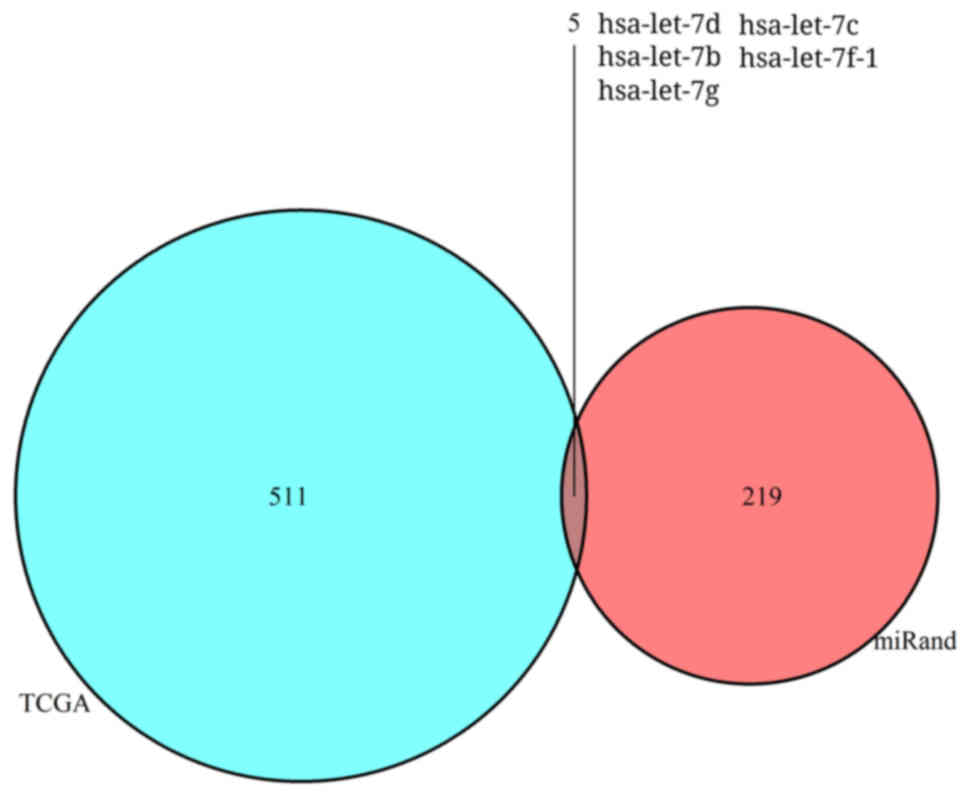
|
1
|
Arnold M, Sierra MS, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global patterns and trends in
colorectal cancer incidence and mortality. Gut. 66:683–691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aran V, Victorino AP, Thuler LC and
Ferreira CG: Colorectal cancer: Epidemiology, disease mechanisms
and interventions to reduce onset and mortality. Clin Colorectal
Cancer. 15:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herman JG, Umar A, Polyak K, Graff JR,
Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW,
et al: Incidence and functional consequences of hMLH1 promoter
hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA.
95:6870–6875. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reya T and Clevers H: Wnt signalling in
stem cells and cancer. Nature. 434:843–850. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wierzbicki PM and Rybarczyk A: The Hippo
pathway in colorectal cancer. Folia Histochem Cytobiol. 53:105–119.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Papadatos-Pastos D, Rabbie R, Ross P and
Sarker D: The role of the PI3K pathway in colorectal cancer. Crit
Rev Oncol Hematol. 94:18–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iwaya T, Yokobori T, Nishida N, Kogo R,
Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, et
al: Downregulation of miR-144 is associated with colorectal cancer
progression via activation of mTOR signaling pathway.
Carcinogenesis. 33:2391–2397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan M, Song M, Bai R, Cheng S and Yan W:
Identification of potential therapeutic targets for colorectal
cancer by bioinformatics analysis. Oncol Lett. 12:5092–5098. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gaudet P, Škunca N, Hu JC and Dessimoz C:
Primer on the gene ontology. Methods Mol Biol. 1446:25–37. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takemasa I, Higuchi H, Yamamoto H,
Sekimoto M, Tomita N, Nakamori S, Matoba R, Monden M and Matsubara
K: Construction of preferential cDNA microarray specialized for
human colorectal carcinoma: Molecular sketch of colorectal cancer.
Biochem Biophys Res Commun. 285:1244–1249. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang
K, Go MY, Ng SC, Chan FK, Sung JJ, et al: Carbonic anhydrase IV
inhibits colon cancer development by inhibiting the Wnt signalling
pathway through targeting the WTAP-WT1-TBL1 axis. Gut.
65:1482–1493. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Farquharson AJ, Steele RJ, Carey FA and
Drew JE: Novel multiplex method to assess insulin, leptin and
adiponectin regulation of inflammatory cytokines associated with
colon cancer. Mol Biol Rep. 39:5727–5736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wood SM, Gill AJ, Brodsky AS, Lu S,
Friedman K, Karashchuk G, Lombardo K, Yang D and Resnick MB: Fatty
acid-binding protein 1 is preferentially lost in microsatellite
instable colorectal carcinomas and is immune modulated via the
interferon γ pathway. Mod Pathol. 30:123–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gaspar C and Fodde R: APC dosage effects
in tumorigenesis and stem cell differentiation. Int J Dev Biol.
48:377–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang
Z, Wang C, Zhang Z, Xia W, et al: γδT17 cells promote the
accumulation and expansion of myeloid-derived suppressor cells in
human colorectal cancer. Immunity. 40:785–800. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zu X, Yan R, Pan J, Zhong L, Cao Y, Ma J,
Cai C, Huang D, Liu J, Chung FL, et al: Aldo-keto reductase 1B10
protects human colon cells from DNA damage induced by electrophilic
carbonyl compounds. Mol Carcinog. 56:118–129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Widegren E, Onnesjö S, Arbman G, Kayed H,
Zentgraf H, Kleeff J, Zhang H and Sun XF: Expression of FXYD3
protein in relation to biological and clinicopathological variables
in colorectal cancers. Chemotherapy. 55:407–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ivanov SV, Panaccione A, Nonaka D, Prasad
ML, Boyd KL, Brown B, Guo Y, Sewell A and Yarbrough WG: Diagnostic
SOX10 gene signatures in salivary adenoid cystic and breast
basal-like carcinomas. Br J Cancer. 109:444–451. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krampitz GW, George BM, Willingham SB,
Volkmer JP, Weiskopf K, Jahchan N, Newman AM, Sahoo D, Zemek AJ,
Yanovsky RL, et al: Identification of tumorigenic cells and
therapeutic targets in pancreatic neuroendocrine tumors. Proc Natl
Acad Sci USA. 113:4464–4469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vincenzi B, Santini D and Tonini G:
Biological interaction between anti-epidermal growth factor
receptor agent cetuximab and magnesium. Expert Opin Pharmacother.
9:1267–1269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wolf FI and Trapani V: Cell
(patho)physiology of magnesium. Clin Sci. 114:27–35. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nasulewicz A, Wietrzyk J, Wolf FI, Dzimira
S, Madej J, Maier JA, Rayssiguier Y, Mazur A and Opolski A:
Magnesium deficiency inhibits primary tumor growth but favors
metastasis in mice. Biochim Biophys Acta. 1739:26–32. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Solinas G, Marchesi F, Garlanda C,
Mantovani A and Allavena P: Inflammation-mediated promotion of
invasion and metastasis. Cancer Metastasis Rev. 29:243–248. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lan FF, Wang H, Chen YC, Chan CY, Ng SS,
Li K, Xie D, He ML, Lin MC and Kung HF: Hsa-let-7g inhibits
proliferation of hepatocellular carcinoma cells by downregulation
of c-Myc and upregulation of p16INK4A. Int J Cancer.
128:319–331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ding Z, Wang X, Schnackenberg L, Khaidakov
M, Liu S, Singla S, Dai Y and Mehta JL: Regulation of autophagy and
apoptosis in response to ox-LDL in vascular smooth muscle cells,
and the modulatory effects of the microRNA hsa-let-7 g. Int J
Cardiol. 168:1378–1385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang S, Feng C, Zhai YZ, Zhou X, Li B,
Wang LL, Chen W, Lv FQ and Li TS: Identification of miRNA
biomarkers of pneumonia using RNA-sequencing and bioinformatics
analysis. Exp Ther Med. 13:1235–1244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Huang D, Chen KY, Cui M, Wang W,
Huang X, Awadellah A, Li Q, Friedman A, Xin WW, et al: Fucosylation
deficiency in mice leads to colitis and adenocarcinoma.
Gastroenterology. 152:193–205.e10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Srinivasan T, Walters J, Bu P, Than EB,
Tung KL, Chen KY, Panarelli N, Milsom J, Augenlicht L, Lipkin SM,
et al: NOTCH signaling regulates asymmetric cell fate of fast- and
slow-cycling colon cancer initiating cells. Cancer Res.
76:3411–3421. 2016. View Article : Google Scholar : PubMed/NCBI
|