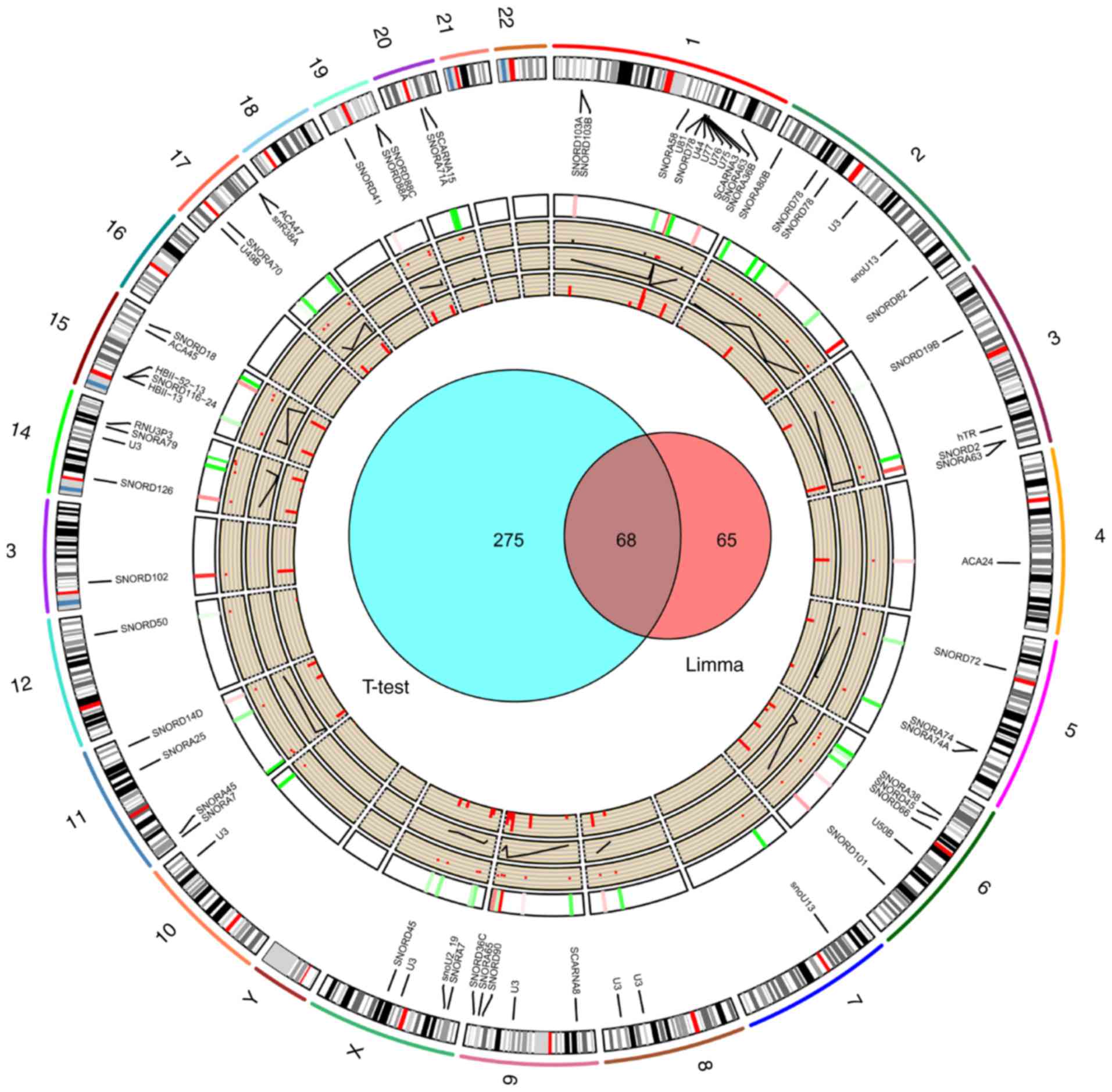
Furthermore, 71 upregulated and 272 downregulated snoRNAs were
identified using the t-test. A total of 68 overlapping snoRNAs (54
upregulated and 14 downregulated snoRNAs) between these methods
were identified (Table I). In
addition, 65 snoRNAs and 275 snoRNAs were identified by either
limma analysis only or the t-test only, respectively. Analysis of
the chromosomal distribution of the genes encoding the
differentially expressed snoRNAs revealed that the genes encoding
these snoRNAs are mostly located on chromosome 1 (Fig. 2).
 | Table I.Detailed information of differentially
expressed snoRNAs in hepatocellular carcinoma. |
Table I.
Detailed information of differentially
expressed snoRNAs in hepatocellular carcinoma.
| Gene symbol | Alias | Ensembl ID | Location | log|FC| | P-value | FDR |
|---|
| SNORD78 | U78 | ENSG00000208317 |
chr1_173834759_173834824 | 2.673130872 | 5.24E-27 | 2.01E-24 |
| SNORA74A | U19 | ENSG00000200959 |
chr5_138614468_138614668 | 2.750097225 | 9.50E-23 | 1.82E-20 |
| SNORD82 | U82 | ENSG00000202400 |
chr2_232325078_232325153 | −1.476302074 | 2.63E-21 | 3.36E-19 |
| hTR |
|
|
chr3_169482397_169482945 | 3.228363337 | 3.51E-21 | 3.37E-19 |
| SNORD72 | HBII-240 |
ENSG00000212296 |
chr5_40832757_40832837 | 1.891459367 | 5.07E-21 | 3.89E-19 |
| SNORD36C | U36C |
ENSG00000252542 |
chr9_136217700_136217768 | −1.053437273 | 2.66E-20 | 1.46E-18 |
| SNORA74 |
|
ENSG00000252213 |
chr5_138611869_138612009 | 3.620879921 | 3.76E-20 | 1.81E-18 |
| U3_chr8-1 |
|
ENSG00000207215 |
chr8_98370493_98370702 | 3.868074921 | 5.31E-20 | 2.26E-18 |
| SNORD103A | U103 |
ENSG00000200154 |
chr1_31408532_31408623 | −1.322045332 | 7.17E-20 | 2.75E-18 |
| SNORA47 | ACA47 |
|
chr17_75085388_75085575 | 2.665034297 | 2.45E-19 | 8.56E-18 |
| SNORD1A | snR38A |
|
chr17_74557714_74557786 | 1.851971297 | 2.89E-19 | 9.05E-18 |
| SNORD103B | U103B |
ENSG00000202107 |
chr1_31421961_31422052 | −1.267507568 | 3.06E-19 | 9.05E-18 |
| U3_chrX |
|
ENSG00000199769 |
chrX_70065931_70066145 | 2.366125925 | 3.43E-19 | 9.41E-18 |
| snoU2_19 |
|
ENSG00000201592 |
chrX_20154424_20154503 | 2.175945961 | 5.13E-19 | 1.31E-17 |
| SNORA7 |
|
ENSG00000200620 |
chrX_15734331_15734469 | 2.606774064 | 1.11E-18 | 2.66E-17 |
| SNORD64 | HBII-13 |
|
chr15_25230246_25230313 | −1.32876822 | 1.35E-18 | 3.06E-17 |
| SCARNA15 |
|
ENSG00000252193 |
chr20_41933195_41933319 | 3.193694106 | 3.48E-18 | 7.43E-17 |
| SNORD78 |
|
ENSG00000212168 |
chr2_57771670_57771734 | 6.087770817 | 4.84E-18 | 9.78E-17 |
| SNORD90 | HBII-295 |
ENSG00000212447 |
chr9_125642491_125642602 | −1.336197987 | 5.78E-18 | 1.11E-16 |
| SNORD126 |
|
ENSG00000238344 |
chr14_20794608_20794685 | −1.171243942 | 3.44E-17 | 5.75E-16 |
| SNORD3P3 |
|
ENSG00000252529 |
chr14_85738276_85738405 | 2.076828048 | 4.65E-17 | 7.44E-16 |
| SNORA58 |
|
ENSG00000201129 |
chr1_154232203_154232338 | 2.491683928 | 6.06E-17 | 9.17E-16 |
| SNORA65 | U65 |
ENSG00000201302 |
chr9_130210779_130210916 | 1.293605056 | 6.21E-17 | 9.17E-16 |
| SNORA7 |
|
ENSG00000206976 |
chr11_3943797_3943933 | 3.723711419 | 6.72E-17 | 9.56E-16 |
| SNORD78 |
|
ENSG00000212378 |
chr2_72987663_72987731 | 3.44943257 | 7.01E-17 | 9.62E-16 |
| SNORD116-24 | HBII-85-24 |
ENSG00000207279 |
chr15_25339182_25339276 | −1.153615572 | 1.05E-16 | 1.38E-15 |
| U3_chr10 |
|
ENSG00000251836 |
chr10_120545264_120545475 | 3.598880515 | 2.34E-16 | 3.00E-15 |
| U3_chr9 |
|
ENSG00000252299 |
chr9_90989184_90989274 | 2.072207774 | 3.65E-16 | 4.53E-15 |
| SNORA45 | ACA45 |
|
chr15_83424696_83424823 | 1.46649302 | 4.78E-16 | 5.56E-15 |
| SNORD76 | U76 |
|
chr1_173835772_173835852 | 1.053351966 | 5.65E-16 | 6.38E-15 |
| SNORD2 | snR39B |
ENSG00000238942 |
chr3_186502584_186502654 | 1.038836932 | 1.59E-15 | 1.65E-14 |
| SNORD44 | U44 |
|
chr1_173835103_173835166 | 1.541593275 | 1.54E-15 | 1.65E-14 |
| U3_chr14 |
|
ENSG00000252792 |
chr14_68212084_68212191 | 3.113376111 | 2.11E-15 | 2.07E-14 |
| SNORD88A | HBII-180A |
ENSG00000221241 |
chr19_51302695_51302792 | 1.543480591 | 4.03E-15 | 3.87E-14 |
| SNORD102 | U102 |
ENSG00000207500 |
chr13_27829200_27829272 | −1.162101322 | 5.98E-15 | 5.47E-14 |
| SNORD18 |
|
ENSG00000200677 |
chr15_91298473_91298542 | 2.653377709 | 1.26E-14 | 1.12E-13 |
| SNORA80B | ACA67B |
ENSG00000206633 |
chr2_10586839_10586975 | 3.131648604 | 2.26E-14 | 1.89E-13 |
| SNORA38 | ACA38 |
ENSG00000200816 |
chr6_31590855_31590987 | 2.842534832 | 3.11E-14 | 2.49E-13 |
| SNORD50 |
|
ENSG00000202335 |
chr12_110934157_110934226 | −2.437841877 | 3.33E-14 | 2.61E-13 |
| SNORD115-13 | HBII-52-13 |
|
chr15_25438467_25438549 | −1.160460557 | 4.18E-14 | 3.15E-13 |
| SNORD49B | U49B |
|
chr17_16342822_16342870 | 1.336562047 | 4.12E-14 | 3.15E-13 |
| SCARNA8 | U92 |
ENSG00000251733 |
chr9_19063653_19063784 | 1.8417627 | 5.69E-14 | 4.12E-13 |
| SNORA71A | U71a |
ENSG00000225091 |
chr20_37055948_37056086 | 2.571597734 | 5.85E-14 | 4.16E-13 |
| SNORD41 | U41 |
ENSG00000209702 |
chr19_12817262_12817332 | 1.180714368 | 1.01E-13 | 7.06E-13 |
| SNORD77 | U77 |
|
chr1_173835438_173835508 | 2.14951456 | 2.32E-13 | 1.59E-12 |
| SNORD88C | HBII-180C |
ENSG00000220988 |
chr19_51305581_51305678 | 1.302824949 | 7.37E-13 | 4.64E-12 |
| SNORD75 | U75 |
|
chr1_173836016_173836076 | 1.982160347 | 1.80E-12 | 1.12E-11 |
| SCARNA3 | HBI-100 |
ENSG00000252906 |
chr1_175937532_175937676 | 1.435536825 | 8.34E-12 | 4.64E-11 |
| SNORA79 | ACA65 |
ENSG00000221303 |
chr14_81669038_81669178 | 2.219512874 | 1.88E-11 | 9.95E-11 |
| SNORA63 |
|
ENSG00000200418 |
chr3_186504112_186504234 | 2.475924302 | 2.23E-11 | 1.14E-10 |
| SNORD81 | U81 |
|
chr1_173833283_173833360 | 1.160446192 | 2.22E-11 | 1.14E-10 |
| SNORA70 |
|
ENSG00000202389 |
chr17_26349356_26349490 | 2.328245545 | 3.11E-11 | 1.55E-10 |
| SNORD66 |
|
ENSG00000212532 |
chr6_51329488_51329563 | 2.023556148 | 7.38E-11 | 3.26E-10 |
| SNORA63 |
|
ENSG00000201791 |
chr1_178722789_178722907 | 2.846016122 | 7.77E-11 | 3.35E-10 |
| snoU13 |
|
ENSG00000238772 |
chr7_37209283_37209384 | 2.608270027 | 1.92E-10 | 7.59E-10 |
| SNORA25 | ACA25 |
ENSG00000207112 |
chr11_93463678_93463812 | 2.42926579 | 2.16E-10 | 8.35E-10 |
| U3_chr8-2 |
|
ENSG00000221461 |
chr8_124192551_124192765 | 1.667436749 | 4.12E-10 | 1.48E-09 |
| SNORD45 |
|
ENSG00000200706 |
chr6_38175050_38175121 | 1.255751287 | 1.17E-09 | 3.92E-09 |
| SNORD45 |
|
ENSG00000200422 |
chrX_86401736_86401807 | 1.31186539 | 1.33E-09 | 4.36E-09 |
| SNORD50B | U50B |
|
chr6_86387306_86387377 | −1.332782809 | 1.98E-09 | 6.38E-09 |
| SNORA45 | ACA3-2 |
ENSG00000212607 |
chr11_8706985_8707116 | 1.063884317 | 2.04E-09 | 6.54E-09 |
| U3_chr2 |
|
ENSG00000212182 |
chr2_114763018_114763232 | 1.593913527 | 2.77E-09 | 8.43E-09 |
| SNORA24 | ACA24 |
|
chr4_119200344_119200475 | 1.357111583 | 8.28E-09 | 2.34E-08 |
| SNORD19B |
|
ENSG00000252787 |
chr3_52722898_52722977 | 1.952644182 | 1.03E-08 | 2.80E-08 |
| SNORD101 | U101 |
ENSG00000206754 |
chr6_133136445_133136518 | 1.224670084 | 3.50E-07 | 7.19E-07 |
| snoU13 |
|
ENSG00000238295 |
chr2_178210586_178210689 | −1.040086867 | 5.72E-07 | 1.11E-06 |
| SNORA36B | ACA36B |
ENSG00000222370 |
chr1_220373887_220374018 | 1.073775965 | 1.95E-06 | 3.41E-06 |
| SNORD14D |
|
ENSG00000207118 |
chr11_122929617_122929703 | −1.025013641 | 0.002356081 | 0.003015784 |
Functional characteristic of snoRNAs
in HCC
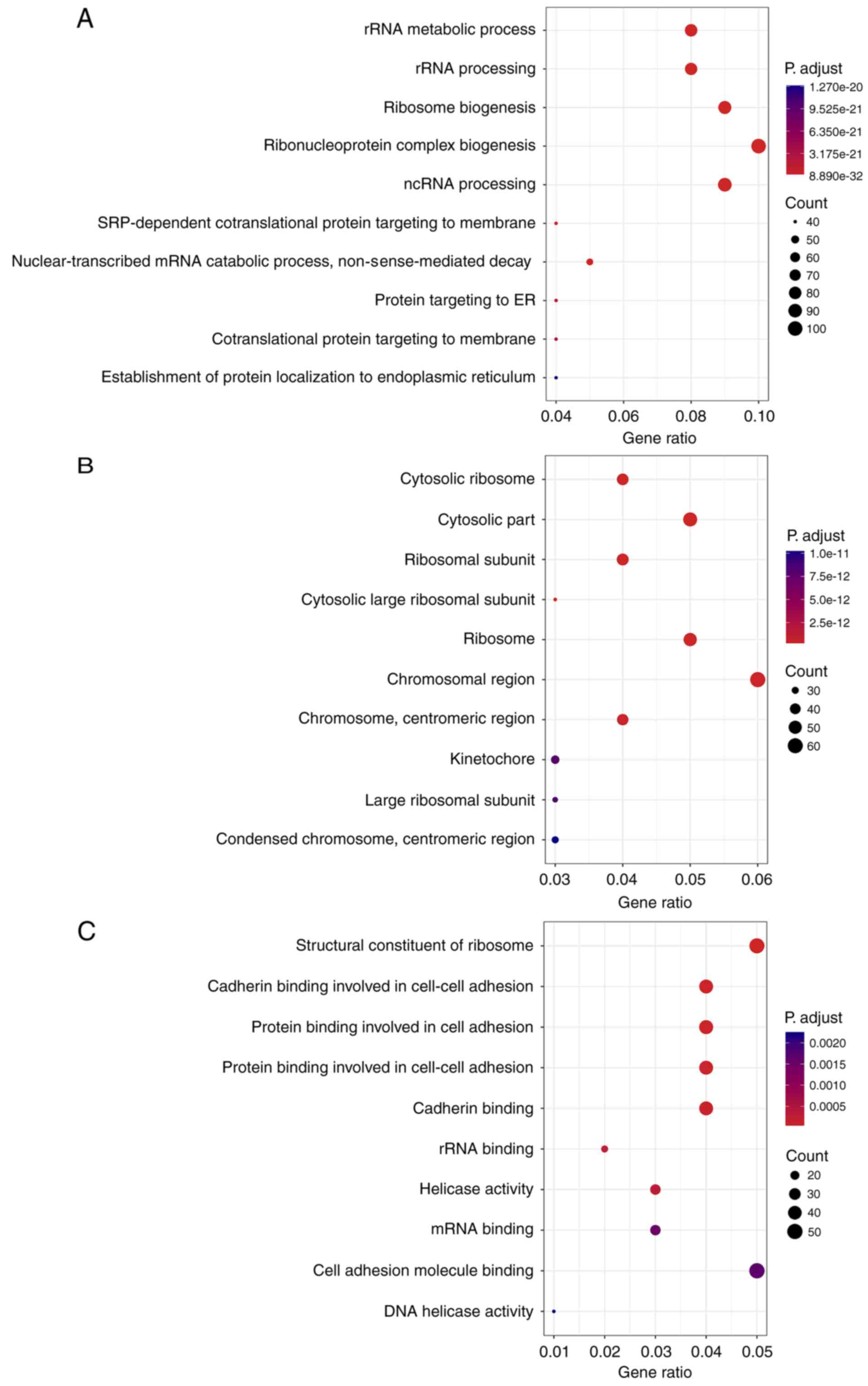
Functional enrichment analysis of 1,149 mRNAs
associated with differentially expressed snoRNAs was performed
using clusterProfiler. Biological processes (BP), cell composition
(CC) and molecular function (MF) were the three categories of GO
terms. In the BP category, the three most enriched items were
‘ribosomal (r)RNA metabolic process’, ‘rRNA processing’ and
‘ribosome biogenesis’ (Fig. 3A). In
the category CC, the mRNAs were mainly concentrated in the terms
‘cytosolic ribosome’, ‘cytosolic part’ and ‘ribosomal subunit’
(Fig. 3B). ‘Structural constituent
of ribosome’, ‘cadherin binding involved in cell-cell adhesion’ and
‘protein binding involved in cell adhesion’ were the more prominent
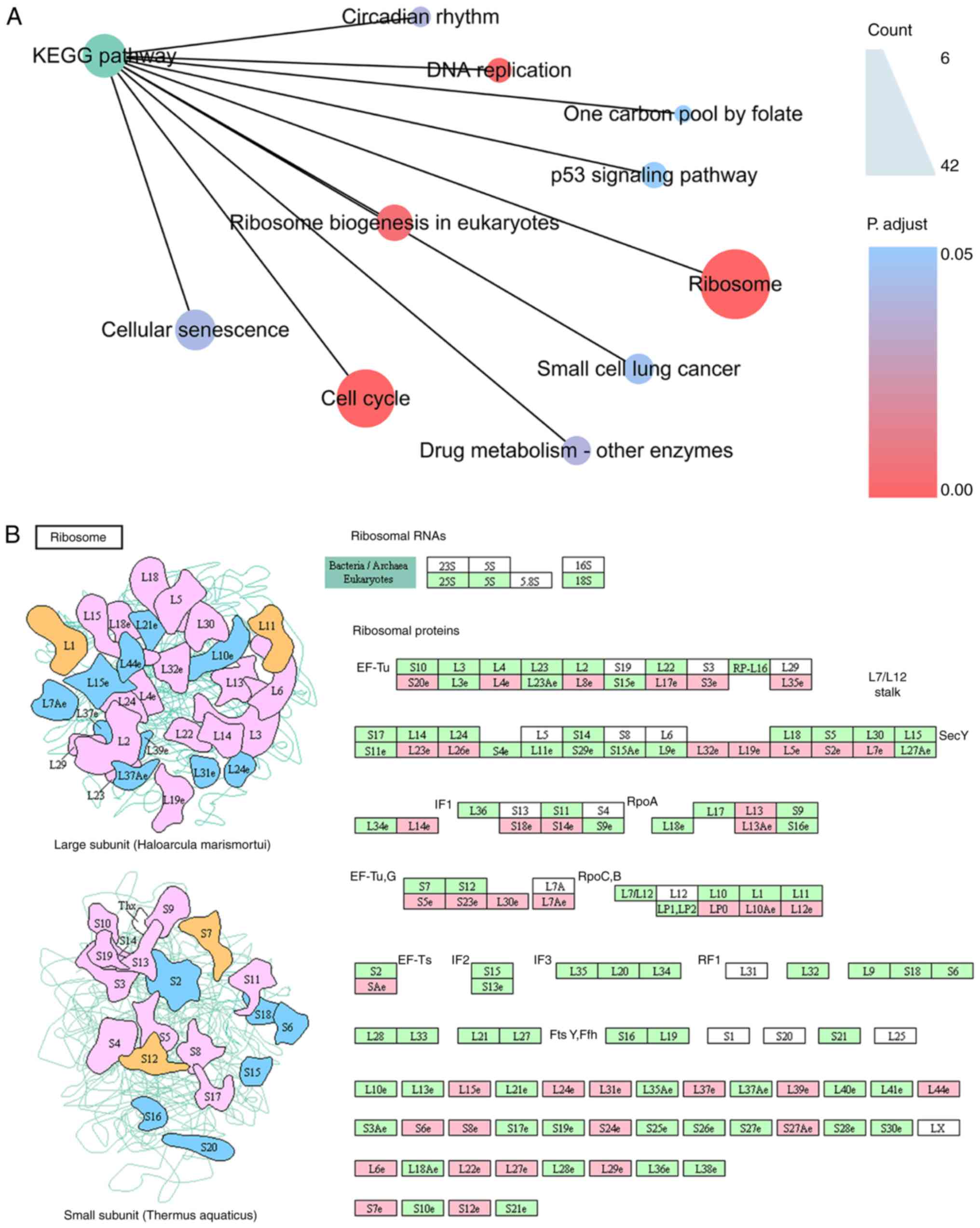
terms enriched by the mRNAs in the MF category (Fig. 3C). More interestingly, KEGG analysis
indicated that the mRNAs associated with the HCC-specific snoRNAs
were most significantly enriched in the pathways ‘Ribosome’, ‘Cell
cycle’ and ‘DNA replication’ (Fig.
4). Among these pathways, ‘Ribosome’ was the most significant
pathway and included 42 genes when the background of the functional
enrichment analysis was set to ‘Homo sapiens’ (https://www.kegg.jp/dbget-bin/www_bget?pathway+hsa03010).
Prognostic predictors for HCC
patients
After removing patients with <90 days of OS, 330
HCC patients were included in the further analysis. The prognostic
value of the differentially expressed snoRNAs was assessed using
univariate Cox regression. A total of 22 snoRNAs with P<0.05
were identified, which were therefore able to predict the survival
of HCC patients. These snoRNAs were then subjected to multivariate
Cox proportional regression analysis, which identified 9 snoRNAs as
independent prognostic indicators for HCC. Finally, the PI was
calculated based on these 9 snoRNAs as follows: [expression of
SNORA (SNOR, H/ACA box)24] × 0.0655 + (expression of SNORA7) ×
0.0991 + (expression of SNORA63) × 0.1196 + (expression of
U3_chr8-2) × 0.2590 + (expression of U3_chr9) × 0.2464 +
[expression of SNOR, C/D box (SNORD)19B] × 0.0613 + (expression of
hTR) × 0.1653 + (expression of SNORD36C) × 0.0830 + (expression of
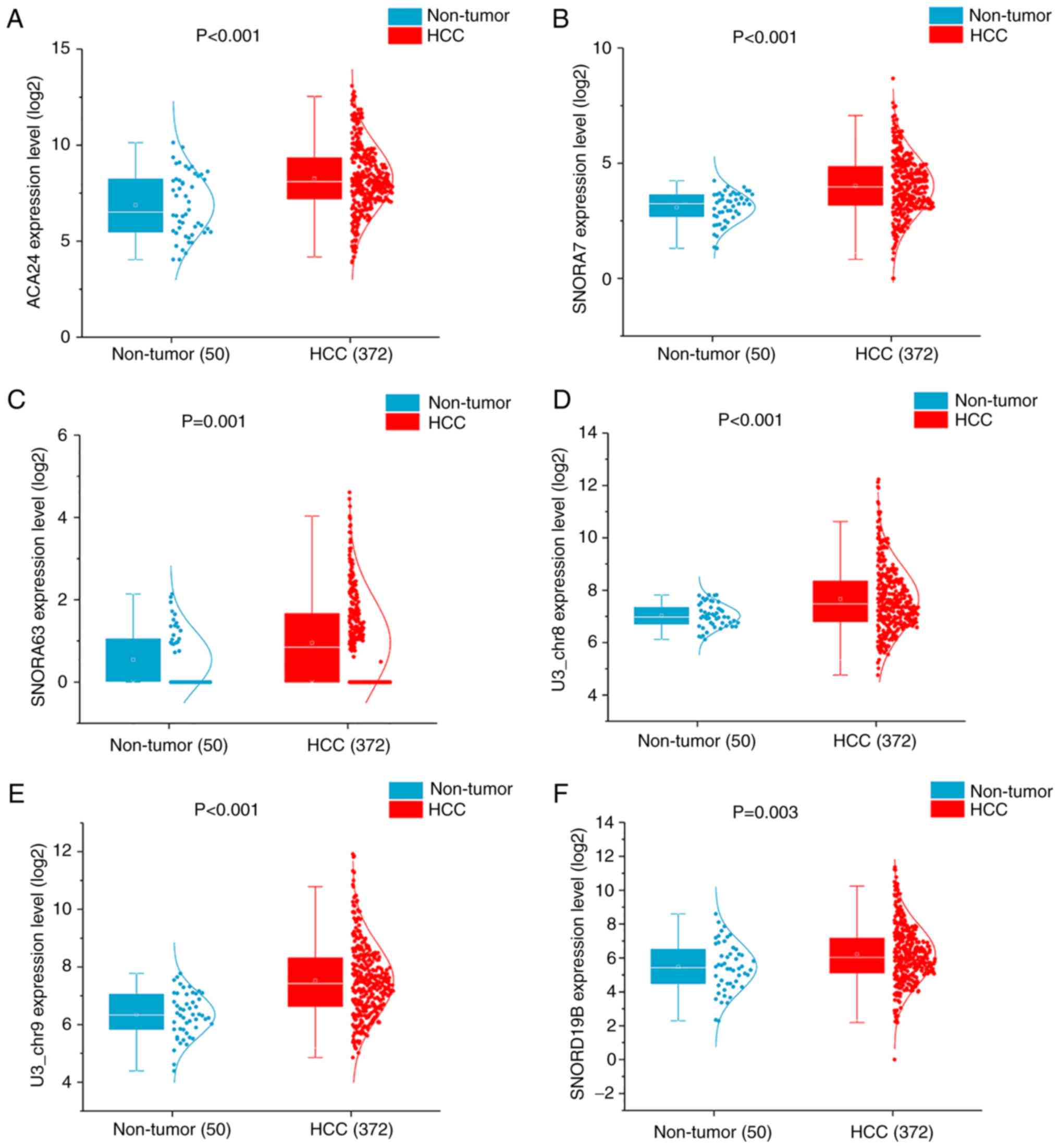
U44) × 0.0964. The expression of SNORD36C was markedly
downregulated in HCC tissues and the remaining snoRNAs were
significantly upregulated in HCC tissues (Fig. 5).
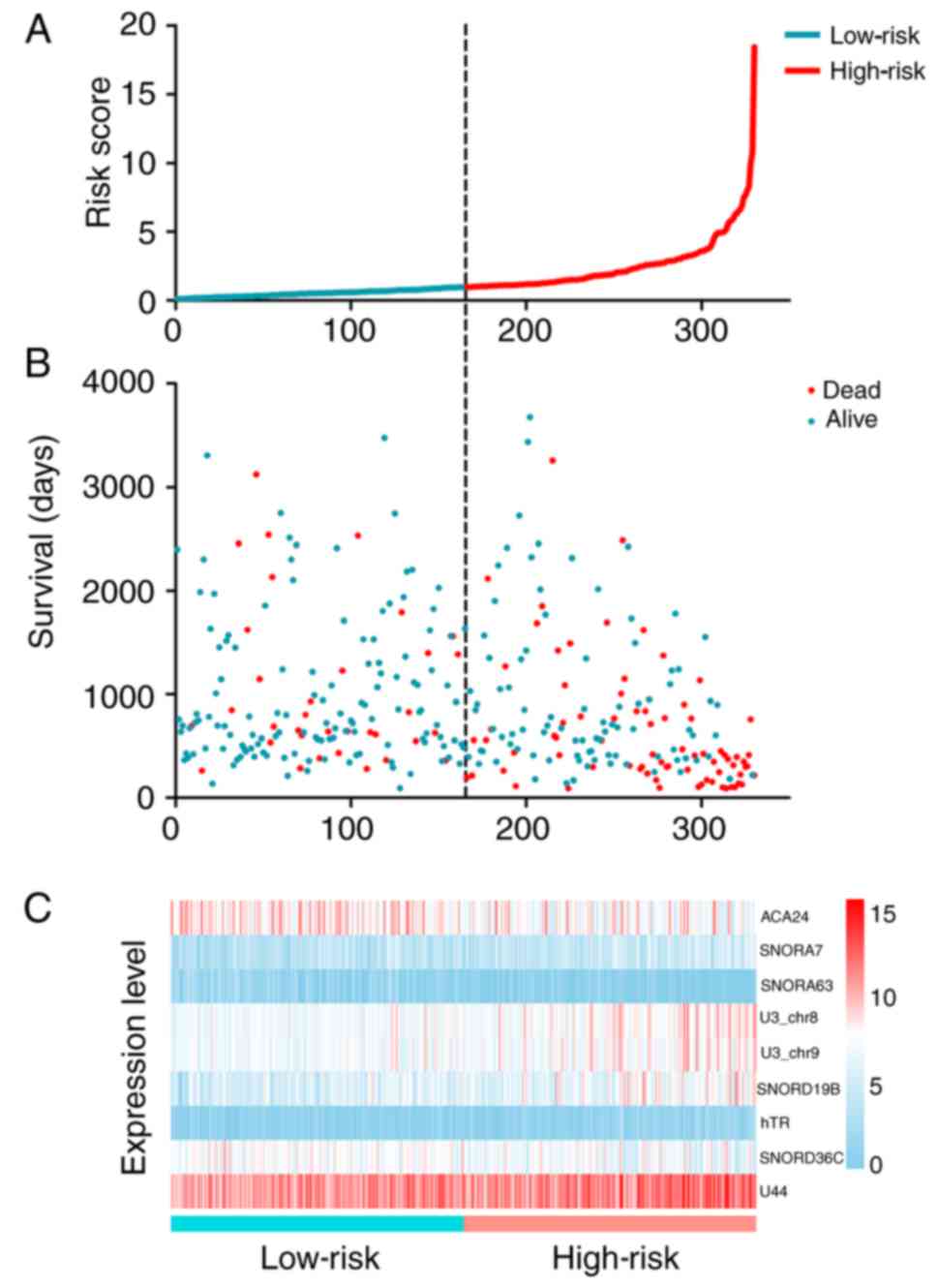
The HCC patients were divided into a high-risk group
(n=165) and a low-risk group (n=165) according to the threshold of
the median PI value (Fig. 6A). The
patients were followed up for a duration of 91–3,675 days. The
dependence of the overall survival status (dead or alive) on the
snoRNA-based risk scores of the HCC patients was also plotted,
displaying inferior survival for patients in the high-risk group
(Fig. 6). Patients in the high-risk
group had a significantly shorter median survival time than those
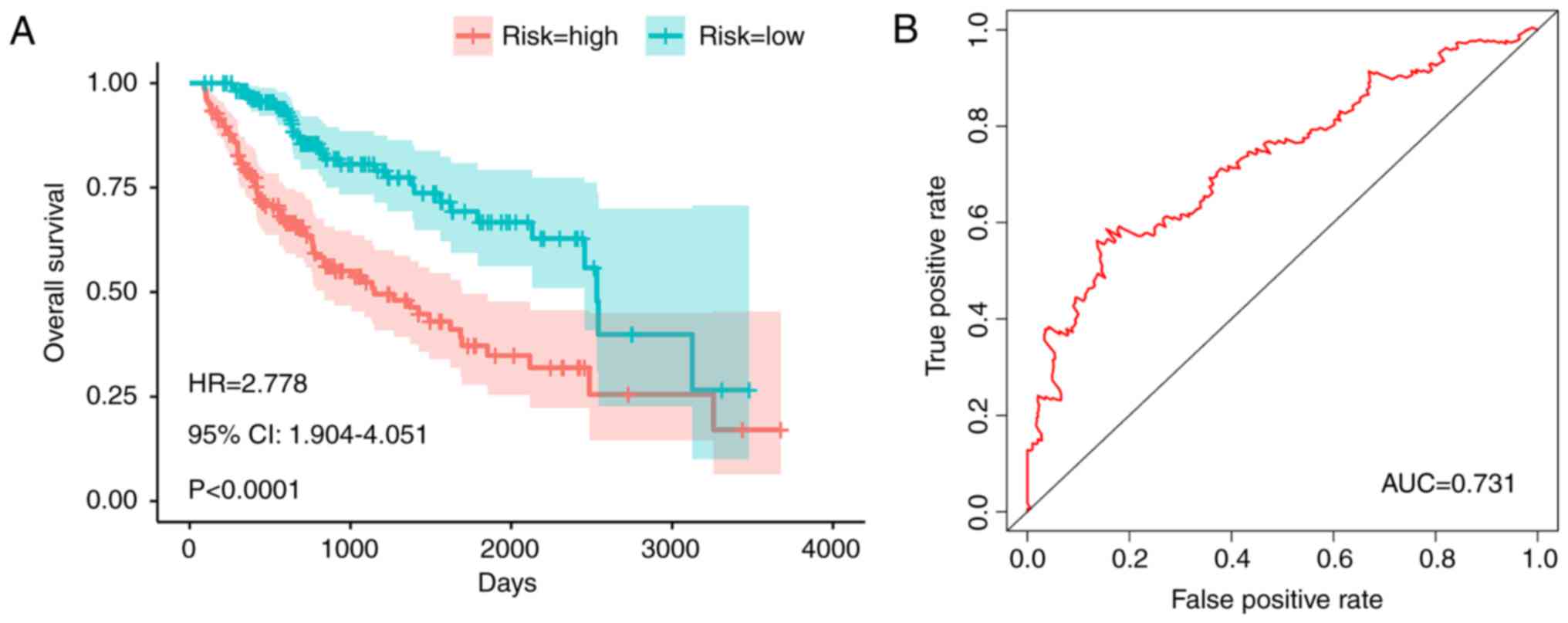
in the low-risk group (hazard ratio=2.778, 95% confidence interval:
1.904–4.051, P<0.001; Fig. 7A).
This result indicated the patients in the high-risk group have a
2.78-fold increased risk of death compared with those in the
low-risk group. The area under the receiver operating
characteristic curve was 0.731, which indicated a moderate survival
prediction ability of the PI (Fig.
7B). In the multivariate analysis (Table II), the risk model/PI that was
proposed was demonstrated to be an independent prognostic factor,
suggesting its independent prognostic value.
 | Table II.Univariate and multivariate analyses
of factors affecting the overall survival of hepatocellular
carcinoma patients from The Cancer Genome Atlas by Cox regression
analysis. |
Table II.
Univariate and multivariate analyses
of factors affecting the overall survival of hepatocellular
carcinoma patients from The Cancer Genome Atlas by Cox regression
analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | Hazard ratio
(95%CI) | P-value | Hazard ratio
(95%CI) | P-value |
|---|
| Age, years |
| (≥60 vs. <60
years) | 1.045
(0.717–1.523) | 0.818 |
|
|
| Gender (male vs.
female) | 0.875
(0.593–1.292) | 0.503 |
|
|
| Pathological stage
(I–II vs. III–IV) | 2.964
(1.978–4.440) | <0.001 | 1.392
(0.188–10.297) | 0.746 |
| Tumor stage (T1-T2
vs. T3-T4) | 3.028
(2.070–4.427) | <0.001 | 1.714
(0.231–12.705) | 0.598 |
| Lymph node
metastasis (positive vs. negative) | 2.507
(0.611–10.287) | 0.202 |
|
|
| Distant metastasis
(positive vs. negative) | 4.911
(1.536–15.701) | 0.007 | 1.395
(0.329–5.909) | 0.652 |
| Histological grade
(G1-G2 vs. G3-G4) | 1.098
(0.743–1.623) | 0.640 |
|
|
| Tumor status (with
tumor vs. tumor free) | 3.610
(2.322–5.613) | <0.001 | 3.313
(1.940–5.659) | <0.001 |
| Vascular tumor cell
type (micro+macro vs. none) | 1.352
(0.852–2.146) | 0.200 |
|
|
| Prognostic index
(high vs. low risk) | 2.853
(1.903–4.278) | <0.001 | 3.023
(1.785–5.119) | <0.001 |
Deregulated pathways between high- and
low-risk groups
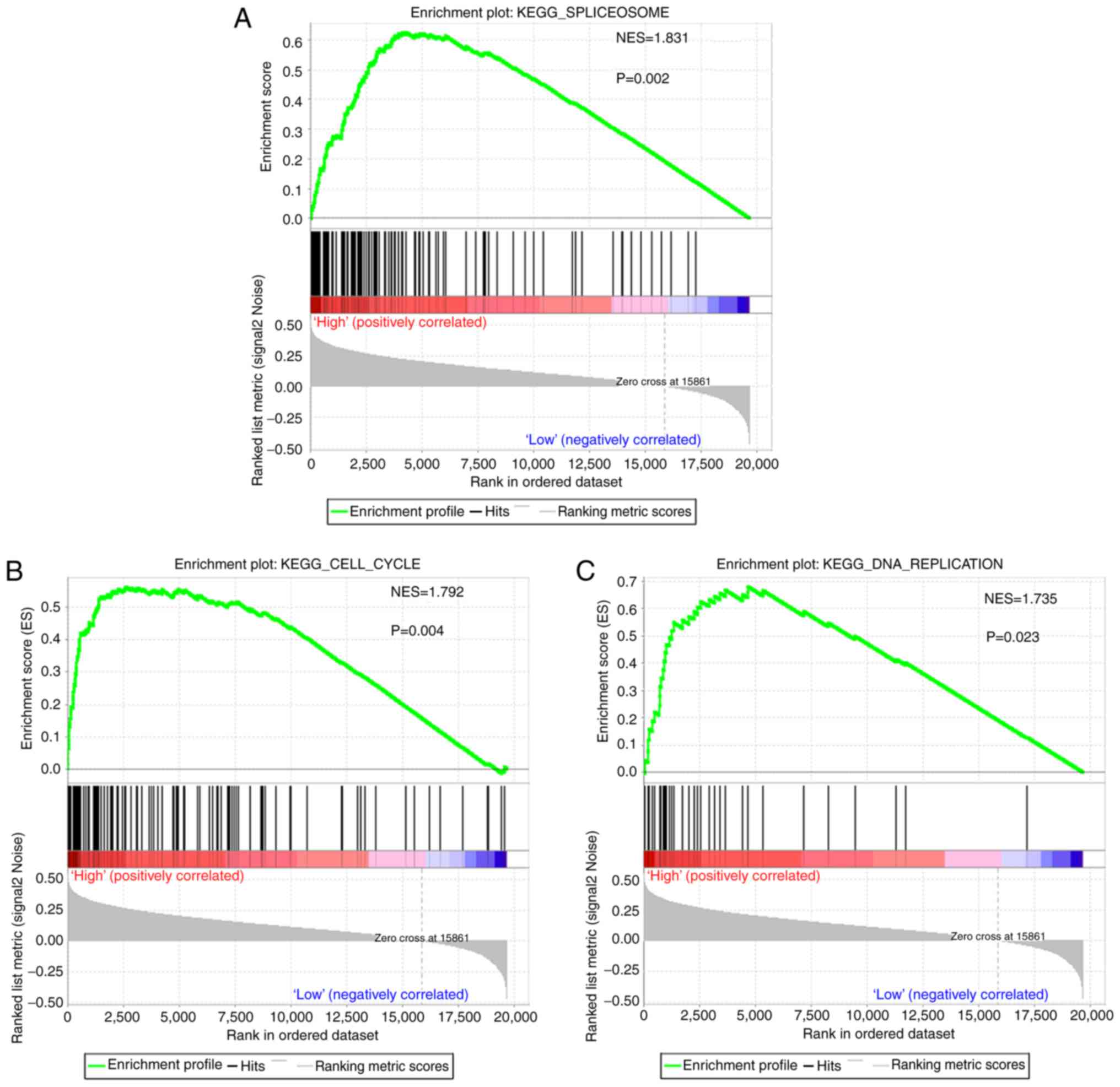
To identify disturbed biological signaling pathways
between the high- and low-risk groups, GSEA analysis was performed.
Among all of the pre-defined KEGG pathway-associated gene sets,
spliceosome, cell cycle and DNA replication signaling pathways were
identified to be significantly linked with the survival risk
estimated by the PI (Fig. 8),
suggesting that patients in the high-risk group may have inferior
survival due to the above cancer-associated signaling pathways.
Discussion
Patients with HCC are at a substantial risk of
metastasis, recurrence and death, although the treatment methods
have markedly improved. A deeper understanding of the molecular
mechanisms is required to develop appropriate treatment protocols
and promote precision medicine. The present study comprehensively
analyzed the expression profiles of snoRNAs in HCC and identified
an overall elevation in the expression of certain snoRNAs.
Furthermore, the potential functional terms and pathways of snoRNAs
were determined, which mainly involved ribosome-associated
processes and the cell cycle. Considering the indispensable
function of certain snoRNAs in HCC, a prognostic method based on 9
snoRNAs was developed to stratify HCC patients into subgroups with
different risks of mortality. Based on the GSEA analysis,
disruption of the spliceosome may be the major contributor to the
poor survival of patients in the high-risk group.
Hepatocarcinogenesis is considered a multi-step
process, with various molecular factors, including snoRNAs,
involved in its development and progression. To date, only few
studies that have delineated the clinical significance and
molecular mechanisms of snoRNAs in HCC. Hence, the present study
comprehensively investigated the expression profiles of snoRNAs in
HCC and observed an overall upregulation of snoRNAs in HCC tissues.
Several HCC-associated oncogenic snoRNAs, which are upregulated in
HCC, have been previously reported, whereas downregulated snoRNAs
may act as tumor suppressors. Several of the dysregulated snoRNAs
identified in the present study were also reported in previous
studies; for instance, Fang et al (14) indicated that SNORD126
(chr14_20794608_20794685) was highly expressed in HCC compared with
non-tumorous samples. They also identified that upregulated
SNORD126 was associated with a shorter survival rate of HCC
patients. However, these results were the opposite of the present
results, according to which SNORD126 was downregulated in HCC
tissues. The limited number of cases in their study (only 30 HCC
tissues) may be the major reason for this difference. Wu et
al (20) reported that the
overexpression of SNORD76 is associated with decreased survival of
HCC patients. In vitro and in vivo functional studies
consistently indicated that SNORD76 promoted HCC cell growth and
tumorigenicity. The high expression levels of another markedly
upregulated snoRNA, SNORD78, has also been validated in HCC, and
knockdown of SNORD78 significantly suppressed the proliferation,
migration and invasion of liver tumor cells (21). These studies have facilitated a
better understanding of the function of snoRNAs in HCC and provided
novel ideas for early diagnosis and the development of precision
medical treatments. The present analysis broadens the scope and
promotes the search for novel snoRNAs as diagnostic and prognostic
markers in HCC.
At present, the rudimentary understanding of the
roles of snoRNAs in HCC limits their clinical application.
Therefore, functional enrichment analysis was performed to
determine the precise biological processes that were deregulated by
the aberrant expression of snoRNAs in HCC. It was identified that
snoRNAs may be involved in the pathways of ribosome structure and
cell cycle, which indicated that snoRNAs significantly affect cell
growth. Indeed, snoRNAs often combine with ribonucleoproteins
(RNPs) to form stable and functional snoRNP particles, which is
necessary for the effective and accurate formation of ribosomes
(22). Ribosomes are considered to
be the processing plants for protein synthesis in cells, but in
tumor cells, this molecular machinery is misaligned and cellular
metabolism is deregulated (23).
Upregulated cell proliferation is usually accompanied by changes in
the ribosome production rate. Perturbations of ribosome and
ribosome-associated pathogenesis have been reported to be
associated with multiple cancer types (24,25).
In HCC, several tumor suppressors and oncogenes have been
identified to either affect the development of the mature ribosome
or to regulate the activity of proteins (26). Therefore, dysregulated snoRNAs
presumably exert an oncogenic or tumor suppressor function and may
regulate the malignant phenotype by altering the ribosome synthesis
machinery.
The highlight of the present study is that it was
the first, to the best of our knowledge, to propose a snoRNA-based
prognostic signature for HCC patients. For a decade, TCGA has
collected large-scale molecular profiles and clinicopathologic
annotation data, which has made it possible to identify key
features that determine the clinical outcome of HCC patients
(17). Identifying the distinct
molecular features of each tumor patient makes it possible to lay a
foundation for the development of personalized medicine (27). Several previous studies have
proposed molecular prognostic signatures based on the expression
levels of lncRNAs (28), miRNAs
(29) and mRNAs (30). However, a snoRNA-based risk score
has not been described to the best of our knowledge. snoRNAs are
stable and measurable in peripheral plasma and serum, which gives
snoRNAs a unique advantage as potential molecular biomarkers for
the diagnosis and prognosis of tumor patients (11,31).
To the best of our knowledge, the present study was
the first to propose a prognostic signature based on snoRNAs, which
had a satisfactory ability to predict survival. The present study
also improved the current understanding of the molecular mechanisms
of HCC. The prospective molecular mechanisms of the key deregulated
snoRNAs were also assessed. Of note, abnormal alternative splicing
events may be the cause for the adverse clinical outcomes for
patients with a high prognostic index in the high-risk group. Of
note, several snoRNAs have been reported to have a role in pre-mRNA
splicing (32). However, the key
snoRNAs identified in the present study have not been reported.
Hence, the specific regulatory mechanisms of snoRNAs in splicing in
HCC should be further explored in the future.
In summary, the present study was the first to
propose a prognostic signature based on 9 snoRNAs in HCC, each of
which is an independent risk factor. Numerous genes with
statistically significant prognostic associations were identified
for further study. These snoRNAs may be utilized as novel
therapeutic targets or molecular markers for HCC with high clinical
significance. The results of the present in silico analysis
should be verified by in vivo and in vitro
experiments in the future. The potential functional terms and
molecular pathways of mRNAs associated with the snoRNAs were also
assessed. The prognostic signature established in the present study
may be a clinically useful tool that is easily incorporated into a
clinical RNA-sequencing program to individualize HCC therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the Fund
of the National Natural Science Foundation of China (grant nos.
NSFC81060202, NSFC81860319 and NSFC81260222), the Guangxi Science
Technology Program (grant no. GuikeAB17195020), the Innovation
Project of Guangxi Graduate Education (grant no. YCSW2018104), the
Guangxi Medical University Training Program for Distinguished Young
Scholars and the Medical Excellence Award Funded by the Creative
Research Development Grant of the First Affiliated Hospital of
Guangxi Medical University.
Availability of data and materials
The datasets used and/or analyzed during this study
are available from the corresponding author on reasonable
request.
Authors' contributions
The study was designed by GC, YWD, YH and HY. PL,
HY, HYW, HYL and YWD performed the statistical analysis. PL and HY
wrote the draft and GC, HY, YH and YWD corrected the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fu J and Wang H: Precision diagnosis and
treatment of liver cancer in China. Cancer Lett. 412:283–288. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bosetti C, Turati F and La Vecchia C:
Hepatocellular carcinoma epidemiology. Best Pract Res Clin
Gastroenterol. 28:753–770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sherman M: Hepatocellular carcinoma:
Epidemiology, surveillance, and diagnosis. Semin Liver Dis.
30:3–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kudo M: Systemic therapy for
hepatocellular carcinoma: 2017 update. Oncology. 93 Suppl
1:S135–S146. 2017. View Article : Google Scholar
|
|
7
|
Costentin CE, Ferrone CR, Arellano RS,
Ganguli S, Hong TS and Zhu AX: Hepatocellular carcinoma with
macrovascular invasion: Defining the optimal treatment strategy.
Liver Cancer. 6:360–374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kissel M, Berndt S, Fiebig L, Kling S, Ji
Q, Gu Q, Lang T, Hafner FT, Teufel M and Zopf D: Antitumor effects
of regorafenib and sorafenib in preclinical models of
hepatocellular carcinoma. Oncotarget. 8:107096–107108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sherman M: Regorafenib for treatment of
hepatocellular carcinoma. Hepatology. 67:1162–1165. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al:
Regorafenib for patients with hepatocellular carcinoma who
progressed on sorafenib treatment (RESORCE): A randomised,
double-blind, placebo-controlled, phase 3 trial. Lancet. 389:56–66.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Romano G, Veneziano D, Acunzo M and Croce
CM: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Samuels DC, Zhao S, Xiang Y, Zhao
YY and Guo Y: Current research on non-coding ribonucleic acid
(RNA). Genes. 8:pii: E3662017. View Article : Google Scholar
|
|
13
|
Xu G, Yang F, Ding CL, Zhao LJ, Ren H,
Zhao P, Wang W and Qi ZT: Small nucleolar RNA 113-1 suppresses
tumorigenesis in hepatocellular carcinoma. Mol Cancer. 13:2162014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fang X, Yang D, Luo H, Wu S, Dong W, Xiao
J, Yuan S, Ni A, Zhang KJ, Liu XY, et al: SNORD126 promotes HCC and
CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J
Mol Cell Biol. 9:243–255. 2017.PubMed/NCBI
|
|
15
|
Gong J, Li Y, Liu CJ, Xiang Y, Li C, Ye Y,
Zhang Z, Hawke DH, Park PK, Diao L, et al: A Pan-cancer analysis of
the expression and clinical relevance of small nucleolar RNAs in
human cancer. Cell Rep. 21:1968–1981. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA Pan-Cancer clinical data resource to
drive High-quality survival outcome analytics. Cell.
173:400–416.e11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jacob H, Stanisavljevic L, Storli KE,
Hestetun KE, Dahl O and Myklebust MP: A four-microRNA classifier as
a novel prognostic marker for tumor recurrence in stage II colon
cancer. Sci Rep. 8:61572018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu L, Chang L, Wang H, Ma W, Peng Q and
Yuan Y: Clinical significance of C/D box small nucleolar RNA U76 as
an oncogene and a prognostic biomarker in hepatocellular carcinoma.
Clin Res Hepatol Gastroenterol. 42:82–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma P, Wang H, Han L, Jing W, Zhou X and
Liu Z: Up-regulation of small nucleolar RNA 78 is correlated with
aggressive phenotype and poor prognosis of hepatocellular
carcinoma. Tumour Biol. Oct 21–2016.(Epub ahead of print).
View Article : Google Scholar
|
|
22
|
Reichow SL, Hamma T, Ferré-D'Amaré AR and
Varani G: The structure and function of small nucleolar
ribonucleoproteins. Nucleic Acids Res. 35:1452–1464. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong Z, Zhu C, Zhan Q and Jiang W: The
roles of RRP15 in nucleolar formation, ribosome biogenesis and
checkpoint control in human cells. Oncotarget. 8:13240–13252.
2017.PubMed/NCBI
|
|
24
|
Su H, Xu T, Ganapathy S, Shadfan M, Long
M, Huang TH, Thompson I and Yuan ZM: Elevated snoRNA biogenesis is
essential in breast cancer. Oncogene. 33:1348–1358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Russo A, Saide A, Smaldone S, Faraonio R
and Russo G: Role of uL3 in multidrug resistance in p53-mutated
lung cancer cells. Int J Mol Sci. 18:pii: E5472017. View Article : Google Scholar
|
|
26
|
Yuan F, Zhang Y, Ma L, Cheng Q, Li G and
Tong T: Enhanced NOLC1 promotes cell senescence and represses
hepatocellular carcinoma cell proliferation by disturbing the
organization of nucleolus. Aging Cell. 16:726–737. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Wu Q, Feng S, Zhao Y and Tao C:
Identification of four prognostic LncRNAs for survival prediction
of patients with hepatocellular carcinoma. PeerJ. 5:e35752017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu G, Wang H, Fu JD, Liu JY, Yan AG and
Guan YY: A five-miRNA expression signature predicts survival in
hepatocellular carcinoma. APMIS. 125:614–622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li B, Feng W, Luo O, Xu T, Cao Y, Wu H, Yu
D and Ding Y: Development and validation of a three-gene prognostic
signature for patients with hepatocellular carcinoma. Sci Rep.
7:55172017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mannoor K, Liao J and Jiang F: Small
nucleolar RNAs in cancer. Biochim Biophys Acta. 1826:121–128.
2012.PubMed/NCBI
|
|
32
|
Warner WA, Spencer DH, Trissal M, White
BS, Helton N, Ley TJ and Link DC: Expression profiling of snoRNAs
in normal hematopoiesis and AML. Blood Adv. 2:151–163. 2018.
View Article : Google Scholar : PubMed/NCBI
|