Introduction
Bladder cancer (BC) is one of the most important
urinary cancers and it remains a difficult cancer to treat due to
metastasis and rapid recurrence (1). The characteristics of BC include rapid
development and high invasion and metastasis potential. Once
distant metastasis is present, ~80% of BC patients will have
succumbed to this disease in the first 5 years (2). Research has focused on studying the
mechanisms of BC, but little progress has been achieved. Therefore,
the definitive mechanisms, as well as novel prognostic markers for
BC need to be discovered.
Studies of RNA networks, especially new patterns of
molecular interactions, are beneficial to the prognosis of patients
with BC (3). Previous studies have
revealed that more than 90% of human genome sequences do not encode
protein. The majority of these sequences are called non-coding RNAs
(ncRNAs). microRNAs (miRNAs) are one type of small nc-RNAs
(4) and play a role in gene
regulation and biological function in numerous cancers. The levels
of miRNAs always change along with cancer progression. By binding
to the target 3′-untranslated region (3′-UTR), a miRNA can induce
mRNA degradation or prevent protein translation (5). miRNAs aberrantly expressed in BC may
play a significant role in cancer metastasis and oncogenesis. Ma
et al (6) revealed that
miR-148a was an independent prognostic marker of BC, and patients
with low miR-148a expression were more likely to have high-grade
tumors. Liu et al (7)
revealed that miR-101-3p inhibited proliferation and metastasis of
BC cells by downregulating EZH2. miR-26a-5p and miR-26b-5p are
deemed cancer-suppressive miRNAs, and both have been revealed to be
significantly downregulated in multiple cancers (6,8).
Additionally, they have been demonstrated to inhibit the
proliferation of BC cells (9).
However, the underlying inhibitory mechanisms still remain largely
unclear. The objective of our study was to investigate the function
of miR-26-5p as a tumor suppressor and identify its target genes in
BC. Thus, discovering the key miRNAs or elucidating miRNA-regulated
mechanisms can provide novel insight and further BC research.
Programmed Cell Death 10 (PDCD10), which has also
been called cerebral cavernous malformation 3 (CCM3), was initially
referred to as TFAR15 (TF-1 cell apoptosis-related gene). Earlier
studies of PDCD10 focused on neural areas. A number of studies have
demonstrated that PDCD10 acts as a critical regulator of neuronal
survival and integrity (10,11).
Subsequently, increasing evidence has indicated a critical role of
PDCD10 in regulating cell death and survival. Zhang et al
(12) observed that PDCD10 was
upregulated in prostate cancer and may play a role in prostate
tumorigenesis. Lauenborg et al demonstrated that PDCD10
enhanced the proliferation of malignant T cells (13). The mechanism by which PDCD10
regulated T-cell proliferation was largely unclear. Therefore, we
hypothesized that miR-26a-5p and miR-26b-5p inhibited BC cell
proliferation by binding to PDCD10 mRNA. In the present study, we
aimed to elucidate the regulatory relationship between
miR-26a-5p/miR-26b-5p and PDCD10 in BC cell proliferation.
Materials and methods
Clinical BC specimens
Fresh BC tissues and corresponding adjacent bladder
tissues were collected from the Urology Department of Shanghai
General Hospital for 8 months from February to October of 2016.
None of the patients (Table I) had
undergone radio- or chemotherapeutic treatment before radical
cystectomy surgery, and all the clinical information of patients is
mentioned in Table I. The present
study was approved by the Medical Ethics Committee of Shanghai
General Hospital, Shanghai Jiao Tong University and informed
consent was obtained before surgery from each patient. All
specimens were staged in accordance with the tumor-node-metastasis
(TNM) classification enacted by the American Joint Committee on
Cancer Union for International Cancer Control enacted by the
American Joint Committee on Cancer (AJCC) (14).
 | Table I.Relationship between miR-26a-5p and
miR-26b-5p expression levels and clinicopathological significance
in BC tissues. |
Table I.
Relationship between miR-26a-5p and
miR-26b-5p expression levels and clinicopathological significance
in BC tissues.
|
Characteristics | Groups | No. | Expression of
miR-26a-5p/U6 | P-value | Expression of
miR-26b-5p/U6 | P-value |
|---|
| Tissue | Cancer tissue |
| 0.076±0.002 (20
pairs) | <0.001 | 0.104±0.002 (21
pairs) |
<0.05a |
|
| Adjacent
tissue |
| 0.152±0.006 (20
pairs) |
| 0.156±0.005 (21
pairs) |
|
| Age, years | ≤60 | 8 | 0.780±0.004 | 0.851 | 0.790±0.041 | 0.172b |
|
| >60 | 12 | 0.393±0.006 |
| 0.619±0.088 |
|
| Sex | Male | 14 | 0.645±0.151 | 0.064 | 0.730±0.078 | 0.342a |
|
| Female | 6 | 0.316±0.031 |
| 0.597±0.063 |
|
| TNM stage | I/II | 8 | 0.823±0.138 | 0.003 | 0.863±0.036 | 0.013b |
|
| III/IV | 12 | 0.362±0.053 |
| 0.570±0.066 |
|
Animals and cell lines
The experiments on animals were approved by the
Ethics Committee for Animal Experimentation of the Shanghai General
Hospital of Shanghai Jiao Tong University and strictly abided by
the Institutional Guidelines for Use and Care of Laboratory
Animals. Forty male pathogen-free 4-week-old nude mice (weight, 15
g) were obtained from Slaccas Animal Laboratory (Shanghai, China).
All experimental animals were maintained at a temperature of 27±1°C
and a humidity of 50±5% under diurnal lighting conditions (12-h
light/dark cycle) with access to food and water ad libitum.
Cell lines of human BC, T24 and 5637, were obtained from the
Typical Culture Preservation Commission Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). Cell lines were cultured in
RPMI-1640 medium supplemented with 1% penicillin/streptomycin and
10% FBS (all from HyClone; GE Healthcare Life Sciences, Logan, UT,
USA) and under a humidified atmosphere of 5% CO2 at 37°C
(15).
In silico analysis of miR-26a-5p and
miR-26b-5p target gene
Candidate target genes of miR-26a-5p and miR-26b-5p
were predicted using the TargetScan database release 7.1
(http://www.targetscan.org/vert_71/).
Genes with cumulative weighed context++ score ≤-0.20 were
included.
Transduction of BC cell lines and RNA
extraction
The short hairpin RNA (shRNA) and GFP-labeled
lentivirus vectors containing the miR-26a-5p/miR-26b-5p mimic
lentivirus (LV-miR-26a-5p) and the corresponding control miRNA
lentivirus (miR-26a-5p/miR-26b-5p-NC), as well as the PDCD10
overexpression lentivirus, the PDCD10 silencing lentivirus, and the
corresponding control lentivirus were obtained from GeneChem
(Shanghai, China). Cells were seeded in 6-well plates
(5×105 cells/well) before transduction. The shRNA
transduction was conducted with HiPerFect Transfection Reagent
according to the manufacturer's instructions (Qiagen, Manchester,
UK). Transduction with the lentiviral vectors was conducted using
transduction reagents and 8 mg/ml Polybrene (Genechem) for 12 h.
For viral transduction, cells were transduced with a multiplicity
of infection (MOI) of 10,100 or 1,000 (16). PDCD10 overexpression, silencing and
the corresponding control stable cell lines were then established,
and the efficiency of transduction was confirmed by quantitative
real-time reverse transcription PCR (RT-qPCR).
RNA extraction and RT-qPCR
Total RNA from the treated cells, including miRNA,
was extracted using RNAsimple Total RNA kit (Tiangen Biotech Co.,
Ltd., Beijing, China) and reverse transcription (RT) was performed
using FastKing RT kit (Tiangen Biotech), following the
manufacturer's protocol. miRNA sample RT was performed using a
miRcute Plus miRNA First-Strand cDNA Synthesis kit (Tiangen
Biotech). All samples were assessed using a Quant One Step RT-qPCR
kit (SYBR-Green) (Tiangen Biotech). The thermocycling conditions
were: 50°C for 30 min for 1 cycle; 95°C for 2 min for 1 cycle;
94°C, 55°C and 68°C for 20 sec for 40 cycles. U6 and GAPDH were
used for normalization, and the relative expression levels of miRNA
and PDCD10 were calculated using the 2−ΔΔCq method
(17). Both of the miRNA primers
used were designed and synthesized by Tiangen Biotech (cat. nos.
miR-26a-5p:CD201-0319 201221000000002 and miR-26b-5p:CD201-0321).
Primers for GAPDH and PDCD10 were designed and synthesized by
Shangon Company (Shanghai, China). The following primers were used:
5′-GGTGAAGGTCGGTGTGAACG-3′ (sense) and 5′-CTCGCTCCTGGAAGATGGTG-3′
(antisense) for GAPDH; and 5′-GACCCAAGCTCCGCCTCTCCC-3′ (sense) and
5′-GGGAGAGGCGGAGCTTGGGTC-3′ (antisense) for PDCD10.
Cell proliferation assays in
vitro
Cells were seeded in 96-well plates at
3×103 cells/well for proliferation assays after
transduction for 48 h. Cell proliferation was determined by Cell
Counting Kit-8 (CCK-8) reagent (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) as previously described (18). CCK-8 is a redox indicator that uses
the highly water-soluble WST-8 to produce a water-soluble formazan
dye, which generated by the activity of dehydrogenases in cells is
directly proportional to the number of living cells. BC cell lines
were collected after 72 h of treatment, and 1 ml of 1X FOXP3
Fix/Perm solution (BioLegend, Inc., San Diego, CA, USA) was added
to each tube for 20 min. According to the manufacturer's
instructions, cells were stained with anti-Ki67 antibodies (1:20;
cat. no. 350514; BioLegend, Inc.) for 30 min, and isotype IgG
antibody (1:20; cat. no. 400141; BioLegend, Inc.) was used as a
control. Ki67 is a nuclear protein which is necessary for cellular
proliferation. The treated samples were analyzed with a FACSCalibur
flow cytometer and CellQuest software (version 5.2.1) (both from BD
Biosciences, Franklin Lakes, NJ, USA). The mean fluorescence
intensity (MFI) of Ki67 was detected with a CBA assay (BD
Biosciences). MFI is the mean of the fluorescence intensity in the
fluorescence channel. The experiments were performed in
triplicate.
Western blot analysis
Treated cells were collected, and lysates were
prepared in lysis buffer (50 mM EDTA, 50 mM NaCl, 1% Triton X-100)
containing a protease inhibitor cocktail (Beyotime Institute of
Biotechnology, Haimen, China). The protein determination method is
BCA. An equal amount of total cell lysate (30 µg) was separated on
12% SDS-PAGE gels and transferred to PVDF membranes. Following
antigen blocking with QuickBlock™ Blocking Buffer for western blot
analysis (P0252; Beyotime Institute of Biotechnology), the
membranes were incubated with primary antibodies against PDCD10 (1
µg/ml; 1:1,000; cat. no. ab110531; Abcam, Cambridge, UK) and GAPDH
(1 µg/ml; 1:1,000; cat. no. ab8245; Abcam)/Tubulin (1 µg/ml;
1:5,000; cat. no. ab7291; Abcam) overnight. Then, the membranes,
which were rinsed with TBST, were incubated with goat anti-rabbit
secondary antibody (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology) and visualized with chemiluminescence (New England
Nuclear, Boston, MA, USA) by Image Lab software (Bio-Rad
Laboratories, Hercules, CA, USA). The experiments were performed in
triplicate.
Immunohistochemistry (IHC)
Tumor specimens and paired adjacent tissues from
bladder patients (n=10) were steeped in 4% paraformaldehyde and
embedded in paraffin (5 µm). After heat-induced epitope retrieval,
immunohistochemical staining was performed using an avidin-biotin
complex method. Subsequently, the endogenous peroxidase activity
was blocked by incubation in 3% H2O2 for 15
min and the samples were incubated at 4°C overnight with primary
anti-PDCD10 antibodies (10 µg/ml; 1:100; cat. no. ab110531; Abcam)
or a goat IgG isotype in a humid chamber. Subsequently, on the
following day, the samples were treated with peroxidase-conjugated
anti-rabbit IgG antibody (1:1,000; cat. no. 101ES60; Golden Bridge
International, Shanghai, China) for 30 min. Until brown granules
appeared, samples were incubated with diaminobenzidine (DAB).
Hematoxylin was used for nuclear staining. Sections were evaluated
blindly by two researchers with an Olympus BX51+DP70 microscope
(Olympus Corp., Tokyo, Japan). The experiments were performed in
triplicate and repeated three times.
Luciferase reporter assay
T24 and 5637 cells were seeded in a 24-well plate,
and then co-transfected with LV-miR-26a-5p or LV-miR-26b-5p,
pMiRTarget-PDCD10-3′-UTR or the mutated 3′-UTR of PDCD10 or an
empty vector as a control. The relative dual-luciferase activity of
cell lysates was normalized to Renilla luciferase activity
and examined with a Dual-Luciferase Reporter Assay system (Promega
Corp., Madison, WI, USA). The experiments were performed in
triplicate.
Xenograft mouse model
The 5637 cells (1×107) stably expressing
LV-miR-26a-5p or LV-miR-26b-5p, LV-PDCD10, LV-PDCD10+miR-26a-5p or
LV-PDCD10+miR-26b-5p were subcutaneously injected into the left
flank area of 4-week-old nude mice (n=6 mice/group). Tumor volumes
were determined every 3 days (0.5 × length × width2).
Three weeks later, the mice were sacrificed by CO2
euthanasia and xenografts were assessed and weighed.
Survival analysis
Clinical data were downloaded by R/Bioconductor
package TCGABiolinks from the TCGA database (https://cancergenome.nih.gov/). Genes or miRNA were
excluded if the expression quantity level in >50% of the samples
was 0. From the entire dataset, 29,627 genes and 627 miRNAs were
brought into the study. The miRNA survival analysis brought in 408
samples and the gene survival analysis brought in 409 samples
because both sets contained complete clinical data.
Statistical analysis
All data are presented as the mean ± SEM and were
analyzed using ANOVA, chi-square test and Student's t-test with
SPSS 21.0 software (IBM Corp., Armonk, NY, USA). Tukey's test was
used to test all pairs of columns. A log-rank test was performed
and Kaplan-Meier survival curves were plotted. The P-values were
two-sided and a value of <0.05 was considered to indicate a
statistically significant difference.
Results
miR-26a-5p and miR-26b-5p inhibit the
proliferation of T24 and 5637 cells in vitro
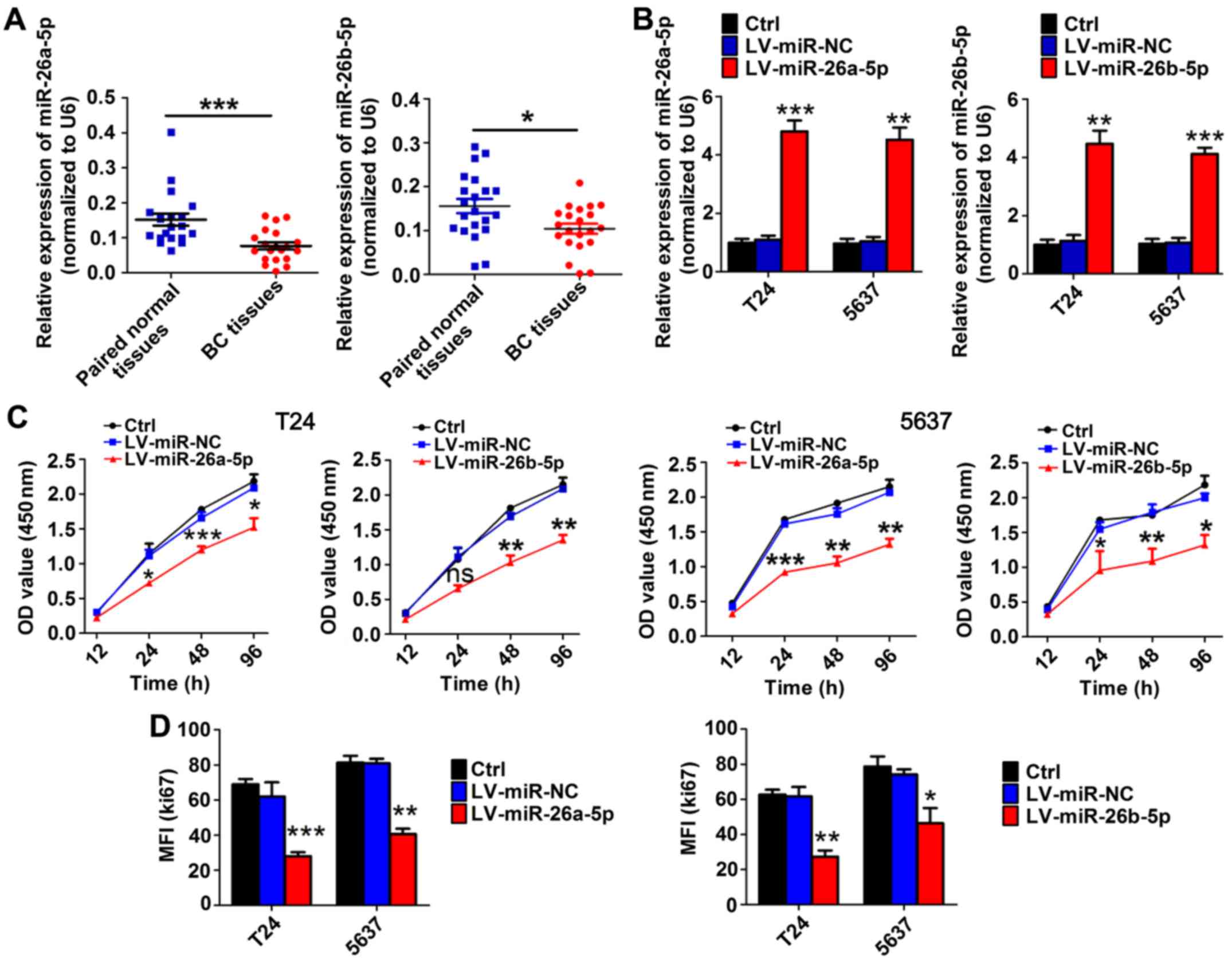
In BC tissues, the expression levels of miR-26a-5p
(n=20) and miR-26b-5p (n=21) were significantly reduced compared
with those in adjacent bladder tissues based on RT-qPCR results
(Fig. 1A). Instead of age and sex,
TNM stage was significantly and negatively associated with the
levels of miR-26-5p (Table I).
To evaluate the role of miR-26a-5p and miR-26b-5p in
BC cell proliferation, we transfected T24 and 5637 cells with
miR-26a-5p or miR-26b-5p overexpression lentivirus and a
high-efficiency of interference was verified by RT-qPCR (Fig. 1B). CCK-8 assays revealed that,
compared with the paired mock or miR-control groups, transduction
with miR-26a-5p and miR-26b-5p inhibited proliferation in T24 and
5637 cell lines (Fig. 1C). In
addition, flow cytometric assays demonstrated that the MFI of Ki67
was significantly reduced in miR-26a-5p and miR-26b-5p
lentivirus-transfected cells compared to mock or miR-control groups
(Fig. 1D). These results indicated
that miR-26a-5p and miR-26b-5p inhibited cell proliferation in BC
cell lines.
PDCD10 is directly modulated by
miR-26a-5p/miR-26b-5p as a target in BC cells
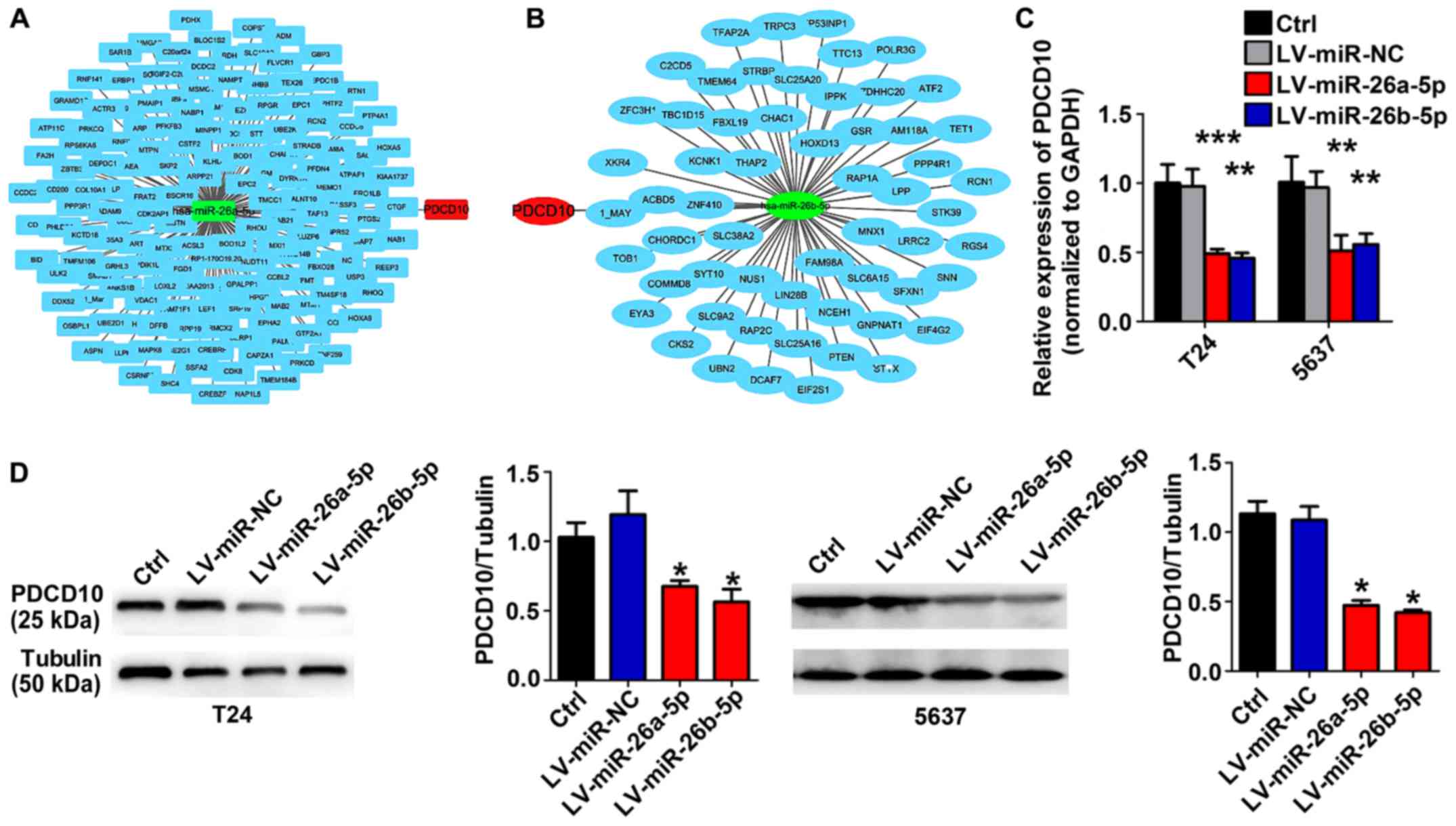
To identify the possible target gene of miR-26-5p in
BC, bioinformatics prediction software was used to analyze
additional information regarding the proliferation-related
molecular mechanisms that are simultaneously regulated by
miR-26a-5p and miR-26b-5p in BC cells. miR-26-5p-regulated
candidate genes were identified using TargetScan database release
7.1 (http://www.targetscan.org/vert_71/). As displayed in
Fig. 2, PDCD10 is the conjunct gene
in the intersection of miR-26a-5p-regulated candidate genes and
miR-26b-5p-regulated candidate genes (Fig. 2A and B). RT-qPCR and western blot
assays revealed that lentivirus-miR-26a-5p or lentivirus-miR-26b-5p
stable BC cell lines expressed a lower level of PDCD10 (Fig. 2C and D).
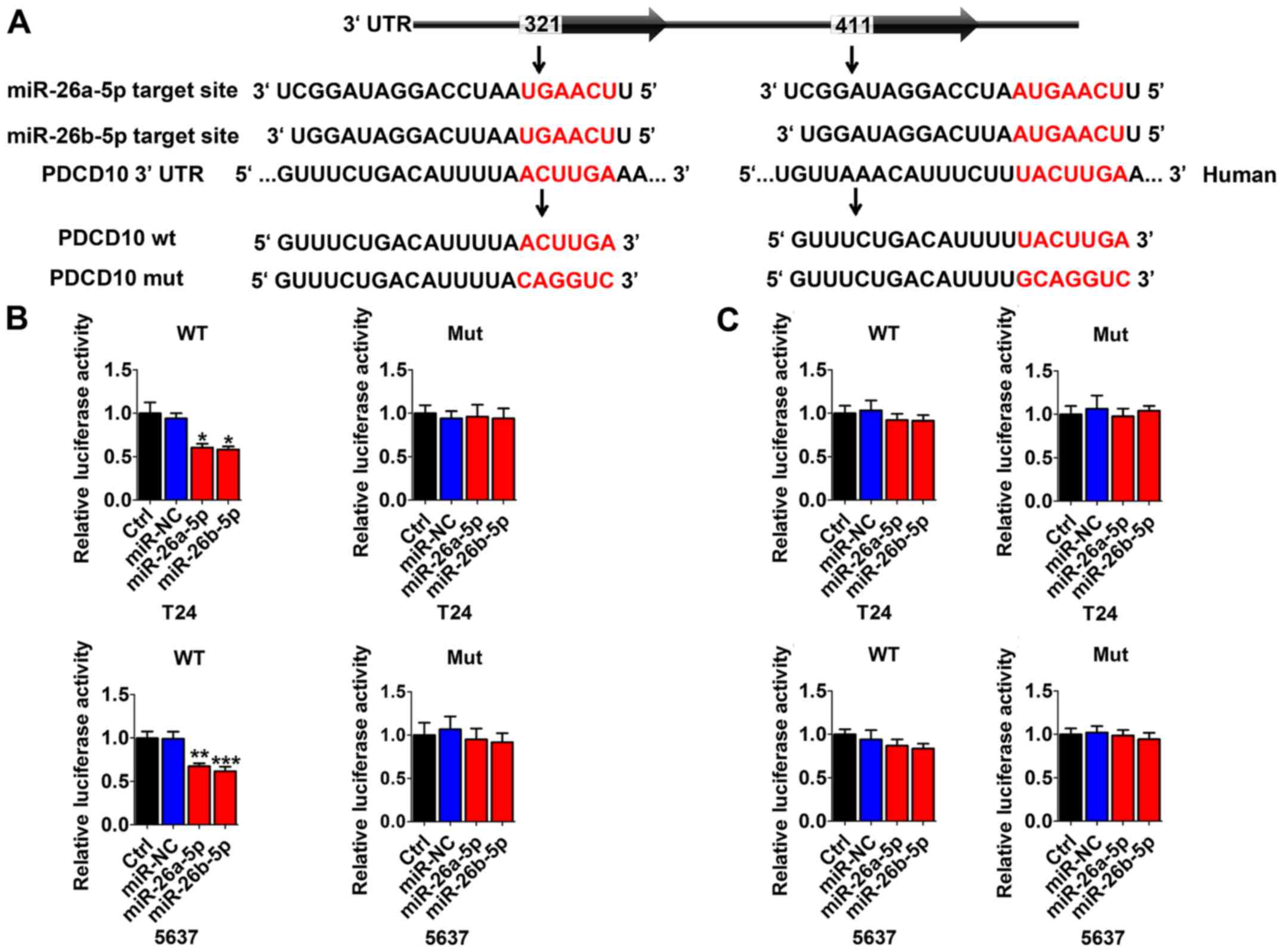
Multiple sequence comparison analysis indicated that
there were two possible miR-26a/b-5p binding sites at positions
321–327 and 411–418 in the PDCD10 3′-UTR sequence (Fig. 3A). Thus, in T24 and 5637 cells,
dual-luciferase reporter assays were performed to identify whether
PDCD10 was directly regulated by miR-26a-5p or miR-26b-5p. We used
vectors encoding the partial 3′-UTR sequence of PDCD10, including
the potential binding miR-26a-5p/miR-26b-5p site. Following
co-transduction with miR-26a-5p/miR-26b-5p and the vector carrying
the wild-type sequence containing the PDCD10 3′-UTR with residues
321–327, the luminescence intensity of PDCD10 was significantly
reduced (Fig. 3B). However,
transduction with the wild-type PDCD10 3′-UTR sequence at positions
411–418 and mutation vector obstructed the decrease in luminescence
(Fig. 3C). These data indicated
that miR-26a-5p/miR-26b-5p directly bind to specific sites at
positions 321–327 of the PDCD10 3′-UTR.
PDCD10 promotes BC cell proliferation
in vitro
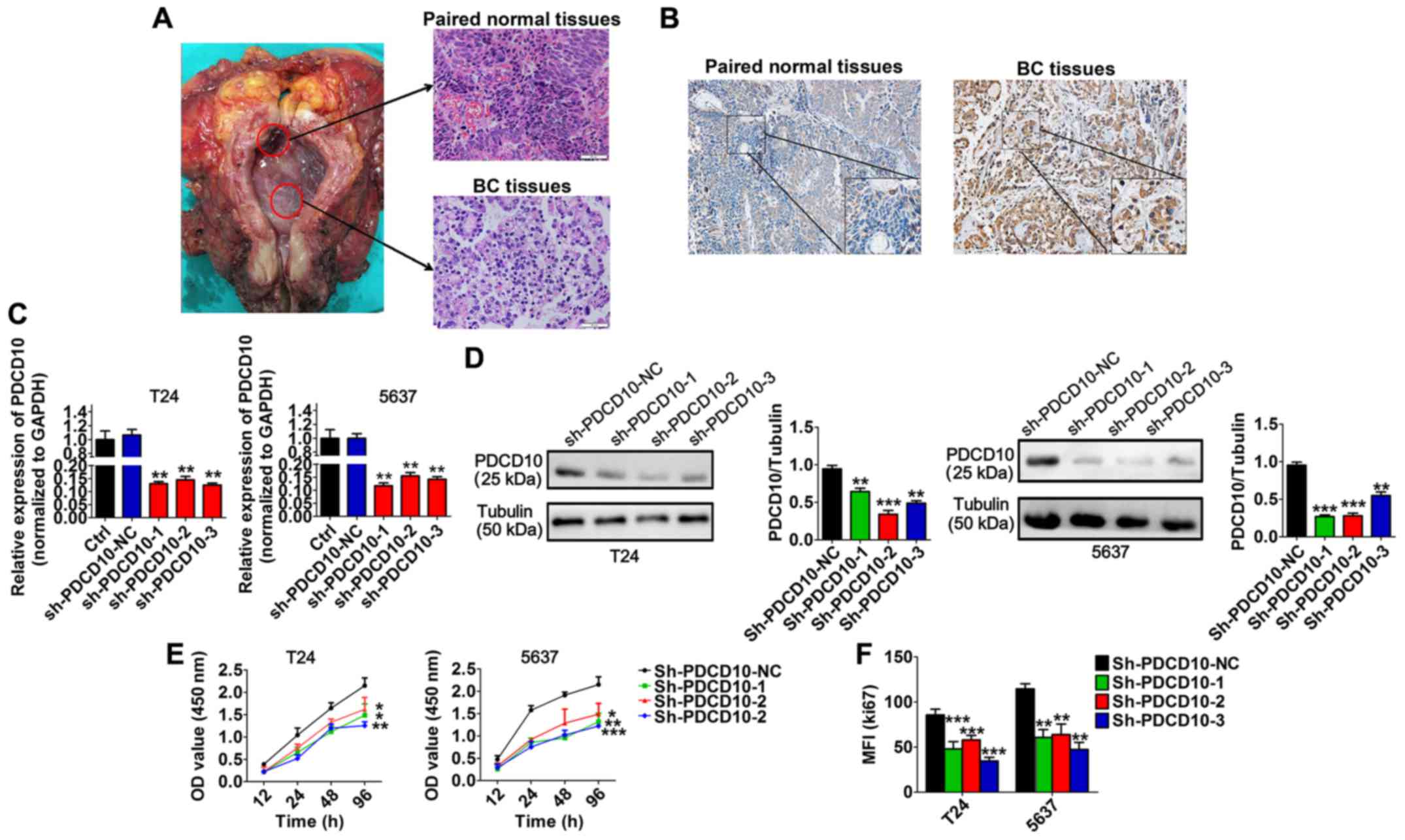
Immunohistochemical results revealed that BC tissues
expressed a higher level of PDCD10 than paired normal tissues
(Fig. 4A and B). We performed
loss-of-function studies by transfecting BC cell lines with three
sh-PDCD10 plasmids (sh-PDCD10-1, sh-PDCD10-2 and sh-PDCD10-3) in
order to explore the function of PDCD10 in BC cell lines and
determined that the mRNA and protein levels of PDCD10 in these cell
lines were effectively downregulated following transfection
(Fig. 4C and D). In addition,
transfection with sh-PDCD10 plasmids significantly inhibited
proliferation of T24 and 5637 cells (Fig. 4E). In a similar situation, the MFI
of Ki67 was significantly lower in T24 and 5637 cells transfected
with sh-PDCD10 plasmids than in cells transfected with sh-PDCD10-NC
(Fig. 4F). These results
ascertained that PDCD10 inhibited the proliferation of T24 and 5637
cell lines in vitro.
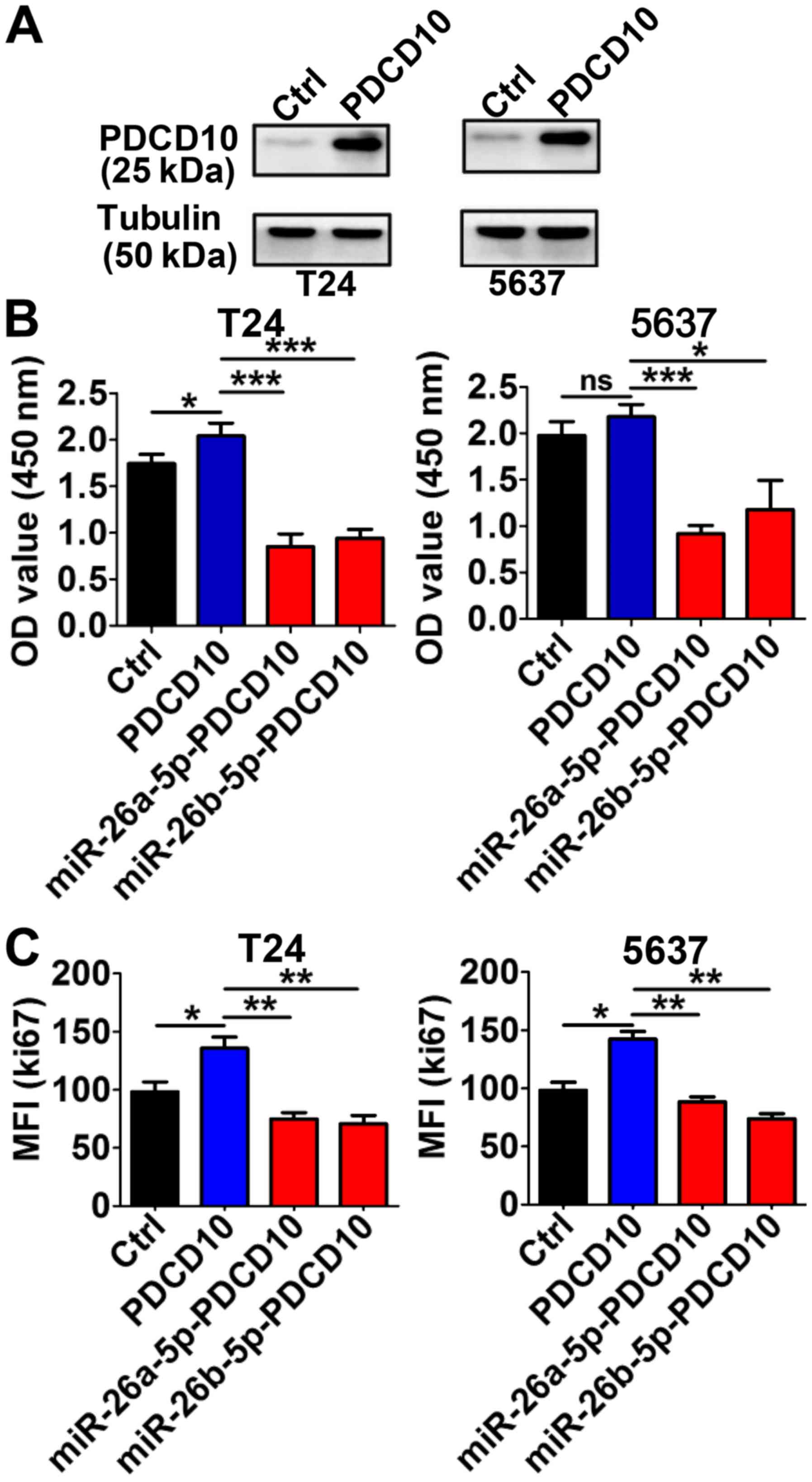
miR-26a-5p and miR-26b-5p inhibit the
proliferation-promoting function of PDCD10
In T24 and 5637 cells, the PDCD10 protein levels
were evidently higher than in the control group cells following
transfection with PDCD10 overexpressing plasmid for 48 h (Fig. 5A). The high level of PDCD10 promoted
proliferation in both T24 and 5637 cells. This effect was partially
blocked by the upregulation of the miRNA level by transduction with
the corresponding miRNA lentivirus (Fig. 5B). Consistent with these results,
the MFI of Ki67 was markedly higher following the upregulation of
PDCD10 via transfection with the plasmid, but when the cells were
simultaneously transfected with miR-26a-5p or miR-26b-5p
lentivirus, the MFI significantly decreased (Fig. 5C). These data indicated that
miR-26-5p inhibited the proliferation-stimulating effect in BC
cells induced by PDCD10.
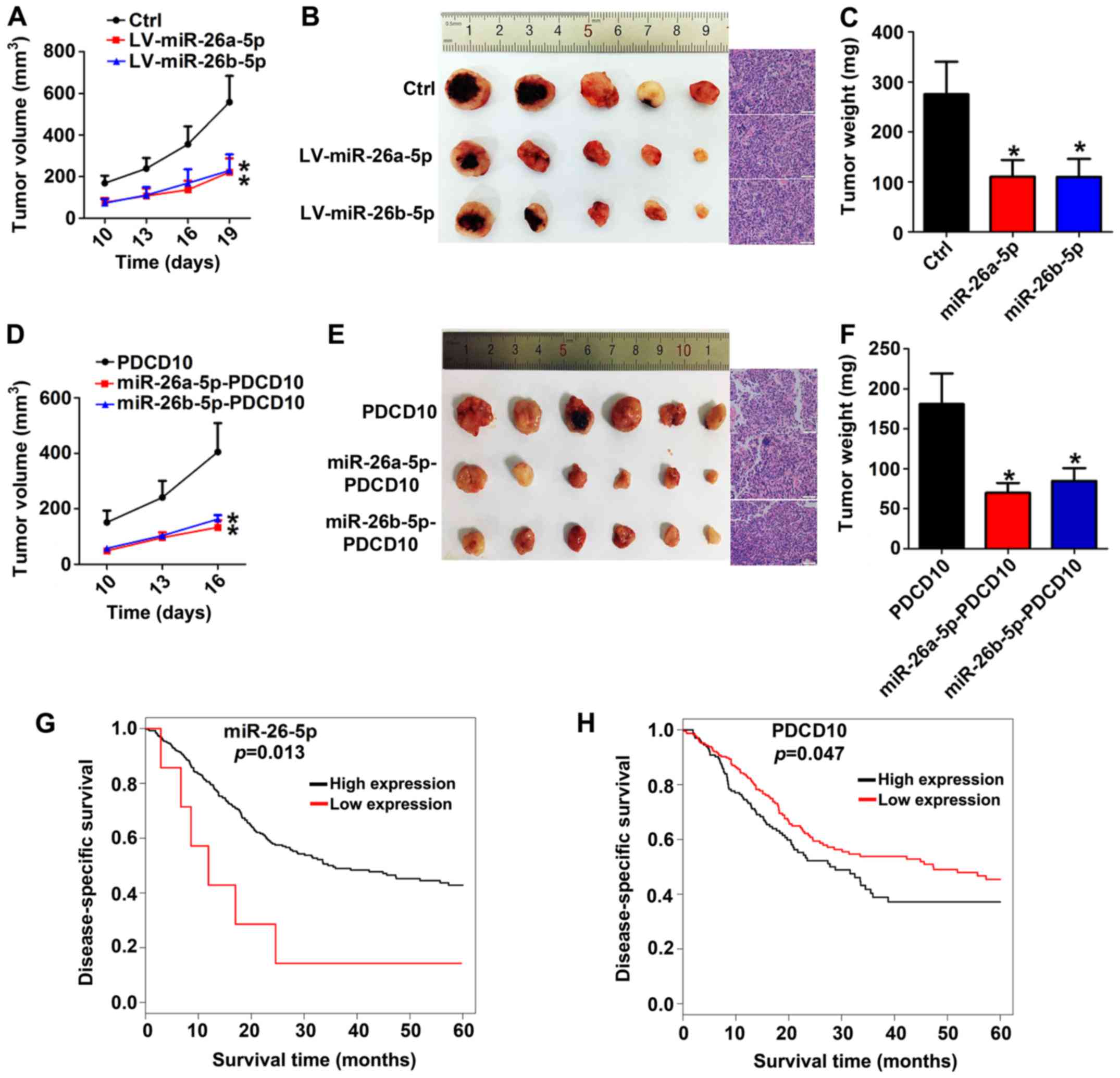
PDCD10 promotes growth and progression
of BC in vivo and is negatively regulated by miR-26a-5p and
miR-26b-5p
Having found that miR-26a-5p/miR-26b-5p inhibited
the ability of PDCD10 to promote proliferation, we then sought to
examine the biological function in vivo. A xenograft mouse
model was constructed after LV-miR-26a-5p or LV-miR-26b-5p or mock
transfected 5637 cells were subcutaneously injected in nude mice
for 7 days. Subsequently, we closely observed the development of
tumors and found that miR-26a-5p and miR-26b-5p overexpression
xenografts developed much slower than those of the mock group
(Fig. 6A-C). To further elucidate
the negative regulation of PDCD10 by miR-26a-5p/miR-26b-5p,
miRNA-PDCD10 co-overexpression 5637 subcutaneous xenografts were
constructed, and they developed more slowly. In addition, growth
tendency, anatomic form and weight assays further indicated that
both miR-26a-5p and miR-26b-5p inhibited the promoting function of
PDCD10 (Fig. 6D-F). As for low and
high miR-26-5p and PDCD10 expression groups, Kaplan-Meier survival
plots revealed that in patients with high miR-26-5p expression or
low PDCD10 expression, overall survival was obviously prolonged
compared with patients with low miR-26-5p expression or high PDCD10
expression (Fig. 6G-H). Animal
experiments and clinical data analysis revealed that regulation
between miR-26a-5p/miR-26b-5p and PDCD10 had clinical value for
evaluating the progression and prognosis of BC patients.
Discussion
miRNAs are transcribed to generate long primary
transcripts (pri-miRNAs) which are then processed into structures
of 60 to 110 nt hairpin precursor miRNAs (pre-miRNAs). These
pre-miRNAs are then cleaved by another RNase III enzyme, Dicer, to
generate ~22 nt miRNAs. Several researchers have reported that
there are many miRNAs that play a part in regulating the
progression of cancer. Yang et al (19) reported that miR-138-5p is a tumor
suppressor that inhibits cell proliferation and invasion by
targeting Survivin. In prostate cancer, papillary thyroid
carcinoma, and head and neck carcinoma, both miR-26a-5p and
miR-26b-5p could inhibit tumor progression (20–22).
miR-26a-5p and miR-26b-5p belong to the miR-26 family, and in
cancer cell lines, these miRNAs can synergistically block G1/S
phase transition. Tian et al (23) reported that both miR-26a-5p and
miR-26b-5p play an important role in invasion, proliferation and
migration abilities of BC cells. In addition, miR-26-5p plays a key
role in cellular growth, development and activation (24). The difference between miR-26a and
miR-26b is just several bases. Actually, both of them belong to the
miR-26 family and have a similar function by targeting the same
mRNAs. The miR-26 family also includes miR-26a-3p and miR-26b-3p.
Wang et al (8) reported that
miR-26b-3p regulated stem cell proliferation by targeting estrogen
receptors. However, the function of miR-26a-3p and miR-26b-3p
warrants further exploration. In the present study, we investigated
whether miR-26-5p is aberrantly expressed in BC by collecting
paired clinical bladder samples, which constitute a set of paired
samples that were perfect for cross-comparison, from 20 patients
who underwent radical cystectomy. Similarly, in BC tissues, we
observed that the expression level of both miR-26a-5p and
miR-26b-5p was significantly lower than in paired normal tissues.
Notably, the trend of the majority of our results was similar to
Miyamoto et al (9), but they
found that aberrant expression of miR-26a-5p did not obviously
impact T24 proliferation. However, unlike the previous study, this
study included transduction with a high-quality lentivirus to
disturb the expression levels of miR-26-5p, which could guarantee a
lasting effect. In addition, CCK-8 assays combined with detection
of Ki67 using flow cytometry provided stronger and more reasonable
results. By analyzing clinical data from TargetScan 7.1 database,
we found that patients with high expression of miR-26-5p tend to
have a better prognosis.
Upregulation of miR-26-5p appears to be an efficient
way to treat BC but the regulatory network of miRNAs is
complicated. An active RNA-induced silencing complex (RISC)
includes mature miRNA, containing Dicer and many associated
proteins. Certainly, coordinated action between Dicer and other
proteins is an efficient method of discarding pre-miRNA (25). Mounting evidence suggests that
lncRNAs can function as miRNA sponges and then compete with
microRNA for a binding site on protein-coding transcripts. Liu
et al revealed that lncRNA SPRY4-IT1 could efficiently
reverse the suppression of Enhancer of zeste homolog 2 (EZH2) by
directly interacting with miR-101-3p, and this ‘sponge’ function
could promote proliferation and metastasis of BC cells (7). Circular RNA (circRNA) is a new type of
ncRNA. Although the roles of circRNAs remain a mystery, insights
into circRNAs have revealed that several circRNAs can function as
microRNA sponges (26). In
addition, Cui et al (27)
revealed that the nuclear factor-κ-B (NF-κB) signaling pathway
could directly regulate transcription of miR-130b. Thus, the
upstream regulation mechanism of miR-26-5p warrants further
study.
Gene silencing may occur either by inhibiting mRNA
translation or mRNA degradation. RISC mediates the translation
inhibition and degradation of the target mRNA by binding to the
3′-UTR in the target mRNA (28). We
used software to assess the intersection of miR-26a-5p and
miR-26a-5p candidate target genes, which includes PDCD10. The
upstream regulatory mechanism of PDCD10 remains unclear at present.
Xu et al (29) found that
miR-181b enhanced angiogenesis of retinoblastoma cells by targeting
PDCD10. Wu et al (30)
demonstrated that miR-613 could regulate cardiomyocyte apoptosis by
targeting the PDCD10 gene. These results indicated that PDCD10 can
be regulated by miRNA. In addition, there are two potential binding
sites in the 3′-UTR of PDCD10 that miR-26a-5p and miR-26a-5p can
simultaneously bind. Our results indicated that
miR-26a-5p/miR-26b-5p directly bound to specific sites at positions
321–327 of the PDCD10 3′-UTR. To gain insight into the regulatory
relationship between miR-26a/b-5p and PDCD10, we used RT-qPCR and
western blotting to demonstrate that miR-26a/b-5p downregulated
PDCD10. We also identified the binding site of miRNA using
luciferase reporter assays. Furthermore, we demonstrated that
upregulation of miR-26a/b-5p weakened the cancer proliferation
function of PDCD10 both in vitro and in vivo.
Several studies have reported that PDCD10 is
aberrantly expressed in some types of human cancers (31). In this study, BC tissues expressed a
higher level of PDCD10 than paired para-carcinoma tissues. PDCD10
is an evolutionarily highly conserved protein that is associated
with cell proliferation and apoptosis. Knockdown of PDCD10 by
sh-RNA inhibited the proliferation of T24 and 5637 cell lines. This
observation was in accordance with previous findings that revealed
that silencing PDCD10 could inhibit the proliferation of
endothelial cells (32). Lambertz
et al (33) proposed that
PDCD10 potentially participated in tumor proliferation, apoptosis,
hyper-angiogenesis and peritumoral edema in human glioblastoma. In
addition, downregulation of PDCD10 inhibited tumor cell
proliferation (33). In addition to
the effect on proliferation, previous studies indicated a positive
correlation between PDCD10 and tumor cell apoptosis (33), and RNA interference indicated that
PDCD10 regulated cell mitogenesis (34). Synthetic biological functions of
PDCD10 warrant more investigation. In contrast to the
well-established mechanism of PDCD10 in vessel permeability and
neo-angiogenesis, limited information about PDCD10 in cancer
research has been reported (35,36).
He et al (37) forecasted
that PDCD10 may play an important role in cancer since it can
influence angiogenesis by targeting vascular endothelial growth
factor receptor 2 (VEGFR2) (37).
He et al revealed that PDCD10 deletion reduced VEGFR2
signaling in embryonic and endothelial cells (37). Notably, VEGF/VEGFR2 signaling could
regulate germ cell proliferation in vitro and promote mouse
testicular regeneration in vivo (38). This indicated that PDCD10 may
regulate proliferation of BC possibly through VEGFR2 signaling. Ma
et al (39) revealed that
PDCD10 interacted with the kinase MST4 and promoted cell growth and
transformation through the extracellular signal-regulated kinase
(ERK) pathway. Riverso et al (40) reported that Krüppel-like factor 4
(KLF4) is regulated by ERK signaling and promoted melanoma cell
growth. Through bioinformatics analysis, clinical data also
indicated that prognosis of patients with low expression of PDCD10
was better than that of patients with high PDCD10 expression.
Further studies on PDCD10 downstream signaling are
necessary. Lauenborg et al (13) reported that PDCD10 regulated
proliferation and survival of malignant T cells by activating Jak3
signaling. In addition, PDCD10 could directly bind to the
phosphatase domain of Fas-associated phosphatase-1 (FAP-1) and
serine/threonine kinase 25 (STK25) in cerebral cavernous
malformations (41). Furthermore,
PDCD10 could form a complex with GM130, a Golgi-resident protein,
and the germinal center kinase III (GCKIII) family to regulate cell
migration (42). The effect of
PDCD10 may be the result of a modulation of one or more of these
processes in our experiments, which warrants further study.
In conclusion, miR-26a-5p and miR-26b-5p play an
antitumor role in BC, and PDCD10 is an important downstream target
gene of miR-26a-5p/miR-26b-5p. Considering that patients with a low
PDCD10 expression level in BC tissues appear to have a better
prognosis than patients with high PDCD10 expression, PDCD10 may be
a new diagnostic biomarker in BC, and the
miR-26a-5p/miR-26b-5p/PDCD10 axis may be a new therapeutic
target.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of China (no. 81672515), the Shanghai Pujiang
Program (no. 16PJD039) and the Commission of Gaofeng Clinical
Medicine Grant (no. 20172019).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZHL and MQL designed this research and provided
research direction. KW and XYM performed the research and collected
the data. JTJ, MYT, RJW and WJZ analyzed the data and finished the
animal experiments. XW and YYH drafted the manuscript and revised
it critically for important content. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Shanghai General Hospital, Shanghai Jiao Tong
University and informed consent was obtained before surgery from
each patient. The animal experiments were approved by the Ethics
Committee for Animal Experimentation of the Shanghai General
Hospital of Shanghai Jiao Tong University and strictly abided by
the Institutional Guidelines for Use and Care of Laboratory
Animals.
Patient consent for publication
Not applicable.
Competing interests
We declare that none of the authors have any
financial and personal relationships with other people or
organizations that can inappropriately influence the quality of the
present study. The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barton MK: High morbidity and mortality
found for high-risk, non-muscle-invasive bladder cancer. CA Cancer
J Clin. 63:371–372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Comprehensive molecular characterization
of urothelial bladder carcinoma. Nature. 507:315–322. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Xu Z, Xu C and Jiang X:
MicroRNA-148a represents an independent prognostic marker in
bladder cancer. Tumour Biol. 37:7915–7920. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X,
Wang M, Huang C, Wang L, Zeng F, et al: LncRNA SPRY4-IT1 sponges
miR-101-3p to promote proliferation and metastasis of bladder
cancer cells through up-regulating EZH2. Cancer Lett. 388:281–291.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Sun B, Zhao X, Zhao N, Sun R, Zhu
D, Zhang Y, Li Y, Gu Q, Dong X, et al: Twist1-related miR-26b-5p
suppresses epithelial-mesenchymal transition, migration and
invasion by targeting SMAD1 in hepatocellular carcinoma.
Oncotarget. 7:24383–24401. 2016.PubMed/NCBI
|
|
9
|
Miyamoto K, Seki N, Matsushita R, Yonemori
M, Yoshino H, Nakagawa M and Enokida H: Tumour-suppressive
miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by
regulating PLOD2 in bladder cancer. Br J Cancer. 115:354–363. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin C, Meng S, Zhu T and Wang X:
PDCD10/CCM3 acts downstream of {gamma}-protocadherins to regulate
neuronal survival. J Biol Chem. 285:41675–41685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stamatovic SM, Sladojevic N, Keep RF and
Andjelkovic AV: PDCD10 (CCM3) regulates brain endothelial barrier
integrity in cerebral cavernous malformation type 3: Role of
CCM3-ERK1/2-cortactin cross-talk. Acta Neuropathol. 130:731–750.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Ma X, Peng S, Nan X and Zhao H:
Differential expression of MST4, STK25 and PDCD10 between benign
prostatic hyperplasia and prostate cancer. Int J Clin Exp Pathol.
7:8105–8111. 2014.PubMed/NCBI
|
|
13
|
Lauenborg B, Kopp K, Krejsgaard T, Eriksen
KW, Geisler C, Dabelsteen S, Gniadecki R, Zhang Q, Wasik MA,
Woetmann A, et al: Programmed cell death-10 enhances proliferation
and protects malignant T cells from apoptosis. APMIS. 118:719–728.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
international union against cancer and the American Joint Committee
on Cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui X, Shen D, Kong C, Zhang Z, Zeng Y,
Lin X and Liu X: NF-κB suppresses apoptosis and promotes bladder
cancer cell proliferation by upregulating survivin expression in
vitro and in vivo. Sci Rep. 7:407232017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sudarshan S, Holman DH, Hyer ML,
Voelkel-Johnson C, Dong JY and Norris JS: In vitro efficacy of Fas
ligand gene therapy for the treatment of bladder cancer. Cancer
Gene Ther. 12:12–18. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin H, Liu W, Fang Z, Liang X, Li J, Bai
Y, Lin L, You H, Pei Y, Wang F, et al: Overexpression of DHX32
contributes to the growth and metastasis of colorectal cancer. Sci
Rep. 5:92472015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang R, Liu M, Liang H, Guo S, Guo X, Yuan
M, Lian H, Yan X, Zhang S, Chen X, et al: miR-138-5p contributes to
cell proliferation and invasion by targeting Survivin in bladder
cancer cells. Mol Cancer. 15:822016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kato M, Goto Y, Matsushita R, Kurozumi A,
Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M,
Ichikawa T, et al: MicroRNA-26a/b directly regulate La-related
protein 1 and inhibit cancer cell invasion in prostate cancer. Int
J Oncol. 47:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu X, Meng Z, Liang W, Tian Y, Wang X, Han
W, Lou G, Wang X, Lou F, Yen Y, et al: miR-26a enhances miRNA
biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth
and metastasis. Oncogene. 33:4296–4306. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fukumoto I, Kikkawa N, Matsushita R, Kato
M, Kurozumi A, Nishikawa R, Goto Y, Koshizuka K, Hanazawa T,
Enokida H, et al: Tumor-suppressive microRNAs (miR-26a/b,
miR-29a/b/c and miR-218) concertedly suppressed
metastasis-promoting LOXL2 in head and neck squamous cell
carcinoma. J Hum Genet. 61:109–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian L, Fang YX, Xue JL and Chen JZ: Four
microRNAs promote prostate cell proliferation with regulation of
PTEN and its downstream signals in vitro. PLoS One. 8:e758852013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Icli B, Dorbala P and Feinberg MW: An
emerging role for the miR-26 family in cardiovascular disease.
Trends Cardiovasc Med. 24:241–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fareh M, Yeom KH, Haagsma AC, Chauhan S,
Heo I and Joo C: TRBP ensures efficient Dicer processing of
precursor microRNA in RNA-crowded environments. Nat Commun.
7:136942016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng X, Li X, Zhang P, Wang J, Zhou Y and
Chen M: Circular RNA: An emerging key player in RNA world. Brief
Bioinform. 18:547–557. 2017.PubMed/NCBI
|
|
27
|
Cui X, Kong C, Zhu Y, Zeng Y, Zhang Z, Liu
X, Zhan B, Piao C and Jiang Z: miR-130b, an onco-miRNA in bladder
cancer, is directly regulated by NF-κB and sustains NF-κB
activation by decreasing Cylindromatosis expression. Oncotarget.
7:48547–48561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nahvi A, Shoemaker CJ and Green R: An
expanded seed sequence definition accounts for full regulation of
the hid 3′ UTR by bantam miRNA. RNA. 15:814–822. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu X, Ge S, Jia R, Zhou Y, Song X, Zhang H
and Fan X: Hypoxia-induced miR-181b enhances angiogenesis of
retinoblastoma cells by targeting PDCD10 and GATA6. Oncol Rep.
33:2789–2796. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu Z, Qi Y, Guo Z, Li P and Zhou D:
miR-613 suppresses ischemia-reperfusion-induced cardiomyocyte
apoptosis by targeting the programmed cell Death 10 gene. Biosci
Trends. 10:251–257. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen PY, Chang WS, Chou RH, Lai YK, Lin
SC, Chi CY and Wu CW: Two non-homologous brain diseases-related
genes, SERPINI1 and PDCD10, are tightly linked by an asymmetric
bidirectional promoter in an evolutionarily conserved manner. BMC
Mol Biol. 8:22007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
You C, Sandalcioglu IE, Dammann P, Felbor
U, Sure U and Zhu Y: Loss of CCM3 impairs DLL4-Notch signalling:
Implication in endothelial angiogenesis and in inherited cerebral
cavernous malformations. J Cell Mol Med. 17:407–418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lambertz N, El HN, Kreitschmann-Andermahr
I, Stein KP, Dammann P, Oezkan N, Mueller O, Sure U and Zhu Y:
Downregulation of programmed cell death 10 is associated with tumor
cell proliferation, hyperangiogenesis and peritumoral edema in
human glioblastoma. BMC Cancer. 15:7592015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kamath RS, Fraser AG, Dong Y, Poulin G,
Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et
al: Systematic functional analysis of the Caenorhabditis elegans
genome using RNAi. Nature. 421:231–237. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stockton RA, Shenkar R, Awad IA and
Ginsberg MH: Cerebral cavernous malformations proteins inhibit Rho
kinase to stabilize vascular integrity. J Exp Med. 207:881–896.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhu Y, Zhao K, Prinz A, Keyvani K,
Lambertz N, Kreitschmann-Andermahr I, Lei T and Sure U: Loss of
endothelial programmed cell death 10 activates glioblastoma cells
and promotes tumor growth. Neuro Oncol. 18:538–548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
He Y, Zhang H, Yu L, Gunel M, Boggon TJ,
Chen H and Min W: Stabilization of VEGFR2 signaling by cerebral
cavernous malformation 3 is critical for vascular development. Sci
Signal. 3:ra262010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tian R, Yang S, Zhu Y, Zou S, Li P, Wang
J, Zhu Z, Huang Y, He Z and Li Z: VEGF/VEGFR2 signaling
regulates germ cell proliferation in vitro and promotes mouse
testicular regeneration in vivo. Cells Tissues Organs. 201:1–13.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma X, Zhao H, Shan J, Long F, Chen Y, Chen
Y, Zhang Y, Han X and Ma D: PDCD10 interacts with Ste20-related
kinase MST4 to promote cell growth and transformation via
modulation of the ERK pathway. Mol Biol Cell. 18:1965–1978. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Riverso M, Montagnani V and Stecca B: KLF4
is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes
melanoma cell growth. Oncogene. 36:3322–3333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Voss K, Stahl S, Schleider E, Ullrich S,
Nickel J, Mueller TD and Felbor U: CCM3 interacts with CCM2
indicating common pathogenesis for cerebral cavernous
malformations. Neurogenetics. 8:249–256. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fidalgo M, Fraile M, Pires A, Force T,
Pombo C and Zalvide J: CCM3/PDCD10 stabilizes GCKIII proteins to
promote Golgi assembly and cell orientation. J Cell Sci.
123:1274–1284. 2010. View Article : Google Scholar : PubMed/NCBI
|