Introduction
Breast cancer (BC) has become the most common
malignancy among women worldwide (1), and ovarian cancer (OC) is the most
lethal gynecological malignancy (2). Extensive evidence has shown that the
etiologies of BC and OC are multifactorial involving hormonal,
reproductive, environmental and genetic factors, and they may share
certain genetic factors (3–5).
Genetic variants that influence the risk of BC and
OC are divided into 3 categories: High-penetrant gene mutations,
intermediate-penetrant gene mutations or variants, and
low-penetrant variants (6).
Approximately 20–30% of BC and OC cases exhibit familial
aggregation, most of which are driven by low-frequency
high-penetrant gene mutations, such as BRCA1, BRCA2, PTEN
and TP53 (7,8). BRCA1 and BRCA2 are
involved in the homologous recombination pathway for double-strand
DNA repair, maintaining genome stability (9). The PTEN gene encodes a
phosphatase that negatively regulates the AKT/PKB signaling
pathway, inducing cell cycle arrest, apoptosis and senescence
(10). P53 is a
tumor-suppressor gene involved in DNA repair, cell cycle arrest and
apoptosis (11). Both family-based
and population-based studies indicate that low-frequency genetic
variants of DNA repair genes CHEK2, ATM, BRIP and
PALB2 are associated with a moderate risk of breast cancer
(12). In particular, the
high-frequency low-penetrance genetic variants such as single
nucleotide polymorphisms (SNPs) are more often associated with
sporadic BC and OC (13).
Although the effect of a single SNP is generally
small, several relevant SNPs may additively or synergistically
contribute to increased breast cancer risk. On average, there is
about one SNP for every 500 nucleotides in the human genome and
each gene is covered by 52 SNPs (14), thus it is costly to genotype all
these known SNPs in the target gene. Fortunately, based on linkage
disequilibrium (LD), applying a much smaller subset of informative
SNPs called haplotype-tagging SNPs (htSNPs) or tagging SNPs (tSNPs)
can capture gene-wide common variations (15). Remarkably, by using these directly
genotyped data of htSNP or tSNPs, many other variants that are not
directly genotyped in laboratoriess can be genotyped on computer,
an approach called genotype imputation (16). It predicts the individual genotypes
at un-typed loci by comparing this individual to other individuals
who shared a common haplotype or haplotypes, usually employing
HapMap data as a reference (17).
Therefore, htSNPs or tSNPs genotype analysis is a cost-effective
strategy to pinpoint the polymorphisms of susceptibility genes in
population association studies (18).
Snail1 (encoded by gene SNAI1) and Twist1
(encoded by gene TWIST1) are 2 key inducers of
epithelial-mesenchymal transition (EMT). The SNAI1 gene is
located on chr20:48,599,513–48,605,420 (human genome hg19 as
reference) and its locus is 20q13.13. TWIST1 is located on
chr7:19,155,091–19,157,295 (human genome hg19 as reference) and its
locus is 7p21.1. Evidence indicates that Snail1 confers tumor cells
with cancer stem cell-like traits, and promotes tumor recurrence,
metastasis and drug resistance (19). It has been demonstrated that Snail1
plays a critical role in tumor growth and metastasis of breast and
ovarian carcinoma by regulating MMP activity (20,21).
Twist1 is reported to be overexpressed in malignant and metastatic
breast cancer (22) and ovarian
cancer (23,24). The positive expression of Twist1 was
found to be associated with poor progression-free survival and
overall survival in epithelial ovarian carcinoma (24). Matsusaka et al analyzed
associations between 7 functional SNPs in EMT-related genes
(TWIST1, ZEB1, SNAI1 and E-cadherin) and outcomes in
metastatic colorectal cancer patients. They found that female
patients who carried the minor allele G of TWIST1 rs2285682
and TWIST1 rs2285681 had improved survival (25). However, little is known concerning
the association between genetic polymorphisms of these 2 genes and
BC/OC susceptibility.
In the present study, we genotyped 7 tagging SNPs,
representatives of all SNPs within the SNAI1 and
TWIST1 genes, in 1,161 BC cases, 286 OC cases and 1,273
cancer-free female controls, and then performed a comprehensive
correlation analysis between genotypes and cancer susceptibility.
To fine-map the potential causal variants, genotype imputation was
conducted to identify more SNPs that contribute to the
susceptibility of the diseases in these two genes. Functional
annotation predicted that SNAI1 rs6125849 and TWIST1
rs4721745 could be causal SNPs, which modulate BC and OC
susceptibility.
Materials and methods
Study population and DNA
isolation
This study included 1,161 BC cases and 978
cancer-free female controls, and 286 ovarian cancer cases and 295
cancer-free female controls. All 1,447 cancer cases were recruited
from Beijing Cancer Hospital, Peking University Third Hospital and
Beijing Hospital between 1995 and 2010, and were pathologically
diagnosed with breast invasive ductal carcinoma or ovarian serous
carcinomas. The epidemiological information was extracted from
their clinical records. The 978 cancer-free female controls and 295
cancer-free female controls, age-matched to BC and OC cases by
5-year age groups respectively, were selected from a
community-based screening program for non-infectious diseases in
Beijing. The epidemiological information of the controls was
collected from the questionnaire. Age, height, weight, age at
menarche and/or menopause, age at the first full-term pregnancy
(FFTP) as well as family history of BC, OC or other cancers in
first-degree relatives and other epidemiologic data were collected
for all patients and controls.
For the 1,161 BC cases and all the 1,273 cancer-free
female controls, genomic DNA was extracted from blood leukocytes as
described by Grimberg et al (26). Briefly, 2 ml of whole blood was
mixed with 8 ml of ice cold Triton lysis buffer (0.32 M sucrose, 10
mM Tris-HCl pH 7.6, 5 mM MgCl2, 1% Triton X-100). After
centrifugation, the nuclear pellet was resuspended in 500 µl of
proteinase K buffer (1 mg/ml proteinase K, 10 mM Tris-HCl pH 8.0,
10 mM NaCl, 10 mM EDTA) and incubated for 2 h at 65°C. Genomic DNA
was subsequently separated from proteins by saturated phenol, and
then with phenol:chloroform:isoamyl alcohol (25:24:1) procedure.
DNA in the supernatant fluid was precipitated by ethanol and
dissolved in 400 µl of TE buffer. DNA concentration was determined
using NanoDrop 2000C (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) at 260 nm.
For the 286 OC cases, genomic DNAs were extracted
from archived formalin-fixed paraffin-embedded (FFPE) non-tumor
tissues as described by Pikor et al (27). Briefly, for each case, 3 pieces of
10-µm-thick paraffin-embedded tissue sections were incubated in
xylene to remove paraffin from the tissues, and then incubated in
proteinase K buffer (5 mg/ml proteinase K, 10 mM Tris-HCl pH 8.0,
10 mM NaCl and 10 mM EDTA) for 2 days until the tissues were fully
dissolved. The DNAs were purified by phenol/chloroform DNA
extraction method, and then precipitated by ethanol and dissolved
in TE buffer. The DNA concentration was determined using NanoDrop
2000C at 260 nm.
Selection of tagging-SNPs
To ensure good marker coverage of the entire genes
and increase the analytic efficiency, tagging SNPs (tSNPs) were
selected by Haploview v.4.2 software program based on the
information of candidate genes in the HapMap database [HapMap Data
Release #27; Chinese Beijing population (CHB)] (www.hapmap.org). The tSNPs met the following
requirements: A 0.8-r2 threshold and a minor allele
frequency (MAF) >0.05, and spanning from 2 kb upstream to 2 kb
downstream for SNAI1 and TWIST1 genes, so that these
tSNPs could capture all known common genetic variants within the
entire gene. In the end, the following tSNPs were chosen, 3 tSNPs
in SNAI1 (rs6125849, rs4647959, rs6020178) and 4 tSNPs in
TWIST1 (rs2285682, rs2285681, rs4721746 and rs4721745).
Genotyping assay and quality
control
Tagging-SNP genotyping was performed using TaqMan
Assay® (Applied Biosystems, Foster City, CA, USA) using
the ABI Step One® or ABI 7900HT® Real-Time
PCR System (Applied Biosystems). Primers and probes (FAM- and
VIC-labeled) were supplied directly by Applied Biosystems as
Assays-by-Design and Assays-on-Demand products. Briefly, all assays
were carried out in 48-well or 384-well plates with positive and
negative controls in each genotyping plate. Plates were sealed and
heated at 95°C for 5 min, and then were subjected to 45–50 cycles
of 92°C for 15 sec and 60°C for 1 min. As for quality control, we
repeated the genotyping on 3% of the samples. The concordance rate
between duplicates was higher than 99%.
Univariate and multivariate
analyses
The distribution of categorical variables and
continuous variables between cases and control groups was compared
by Pearson's χ2 test and Student's t-test, respectively.
For each SNP, Hardy-Weinberg equilibrium was evaluated using a
one-degree of freedom goodness-of-fit χ2 test to compare
the observed with the expected frequency of genotypes among the
female controls. LD plots of the D' values for these SNPs were
produced using the Haploview program (28). The most probable haplotypes of each
participant were estimated using the expectation-maximization (EM)
algorithm. For SNP analysis, we tested 3 different genetic models,
namely dominant model, recessive model and codominant model, to
identify the best-fitting one with the smallest P-value (29). Two-sided chi-square test was also
used to investigate the differences in the distributions of
genotypes between cases and controls. To analyze the associations
of an individual SNP with BC and OC risk, univariate and
multivariate logistic regression models were used to estimate the
odds ratios (ORs) and 95% confidence intervals (CIs). The above
statistical analyses were performed by Statistic Analysis System
software (SAS v9.1; SAS Institute, Cary, NC, USA).
False-positive report probability and
statistical power
We calculated the false-positive report probability
(FPRP) values for the statistically significant associations. We
set 0.2 as the FPRP threshold and adopted a prior probability of
0.1 to detect OR of 1.50/0.67 (risk/protective effects) as
described previously (30,31). The association that reached the FPRP
threshold of <0.2 was considered noteworthy.
Genotype imputation
Genotype imputation is used to predict the genotypes
at the variants that are not directly assayed in the study sample
(32). In this study, we used MACH
software (http://csg.sph.umich.edu/abecasis/MACH/tour/imputation.html)
to complete genotype imputation. To obtain more reliable results,
we constructed reference haplotypes from the CHB population
according to 1000 Genomes datasets. What's more, the module of
imputation helper in GenGen software tool (http://gengen.openbioinformatics.org) was used to
facilitate the analysis of high-throughput genomics datasets.
Imputed variants that had an R-square measure <0.3 were
excluded. Well-imputed variants were then genotyped and used to
analyze the associations between the variants and BC/OC
susceptibility.
Multifactor dimensionality reduction
(MDR) analysis
MDR analysis was conducted to identify higher-order
interactions that were associated with cancer risk using MDR
software (v.2.0 beta 8.4) (http://sourceforge.net/projects/mdr/). MDR analysis
collapsed multi-dimensional data into a single independent
dimensional variable with two levels (high and low risk) using the
ratio of the number of cases to the number of controls, and thereby
reduced multiple dimensional data into one dimension and permitted
detection of interactions in relatively small sample sizes
(33). MDR was also performed in
cross-validation and permutation test to classify and predict
disease status. The best candidate interaction model was selected
to maximize testing accuracy and cross-validation consistency (CVC)
(34).
Functional annotation tools
To explore the functional relevance of the selected
SNPs, we annotated each variant using publicly available
bioinformatics data and existing functional annotation software.
HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)
and RegulomeDB (http://regulome.stanford.edu/) provide annotation for
variations, including the ENCODE project data, DNase
hypersensitivity data, histone modification data, transcription
factor ChIP-seq data, and eQTL datasets. Furthermore, we analyzed
the ENCODE project (https://www.encodeproject.org/) data very closely in
the breast cancer cell lines (HMEC and MCF-7) by using the UCSC
Genome Browser (http://genome.ucsc.edu), which may indicate similar
roles in ovarian cancer.
Ethics statement
This study was approved by the Peking University IRB
(reference no. IRB00001052-11029). We obtained written informed
consents from all BC cases and control women. Because we could not
obtain the contact information of ovarian cancer patients treated
before 2011, PKU IRB approved our application to waive informed
consent for the archived samples collected before April 2011. This
study only used these samples. We used all the data/samples
anonymously.
Results
Characteristics of the study
population
This study included 1,161 BC cases and 978
cancer-free female controls, 286 ovarian cancer cases and 295
cancer-free female controls. Since the etiology of BC and OC has
similarity, we merged the BC cases and OC cases (designated as
BC/OC) as well as control groups to increase the sample size. As
shown in Table I, the 1,447 BC/OC
patients and 1273 controls appeared to be adequately matched in age
(P=0.1673). The patients had a much younger age at menarche
(P<0.0001) and an elder age at first full-term pregnancy
(P<0.0001) compared with the controls. In addition, a higher
proportion of women were in pre-menopause status in the case group
than that noted in the controls. For other characteristics such as
body mass index (BMI), age at menopause and family history of
cancer in first-degree relatives, no statistically differences were
found (P>0.05). These variables were further used in
multivariate logistic regression to adjust for any possible
confounding effect on BC/OC risk.
 | Table I.Characteristics of the BC/OC patients
and cancer-free controls |
Table I.
Characteristics of the BC/OC patients
and cancer-free controls
| Variable | Case, n=1,447 | Control,
n=1,273 | P-value |
|---|
| Age, years (mean ±
SD) | 50.33±11.00 | 49.79±9.05 | 0.1673 |
| Age, n (%),
years |
|
| 0.0742 |
|
<50 | 753 (52.04) | 706 (55.46) |
|
|
≥50 | 694 (47.96) | 567 (44.54) |
|
| Body mass index
(BMI), (mean ± SD) | 24.48±3.32 | 24.65±3.49 | 0.1915 |
| Age at menarche,
years (mean ± SD) | 14.58±1.88 | 15.12±1.91 |
<0.0001 |
| Age at menopause,
years (mean ± SD) | 49.28±4.20 | 49.22±3.80 | 0.8150 |
| Age of first
full-term pregnancy (FFTP), years (mean ± SD) | 26.03±2.91 | 25.40±2.79 |
<0.0001 |
| Menopause status, n
(%) |
|
| 0.0001 |
|
Premenopause | 827 (57.75) | 633 (50.36) |
|
|
Postmenopause | 605 (42.25) | 624 (49.64) |
|
| Family history of
cancer in first-degree relatives, n (%) |
|
| 0.1374 |
|
Yes | 310 (21.47) | 244 (19.17) |
|
| No | 1134 (78.53) | 1029 (80.83) |
|
Similarly, the selected characteristics were
separately analyzed in the BC and OC cases. The mean ages of the
1,161 BC cases at diagnosis and the 978 cancer-free female controls
were 49.28±10.53 and 48.61±8.25, respectively. They were adequately
matched in age (P=0.1037). BC patients had a much younger age at
menarche (P<0.0001) and an elder age at primiparity
(P<0.0001) compared with controls.
The mean ages of the 286 BC cases at diagnosis and
the 295 cancer-free female controls were 54.57±11.85 and
53.71±10.40, respectively. They were adequately matched in age
(P=0.3517). The OC cases had a lower BMI (P=0.0309), younger age at
menarche (P=0.0020) and higher proportion of post-menopausal
individuals (P<0.0001) than controls.
Association of individual tSNPs in
SNAI1 and TWIST1 with BC and OC risk
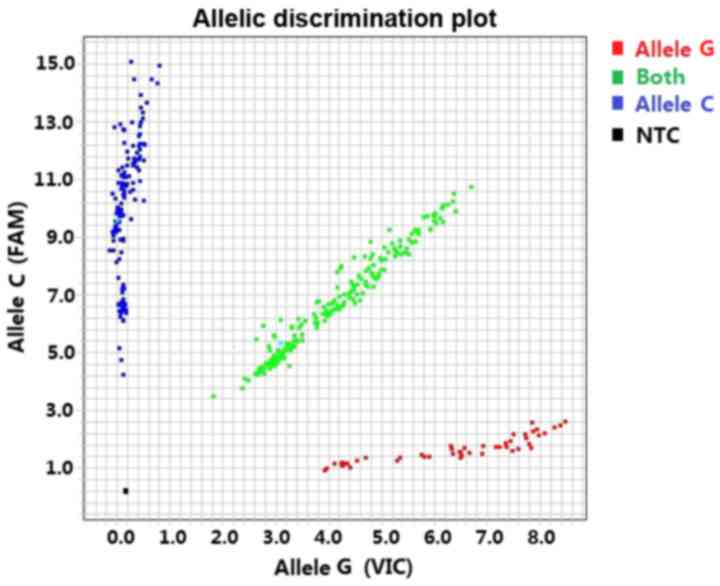
SNPs were genotyped by TaqMan Assay®
(Applied Biosystems) using the Real-Time PCR System ABI Step One
(48-well plate) or ABI 7900HT (384-well plates) (Fig. 1).
The 7 tSNPs in SNAI1 and TWIST1 were
all in agreement with Hardy-Weinberg equilibrium (HWE) in the
control population (P>0.05).
As shown in Table
II, there were significant differences in genotype distribution
between BC/OC cases and controls in polymorphisms SNAI1
rs6125849 (G>A) and TWIST1 rs4721745 (C>G) by
χ2 test (P=0.0128 and 0.0007 respectively). Multivariate
logistic regression showed that SNAI1 rs6125849,
TWIST1 rs4721746 and TWIST1 rs4721745 were all
associated with decreased BC/OC risk in the dominant model after
adjusting for body mass index (BMI), age at menarche, age of first
full-term pregnancy and menopause status (adjusted odds ratio, aOR
=0.77, P=0.0087; aOR=0.83, P=0.0434; aOR=0.73, P=0.0002,
respectively). Moreover, the cancer risk was further decreased with
the increase in minor allele dose for the 3 tagging SNPs
(Ptrend=0.0038, 0.0492 and 0.0006, respectively), which
suggested those loci were strongly associated with BC/OC
susceptibility. The other tSNPs did not show statistical
significance in the multivariate logistic analysis.
 | Table II.Genotype and allele frequencies of
the tagging-SNPs and their associations with the risk of BC/OC. |
Table II.
Genotype and allele frequencies of
the tagging-SNPs and their associations with the risk of BC/OC.
| Gene | SNPs | Genotype | Cases (%) | Controls (%) |
P-valuea |
P-valueb |
P-valuetrend | OR (95% CI) | P-value | aORc (95% CI) |
P-valued |
|---|
| SNAI1 | rs6125849 | GG | 457 (31.58) | 337 (26.47) | 0.0128 |
| 0.0038 |
|
|
|
|
|
|
| AG | 703 (48.58) | 657 (51.61) |
|
|
| 0.79
(0.66–0.94) | 0.0085 | 0.79
(0.66–0.94) | 0.0087 |
|
|
| AA | 287 (19.83) | 279 (21.92) |
|
|
| 0.76
(0.61–0.94) | 0.0124 | 0.75
(0.60–0.93) | 0.0093 |
|
|
| A allele
frequency | 1,277 (44.13) | 1,215 (47.72) |
| 0.0079 |
|
|
|
|
|
|
|
| AG/AA vs. GG
(dominant model) |
|
|
|
|
| 0.78
(0.66–0.92) | 0.0035 | 0.77
(0.65–0.92) | 0.0031 |
|
|
| AA vs. GG/AG
(recessive model) |
|
|
|
|
| 0.88
(0.73–1.06) | 0.1813 | 0.87
(0.72–1.05) | 0.1458 |
|
| rs6020178 | TT | 1,195 (82.58) | 1,064 (83.58) | 0.7862 |
| 0.2538 |
|
|
|
|
|
|
| TC | 239 (16.52) | 198 (15.55) |
|
|
| 1.08
(0.88–1.32) | 0.4929 | 1.11
(0.90–1.37) | 0.3163 |
|
|
| CC | 13 (0.90) | 11 (0.86) |
|
|
| 1.05
(0.47–2.36) | 0.9016 | 1.13
(0.49–2.63) | 0.7757 |
|
|
| C allele
frequency | 265 (9.16) | 220 (8.64) |
| 0.5053 |
|
|
|
|
|
|
|
| TC/CC vs. TT
(dominant model) |
|
|
|
|
| 1.07
(0.88–1.31) | 0.4898 | 1.11
(0.91–1.37) | 0.3026 |
|
|
| CC vs. TT/TC
(recessive model) |
|
|
|
|
| 1.04
(0.46–2.33) | 0.9240 | 1.11
(0.48–2.59) | 0.8066 |
|
| rs4647959 | TT | 1,044 (72.15) | 939 (73.76) | 0.3773 |
| 0.2697 |
|
|
|
|
|
|
| TC | 374 (25.85) | 303 (23.80) |
|
|
| 1.11
(0.93–1.32) | 0.2436 | 1.12
(0.93–1.33) | 0.2296 |
|
|
| CC | 29 (2.00) | 31 (2.44) |
|
|
| 0.84
(0.50–1.41) | 0.5088 | 0.80
(0.47–1.36) | 0.4018 |
|
|
| C allele
frequency | 432 (14.93) | 365 (14.34) |
| 0.5383 |
|
|
|
|
|
|
|
| TC/CC vs. TT
(dominant model) |
|
|
|
|
| 1.09
(0.92–1.29) | 0.3455 | 1.09
(0.91–1.29) | 0.3496 |
|
|
| CC vs. TT/TC
(recessive model) |
|
|
|
|
| 0.82
(0.49–1.37) | 0.4444 | 0.77
(0.46–1.32) | 0.3456 |
| TWIST1 | rs2285682 | TT | 1,113 (76.92) | 969 (76.12) | 0.7337 |
| 0.3774 |
|
|
|
|
|
|
| TG | 309 (21.35) | 285 (22.39) |
|
|
| 0.94
(0.79–1.13) | 0.5352 | 0.93
(0.78–1.12) | 0.4662 |
|
|
| GG | 25 (1.73) | 19 (1.49) |
|
|
| 1.14
(0.63–2.09) | 0.6607 | 1.01
(0.54–1.88) | 0.9825 |
|
|
| G allele
frequency | 359 (12.40) | 323 (12.69) |
| 0.7543 |
|
|
|
|
|
|
|
| TG/TT vs. TT
(dominant model) |
|
|
|
|
| 0.96
(0.80–1.14) | 0.6237 | 0.94
(0.78–1.12) | 0.4879 |
|
|
| GG vs. TT/TG
(recessive model) |
|
|
|
|
| 1.16
(0.64–2.12) | 0.6300 | 1.02
(0.55–1.90) | 0.9438 |
| TWIST1 | rs2285681 | GG | 795 (54.94) | 673 (52.87) | 0.1907 |
| 0.3375 |
|
|
|
|
|
|
| CG | 540 (37.32) | 515 (40.46) |
|
|
| 0.89
(0.76–1.04) | 0.1404 | 0.89
(0.76–1.05) | 0.1562 |
|
|
| CC | 112 (7.74) | 85 (6.68) |
|
|
| 1.12
(0.83–1.51) | 0.4755 | 1.08
(0.80–1.46) | 0.6272 |
|
|
| C allele
frequency | 764 (26.40) | 685 (26.90) |
| 0.6739 |
|
|
|
|
|
|
|
| CG/CC vs. GG
(dominant model) |
|
|
|
|
| 0.92
(0.79–1.07) | 0.2789 | 0.92
(0.79–1.07) | 0.2696 |
|
|
| CC vs. GG/CG
(recessive model) |
|
|
|
|
| 1.17
(0.88–1.57) | 0.2863 | 1.13
(0.84–1.53) | 0.4116 |
|
| rs4721746 | CC | 1,106 (76.43) | 933 (73.29) | 0.1475 |
| 0.0492 |
|
|
|
|
|
|
| AC | 307 (21.22) | 310 (24.35) |
|
|
| 0.84
(0.70–1.00) | 0.0506 | 0.83
(0.69–1.00) | 0.0478 |
|
|
| AA | 34 (2.35) | 30 (2.36) |
|
|
| 0.96
(0.58–1.57) | 0.8598 | 0.86
(0.52–1.43) | 0.5590 |
|
|
| A allele
frequency | 375 (12.96) | 370 (14.53) |
| 0.0918 |
|
|
|
|
|
|
|
| AC/AA vs. CC
(dominant model) |
|
|
|
|
| 0.85
(0.71–1.01) | 0.0592 | 0.83
(0.70–1.00) | 0.0434 |
|
|
| AA vs. CC/AC
(recessive model) |
|
|
|
|
| 1.00
(0.61–1.64) | 0.9905 | 0.90
(0.54–1.49) | 0.6757 |
|
| rs4721745 | CC | 558 (38.56) | 402 (31.58) | 0.0007 |
| 0.0006 |
|
|
|
|
|
|
| CG | 650 (44.92) | 638 (50.12) |
|
|
| 0.73
(0.62–0.87) | 0.0003 | 0.74
(0.62–0.87) | 0.0005 |
|
|
| GG | 239 (16.52) | 233 (18.30) |
|
|
| 0.74
(0.60–0.92) | 0.0074 | 0.73
(0.58–0.91) | 0.0057 |
|
|
| G allele
frequency | 1,128 (38.98) | 1,104 (43.36) |
| 0.0010 |
|
|
|
|
|
|
|
| CG/GG vs. CC
(dominant model) |
|
|
|
|
| 0.74
(0.63–0.86) | 0.0001 | 0.73
(0.62–0.86) | 0.0002 |
|
|
| GG vs. GG/CC
(recessive model) |
|
|
|
|
| 0.88
(0.72–1.08) | 0.2190 | 0.87
(0.71–1.06) | 0.1712 |
Similarly, we analyzed the associations of
individual tSNPs in SNAI1 and TWIST1 with BC and OC
susceptibility respectively. The three variants (SNAI1
rs6125849, TWIST1 rs4721746, TWIST1 rs4721745) were
strongly associated with decreased breast cancer risk (P<0.05).
For ovarian cancer, no SNP reached statistical significance.
Fine-scale genetic mapping by genotype
imputation
Based on the 1000 Genomes dataset, there are 44
variants (MAF>5%) in SNAI1 and 13 variants (MAF>5%) in
TWIST1 in the Chinese Han Beijing population. Using our
directly genotyped 7 tSNPs data, 28 variants (22 variants in
SNAI1 and 6 variants in TWIST1) were well-imputed in
cases and controls by ‘in silico’ genotype imputation. Then,
we examined the 35 variants for their allelic associations with
BC/OC susceptibility using allelic association tests in PLINK
software (http://www.cog-genomics.org/plink2/). As shown in
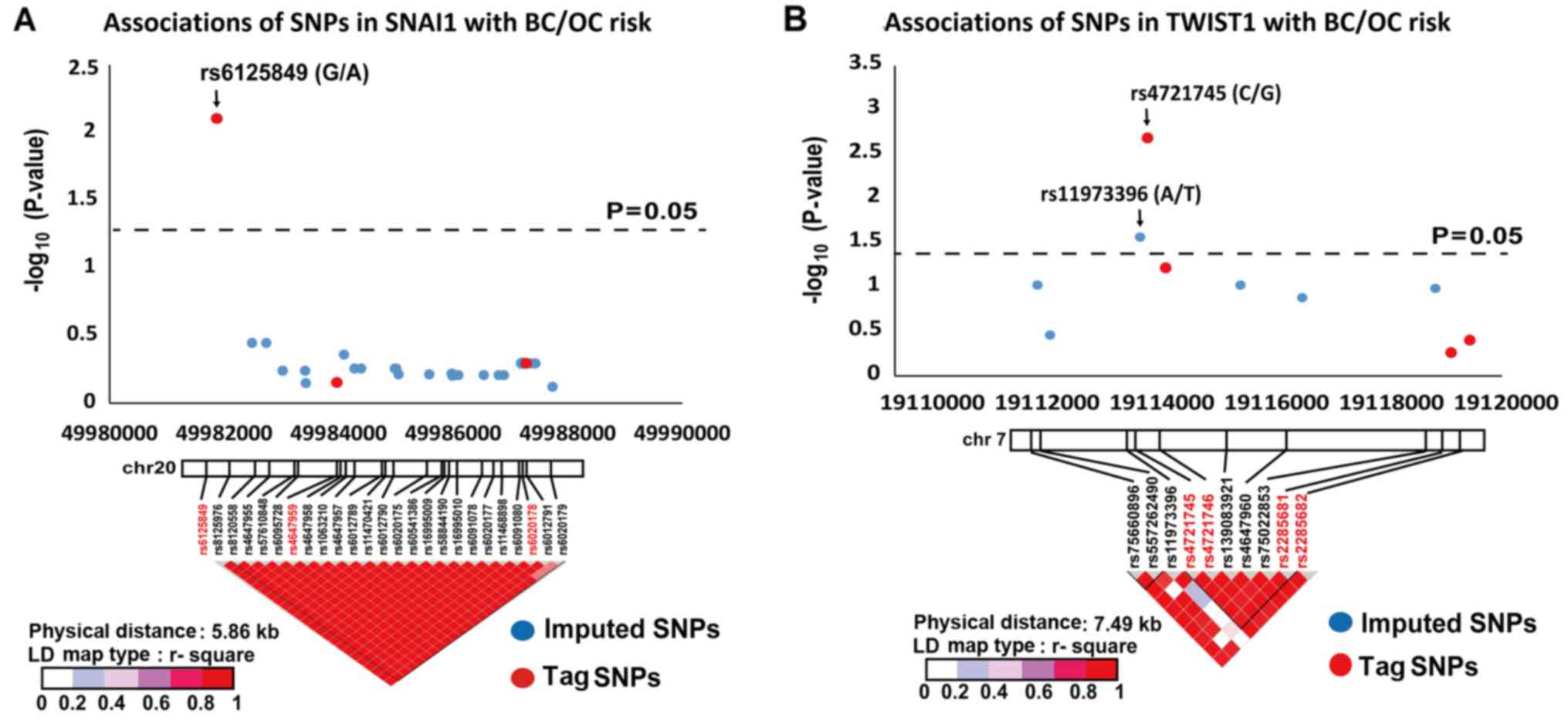
Fig. 2, three variants
(SNAI1 rs6125849, TWIST1 rs4721745 and TWIST1
rs11973396) demonstrated strong allelic associations with BC/OC
susceptibility (OR=0.87, P=0.0079; OR=0.83, P=0.0011; OR=0.87,
P=0.0270, respectively).
For breast cancer alone, SNAI1 rs6125849 and
TWIST1 rs4721745 were associated with decreased BC risk
(OR=0.86, P=0.0132; OR=0.83, P=0.0029). For ovarian cancer,
however, no SNP reached statistical significance in the gene-wide
association study.
Multivariate logistic analysis to
identify independently related SNPs
As previously shown in the BC/OC case-control study,
we identified four protective SNPs by tSNP direct genotyping and
genotype imputation, these being SNAI1 rs6125849,
TWIST1 rs4721746, TWIST1 rs4721745 and TWIST1
rs11973396. Here, we performed multivariate logistic regression
analysis to identify which SNPs are related with BC/OC cancer risk
independently. As shown in Table
III, all 4 SNPs decreased the BC/OC risk independently.
 | Table III.Multivariate logistic analysis of the
BC/OC cases to identify independently related SNPs. |
Table III.
Multivariate logistic analysis of the
BC/OC cases to identify independently related SNPs.
| Gene | SNPs | Genotype | Cases (%) | Controls (%) | aORa (95% CI) |
P-valueb |
|---|
| SNAI1 | rs6125849 | GG | 457 (31.58) | 337 (26.47) |
|
|
|
|
| AG | 703 (48.58) | 657 (51.61) | 0.80
(0.66–0.96) | 0.0140 |
|
|
| AA | 287 (19.83) | 279 (21.92) | 0.77
(0.62–0.96) | 0.0200 |
|
|
| AG/AA vs. GG
(dominant model) |
|
| 0.79
(0.66–0.94) | 0.0065 |
|
|
| AA vs. GG/AG
(recessive model) |
|
| 0.89
(0.74–1.07) | 0.2183 |
| TWIST1 | rs4721746 | CC | 1106 (76.43) | 933 (73.29) |
|
|
|
|
| AC | 307 (21.22) | 310 (24.35) | 0.79
(0.65–0.96) | 0.0150 |
|
|
| AA | 34 (2.35) | 30 (2.36) | 0.75
(0.44–1.26) | 0.2796 |
|
|
| AC/AA vs. CC
(dominant model) |
|
| 0.79
(0.65–0.95) | 0.0107 |
|
|
| AA vs. CC/AC
(recessive model) |
|
| 1.04
(0.61–1.75) | 0.8971 |
| TWIST1 | rs4721745 | CC | 558 (38.56) | 402 (31.58) |
|
|
|
|
| CG | 650 (44.92) | 638 (50.12) | 0.72
(0.61–0.86) | 0.0002 |
|
|
| GG | 239 (16.52) | 233 (18.30) | 0.73
(0.58–0.91) | 0.0060 |
|
|
| CG/GG vs. CC
(dominant model) |
|
| 0.73
(0.62–0.85) | 0.0001 |
|
|
| GG vs. GG/CC
(recessive model) |
|
| 0.88
(0.71–1.07) | 0.2004 |
| TWIST1 | rs11973396 | AA | 799 (55.22) | 641 (50.35) |
|
|
|
|
| AT | 526 (36.35) | 518 (40.69) | 0.97
(0.79–1.20) | 0.7918 |
|
|
| TT | 122 (8.43) | 114 (8.96) | 1.17
(0.79–1.73) | 0.4264 |
|
|
| AT/TT vs. AA
(dominant model) |
|
| 0.78
(0.67–0.92) | 0.0024 |
|
|
| TT vs. AA/AT
(recessive model) |
|
| 1.21
(0.88–1.67) | 0.2473 |
Similarly, multivariate logistic regression was used
to analyze whether the three aforementioned protective SNPs for
breast cancer (SNAI1 rs6125849, TWIST1 rs4721745 and
TWIST1 rs4721746) could affect breast cancer risk
independently. It was demonstrated that SNAI1 rs6125849 and
TWIST1 rs4721745 were still strongly associated with breast
cancer susceptibility between the 1,161 BC cases and 978 controls,
whereas TWIST1 rs4721746 was not further associated with BC
susceptibility.
Gene-gene interactions between genetic
variants
In the above analyses, the four SNPs, SNAI1
rs6125849, TWIST1 rs4721746, TWIST1 rs4721745 and
TWIST1 rs11973396, were demonstrated to be independent
genetic variants associated with decreased BC/OC risk. Therefore,
we performed logistic regression analyses to assess their joint
effects on BC/OC susceptibility. When all the women carrying none
of the 4 protective genotypes at the 4 loci were pooled together to
serve as a reference, those harboring three protective genotypes
showed significantly lower risk (aOR=0.69, P=0.0061), and those
harboring 4 protective genotypes demonstrated the lowest risk
(aOR=0.62, P=0.0220) (Table IV).
In subsequent trend test analysis, the dose-dependent effect of the
four protective loci was observed with Ptrend<0.0001,
indicating a synergistic effect of the four loci on BC/OC
susceptibility.
 | Table IV.Combination effects of rs6125849,
rs4721746, rs4721745 and rs11973396 in the dominant model on BC/OC
susceptibility. |
Table IV.
Combination effects of rs6125849,
rs4721746, rs4721745 and rs11973396 in the dominant model on BC/OC
susceptibility.
| Number of
protective genotypesa | Cases (%) | Controls (%) |
Ptrend | OR (95% CI) | P-value | aORb (95% CI) |
P-valuec |
|---|
| 0 | 162 (11.20) | 115 (9.03) |
<0001 |
Reference |
|
Reference |
|
| 1 | 392 (27.09) | 281 (22.07) |
| 0.99
(0.75–1.32) | 0.9463 | 1.03
(0.78–1.38) | 0.8204 |
| 2 | 269 (18.59) | 210 (16.50) |
| 0.91
(0.67–1.23) | 0.5339 | 0.92
(0.68–1.25) | 0.6051 |
| 3 | 558 (38.56) | 590 (46.35) |
| 0.67
(0.52–0.88) | 0.0033 | 0.69
(0.52–0.90) | 0.0061 |
| 4 | 66 (4.56) | 77 (6.05) |
| 0.61
(0.41–0.91) | 0.0166 | 0.62
(0.41–0.93) | 0.0220 |
Similarly, logistic regression demonstrated that
SNAI1 rs6125849 and TWIST1 rs4721745 could jointly
affect breast cancer susceptibility as well. Those harboring 2
protective genotypes of SNAI1 rs6125849 and TWIST1
rs4721745 showed the lowest risk (aOR=0.60, P=0.0011). In
subsequent trend test analysis, the dose-dependent effect of the
two loci was observed with Ptrend<0.0001.
Association of high-order interactions
with cancer risk by MDR analysis
MDR is a nonparametric and genetic model-free
alternative to traditional logistic regression analysis. Since the
7 tSNPs can cover the genetic variants in SNAI1 and
TWIST1 gene, we analyzed the interactions among these SNPs
by MDR. Table V shows the best
interaction model by MDR analysis for BC/OC. The best one-factor
model for predicting cancer risk was TWIST1 rs4721745
(C>G). It yielded a high testing accuracy of 0.5349 and CVC of
10/10 with permutation P=0.035, suggesting that this SNP is the
primary factor that contributes to BC/OC susceptibility compared
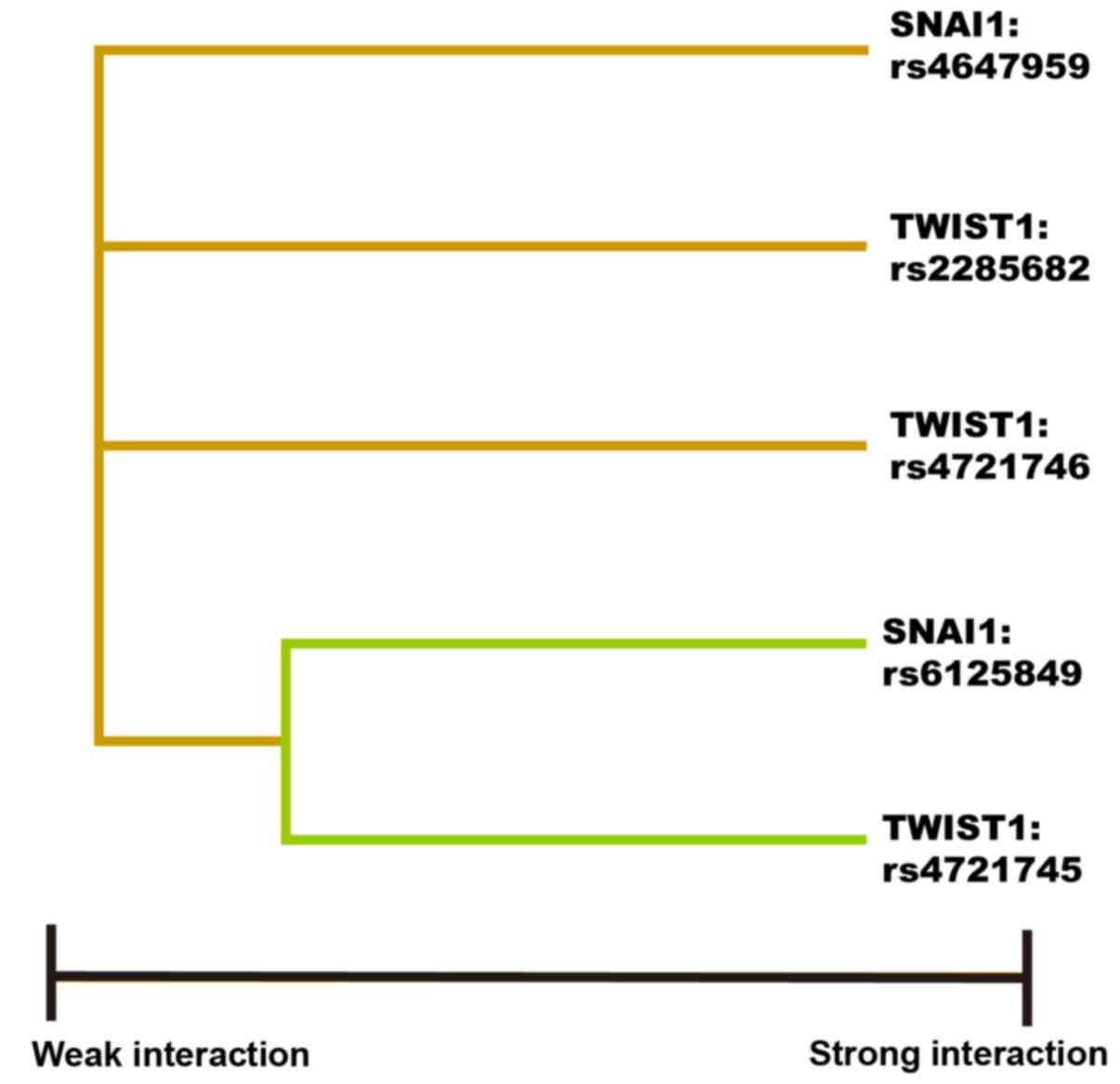
with the other genetic factors. The best three-factor model for
predicting cancer risk included SNAI1 rs6125849,
TWIST1 rs4721745 and TWIST1 rs2285682 (Table V). The interaction dendrogram showed
that SNAI1 rs6125849 and TWIST1 rs4721745 had the
strongest synergistic interaction (Fig.
3).
 | Table V.Association of higher-order gene-gene
interactions with BC/OC risk by MDR analysis. |
Table V.
Association of higher-order gene-gene
interactions with BC/OC risk by MDR analysis.
| No. of loci | Best interaction
modelsa | Testing
accuracy | CVC | P-value for
permutation test |
|---|
| 1 | TWIST1
rs4721745 | 0.5349 | 10/10 | 0.035 |
| 2 | SNAI1
rs6125849 + TWIST1 rs4721745 | 0.5340 | 6/10 | 0.042 |
| 3 | SNAI1
rs6125849 + TWIST1 rs2285682 + TWIST1 rs4721745 | 0.5440 | 7/10 | 0.005 |
| 4 | SNAI1
rs6125849 + SNAI1 rs4647959 + TWIST1 rs4721746 +
TWIST1 rs4721745 | 0.5350 | 5/10 | 0.035 |
Similarly, MDR analysis was performed in BC and OC
separately, and the interaction between SNAI1 rs6125849 and
TWIST1 rs4721745 remained in both breast cancer and ovarian
cancer (data not shown).
False-positive report probability and
statistical power analysis
All the above analyses indicated that SNAI1
rs6125849, TWIST1 rs4721745 and rs4721746 were strongly
associated with BC/OC susceptibility. To decide whether these
findings deserve attention or are ‘noteworthy’, we calculated
false-positive report probability (FPRP) and statistical power. As
shown in Table VI, at the FPRP
threshold of 0.2 and prior probability level of 0.1, the
associations of SNAI1 rs6125849 and TWIST1 rs4721745
with BC/OC susceptibility remained noteworthy.
 | Table VI.False-positive report probability
(FPRP) values for positive results on associations between
tagging-SNPs and BC/OC. |
Table VI.
False-positive report probability
(FPRP) values for positive results on associations between
tagging-SNPs and BC/OC.
|
|
|
|
| Prior
probability |
|---|
|
|
|
|
|
|
|---|
| Gene/SNP | Crude OR (95%
CI) |
P-valuea | Statistical
powerb | 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 |
|---|
| SNAI1
rs6125849 (G>A) |
| AG vs.
GG | 0.79
(0.66–0.94) | 0.0085 | 0.9999 | 0.0249 | 0.0711 | 0.4570 | 0.8947 | 0.9884 |
| AA vs.
GG | 0.76
(0.61–0.94) | 0.0124 | 0.9988 | 0.0359 | 0.1005 | 0.5514 | 0.9254 | 0.9920 |
| AG/AA
vs. GG | 0.78
(0.66–0.92) | 0.0035 | 0.8402 | 0.0123 | 0.0361 | 0.2920 | 0.8063 | 0.9766 |
| TWIST1
rs4721746 (C>A) |
| AC vs.
CC | 0.84
(0.70–1.00) | 0.0506 | 0.5917 | 0.2042 | 0.4349 | 0.8944 | 0.9884 | 0.9988 |
| AC/AA
vs. CC | 0.85
(0.71–1.01) | 0.0592 | 0.4526 | 0.2818 | 0.5407 | 0.9283 | 0.9924 | 0.9992 |
| TWIST1
rs4721745 (C>G) |
| CG vs.
CC | 0.73
(0.62–0.87) | 0.0003 | 0.9890 | 0.0009 | 0.0027 | 0.0292 | 0.2326 | 0.7521 |
| GG vs.
CC | 0.74
(0.60–0.92) | 0.0074 | 0.9829 | 0.0221 | 0.0635 | 0.4270 | 0.8826 | 0.9869 |
| CG/GG
vs. CC | 0.74
(0.63–0.86) | 0.0001 | 0.9483 | 0.0003 | 0.0009 | 0.0103 | 0.0953 | 0.5132 |
Functional annotation
Functional annotations were conducted for these
‘noteworthy’ SNPs SNAI1 rs6125849 and TWIST1
rs4721745 using HaploReg (http://www.broadinstitute.org/mammals/haploreg/haploreg.php)
and RegulomeDB (http://regulome.stanford.edu/). In ovarian tissues,
breast cancer cell line MCF-7, and human mammary epithelial cell
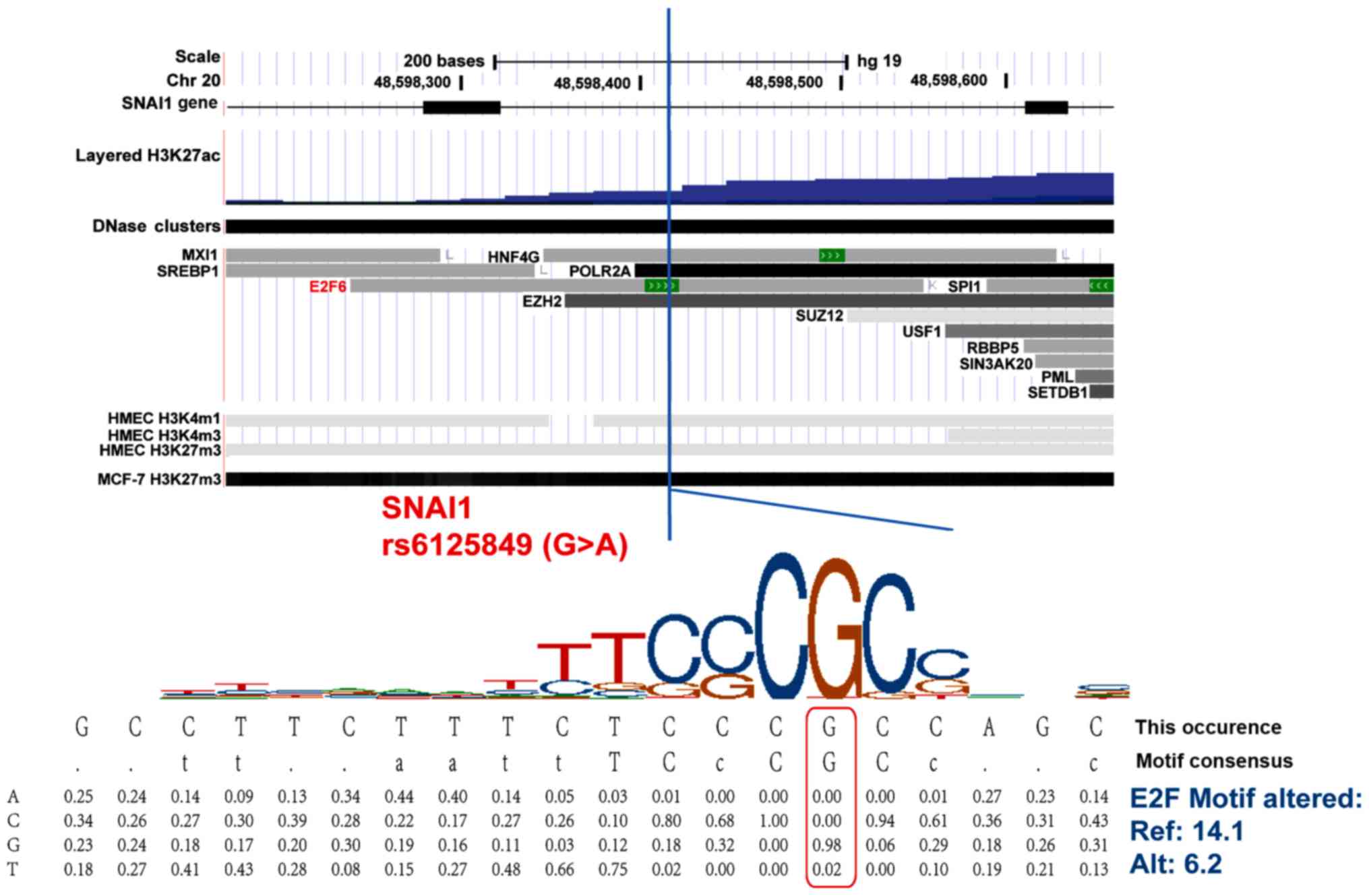
line HMEC, SNAI1 rs6125849 (G>A) is located in the region
with histone methylation, acetylation modifications and many
transcriptional factor binding sites, indicating a role of
expressional regulation in this region (Fig. 3). In addition, the minor allele A of
SNAI1 rs6125849 (G>A) alters its binding affinity with
transcription factor E2F6 based on the HaploReg database (Fig. 4). The TWIST1 rs4721745 is
located in the region with histone modifications in human mammary
epithelial cell line HMEC. Using Jaspar database, it predicted that
the minor allele G of TWIST1 rs4721745 (C>G) altered its
binding affinity with transcription factor TCF11-MafG.
Overall, SNAI1 rs6125849 and TWIST1 rs4721746 may be
functional loci.
Discussion
To the best of our knowledge, this is the first
gene-wide SNP study to comprehensively evaluate the association of
genetic variants in SNAI1 and TWIST1 with the risk of
breast cancer (BC) and ovarian cancer (OC) in Chinese Han
women.
In this study, we applied multiple strategies
including tagging-SNP genotyping, genotype imputation, logistic
regression (LR), MDR and functional annotation to systematically
evaluate the association of BC/OC susceptibility with germline
variants in SNAI1 and TWIST1 among Chinese Han women.
By LR, we discovered that three tSNPs (SNAI1 rs6125849,
TWIST1 rs4721746, and TWIST1 rs4721745) were
associated with decreased risks of BC/OC. Since a fine-mapping
study with high-density SNPs within the target region may be
helpful in identifying the causal variants, we used genotype
imputation to analyze more variants among the 2 genes, and revealed
an imputed SNP TWIST1 rs11973396 to be associated with
decreased BC/OC risk as well. Moreover, we demonstrated the joint
effects of these protective loci by LR and MDR analyses, and found
that women carrying minor alleles of both SNAI1 rs6125849
and TWIST1 rs4721745 could decrease their risk of BC/OC by
40%.
According to our single-locus analysis,
TWIST1 rs4721745 (C>G), located in the 3′ flanking region
of the gene, might be the strongest protective locus against BC/OC
in our population. A recently published study on endometrial cancer
susceptibility from our laboratory also showed rs4721745 to be a
protective locus in the CHB population, which is consistent with
our results (35). Using web-based
software F-SNP (http://compbio.cs.queensu.ca/F-SNP/) (36), this polymorphism is predicted to
have transcriptional regulation function by TFSearch and Consite
tools (FS score 0.208). In addition, by using miRNA prediction
module PicTar and TargetScan from UCSC genome browser, miR-33/381
is predicted to be the twist-specific target regulatory elements
(37). Zhou et al found that
the miR-33a level was negatively correlated with the Twist level in
Saos-2 cells, and inhibition of miR-33a increased cisplatin-induced
cell apoptosis, which was reversed by knockdown of Twist (38). Our functional annotation also
indicated that rs4721745 was located in a region with histone
methylation modifications such as H3K4me3, a modification usually
associated with active transcription of nearby genes, suggesting
that the variation of this locus may alter TWIST1 expression
by influencing its transcription level. Jaspar database predicted
that the minor allele G of TWIST1 rs4721745 (C>G) altered
its binding affinity with transcription factor TCF11-MafG.
Based on these findings, we predict that this protective locus may
be involved in TWIST1 transcription and alter its biological
functions.
SNAI1 inhibits the expression of epithelial
markers and activates mesenchymal molecules, thus participating in
tumor epithelial-mesenchymal transition and metastasis (39). Our results showed that SNAI1
rs6125849 (located near 5′ flanking region) decreased BC risk. The
3DSNP (http://cbportal.org/3dsnp/) predicted
that this locus was located in the loop between the promoter and
enhancer with histone modifications and many transcriptional factor
binding sites. HaploReg database predicted that the minor allele A
of SNAI1 rs6125849 (G>A) altered its binding affinity
with transcription factor E2F6. We presumed this protective locus
might affect SNAI1 transcription, thereby inhibiting tumor
development. Regarding SNAI1 rs4647958 (Val118Ala), also
known as c.353T>C, Lei et al genotyped this missense
variation in 2,072 lung cancer cases and 2,077 control subjects,
and demonstrated a protective effect of rs4647958 on lung cancer
susceptibility (OR=0.76, 95% CI=0.65–0.90, P=0.018) (40). In this study, however, we did not
find any association between rs4647958 and BC/OC risk. It is
possible that rs4647958 plays a different role in lung cancer and
BC/OC.
Our study inevitably has limitations. First, we
only analyzed the association of common genetic variants (MAF
≥0.05) with BC/OC susceptibility, but did not analyze the
association of low frequency or very low frequency of genetic
variants (MAF <0.05), which might have led to the omission of
important variants. Also, due to the small sample size of the
subgroups, various results may be chance findings, although we used
various statistical methods to control the false positives.
In summary, this study suggests that genetic
variants in SNAI1 and TWIST1 are associated with
BC/OC susceptibility. Our data also suggest a synergistic effect of
those related loci on BC/OC risk. Nevertheless, further validation
studies are needed for further determination of the truly causal
SNPs and their exact functional mechanisms. These cancer-associated
SNPs have the potential to improve our future ability of
personalized evaluation of BC/OC susceptibility and facilitate the
identification of high-risk subgroups in the general
population.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81672790 and 81621063)
and the Open Project of the Key Laboratory of Genomic and Precision
Medicine, Chinese Academy of Sciences.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YJW, XXT and WGF conceived and designed the study.
YJW, LYZ and LY performed the experiments. YJW, ZFW, RS, LY and YHG
analyzed the data. FM, YTX, LC, DGL, XHL and YLS designed the study
and contributed with reagents/materials/analysis tools. YJW, ZFW,
YHG and RS wrote the paper. XXT, WGF and YHG revised the paper. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Peking
University IRB (reference no. IRB00001052-11029). We obtained
written informed consents from all BC cases and control women.
Since we could not obtain the contact information of ovarian cancer
patients treated before 2011, PKU IRB approved our application to
waive informed consent for the archived samples collected before
April 2011. This study only used these samples. We used all the
data/samples anonymously.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H
and Zou X: Report of cancer incidence and mortality in China, 2010.
Ann Transl Med. 2:61–85. 2014.PubMed/NCBI
|
|
2
|
Jordan SJ, Cushing-Haugen KL, Wicklund KG,
Doherty JA and Rossing MA: Breast-feeding and risk of epithelial
ovarian cancer. Cancer Causes Control. 23:919–927. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng L, Song A, Ruan Y, Chen L, Liu D, Li
X, Guo H, Han J, Li Y, Tian X, et al: Genetic polymorphisms in
AURKA, BRCA1, CCNE1 and CDK2 are associated with ovarian cancer
susceptibility among Chinese Han women. Cancer Epidemiol.
37:639–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Steiner E, Klubert D and Knutson D:
Assessing breast cancer risk in women. Am Fam Physician.
78:1361–1366. 2008.PubMed/NCBI
|
|
5
|
Bodelon C, Wentzensen N, Schonfeld SJ,
Visvanathan K, Hartge P, Park Y and Pfeiffer RM: Hormonal risk
factors and invasive epithelial ovarian cancer risk by parity. Br J
Cancer. 109:769–776. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scollen S, Luccarini C, Baynes C, Driver
K, Humphreys MK, Garcia-Closas M, Figueroa J, Lissowska J, Pharoah
PD, Easton DF, et al: TGF-β signaling pathway and breast cancer
susceptibility. Cancer Epidemiol Biomarkers Prev. 20:1112–1119.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Antoniou A, Pharoah PD, Narod S, Risch HA,
Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et
al: Average risks of breast and ovarian cancer associated with
BRCA1 or BRCA2 mutations detected in case Series unselected for
family history: A combined analysis of 22 studies. Am J Hum Genet.
72:1117–1130. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Antoniou AC and Easton DF: Models of
genetic susceptibility to breast cancer. Oncogene. 25:5898–5905.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yoshida K and Miki Y: Role of BRCA1 and
BRCA2 as regulators of DNA repair, transcription, and cell cycle in
response to DNA damage. Cancer Sci. 95:866–871. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rahdar M, Inoue T, Meyer T, Zhang J,
Vazquez F and Devreotes PN: A phosphorylation-dependent
intramolecular interaction regulates the membrane association and
activity of the tumor suppressor PTEN. Proc Natl Acad Sci USA.
106:480–485. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vousden KH: p53: Death star. Cell.
103:691–694. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hollestelle A, Wasielewski M, Martens JW
and Schutte M: Discovering moderate-risk breast cancer
susceptibility genes. Curr Opin Genet Dev. 20:268–276. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schork NJ, Murray SS, Frazer KA and Topol
EJ: Common vs. rare allele hypotheses for complex diseases. Curr
Opin Genet Dev. 19:212–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu CT and Lin H and Lin H: Functional
analysis of HapMap SNPs. Gene. 511:358–363. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stram DO, Haiman CA, Hirschhorn JN,
Altshuler D, Kolonel LN, Henderson BE and Pike MC: Choosing
haplotype-tagging SNPS based on unphased genotype data using a
preliminary sample of unrelated subjects with an example from the
Multiethnic Cohort Study. Hum Hered. 55:27–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Willer C, Sanna S and Abecasis G:
Genotype imputation. Annu Rev Genomics Hum Genet. 10:387–406. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nothnagel M, Ellinghaus D, Schreiber S,
Krawczak M and Franke A: A comprehensive evaluation of SNP genotype
imputation. Hum Genet. 125:163–171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clark AG: The role of haplotypes in
candidate gene studies. Genet Epidemiol. 27:321–333. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Shi J, Chai K, Ying X and Zhou BP:
The role of snail in EMT and tumorigenesis. Curr Cancer Drug
Targets. 13:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin H, Yu Y, Zhang T, Zhou X, Zhou J, Jia
L, Wu Y, Zhou BP and Feng Y: Snail is critical for tumor growth and
metastasis of ovarian carcinoma. Int J Cancer. 126:2102–2111.
2010.PubMed/NCBI
|
|
21
|
Moody SE, Perez D, Pan TC, Sarkisian CJ,
Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD and
Chodosh LA: The transcriptional repressor Snail promotes mammary
tumor recurrence. Cancer Cell. 8:197–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang X, Guo D, Li W, Yu T, Zhou J and
Gong J: Combination Twist1 and CA15-3 in axillary lymph nodes for
breast cancer prognosis. Mol Med Rep. 15:1123–1134. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Terauchi M, Kajiyama H, Yamashita M, Kato
M, Tsukamoto H, Umezu T, Hosono S, Yamamoto E, Shibata K, Ino K, et
al: Possible involvement of TWIST in enhanced peritoneal metastasis
of epithelial ovarian carcinoma. Clin Exp Metastasis. 24:329–339.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hosono S, Kajiyama H, Terauchi M, Shibata
K, Ino K, Nawa A and Kikkawa F: Expression of Twist increases the
risk for recurrence and for poor survival in epithelial ovarian
carcinoma patients. Br J Cancer. 96:314–320. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matsusaka S, Zhang W, Cao S, Hanna DL,
Sunakawa Y, Sebio A, Ueno M, Yang D, Ning Y, Parekh A, et al:
TWIST1 polymorphisms predict survival in patients with metastatic
colorectal cancer receiving first-line bevacizumab plus
oxaliplatin-based chemotherapy. Mol Cancer Ther. 15:1405–1411.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grimberg J, Nawoschik S, Belluscio L,
McKee R, Turck A and Eisenberg A: A simple and efficient
non-organic procedure for the isolation of genomic DNA from blood.
Nucleic Acids Res. 17:83901989. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pikor LA, Enfield KSS, Cameron H and Lam
WL: DNA extraction from paraffin embedded material for genetic and
epigenetic analyses. J Vis Exp. 49:e27632011.
|
|
28
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lewis CM: Genetic association studies:
Design, analysis and interpretation. Brief Bioinform. 3:146–153.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He J, Wang F, Zhu J, Zhang R, Yang T, Zou
Y and Xia H: Association of potentially functional variants in the
XPG gene with neuroblastoma risk in a Chinese population. J Cell
Mol Med. 20:1481–1490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wacholder S, Chanock S, Garcia-Closas M,
El Ghormli L and Rothman N: Assessing the probability that a
positive report is false: An approach for molecular epidemiology
studies. J Natl Cancer Inst. 96:434–442. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Browning BL and Browning SR: A unified
approach to genotype imputation and haplotype-phase inference for
large data sets of trios and unrelated individuals. Am J Hum Genet.
84:210–223. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu L, Wu C, Wang Y, Zhong R, Wang F,
Zhang X, Duan S, Lou J, Yu D, Tan W, et al: Association of
candidate genetic variations with gastric cardia adenocarcinoma in
Chinese population: A multiple interaction analysis.
Carcinogenesis. 32:336–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ghosh J, Pradhan S and Mittal B:
Multilocus analysis of hormonal, neurotransmitter, inflammatory
pathways and genome-wide associated variants in migraine
susceptibility. Eur J Neurol. 21:1011–1020. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang L, Wang YJ, Zheng LY, Jia YM, Chen
YL, Chen L, Liu DG, Li XH, Guo HY, Sun YL, et al: Genetic
polymorphisms of TGFB1, TGFBR1, SNAI1 and TWIST1 are associated
with endometrial cancer susceptibility in chinese Han women. PLoS
One. 11:e01552702016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee PH and Shatkay H: F-SNP:
Computationally predicted functional SNPs for disease association
studies. Nucleic Acids Res. 36:D820–D824. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Huang Z, Wu S, Zang X, Liu M and
Shi J: miR-33a is up-regulated in chemoresistant osteosarcoma and
promotes osteosarcoma cell resistance to cisplatin by
down-regulating TWIST. J Exp Clin Cancer Res. 33:122014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dhasarathy A, Phadke D, Mav D, Shah RR and
Wade PA: The transcription factors Snail and Slug activate the
transforming growth factor-beta signaling pathway in breast cancer.
PLoS One. 6:e265142011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang L, Yang X, Ji W, Deng J, Qiu F, Yang
R, Fang W, Zhang L, Huang D, Xie C, et al: Effects of a functional
variant c.353T>C in snai1 on risk of two contextual diseases.
Chronic obstructive pulmonary disease and lung cancer. Am J Respir
Crit Care Med. 189:139–148. 2014.PubMed/NCBI
|