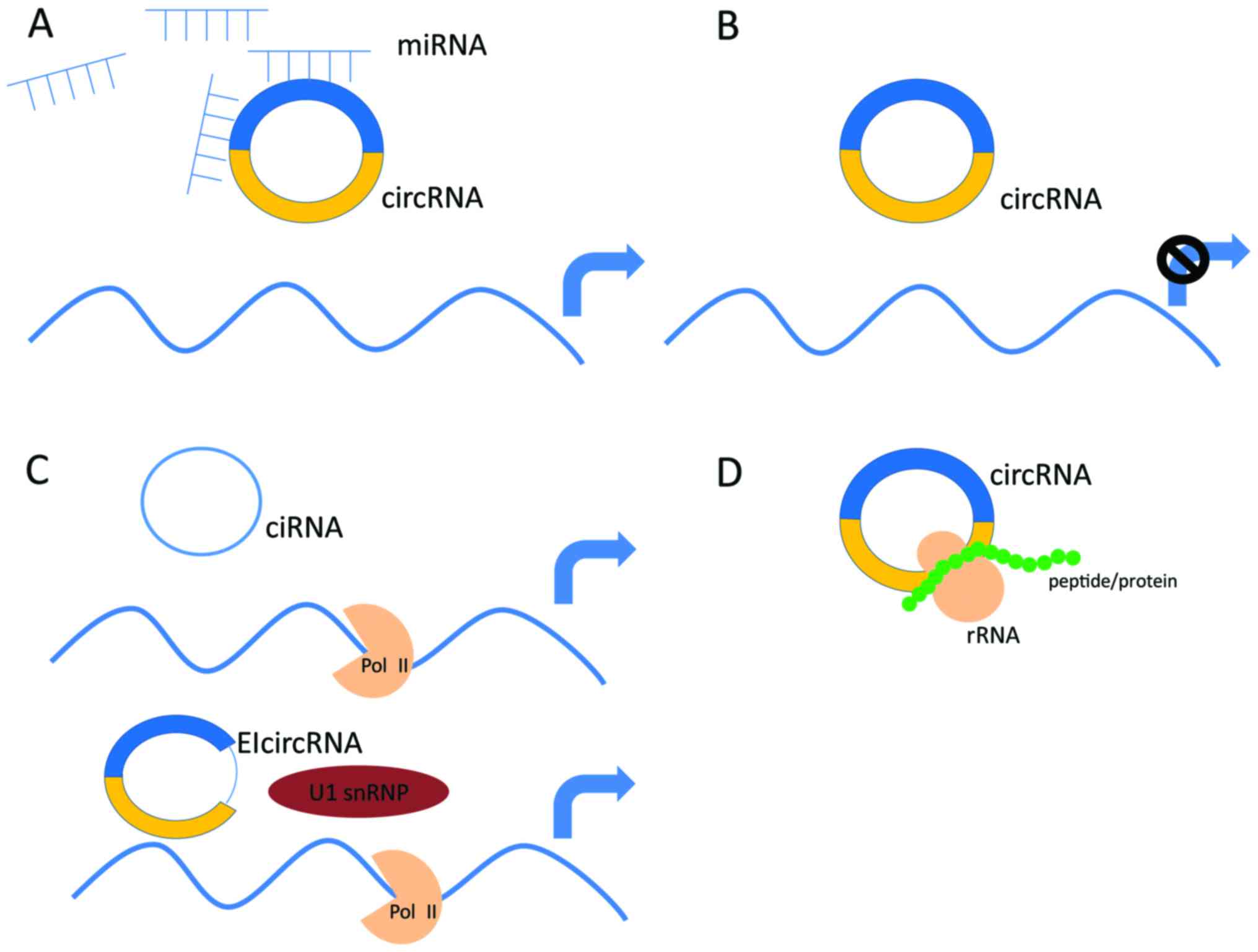
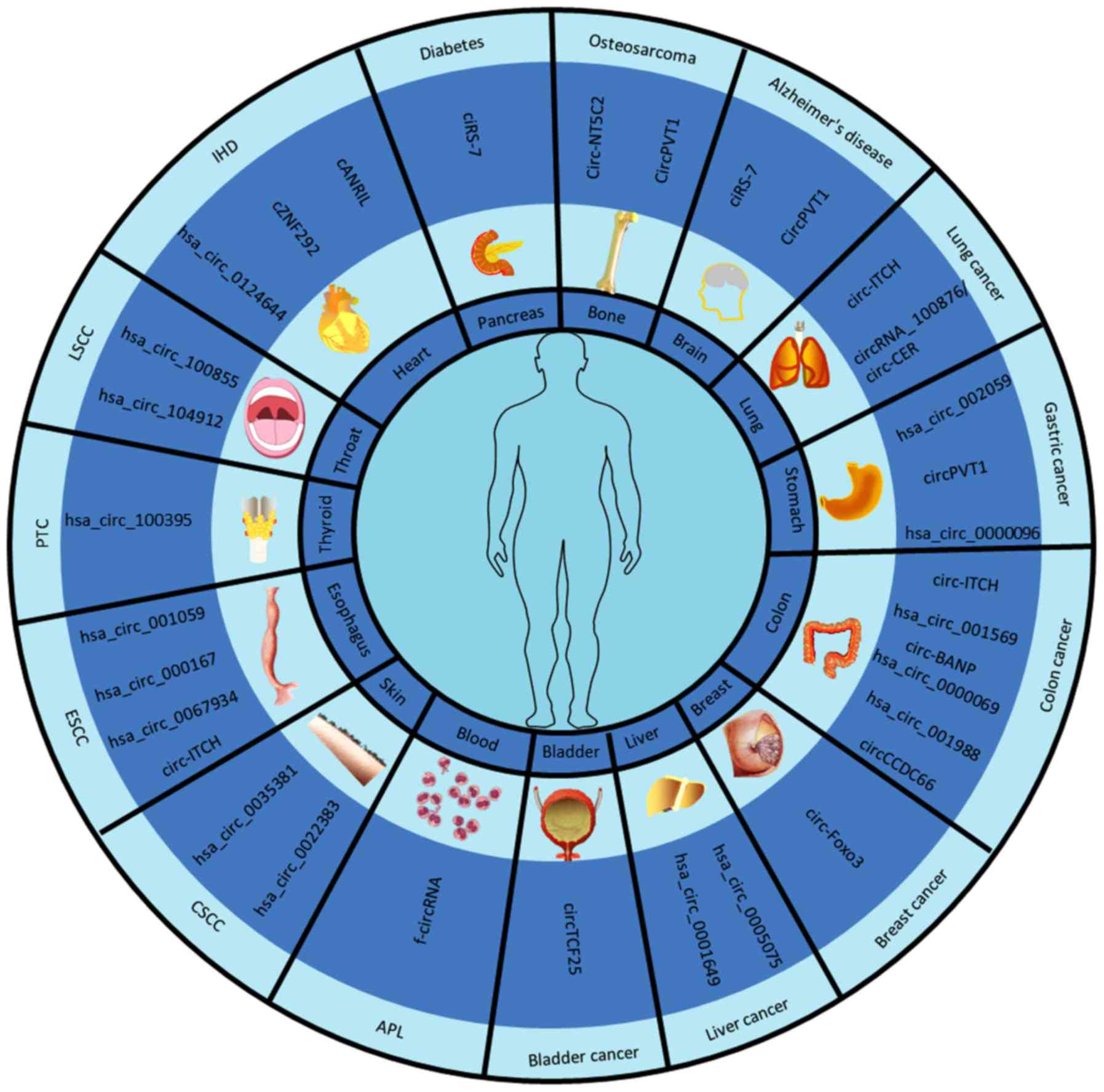
|
1
|
Amodio N, Raimondi L, Juli G, Stamato MA,
Caracciolo D, Tagliaferri P and Tassone P: MALAT1: A druggable long
non-coding RNA for targeted anti-cancer approaches. J Hematol
Oncol. 11:632018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Glasgow AMA, De Santi C and Greene CM:
Non-coding RNA in cystic fibrosis. Biochem Soc Trans. 46:619–630.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zong L, Sun Q, Zhang H, Chen Z, Deng Y, Li
D and Zhang L: Increased expression of circRNA_102231 in lung
cancer and its clinical significance. Biomed Pharmacother.
102:639–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soslau G: Circular RNA (circRNA) was an
important bridge in the switch from the RNA world to the DNA world.
J Theor Biol. 447:32–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang XM, Li ZL, Li JL, Xu Y, Leng KM, Cui
YF and Sun DJ: A novel prognostic biomarker for cholangiocarcinoma:
circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 22:365–371.
2018.PubMed/NCBI
|
|
6
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao Y, Zhang J and Zhao F: Circular RNA
identification based on multiple seed matching. Brief Bioinform.
Feb 28–2017.(Epub ahead of print). doi: 10.1093/bib/bbx014.
|
|
9
|
Lei K, Bai H, Wei Z, Xie C, Wang J, Li J
and Chen Q: The mechanism and function of circular RNAs in human
diseases. Exp Cell Res. 368:147–158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li D, Chen Y, Mei H, Jiao W, Song H, Ye L,
Fang E, Wang X, Yang F, Huang K, et al: Ets-1 promoter-associated
noncoding RNA regulates the NONO/ERG/Ets-1 axis to drive gastric
cancer progression. Oncogene. 37:4871–4886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao ZJ and Shen J: Circular RNA
participates in the carcinogenesis and the malignant behavior of
cancer. RNA Biol. 14:514–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Militello G, Weirick T, John D, Döring C,
Dimmeler S and Uchida S: Screening and validation of lncRNAs and
circRNAs as miRNA sponges. Brief Bioinform. 18:780–788.
2017.PubMed/NCBI
|
|
16
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim J, Abdelmohsen K, Yang X, De S,
Grammatikakis I, Noh JH and Gorospe M: LncRNA OIP5-AS1/cyrano
sponges RNA-binding protein HuR. Nucleic Acids Res. 44:2378–2392.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dai X, Zhang N, Cheng Y, Yang T, Chen Y,
Liu Z, Wang Z, Yang C and Jiang Y: RNA-binding protein
trinucleotide repeat-containing 6A regulates the formation of
circular RNA 0006916, with important functions in lung cancer
cells. Carcinogenesis. May 3–2018.(Epub ahead of print). doi:
10.1093/carcin/bgy061. View Article : Google Scholar
|
|
19
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J,
Wang Z, Wang Q, Li A, Marks JR, et al: CircIRAK3 sponges miR-3607
to facilitate breast cancer metastasis. Cancer Lett. 430:179–192.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan Y, Liu W, Zhang Y, Zhang Y and Sun S:
CircRNA circ_0026344 as a prognostic biomarker suppresses
colorectal cancer progression via microRNA-21 and microRNA-31.
Biochem Biophys Res Commun. 503:870–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou J, Wang H, Chu J, Huang Q, Li G, Yan
Y, Xu T, Chen J and Wang Y: Circular RNA hsa_circ_0008344 regulates
glioblastoma cell proliferation, migration, invasion, and
apoptosis. J Clin Lab Anal. e224542018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
24
|
Hou LD and Zhang J: Circular RNAs: An
emerging type of RNA in cancer. Int J Immunopathol Pharmacol.
30:1–6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jakobi T and Dieterich C: Deep
computational circular RNA analytics from RNA-seq data. Methods Mol
Biol. 1724:9–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen BJ, Byrne FL, Takenaka K, Modesitt
SC, Olzomer EM, Mills JD, Farrell R, Hoehn KL and Janitz M:
Analysis of the circular RNA transcriptome in endometrial cancer.
Oncotarget. 9:5786–5796. 2018.PubMed/NCBI
|
|
27
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fumagalli MR, Zapperi S and La Porta CAM:
Impact of the cross-talk between circular and messenger RNAs on
cell regulation. J Theor Biol. 454:386–395. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siede D, Rapti K, Gorska AA, Katus HA,
Altmüller J, Boeckel JN, Meder B, Maack C, Völkers M, Müller OJ, et
al: Identification of circular RNAs with host gene-independent
expression in human model systems for cardiac differentiation and
disease. J Mol Cell Cardiol. 109:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Y and Wang Z: Efficient backsplicing
produces translatable circular mRNAs. RNA. 21:172–179. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N,
Kadener S, et al: circRNA biogenesis competes with pre-mRNA
splicing. Mol Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wilusz JE: Circular RNAs: Unexpected
outputs of many protein-coding genes. RNA Biol. 14:1007–1017. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsiao KY, Sun HS and Tsai SJ: Circular
RNA-new member of noncoding RNA with novel functions. Exp Biol Med.
242:1136–1141. 2017. View Article : Google Scholar
|
|
36
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Petkovic S and Müller S: Synthesis and
engineering of circular RNAs. Methods Mol Biol. 1724:167–180. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen L, Huang C, Wang X and Shan G:
Circular RNAs in eukaryotic cells. Curr Genomics. 16:312–318. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang Y and Wang Z: Constructing GFP-based
reporter to study back splicing and translation of circular RNA.
Methods Mol Biol. 1724:107–118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schmitz KM, Mayer C, Postepska A and
Grummt I: Interaction of noncoding RNA with the rDNA promoter
mediates recruitment of DNMT3b and silencing of rRNA genes. Genes
Dev. 24:2264–2269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Beckedorff FC, Ayupe AC, Crocci-Souza R,
Amaral MS, Nakaya HI, Soltys DT, Menck CF, Reis EM and
Verjovski-Almeida S: The intronic long noncoding RNA ANRASSF1
recruits PRC2 to the RASSF1A promoter, reducing the expression of
RASSF1A and increasing cell proliferation. PLoS Genet.
9:e10037052013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Müller S and Appel B: In vitro
circularization of RNA. RNA Biol. 14:1018–1027. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao F, Han Y, Liu Z, Zhao Z, Li Z and Jia
K: circFADS2 regulates lung cancer cells proliferation and invasion
via acting as a sponge of miR-498. Biosci Rep. 38:pii:
BSR201805702018. View Article : Google Scholar
|
|
49
|
Lin X and Chen Y: Identification of
potentially functional CircRNA-miRNA-mRNA regulatory network in
hepatocellular carcinoma by integrated microarray analysis. Med Sci
Monit Basic Res. 24:70–78. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang M, Jia L and Zheng Y: circRNA
expression profiles in human bone marrow stem cells undergoing
osteoblast differentiation. Stem Cell Rev. Jul 25–2018.(Epub ahead
of print). doi: 10.1007/s12015-018-9841-x. View Article : Google Scholar
|
|
51
|
Li S, Sun X, Miao S, Lu T, Wang Y, Liu J
and Jiao W: hsa_circ_0000729, a potential prognostic biomarker in
lung adenocarcinoma. Thorac Cancer. 9:924–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Haque S and Harries LW: Circular RNAs
(circRNAs) in health and disease. Genes. 8:E3532017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schneider T, Hung LH, Schreiner S, Starke
S, Eckhof H, Rossbach O, Reich S, Medenbach J and Bindereif A:
CircRNA-protein complexes: IMP3 protein component defines subfamily
of circRNPs. Sci Rep. 6:313132016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gehring NH, Kunz JB, Neu-Yilik G, Breit S,
Viegas MH, Hentze MW and Kulozik AE: Exon-junction complex
components specify distinct routes of nonsense-mediated mRNA decay
with differential cofactor requirements. Mol Cell. 20:65–75. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Perriman R and Ares M Jr: Circular mRNA
can direct translation of extremely long repeating-sequence
proteins in vivo. RNA. 4:1047–1054. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
AbouHaidar MG, Venkataraman S, Golshani A,
Liu B and Ahmad T: Novel coding, translation, and gene expression
of a replicating covalently closed circular RNA of 220 nt. Proc
Natl Acad Sci USA. 111:14542–14547. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen X, Han P, Zhou T, Guo X, Song X and
Li Y: circRNADb: A comprehensive database for human circular RNAs
with protein-coding annotations. Sci Rep. 6:349852016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lasda E and Parker R: Circular RNAs
co-precipitate with extracellular vesicles: A possible mechanism
for circRNA clearance. PLoS One. 11:e01484072016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44:D209–D215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He J, Xie Q, Xu H, Li J and Li Y: Circular
RNAs and cancer. Cancer Lett. 396:138–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
69
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Peng Y and Croce CM: The role of MicroRNAs
in human cancer. Signal Transduct Target Ther. 1:150042016.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Scotti MM and Swanson MS: RNA mis-splicing
in disease. Nat Rev Genet. 17:19–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li P, Chen H, Chen S, Mo X, Li T, Xiao B,
Yu R and Guo J: Circular RNA 0000096 affects cell growth and
migration in gastric cancer. Br J Cancer. 116:626–633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yao JT, Zhao SH, Liu QP, Lv MQ, Zhou DX,
Liao ZJ and Nan KJ: Over-expression of CircRNA_100876 in non-small
cell lung cancer and its prognostic value. Pathol Res Pract.
213:453–456. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation - exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ, et al: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
81
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Jiang Y, Yim SH, Xu HD, Jung SH, Yang SY,
Hu HJ, Jung CK and Chung YJ: A potential oncogenic role of the
commonly observed E2F5 overexpression in hepatocellular carcinoma.
World J Gastroenterol. 17:470–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Annunziata CM, Kleinberg L, Davidson B,
Berner A, Gius D, Tchabo N, Steinberg SM and Kohn EC: BAG-4/SODD
and associated antiapoptotic proteins are linked to aggressiveness
of epithelial ovarian cancer. Clin Cancer Res. 13:6585–6592. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ozawa F, Friess H, Zimmermann A, Kleeff J
and Büchler MW: Enhanced expression of Silencer of death domains
(SODD/BAG-4) in pancreatic cancer. Biochem Biophys Res Commun.
271:409–413. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Li Y, Zhu X, Zeng Y, Wang J, Zhang X, Ding
YQ and Liang L: FMNL2 enhances invasion of colorectal carcinoma by
inducing epithelial-mesenchymal transition. Mol Cancer Res.
8:1579–1590. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liang L, Li X, Zhang X, Lv Z, He G, Zhao
W, Ren X, Li Y, Bian X, Liao W, et al: MicroRNA-137, an HMGA1
target, suppresses colorectal cancer cell invasion and metastasis
in mice by directly targeting FMNL2. Gastroenterology.
144:624–635.e4. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Guo JN, Li J, Zhu CL, Feng WT, Shao JX,
Wan L, Huang MD and He JD: Comprehensive profile of differentially
expressed circular RNAs reveals that hsa_circ_0000069 is
upregulated and promotes cell proliferation, migration, and
invasion in colorectal cancer. Onco Targets Ther. 9:7451–7458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
90
|
Cortés-López M and Miura P: Emerging
functions of circular RNAs. Yale J Biol Med. 89:527–537.
2016.PubMed/NCBI
|
|
91
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
93
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pinder SE: Ductal carcinoma in situ
(DCIS): Pathological features, differential diagnosis, prognostic
factors and specimen evaluation. Mod Pathol. 23 Suppl 2:S8–S13.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hernandez L, Wilkerson PM, Lambros MB,
Campion-Flora A, Rodrigues DN, Gauthier A, Cabral C, Pawar V,
Mackay A, A'Hern R, et al: Genomic and mutational profiling of
ductal carcinomas in situ and matched adjacent invasive breast
cancers reveals intra-tumour genetic heterogeneity and clonal
selection. J Pathol. 227:42–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Galasso M, Costantino G, Pasquali L,
Minotti L, Baldassari F, Corrà F, Agnoletto C and Volinia S:
Profiling of the predicted circular RNAs in ductal in situ and
invasive breast cancer: A pilot study. Int J Genomics.
2016:45038402016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing
C, Gao K, Liu ZH and Yu SJ: miR-429 inhibits migration and invasion
of breast cancer cells in vitro. Int J Oncol. 46:531–538. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Yang W, Du WW, Li X, Yee AJ and Yang BB:
Foxo3 activity promoted by non-coding effects of circular RNA and
Foxo3 pseudogene in the inhibition of tumor growth and
angiogenesis. Oncogene. 35:3919–3931. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Du WW, Ling F, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Caiment F, Gaj S, Claessen S and Kleinjans
J: High-throughput data integration of RNA-miRNA-circRNA reveals
novel insights into mechanisms of benzo[a]pyrene-induced
carcinogenicity. Nucleic Acids Res. 43:2525–2534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular rna biomarker, is involved in
hepatocellular crcinoma development. Medicine. 95:e38112016.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dong L, Deng J, Sun ZM, Pan AP, Xiang XJ,
Zhang L, Yu F, Chen J, Sun Z, Feng M and Xiong JP: Interference
with the β-catenin gene in gastric cancer induces changes to the
miRNA expression profile. Tumour Biol. 36:6973–6983. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Warnecke-Eberz U, Chon SH, Hölscher AH,
Drebber U and Bollschweiler E: Exosomal onco-miRs from serum of
patients with adenocarcinoma of the esophagus: Comparison of miRNA
profiles of exosomes and matching tumor. Tumour Biol. 36:4643–4653.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA Cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Xu L, Zhang M, Zheng X, Yi P, Lan C and Xu
M: The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of
hepatic microvascular invasion in hepatocellular carcinoma. J
Cancer Res Clin Oncol. 143:17–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circrnas derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Alhasan AA, Izuogu OG, Al-Balool HH, Steyn
JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L,
Elliott DJ, et al: Circular RNA enrichment in platelets is a
signature of transcriptome degradation. Blood. 127:e1–e11. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Best MG, Sol N, Kooi I, Tannous J,
Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E,
Koster J, et al: RNA-Seq of tumor-educated platelets enables
blood-based pan-cancer, multiclass, and molecular pathway cancer
diagnostics. Cancer Cell. 28:666–676. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu X, Zhong Y, Li J and Shan A: Circular
RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation
and metastasis through targeting miR-448. Oncotarget.
8:114829–114838. 2017.PubMed/NCBI
|
|
115
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genom
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Sand M, Bechara FG, Gambichler T, Sand D,
Bromba M, Hahn SA, Stockfleth E and Hessam S: Circular RNA
expression in cutaneous squamous cell carcinoma. J Dermatol Sci.
83:210–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Peng N, Shi L, Zhang Q, Hu Y, Wang N and
Ye H: Microarray profiling of circular RNAs in human papillary
thyroid carcinoma. PLoS One. 12:e01702872017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T,
Wang X, Li Q, Tian L, Liu M and Sun Y: Circular RNA: A novel
biomarker for progressive laryngeal cancer. Am J Transl Res.
8:932–939. 2016.PubMed/NCBI
|
|
120
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Melo SA, Luecke LB, Kahlert C, Fernandez
AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari
N, et al: Glypican-1 identifies cancer exosomes and detects early
pancreatic cancer. Nature. 523:177–182. 2015. View Article : Google Scholar : PubMed/NCBI
|