Introduction
Esophageal squamous cell carcinoma (ESCC) is
associated with a considerable decline in quality of life, in
addition to a poor prognosis (1,2). A
primary goal of efforts to improve the management of the disease
and patient outcomes is establishing methods to accurately predict
the risk of recurrence, in addition to survival, following curative
esophageal resection (3). Such
information is urgently required to provide appropriate
individualized perioperative follow-up and treatment (4,5).
Furthermore, a better understanding of the molecular mechanisms of
disease progression is essential, and identification of molecules
that contribute to the pathogenesis of ESCC may lead to the
development of novel biomarkers that facilitate precise risk
stratification and monitoring of recurrence following esophagectomy
(6,7).
The copine 5 (CPNE5) gene located on human
chromosome 6p21.2 (8) belongs to
the copine gene family, encoding calcium-dependent lipid-binding
proteins comprising two N-terminal C2 domains (C2Ds) and a
C-terminal A domain (9). CPNE5
localizes to the cytosol and is expressed at high levels in the
brain, lymph nodes, testes and heart (10). CPNE5 is expressed by
differentiated neurons during neural development, suggesting that
CPNE5 function is important for the function of the central nervous
system (11). Furthermore,
CPNE5 expression is associated with alcohol dependence and
obesity in Caucasians (12).
CPNE5 expression may be associated with the progression of
ESCC, since copine proteins interact with diverse target proteins,
including dual specificity mitogen-activated protein kinase kinase
1 (13), protein phosphatase 5
(14), and CDC42-regulated kinase
(15), which are components of
intracellular signaling pathways that influence the malignant
phenotype (16). However, the role
of CPNE5 in cancer is unknown.
The present study assessed whether CPNE5 may
serve as a predictive marker of ESCC outcomes following curative
resection. To answer this question, the expression of CPNE5
mRNA and the methylation of the CPNE5 promoter was measured
in ESCC cell lines and in surgically-resected ESCC tissues.
Materials and methods
Ethics approval and consent to
participate
The present study rigidly adhered to the ethical
guidelines of the World Medical Association Declaration of Helsinki
Ethical Principles for Medical Research Involving Human Subjects.
Written informed consent for the use of clinical samples and data
was obtained from all patients, as required by the Institutional
Review Board of Nagoya University (Nagoya, Japan; approval no.
2014-0044).
Sample collection
ESCC cell lines (TE1, TT and TTn) and a
non-tumorigenic epithelial cell line (Het-1A) were obtained from
the American Type Culture Collection (Manassas, VA, USA). NUEC2 and
WSSC cell lines were established at Nagoya University (17). KYSE510, KYSE590, KYSE890, KYSE1170,
KYSE1260 and KYSE1440 cells were purchased from the Japanese
Collection of Research Bioresources Cell Bank (Osaka, Japan)
(18). Cells were stored at −80°C
in a cell preservative (Cell Banker; LSI Medience Corporation,
Tokyo, Japan) and cultured in RPMI-1640 medium (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) supplemented with 10% fetal bovine
serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA) in an
atmosphere containing 5% CO2 at 37°C. A total of 106
primary ESCC tissues and adjacent normal tissues were acquired from
patients who underwent radical esophageal resection at Nagoya
University Hospital between October 2001 and January 2016 (19). The tumors were determined to be
radically resected when pathologically diagnosed as stage I to III.
All tissue samples were histologically diagnosed as ESCC,
immediately frozen following resection and stored at −80°C.
Specimens were histologically classified using the 7th edition of
the UICC staging system for esophageal cancer (20). Patients who received neoadjuvant
chemotherapy were excluded. Postoperative follow-up included
physical examination, measurement of serum tumor markers every 3
months, and enhanced computed tomography of the chest and abdominal
cavity every 6 months. Adjuvant chemotherapy was administered to
selected patients according to their condition and at the
discretion of the physician.
Analysis of CPNE5 mRNA expression
levels
The expression levels of CPNE5 mRNA were
measured using a reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) assay. Total RNA isolated using RNeasy
Mini Kit (Qiagen GmbH, Hilden, Germany) from cell lines and 106
pairs of surgically-resected primary ESCCs and adjacent normal
tissues served as template for cDNA synthesis. Reverse
transcription was performed as follows: 10.5 µl 1 µg/µl RNA, 4 µl
of 5X first strand buffer (Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 2 µl 100 mM dithiothreitol (Thermo Fisher Scientific,
Inc.), 1 µl 10 mM dNTP mix (Promega Corporation, Madison, WI, USA),
1 µl random primer (Roche Diagnostics, Basel, Switzerland), 1 µl
200 U/µl Moloney murine leukemia virus reverse transcriptase
(Thermo Fisher Scientific, Inc.) and 0.5 µl RNase inhibitor (Roche
Diagnostics) were mixed and incubated for 60 min at 37°C.
GAPDH mRNA expression levels (TaqMan; GAPDH control
reagents; Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) were quantified, and the data were used to
normalize the expression levels. RT-qPCR was performed using the
SYBR Green PCR Core Reagents kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) as follows: One cycle at 95°C for 10 min; 40
cycles at 95°C for 5 sec and 60°C for 60 sec without a final
extension step. The samples were tested in triplicate, and samples
without a template were included in each PCR plate as a negative
control (21). Real-time SYBR Green
fluorescence was detected using an ABI StepOnePlus Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.)
(22) and the 2−ΔΔCq
method was used for PCR quantification (23). The expression level of each sample
is expressed as the value of the CPNE5 amplicon divided by
that of GAPDH. The sequences of the specific primers are
listed in Table I.
 | Table I.Primers and annealing
temperatures. |
Table I.
Primers and annealing
temperatures.
| Gene | Experiment | Type | Sequence
(5′-3′) | Product size
(bp) | Annealing
temperature |
|---|
| CPNE5 | RT-qPCR | Forward |
CATGTTTTCCAAGTCCGACC | 106 | 60°C |
|
|
| Reverse |
ATTGAGCGTGTTGTCGATGA |
|
|
|
| Bisulfite
sequencing | Forward |
GGTAGGAGTTTTTAGATTTGGAGGT | 172 | 56°C |
|
|
| Reverse |
ATTTCCCAATAACCCAAATAAAATC |
|
|
| GAPDH | RT-qPCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 | 60°C |
|
|
| Probe |
CAAGCTTCCCGTTCTCAGCC |
|
|
|
|
| Reverse |
GAAGATGGTGATGGGATTTC |
|
|
Western blot analysis
The protein was extracted from each cell line using
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) and protein concentration was determined using a
bicinchoninic acid protein assay kit (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). For SDS-PAGE, 20 µg protein was added to a
NuPAGE 4–12% Bis-Tris Gel (Thermo Fisher Scientific, Inc.) and
electrophoresed for 35 min a 200 V. A polyvinylidene difluoride
membrane was used for blotting and the membrane was blocked with 5%
skim milk (Wako Pure Chemical Industries, Ltd., Osaka, Japan) for
60 min at room temperature. The CPNE5 protein expression levels in
ESCC cell lines were evaluated with a rabbit anti-CPNE5 polyclonal
antibody (1:100 dilution and overnight incubation at 4°C; cat. no.
HPA031369; Atlas Antibodies AB, Bromma, Sweden) as a primary
antibody and anti-rabbit IgG HRP-linked antibody (1:1,000 dilution
and 60 min incubation at room temperature; cat. no. 7074S; Cell
Signaling Technology, Inc., Danvers, MA, USA) as a secondary
antibody. As an internal control, β-actin protein expression was
detected with a mouse anti-β-actin polyclonal antibody (1:10,000
dilution and incubated for 60 min at room temperature; cat. no.
ab6276; Abcam, Cambridge, UK) as a primary antibody and anti-mouse
IgG HRP-linked antibody (1:1,000 dilution and 60 min incubation at
room temperature; cat. no. 7076S; Cell Signaling Technology, Inc.)
as a secondary antibody. Enhanced Chemiluminescence Western Blot
Analysis System (GE Healthcare, Chicago, IL, USA) was used for
visualization of the secondary antibody. An ESCC cell line with
relatively high CPNE5 mRNA expression (KYSE590) and a
low-expression ESCC cell line (TT) were evaluated.
Bisulfite nucleotide sequencing
Genomic DNA was isolated from the cell lines using a
QIAamp DNA Mini kit (Qiagen GmbH) and treated with bisulfite (2
cycles at 95°C for 5 min and 60°C for 10 min). Bisulfite-modified
DNA from ESCC cell lines and a non-cancerous esophageal mucosa cell
line (Het-1A) were amplified as follows: One cycle at 94°C for 2
min; and 50 cycles at 94°C for 15 sec, 56°C for 15 sec and 72°C for
30 sec, using specific primers (Table
I). Sequencing was performed by Eurofins Genomics Tokyo Co.,
Ltd. (Tokyo, Japan), using a Big Dye Terminator v3.1 Cycle
Sequencing kit (Thermo Fisher Scientific, Inc.) and a 3730× l DNA
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.)
(24).
Immunohistochemistry
Immunohistochemical staining was performed to
determine the difference in CPNE5 protein expression between
cancerous tissue and non-cancerous tissues in 45 representative
clinical cases. Sections were incubated for 16 h at 4°C with a
rabbit polyclonal antibody raised against CPNE5 (cat. no.
HPA031369; Atlas Antibodies AB) diluted 1:100 in Antibody Diluent
(Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Sections
were incubated with secondary antibody (SignalStain®
Boost IHC Detection Reagent labelled with HRP; Cell Signaling
Technology, Inc.) for 30 min at room temperature. Antigen antibody
complexes were visualized by exposure with liquid
3,3′-diaminobenzidine (Nichirei, Tokyo, Japan) for 2 min. A total
of two independent observers evaluated the specimens using an
optical microscope with ×400 magnification as follows: Cancerous
tissue >non-cancerous tissue; equivalent; or cancerous tissue
<non-cancerous tissue.
Statistical analysis
Quantitative data are presented as the mean ±
standard deviation. Patients were divided into low and high
CPNE5 groups according to the median levels of CPNE5
mRNA expression in the cancerous tissues. The differences between
CPNE5 mRNA expression values in the two groups
(differentiated cell lines vs. undifferentiated cell lines, or
cancerous tissues vs. non-cancerous tissues) were compared using
the Mann-Whitney test. The χ2 test was used to analyze
the association between CPNE5 mRNA expression levels and
clinicopathological characteristics. Overall survival (OS) and
disease-free survival (DFS) rates were calculated using the
Kaplan-Meier method. The Cox proportional hazards model was used to
compare survival rates, and multivariable regression analysis was
used to identify prognostic factors. Statistical analysis was
performed using JMP 10 software (SAS Institute Inc., Cary, NC,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of CPNE5 and
promoter methylation in ESCC cell lines
CPNE5 mRNA expression levels differed among
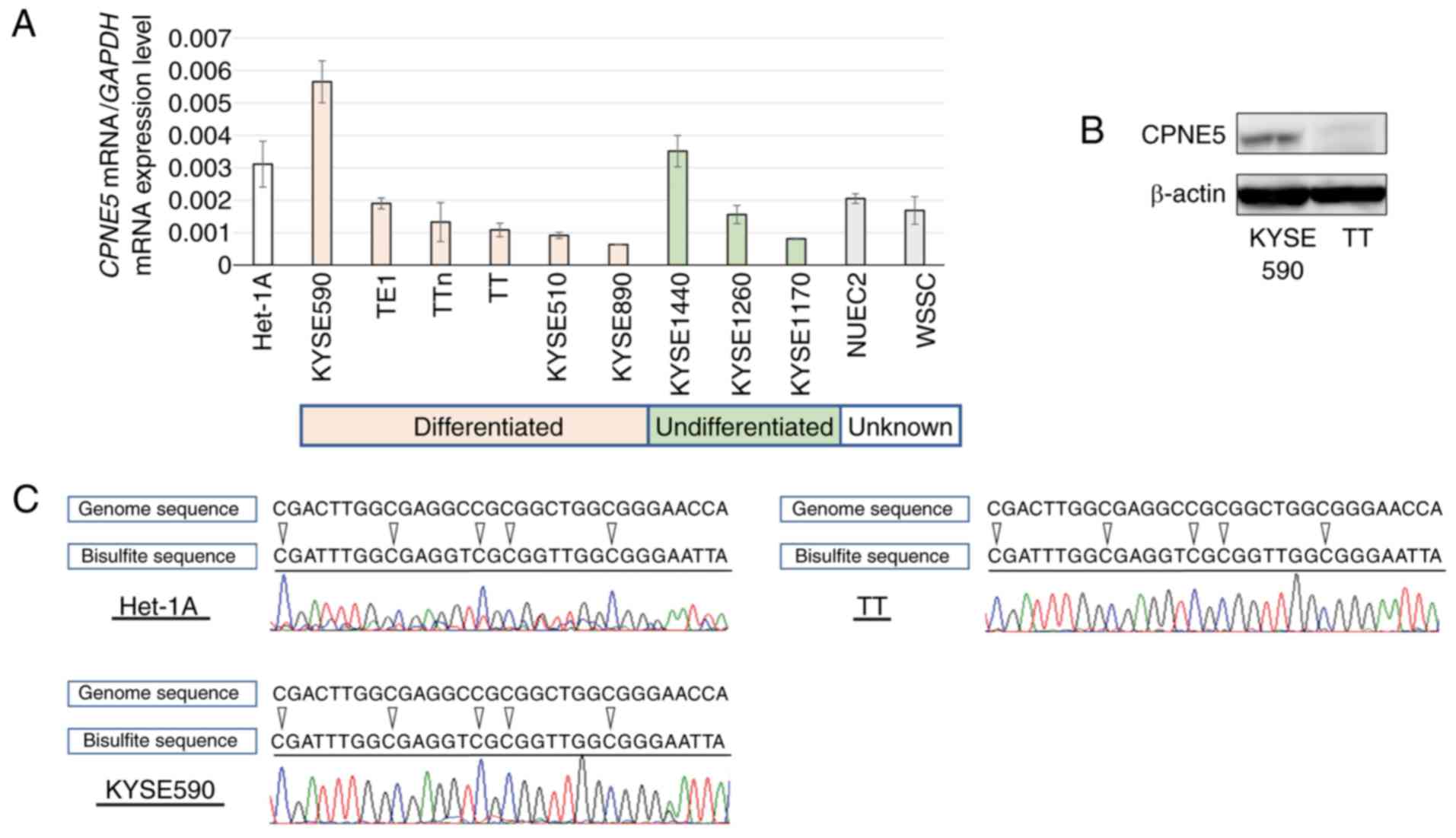
the 11 ESCC cell lines (Fig. 1A),
and were lower compared with those of Het-1A cells, except for
KYSE590 and KYSE1440. There were no significant differences in
CPNE5 mRNA expression levels between differentiated
(0.00197±0.00172) and undifferentiated (0.00161±0.00133) cell lines
(P=0.897). ESCC cell lines established from metastatic sites,
including TT and TTn, and those established from lymph node
metastases, including KYSE1170 and KYSE1260, expressed low levels
of CPNE5 mRNA. Western blot analysis using an anti-CPNE5
antibody illustrated high CPNE5 protein expression in an ESCC cell
line with high CPNE5 mRNA expression (KYSE590) and low CPNE5
protein expression in a low-expression ESCC cell line (TT)
(Fig. 1B). Bisulfite sequencing
analysis of CPNE5 revealed that the CpG sites in the
CPNE5 DNA promotor region in all ESCC cell lines and Het-1A
were completely methylated. The bisulfite sequencing results for
Het-1A, KYSE590 and TT cells are presented as representative cell
lines expressing high and low levels of CPNE5 mRNA,
respectively (Fig. 1C).
Characteristics of patients with
ESCC
The median age of the 106 patients was 65 years
(range, 44–84 years), and the female: Male ratio was 20:86.
According to the UICC staging system (7th edition), 24, 29 and 53
patients were diagnosed with disease at pathological stages I, II
and III, respectively. Adjuvant chemotherapy was administered to 36
patients (34%). The median duration of follow-up was 34.1 months,
during which 44 patients (42%) experienced recurrence and 39
patients (37%) succumbed to the disease.
CPNE5 mRNA expression levels in
clinical samples
The mean normalized CPNE5 mRNA expression
level was significantly lower in ESCC tissues (0.0107±0.0138)
compared with the corresponding non-cancerous adjacent mucosal
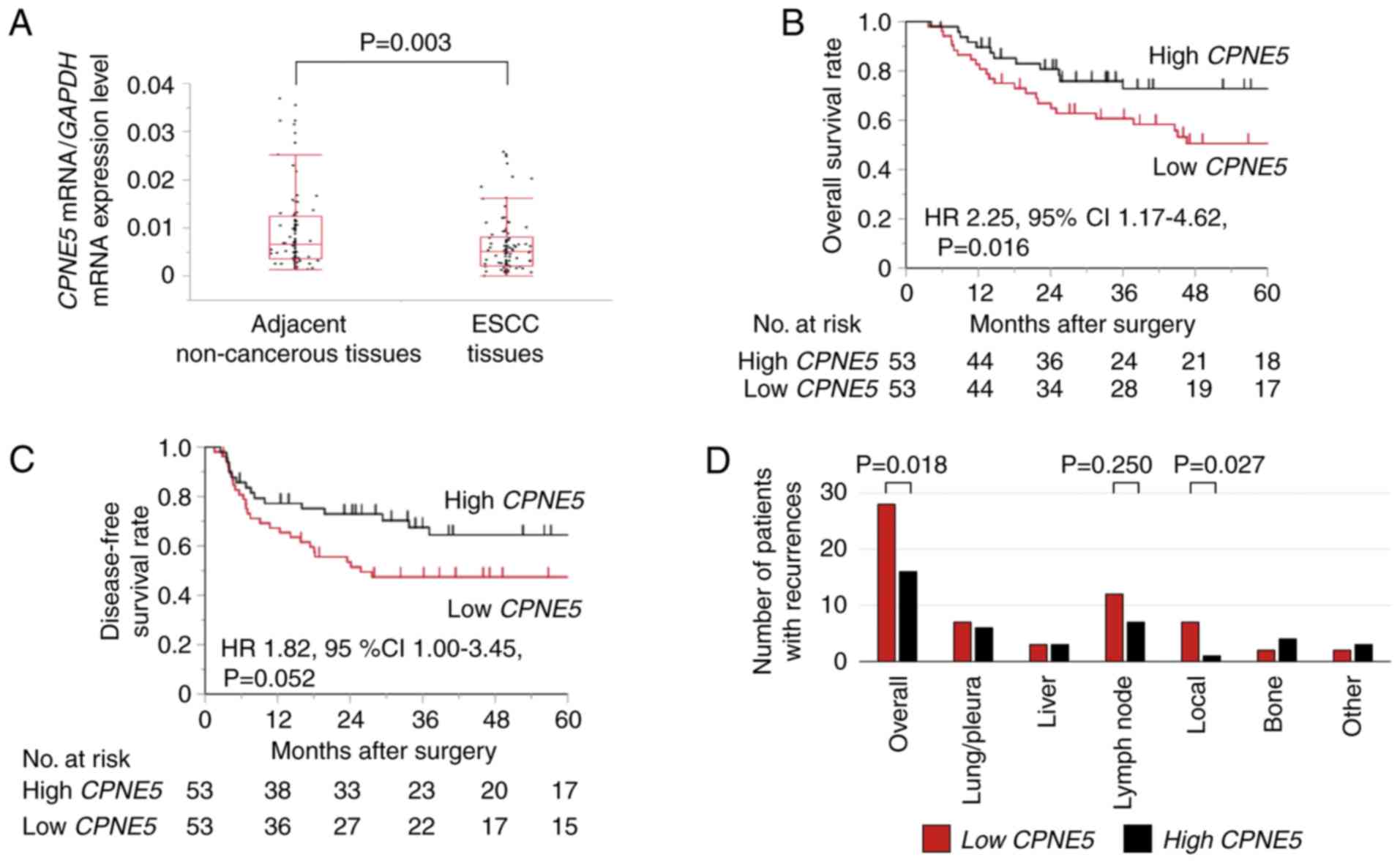
tissues (0.00829±0.0123; P=0.003; Fig.
2A).
Association between levels of CPNE5
mRNA and the clinicopathological characteristics of patients who
underwent resection
There was no significant association between the low
and high CPNE5 expression groups with their
clinicopathological characteristics (sex, tumor size and depth,
lymphatic involvement, vascular invasion and pathological stage).
By contrast, the percentage of patients who received adjuvant
chemotherapy was significantly higher in the low CPNE5 group
(Table II).
 | Table II.Association between the expression
level of CPNE5 mRNA and clinicopathological parameters in
106 patients with resected esophageal cancer. |
Table II.
Association between the expression
level of CPNE5 mRNA and clinicopathological parameters in
106 patients with resected esophageal cancer.
| Clinicopathological
parameters | Low CPNE5 in
ESCC tissue, no. of patients | High CPNE5
in ESCC tissue, no. of patients | P-value |
|---|
| Age, years |
|
| 0.437 |
|
<65 | 23 | 28 |
|
|
≥65 | 30 | 25 |
|
| Sex |
|
| 1.000 |
|
Male | 43 | 43 |
|
|
Female | 10 | 10 |
|
| Smoking
history |
|
| 0.487 |
|
Yes | 39 | 43 |
|
| No | 14 | 10 |
|
| Double cancer |
|
| 0.698 |
|
Present | 7 | 10 |
|
|
Absent | 46 | 43 |
|
| Tumor location |
|
| 0.111 |
| Ce | 0 | 1 |
|
| Ut | 3 | 5 |
|
| Mt | 24 | 27 |
|
| Lt | 25 | 15 |
|
| Ae | 1 | 5 |
|
| Tumor
multiplicity |
|
| 1.000 |
|
Present | 7 | 8 |
|
|
Absent | 46 | 45 |
|
| Tumor size, mm |
|
| 0.151 |
|
<50 | 31 | 39 |
|
|
≥50 | 22 | 14 |
|
| CEA, ng/ml |
|
| 0.761 |
| ≤5 | 48 | 46 |
|
|
>5 | 5 | 7 |
|
| SCC, IU/ml |
|
| 0.531 |
|
≤1.5 | 36 | 33 |
|
|
>1.5 | 15 | 19 |
|
| pT |
|
| 0.116 |
| T1 or
2 | 18 | 27 |
|
| T3 | 35 | 26 |
|
| Lymph node
metastasis |
|
| 0.843 |
|
Present | 31 | 33 |
|
|
Absent | 22 | 20 |
|
|
Differentiation |
|
| 0.267 |
|
Differentiated | 43 | 45 |
|
|
Undifferentiated | 10 | 5 |
|
| Lymphatic
involvement |
|
| 0.822 |
|
Present | 39 | 41 |
|
|
Absent | 14 | 12 |
|
| Vascular
invasion |
|
| 1.000 |
|
Present | 21 | 22 |
|
|
Absent | 32 | 31 |
|
| Intraepithelial
progress |
|
| 0.057 |
|
Present | 18 | 22 |
|
|
Absent | 23 | 10 |
|
| Pathological UICC
stage |
|
| 0.466 |
| I | 10 | 14 |
|
| II | 17 | 12 |
|
|
III | 26 | 27 |
|
| Postoperative
adjuvant chemotherapy |
|
| 0.007 |
|
Present | 25 | 11 |
|
|
Absent | 28 | 42 |
|
Ability of CPNE5 mRNA expression
levels to predict prognosis
Patients in the low CPNE5 expression group
experienced a significantly shorter OS time compared with those in
the high CPNE5 expression group (the 5-year OS rates were
55.6 and 77.3% for the low and high expression groups,
respectively; P=0.016; Fig. 2B).
Though the difference was not statistically significant, there was
a similar trend observed in DFS (P=0.052; Fig. 2C). Univariate analysis revealed that
tumor depth, lymph node metastasis, undifferentiated tumor
phenotype, lymphatic involvement, postoperative chemotherapy and
low CPNE5 expression were significantly associated with
lower survival rates. Multivariable analysis identified low
CPNE5 expression and lymphatic involvement as independent
prognostic factors for OS (hazard ratio, 2.55; 95% confidence
interval, 1.21–5.68; P=0.014; and hazard ratio, 6.28; 95%
confidence interval, 1.70–40.6; P=0.004, respectively; Table III).
 | Table III.Prognostic factors for overall
survival of 106 patients. |
Table III.
Prognostic factors for overall
survival of 106 patients.
|
| Univariate | Multivariable |
|---|
|
|
|
|
|---|
|
| Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age, ≥65 years | 1.45 | 0.75–2.79 | 0.259 | – | – | – |
| Sex, male | 2.19 | 0.94–6.43 | 0.072 | – | – | – |
| Smoking | 0.98 | 0.49–2.20 | 0.963 | – | – | – |
| Double cancer | 1.29 | 0.52–2.76 | 0.559 | – | – | – |
| Tumor
multiplicity | 0.70 | 0.21–1.74 | 0.470 | – | – | – |
| Tumor size, ≥50
mm | 1.28 | 0.66–2.41 | 0.462 | – | – | – |
| CEA, >5
ng/ml | 1.34 | 0.50–2.97 | 0.528 | – | – | – |
| SCC, >1.5
IU/ml | 1.11 | 0.55–2.14 | 0.769 | – | – | – |
| Tumor depth,
pT3 | 2.75 | 1.38–5.96 | 0.003 | 1.48 | 0.72–3.27 | 0.293 |
| Lymph node
metastasis | 2.36 | 1.19–5.09 | 0.013 | 1.53 | 0.64–3.92 | 0.349 |
| Tumor
differentiation, undifferentiated | 2.75 | 1.22–5.62 | 0.002 | 2.10 | 0.92–4.37 | 0.075 |
| Lymphatic
involvement | 8.61 | 2.63–53.0 | <0.001 | 6.28 | 1.70–40.6 | 0.004a |
| Vascular
invasion | 1.39 | 0.73–2.61 | 0.304 | – | – | – |
| Intraepithelial
progress | 0.64 | 0.32–1.27 | 0.204 | – | – | – |
| Postoperative
adjuvant chemotherapy | 2.08 | 1.11–3.93 | 0.023 | 0.92 | 0.42–2.04 | 0.837 |
| Low CPNE5
expression | 2.25 | 1.17–4.62 | 0.015 | 2.55 | 1.21–5.68 | 0.014a |
Analysis of patients with
recurrence
Differences between recurrence patterns were
predicted, since the Kaplan-Meier analysis revealed
fold-differences between OS and DFS. Among patients with
recurrence, a significant number were members of the low
CPNE5 group (P=0.018). Among patients with local recurrence,
significantly more were members of the low CPNE5 group
(P=0.027; Fig. 2D). A similar
tendency was observed in patients with lymph node recurrence
(P=0.250; Fig. 2D). By contrast,
the rates of distant metastasis to tissues including the
lung/pleura and liver were not significantly different between
groups (Fig. 2D).
CPNE5 protein expression levels in
clinical samples
The expression patterns of CPNE5 protein were
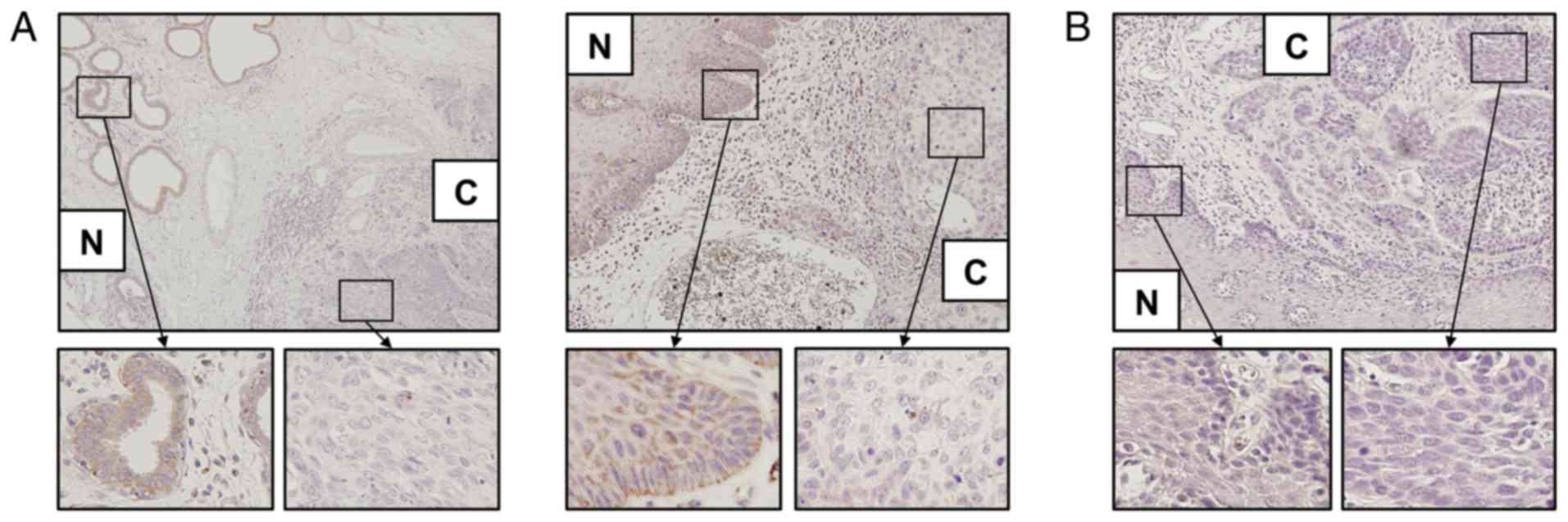
evaluated using immunohistochemical staining. Among the 45 clinical
samples, CPNE5 protein expression was suppressed in the cancerous
tissue in 14 (31%) samples, equally expressed in cancerous and
non-cancerous tissue in 19 (42%) samples and overexpressed in
cancerous tissue in 12 (27%) samples. Representative examples of
suppression in cancerous tissue, and an example of equivalent
expression, are presented (Fig.
3).
Discussion
The results of the present study provided evidence
to support the predictive value of CPNE5 expression levels
in ESCC tissues following curative resection. Specifically, the
CPNE5 mRNA expression levels of the majority ESCC cell lines
were lower compared with those of a non-tumorigenic esophageal cell
line. Bisulfite sequencing analysis was performed to reveal the
mechanism of downregulation of CPNE5 transcription, as the
promoter region harbors CpG islands (25,26).
Methylation of the CPNE5 promoter region was detected in all
ESCC cell lines and in a non-tumorigenic esophageal cell line,
indicating that promoter hypermethylation did not contribute to the
regulation of CPNE5 transcription. Acetylation of histones
(27), copy-number alterations
(28), microRNAs (27) and genomic mutations (29) may therefore contribute to the
downregulation of CPNE5 transcription.
The expression levels of CPNE5 mRNA in ESCC
tissues were lower compared with those of adjacent non-cancerous
tissue, revealing an association between CPNE5 expression
and the pathology of ESCC. CPNE5 mRNA expression levels were
not significantly associated with clinicopathological
characteristics known to be associated with an unfavorable
prognosis of ESCC [including tumor multiplicity, tumor size, tumor
markers, lymphatic involvement, vascular invasion, and UICC stage
(30)], although low levels of
CPNE5 mRNA were associated with shorter OS. Furthermore,
multivariable analysis identified low levels of CPNE5 mRNA
as an independent risk factor of shorter OS. Therefore,
CPNE5 may serve as an effective marker for predicting
prognosis compared with the tumor-node-metastasis classification of
esophageal cancer.
CPNE5 mRNA expression levels were lower in
patients who received adjuvant chemotherapy, although the
administration of chemotherapy was at the physician's discretion.
The association between adjuvant chemotherapy and CPNE5
expression may be explained by the association, albeit not
statistically significant, of CPNE5 mRNA expression levels
with tumor size and depth, in addition to intraepithelial
progression, and physicians may therefore decide to administer
adjuvant chemotherapy according to their interpretations of the
totality of pathological findings. However, it was noted that
patients in the low CPNE5 expression group had poor
prognoses despite adjuvant chemotherapy (31), suggesting an association between
CPNE5 expression and resistance to chemotherapy.
OS and DFS rates following curative resection of
ESCC were lower in the low CPNE5 group compared with the
high CPNE5 group. The difference in OS was statistically
significant, although that for DFS was not. The Kaplan-Meier
analysis demonstrated that the primary curves for DFS of the two
groups overlapped. By contrast, the primary curves of OS were
separated, and this difference may reflect the statistical
difference. These results demonstrated that the low CPNE5
group experienced shorter survival following recurrence.
Accordingly, two hypotheses were developed to explain the data as
follows: i) The clinicopathological findings indicate resistance to
chemotherapy; thus, patients in the low CPNE5 group may
succumb following recurrence, as they did not benefit from
chemotherapy; and ii) the differences in recurrence patterns may
explain survival patterns; it was expected that recurrence in the
low CPNE5 group may arise in a site that is difficult to
treat, for example the lung or liver, which is associated with poor
prognosis following recurrence (32). The analysis of recurrence patterns,
which was conducted to evaluate these hypotheses, revealed that
significantly more patients in the low CPNE5 group
experienced more frequent recurrence compared with those in the
high CPNE5 group. In contrast to expectations, the
difference in the numbers of patients in the low and high
CPNE5 groups was not significant for patients with lung and
hepatic recurrences, which are associated with poor prognosis
following recurrence.
However, it was noted that the differences in
recurrence rates between the low and high CPNE5 groups was
explained by local and lymphatic recurrences, indicating that
CPNE5 expression may be associated with the local growth of
esophageal cancer. To translate these findings into clinical
practice, patients with low CPNE5 expression ought to
undergo surgery with rigorous lymph node dissection (33) and adequate surgical margins
(34). Frequent follow-up,
including esophagoscopy (35) and
computed tomography (36), may
enhance the detection of local recurrence.
There are certain limitations to the present study.
First, an association between CPNE5 expression and
resistance to chemotherapy was identified. Unfortunately, it was
not possible to obtain details of patients who received
chemotherapy following recurrence. The analysis of biopsied or
micro-dissected resected tissues from patients who have received
neoadjuvant chemotherapy may help to investigate this further.
Second, this was a retrospective study of a small number patients
treated at a single center. External validation using large cohorts
from multiple institutions is required to validate the present
findings. Third, the function of CPNE5 in esophageal cancer
and its mechanism of regulation remain unexplained. Functional
analysis of CPNE5-knockdown cell lines, protein expression
and animal tumor xenograft models are required to overcome these
limitations.
In conclusion, the present data suggested that
CPNE5 expression in ESCC tissues may represent a promising
biomarker for predicting ESCC recurrence, particularly for patients
with local recurrence, and may help ensure that patients receive
optimal treatment and follow-up.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SU, MKa, TM and HT performed the experiments and the
data analysis. SU, MKa, TM, HT, CT, DK, MKo, MS, MH, SY, GN and YK
collected the cases and the clinical data. SU and MKa conceived and
designed the study and prepared the initial manuscript. YK
supervised the project. All authors contributed to the final
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study conformed to the ethical
guidelines of the World Medical Association Declaration of
Helsinki, Ethical Principles for Medical Research Involving Human
Subjects, and was approved by the Institutional Review Board of
Nagoya University (Nagoya, Japan). Written informed consent for the
use of clinical samples and data, as required by the institutional
review board, was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lagergren J, Smyth E, Cunningham D and
Lagergren P: Oesophageal cancer. Lancet. 390:2383–2396. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000–14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tanaka H, Kanda M, Koike M, Iwata N,
Shimizu D, Ezaka K, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et
al: Adherens junctions associated protein 1 serves as a predictor
of recurrence of squamous cell carcinoma of the esophagus. Int J
Oncol. 47:1811–1818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xi M, Yang Y, Zhang L, Yang H, Merrell KW,
Hallemeier CL, Shen RK, Haddock MG, Hofstetter WL, Maru DM, et al:
Multi-institutional analysis of recurrence and survival after
neoadjuvant chemoradiotherapy of esophageal cancer: Impact of
histology on recurrence patterns and outcomes. Ann Surg. 2018.
View Article : Google Scholar
|
|
5
|
Chen HS, Hsu PK, Liu CC and Wu SC: Upfront
surgery and pathological stage-based adjuvant chemoradiation
strategy in locally advanced esophageal squamous cell carcinoma.
Sci Rep. 8:21802018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hibino S, Kanda M, Oya H, Takami H,
Shimizu D, Nomoto S, Hishida M, Niwa Y, Koike M, Yamada S, et al:
Reduced expression of DENND2D through promoter hypermethylation is
an adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kunzmann AT, McMenamin ÚC, Spence AD, Gray
RT, Murray LJ, Turkington RC and Coleman HG: Blood biomarkers for
early diagnosis of oesophageal cancer: A systematic review. Eur J
Gastroenterol Hepatol. 30:263–273. 2018.PubMed/NCBI
|
|
8
|
Tripodis N, Mason R, Humphray SJ, Davies
AF, Herberg JA, Trowsdale J, Nizetic D, Senger G and Ragoussis J:
Physical map of human 6p21.2-6p21.3: Region flanking the
centromeric end of the major histocompatibility complex. Genome
Res. 8:631–643. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Creutz CE, Tomsig JL, Snyder SL, Gautier
MC, Skouri F, Beisson J and Cohen J: The copines, a novel class of
C2 domain-containing, calcium-dependent, phospholipid-binding
proteins conserved from Paramecium to humans. J Biol Chem.
273:1393–1402. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fagerberg L, Hallström BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ding X, Jin Y, Wu Y, Wu Y, Wu H, Xiong L,
Song X, Liu S, Fan W and Fan M: Localization and cellular
distribution of CPNE5 in embryonic mouse brain. Brain Res.
1224:20–28. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang KS, Zuo L, Pan Y, Xie C and Luo X:
Genetic variants in the CPNE5 gene are associated with alcohol
dependence and obesity in Caucasian populations. J Psychiatr Res.
71:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kidger AM, Sipthorp J and Cook SJ: ERK1/2
inhibitors: New weapons to inhibit the RAS-regulated
RAF-MEK1/2-ERK1/2 pathway. Pharmacol Ther. 187:45–60. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsieh FS, Hung MH, Wang CY, Chen YL, Hsiao
YJ, Tsai MH, Li JR, Chen LJ, Shih CT, Chao TI, et al: Inhibition of
protein phosphatase 5 suppresses non-small cell lung cancer through
AMP-activated kinase activation. Lung Cancer. 112:81–89. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schlessinger K, McManus EJ and Hall A:
Cdc42 and noncanonical Wnt signal transduction pathways cooperate
to promote cell polarity. J Cell Biol. 178:355–361. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tomsig JL, Snyder SL and Creutz CE:
Identification of targets for calcium signaling through the copine
family of proteins. Characterization of a coiled-coil
copine-binding motif. J Biol Chem. 278:10048–10054. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsunoo H, Komura S, Ohishi N, Yajima H,
Akiyama S, Kasai Y, Ito K, Nakao A and Yagi K: Effect of
transfection with human interferon-beta gene entrapped in cationic
multilamellar liposomes in combination with 5-fluorouracil on the
growth of human esophageal cancer cells in vitro. Anticancer Res.
22:1537–1543. 2002.PubMed/NCBI
|
|
18
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miwa T, Kanda M, Koike M, Iwata N, Tanaka
H, Umeda S, Tanaka C, Kobayashi D, Hayashi M, Yamada S, et al:
Identification of NCCRP1 as an epigenetically regulated tumor
suppressor and biomarker for malignant phenotypes of squamous cell
carcinoma of the esophagus. Oncol Lett. 14:4822–4828. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: International Union Against CancerTNM Classification of
Malignant Tumours. 7th edition. Wiley-Blackwell; New York, NY: pp.
66–72. 2009
|
|
21
|
Kanda M, Shimizu D, Sueoka S, Nomoto S,
Oya H, Takami H, Ezaka K, Hashimoto R, Tanaka Y, Kobayashi D, et
al: Prognostic relevance of SAMSN1 expression in gastric cancer.
Oncol Lett. 12:4708–4716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shimizu D, Kanda M, Tanaka H, Kobayashi D,
Tanaka C, Hayashi M, Iwata N, Niwa Y, Takami H, Yamada S, et al:
GPR155 serves as a predictive biomarker for hematogenous metastasis
in patients with gastric cancer. Sci Rep. 7:420892017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanda M, Tanaka C, Kobayashi D, Tanaka H,
Shimizu D, Shibata M, Takami H, Hayashi M, Iwata N, Niwa Y, et al:
Epigenetic suppression of the immunoregulator MZB1 is associated
with the malignant phenotype of gastric cancer. Int J Cancer.
139:2290–2298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Beck S, Rhee C, Song J, Lee BK, LeBlanc L,
Cannon L and Kim J: Implications of CpG islands on chromosomal
architectures and modes of global gene regulation. Nucleic Acids
Res. 46:4362–4391. 2018. View Article : Google Scholar
|
|
26
|
Pu W, Wang C, Chen S, Zhao D, Zhou Y, Ma
Y, Wang Y, Li C, Huang Z, Jin L, et al: Targeted bisulfite
sequencing identified a panel of DNA methylation-based biomarkers
for esophageal squamous cell carcinoma (ESCC). Clin Epigenetics.
9:1292017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kailasam A, Mittal SK and Agrawal DK:
Epigenetics in the pathogenesis of esophageal adenocarcinoma. Clin
Transl Sci. 8:394–402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong G, Mao Q, Yu D, Zhang Y, Qiu M, Dong
G, Chen Q, Xia W, Wang J, Xu L, et al: Integrative analysis of copy
number and transcriptional expression profiles in esophageal cancer
to identify a novel driver gene for therapy. Sci Rep. 7:420602017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen XX, Zhong Q, Liu Y, Yan SM, Chen ZH,
Jin SZ, Xia TL, Li RY, Zhou AJ, Su Z, et al: Genomic comparison of
esophageal squamous cell carcinoma and its precursor lesions by
multi-region whole-exome sequencing. Nat Commun. 8:5242017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang D, Zheng Y, Wang Z, Huang Q, Cao X,
Wang F and Liu S: Comparison of the 7th and proposed 8th editions
of the AJCC/UICC TNM staging system for esophageal squamous cell
carcinoma underwent radical surgery. Eur J Surg Oncol.
43:1949–1955. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang SS, Yang H, Xie X, Luo KJ, Wen J,
Bella AE, Hu Y, Yang F and Fu JH: Adjuvant chemotherapy versus
surgery alone for esophageal squamous cell carcinoma: A
meta-analysis of randomized controlled trials and nonrandomized
studies. Dis Esophagus. 27:574–584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu SG, Zhang WW, He ZY, Sun JY, Chen YX
and Guo L: Sites of metastasis and overall survival in esophageal
cancer: A population-based study. Cancer Manag Res. 9:781–788.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo JC, Lin CC, Huang TC, Huang PM, Kuo
HY, Chang CH, Wang CC, Cheng JC, Yeh KH, Hsu CH, et al: Number of
resected lymph nodes and survival of patients with locally advanced
esophageal squamous cell carcinoma receiving preoperative
chemoradiotherapy. Anticancer Res. 38:1569–1577. 2018.PubMed/NCBI
|
|
34
|
Tam PC, Siu KF, Cheung HC, Ma L and Wong
J: Local recurrences after subtotal esophagectomy for squamous cell
carcinoma. Ann Surg. 205:189–194. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Catalano MF, Sivak MV Jr, Rice TW and Van
Dam J: Postoperative screening for anastomotic recurrence of
esophageal carcinoma by endoscopic ultrasonography. Gastrointest
Endosc. 42:540–544. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim TJ, Lee KH, Kim YH, Sung SW, Jheon S,
Cho SK and Lee KW: Postoperative imaging of esophageal cancer: What
chest radiologists need to know. Radiographics. 27:409–429. 2007.
View Article : Google Scholar : PubMed/NCBI
|