Introduction
Hepatocellular carcinoma (HCC) is clinically
considered a lethal cancer that usually results in intra- and
extrahepatic metastasis and has a poor prognosis. Although a
majority of HCC cases occur in underdeveloped nations, HCC is the
most frequent cancer as well as the second leading cause of
mortality in China (1). Although
earlier studies have revealed links between several genes and HCC,
the underlying mechanisms remain unexplored. Therefore, an
understanding of the underlying pathophysiological factors
associated with HCC is vital for uncovering novel prognostic
biomarkers and developing therapeutic strategies.
The transcription of RNA from non-protein-coding
segments of DNA is one of the most crucial discoveries of the
postgenomic era (2). Protein-coding
genes constitute merely ~2% of the human genome, while a greater
proportion of the genome includes non-coding RNAs (ncRNAs) such as
long ncRNAs (lncRNAs). Recent research has revealed the roles of
lncRNAs in cancer; lncRNAs can act either as oncogenes or as tumor
suppressors. Several research groups have established that
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)
positively modulates growth, apoptosis, and migration in many
cancers (3,4). On the other hand, maternally expressed
gene 3 (MEG3) was revealed to function as a tumor inhibitor that
promoted p53-mediated transactivation (5,6).
lncRNA activated by transforming growth factor β (TGF-β)
(lncRNA-ATB), which is an important regulator of the
invasion-metastasis cascade, was revealed to promote cell invasion
through its role as a competing endogenous RNA (ceRNA) for zinc
finger E-box-binding homeobox (ZEB) genes and facilitated the
colonization of disseminated HCC cells in distant organs by binding
to interleukin-11 (IL-11) mRNA (7).
A previous study found that FOXD2 adjacent opposite strand RNA 1
(FOXD2-AS1) regulated tumor development in lung cancer (8). In terms of microRNAs (miRNAs),
endogenous transcripts containing miRNA response elements (MREs)
have been proposed to interact with each other by acting as miRNA
sponges or as ceRNAs, thus forming large-scale regulatory networks
across the transcriptome (9). For
example, Tan et al demonstrated that the double-negative
feedback loop between lncRNA taurine upregulated 1 (TUG1) and
miR-145 promoted epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells (10). Nevertheless, the relationship
between aberrant FOXD2-AS1 expression and malignant behavior in HCC
remains unclear, and the mechanism underlying the oncogenic
activity of FOXD2-AS1 warrants elucidation.
We hypothesized that lncRNA FOXD2-AS1 contributes to
HCC progression by acting as a miRNA sponge and, moreover, that
FOXD2-AS1 expression is increased in HCC tissues and is correlated
with inferior prognosis. Additional investigations revealed that
when overexpressed, FOXD2-AS1 functioned like a ceRNA that enhanced
cell viability and migration, thus resulting in the upregulation of
the annexin A2 (ANXA2) protein, which was regulated by the
targeting of miR-206 by FOXD2-AS1. As a whole, this study
established that the upregulation of FOXD2-AS1 promoted the
viability of HCC cells in part through the role of FOXD2-AS1 as a
miR-206 sponge.
Materials and methods
Patients
During 2008–2010, a total of 140 HCC tissues and the
corresponding neighboring healthy hepatic tissues from patients
subjected to resection were acquired from the First Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China). The patients
were monitored via phone calls or visits every 6 months until their
death or the completion of the study. The patients had not
undergone chemotherapy or radiotherapy prior to surgery, and the
tissue samples were subjected to pathological examination. Prior to
the the study, the patients signed written informed consent forms.
In addition, the present study was approved by the Ethics Committee
of the First Affiliated Hospital of Xi'an Jiaotong University
(Xi'an, China).
Cell culture, cell transfection and
MTT assay
Human liver cancer cell lines (Hep3B, MHCC97-L,
MHCC97-H, SK-HEP1 and HCCLM3) and a cultured normal liver cell line
(HL7702) were obtained from the Cell Bank of the Chinese Academy of
Sciences. These cells were cultured in RPMI-1640 medium containing
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.
Waltham, MA, USA) at 37°C in 5% CO2. Cells in the
logarithmic growth phase were monitored after being passaged every
2–3 days. The miRNA mimics, inhibitors and small interfering RNAs
(siRNAs) were obtained from GenePharma Co., Ltd. (Shanghai, China).
Oligonucleotide and plasmid transfections were carried out with
Lipofectamine™ 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
Following transfection, the cells were collected and used for
subsequent studies. The siRNA oligo sequences were as follows:
siFOXD2-AS1 sense, 5′-GCGCGGUUGUUGAGACCAAGG-3′ and siFOXD2-AS1
antisense, 5′-UUGGUCUCAACAACCGCGCAG-3′.
After transfection, cell proliferation was analyzed
via a
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium
bromide (MTT) assay (Promega Corp., Madison, WI, USA) according to
the manufacturer's instructions. Cells (5×103
cells/well) were seeded in 96-well plates and treated 24 h later.
After a further 24 h, 20 µl of 5 mg/ml MTT was added, and the
plates were placed in an incubator for 4 h. Next, the supernatant
was aspirated and 200 µl of dimethyl sulfoxide (DMSO) was added to
the wells to dissolve the formazan. The optical density (OD) was
determined at 450 nm.
RNA isolation and quantitative
real-time PCR (qRT-PCR)
RNA was extracted with TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). After RNA extraction, first strand
cDNA was synthesized with a High Capacity cDNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The mRNA and lncRNA expression analyses were carried out by
RT-PCR (11). qRT-PCR was carried
out with SYBR® Premix Ex Taq™ II (Takara Biotechnology
Co., Ltd., Dalian, China) on a StepOne Plus Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) following the
supplier's mRNA analysis protocols. GAPDH and U6 were used as the
endogenous controls for lncRNAs and miRNAs, respectively. The
qRT-PCR experiments were carried out in triplicate, whereas the
changes in the expression of the candidate genes were analyzed via
the 2−ΔΔCq method (12).
The primer sequences were as follows: FOXD2-AS1 forward,
5′-TGTTCGTGGGAAGAGGGTTG-3′ and reverse, 5′-TACCACTCCGGGAACTCTGT-3′;
and GAPDH forward, 5′-ACTGCCACCCAGAAGACT-3′ and reverse,
5′-GCTCAGTGTAGCCCAGGAT-3′. qRT-PCR included an initial denaturation
cycle at 95°C for 2 min, followed by 35 cycles of denaturation at
98°C for 10 sec and annealing and extension at 60°C for 45 sec.
Vector construction and
transfection
Full-length lncRNA FOXD2-AS1 was amplified using
Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher
Scientific, Inc.) and cloned into pcDNA3.1 (Invitrogen; Thermo
Fisher Scientific, Inc.); the resulting vector was named
pcDNA3.1-FOXD2-AS1. FOXD2-AS1 constructs mutated at the putative
miR-206 target site were generated with the appropriate primers.
The resulting vectors were sequenced and were named FOXD2-AS1-WT
(wild-type FOXD2-AS1) and FOXD2-AS1-Mut (mutated FOXD2-AS1). siRNA
targeting ANXA2 was obtained from GenePharma Co., Ltd. The
double-stranded miRNA mimic (miR-206) and the control miRNA
(miR-con) were obtained from GenePharma Co., Ltd. The vectors and
miRNAs were transfected into HCC cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). The primers used for
cloning and plasmid assembly were as follows: pCDNA/FOXD2-AS1-F,
5′-GGATCCCCTGTTCGGCGTCTTGCAGCAGTGC-3′) and pCDNA/FOXD2-AS1-R,
5′-CTCGAGGGAATGAATAACTTCAGTC-3′.
Wound healing assay
The wound healing assay was carried out to verify
cell metastasis. First, cells (5×105 cells/well)
suspended in DMEM were cultured in 6-well plates as confluent
monolayers for 24 h. Subsequently, scratches were made on the
monolayer surface with a 200-µl pipette tip. Images depicting cell
migration across the scratches were captured in 5 randomly selected
fields at 0 and 24 h following treatment.
Cell migration assay
To study the impact of lncRNA FOXD2-AS1 on cell
migration, Transwell assays were performed with 8-µm pore chambers
(BD Biosciences, Franklin Lakes, NJ, USA). For the Transwell
assays, cells in which lncRNA FOXD2-AS1 was either overexpressed or
knocked down were added to serum-free medium and permitted to
migrate in the direction of medium supplemented with 10% FBS at
37°C for 24 h. The migrated cells were treated with 100% methanol
for 30 min, whereas the non-migrated cells were removed.
Subsequently, the cells on the underside of the membrane were
stained with 0.1% crystal violet for 20 min. The number of cells in
5 arbitrary fields of each replicate was determined with a light
microscope (Nikon ECLIPSE 80i; ×100 magnification). All experiments
were repeated three times.
RNA immunoprecipitation (RIP)
assay
Hep3B cells were used in the RIP assay along with an
anti-Ago2 antibody (Cell Signaling Technology, Inc., Danvers, MA,
USA) and a Magna RIP™ RNA-Binding Protein Immunoprecipitation kit
(EMD Millipore, Billerica, MA, USA). After antibody recovery with
protein A/G beads, qRT-PCR was carried out to quantify FOXD2-AS1
and the target miRNAs in the precipitate.
Bioinformatics analysis
The putative miRNA binding sites on lncRNA FOXD2-AS1
were identified by DIANA software (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php)
with the minimum cutoff score set at 0.802.
Luciferase reporter assay
The 3′ untranslated regions (3′-UTRs) of
FOXD2-AS1-WT and FOXD2-AS1-Mut were cloned into the pGL3-basic
luciferase reporter vector (Promega Corp.). For the luciferase
reporter assay, 100 ng of either the FOXD2-AS1-WT vector or the
FOXD2-AS1-Mut vector, along with 100 nM miR-206 mimic and 20 ng of
a Renilla luciferase vector (Promega Corp.) as the control,
were transfected into cells by Lipofectamine 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). The relative luciferase activity
was determined via a Dual-Luciferase reporter gene assay system by
normalization to the Renilla luciferase activity at 48 h
after transfection. Transfections were carried out in duplicate and
repeated three times.
Western blot analysis
Cell lysis was carried out in RIPA buffer
supplemented with a protease inhibitor (Roche Diagnostics,
Indianapolis, IN, USA) and heated at 95°C for 5 min. The protein
concentration was quantified with a BCA Protein Assay kit (Qiagen,
Valencia, CA, USA), according to the manufacturer's protocol.
Lysates (40 µg protein) were purified by 10% SDS-PAGE gels and
transferred to nitrocellulose membranes over a subsequent 2-h time
period. Membranes were blocked in 5% skim milk in 1X TBST for 2 h
at room temperature and incubated with ANXA2 (1:1,000 dilution;
cat. no. 8235; Cell Signaling Technology, Inc.) and GAPDH (1:2,000
dilution; cat. no. 5174; Cell Signaling Technology, Inc.) primary
antibodies at 4°C overnight. Next, the membranes were rinsed three
times with 1% TBST, incubated with secondary antibodies (goat
anti-rabbit IgG-HRP;0 1:10,000 dilution; cat. no. ab6721; Abcam,
Cambridge, MA, USA) at room temperature for 1 h and detected with
enhanced chemiluminescence (ECL) reagents (Pierce; Thermo Fisher
Scientific, Inc.). In addition, densitometry was employed to
quantify the intensity of the protein bands (Image Lab software
4.1; Bio-Rad Laboatories, Inc., Hercules, CA, USA) and normalize
them to their respective GAPDH bands.
Xenograft tumor model
Twelve 4- to 6-week-old female BALB/c nude mice were
obtained from Shanghai SLAC Laboratory Animal Co., Ltd. and
maintained in a sterile pathogen-free environment. The specific
housing conditions were as follows: Temperature, 21±2°C; humidity,
30–70%; 12-h light/dark cycle; the ingested food and water were
sterile feed and sterilized bottled water. Animals were fed ad
libitum mouse chow and given free access to water. All animal
experiments were performed with stringent adherence to the Guide
for the Care and Use of Laboratory Animals and were approved by the
Laboratory Animal Care Committee of the Xi'an Jiaotong University
(no. XJTULAC2018-462). We used 6 mice per group. Control (LV-CON)
and FOXD2-AS1-overexpressing (LV-FOXD2-AS1) HCC cells were
trypsinized and harvested in serum-free DMEM; 0.1 ml serum-free
DMEM containing 3×106 cells was then administered by
subcutaneous injection into the right flank of the mice. Tumor
growth was evaluated by determining tumor diameters with a digital
caliper, whereas the tumor volume was determined using the
following formula: tumor volume = ab2/2, where
a is the larger and b is the smaller of the two
dimensions. According to the Institutional Animal Care and Use
Committee guidelines, the maximum allowable tumor size for mice is
4.2 cm3. The mice were sacrificed via CO2
inhalation 24 days after injection, and the tumors were harvested
by excision for further analysis.
Statistical analysis
Differences between the groups were determined by
one way ANOVA followed by LSD post hoc test, the Wilcoxon
signed-rank test, or Pearson's χ2 test as appropriate.
Differences were regarded as statistically significant for P-values
<0.05. Pearson correlation analysis was used to examine the
association between FOXD2-AS1 expression and miR-206. Overall
survival (OS) curves were estimated using the Kaplan-Meier method
and the log-rank test. Cox proportional hazard models were used,
and P-values were determined by SPSS 20.0 software (IBM Corp.,
Armonk, NY, USA).
Results
FOXD2-AS1 expression is increased in
HCC and is linked to inferior prognosis
To assess the involvement of FOXD2-AS1 in
oncogenesis, we initially examined FOXD2-AS1 expression levels in
360 HCC tissue specimens in TCGA. To determine the correlation
between FOXD2-AS1 expression and clinicopathological
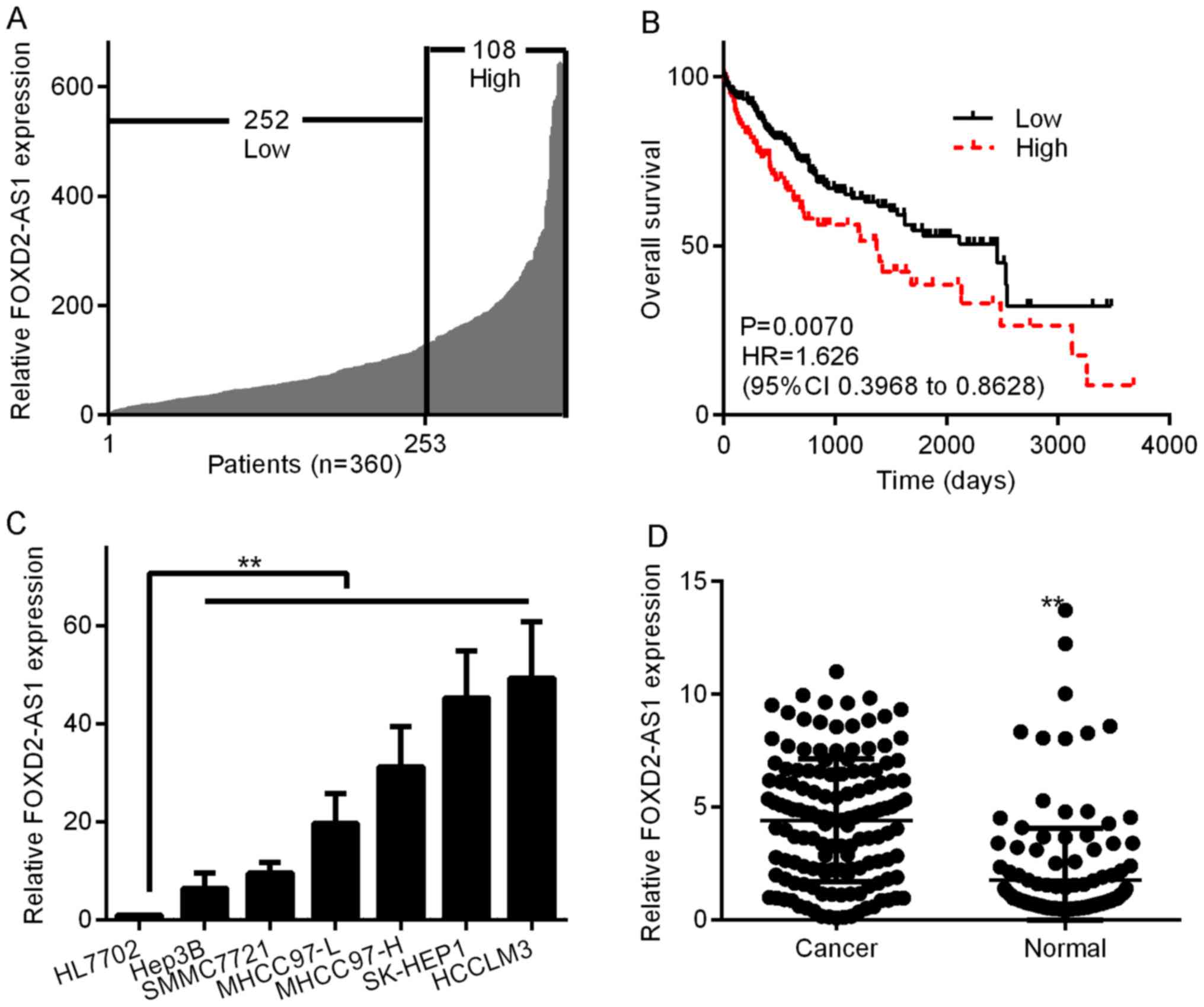
characteristics, the FOXD2-AS1 expression levels were classified as
low (n=252) or high (n=108) according to the Youden index (Fig. 1A). Then, we analyzed the OS curves
to determine whether the FOXD2-AS1 expression level was related to
prognosis. As shown in Fig. 1B,
high FOXD2-AS1 expression in HCC tissues was linked to shorter OS
(P=0.0070, log-rank test). Additionally, we determined the levels
of FOXD2-AS1 in six HCC cell lines (Hep3B, MHCC97-L, MHCC97-H,
SK-HEP1 and HCCLM3); FOXD2-AS1 expression was considerably higher
in all six HCC cell lines than in the normal human liver cell line
HL7702 (Fig. 1C). Of the six HCC
cell lines, SK-HEP1 and HCCLM3 cells had the highest FOXD2-AS1
expression, while Hep3B exhibited the lowest FOXD2-AS1 expression.
To explore whether FOXD2-AS1 is detectable in HCC, we studied its
expression in 140 HCC tissues and neighboring healthy hepatic
tissues. As shown in Fig. 1D,
greater expression of FOXD2-AS1 was observed in tumor tissues than
in the neighboring healthy tissues (P<0.01). These results
indicated that FOXD2-AS1 likely plays a key role in tumor
progression in human HCC.
FOXD2-AS1 enhances the viability and
migration of HCC cells
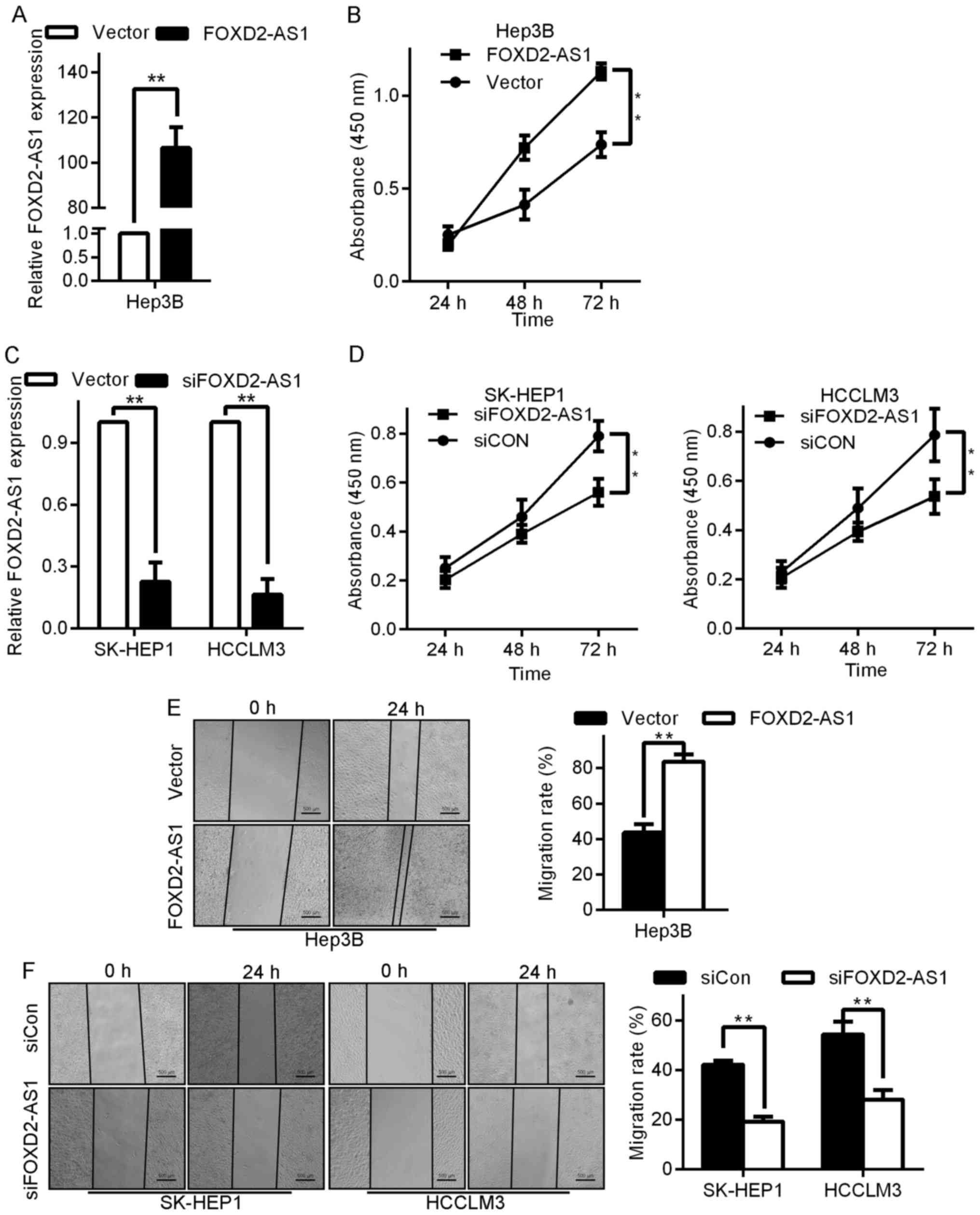
To explore the involvement of FOXD2-AS1 in HCC
pathogenesis, Hep3B cells were selected for FOXD2-AS1
overexpression, while SK-HEP1 and HCCLM3 were selected for
FOXD2-AS1 silencing. The expression levels of FOXD2-AS1 were
significantly higher in FOXD2-AS1-transfected cells than in their
vector-transfected counterparts (Fig.
2A), whereas FOXD2-AS1 expression was lower in
siFOXD2-AS1-transfected cells (Fig.
2C). Subsequently, a cell counting assay was conducted to
examine the influence of FOXD2-AS1 on cancer cell viability. The
MTT assay revealed that compared with that of the control cells,
the growth of the FOXD2-AS1-overexpressing Hep3B cells was notably
enhanced (Fig. 2B), whereas the
downregulation of FOXD2-AS1 by siRNA effectively inhibited the
viability of SK-HEP1 and HCCLM3 cells (Fig. 2D).
Next, to determine whether FOXD2-AS1 alters cell
motility, we studied cell migration using a wound healing assay and
Transwell chamber assay after the transfection of FOXD2-AS1 or
siFOXD2-AS1 into the indicated cells. The ectopic expression of
FOXD2-AS1 markedly increased the migration of Hep3B cells (Fig. 2G, left upper image). To quantify
this effect, we calculated the number of cells that migrated to the
underside of the Transwell chambers following transfection with
either the FOXD2-AS1-expressing vector or the control vector. The
migration of FOXD2-AS1-transfected cells was significantly higher
than that of the control vector-transfected cells (P<0.05;
Fig. 2G, left lower image), whereas
the migration of siFOXD2-AS1-transfected cells was considerably
lower than that of siRNA control (siCon)-transfected cells
(Fig. 2G, right images). Similarly,
in the wound healing assay, FOXD2-AS1 promoted the migration of
Hep3B cells while siFOXD2-AS1 decreased the migration of SK-HEP1
and HCCLM3 cells (Figs. 2E and
2F). Overall, these results
indicated that the overexpression of FOXD2-AS1 enhanced biological
behaviors related to tumor progression in HCC cells in
vitro.
FOXD2-AS1 functions as a molecular
sponge of miR-206
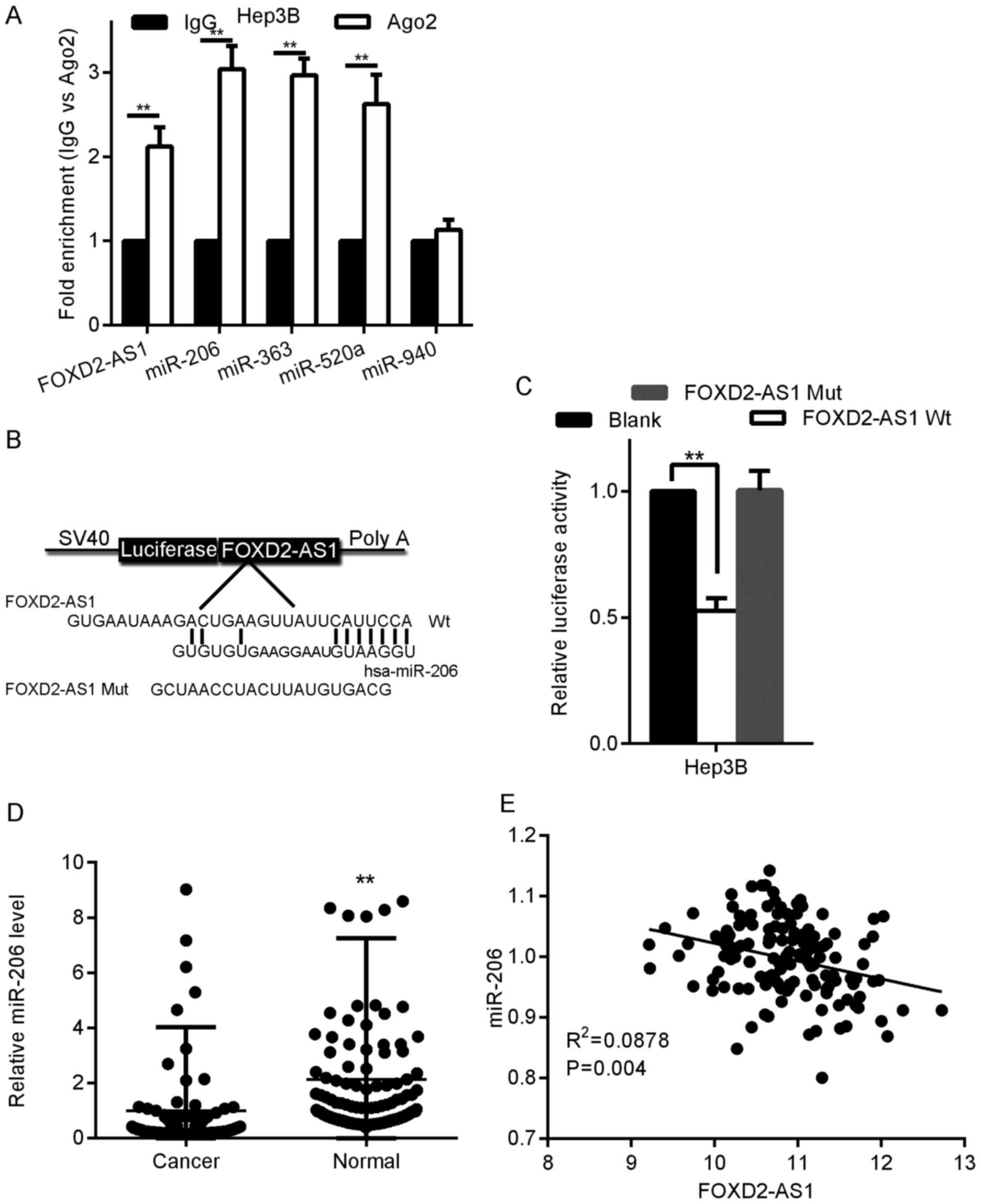
Recently, FOXD2-AS1 has been reported to play a role
in cancer development by acting as a ceRNA for miRNAs (13). To determine whether FOXD2-AS1 has an
analogous role in HCC, we predicted miRNA binding sites using DIANA
software, which is a miRNA target identification tool. We
identified 10 typical miRNAs and their corresponding target
sequences in FOXD2-AS1. The miRNA expression levels were determined
in pcDNA-FOXD2-AS1-transfected Hep3B cells via qRT-PCR. Compared
with their corresponding expression levels in pcDNA-NC-transfected
cells, the miR-363, miR-520a, miR-940, and miR-206 expression
levels displayed a considerable decrease in
pcDNA-FOXD2-AS1-transfected Hep3B cells (data not shown).
miRNA acts by tethering to Ago2, an essential part
of the RNA-induced silencing complex (RISC) that is vital to gene
silencing. Furthermore, miRNA targets can be separated from the
RISC by Ago2 coimmunoprecipitation (14). To determine whether FOXD2-AS1
associates with the RISC, we identified miR-363, miR-520a, miR-940,
miR-206, and FOXD2-AS1 in the Ago2 pellet. As revealed in Fig. 3A, miR-520a, miR-363 and FOXD2-AS1
expression was increased ~3-fold, whereas miR-206 expression was
increased by >3-fold. Therefore, FOXD2-AS1 may be associated
with the dysregulation of miR-206. For further confirmation, we
generated luciferase reporter vectors expressing FOXD2-AS1 with
either WT (FOXD2-AS1-WT) or mutant (FOXD2-AS1-Mut) miR-206 target
sites (Fig. 3B). We established
that the corresponding FOXD2-AS1-Mut construct did not suppress
miR-206 expression (Fig. 3C), thus
indicating that miR-206 is a FOXD2-AS1-specific miRNA.
Subsequently, we quantified the expression levels of miR-206 in 120
HCC tissues from the same set of patients described in Fig. 1D; miR-206 expression was notably
lower in the HCC tissues than in the paired neighboring healthy
liver tissues (Fig. 3D), and the
miR-206 level was significantly negatively correlated with the
FOXD2-AS1 level (Fig. 3E).
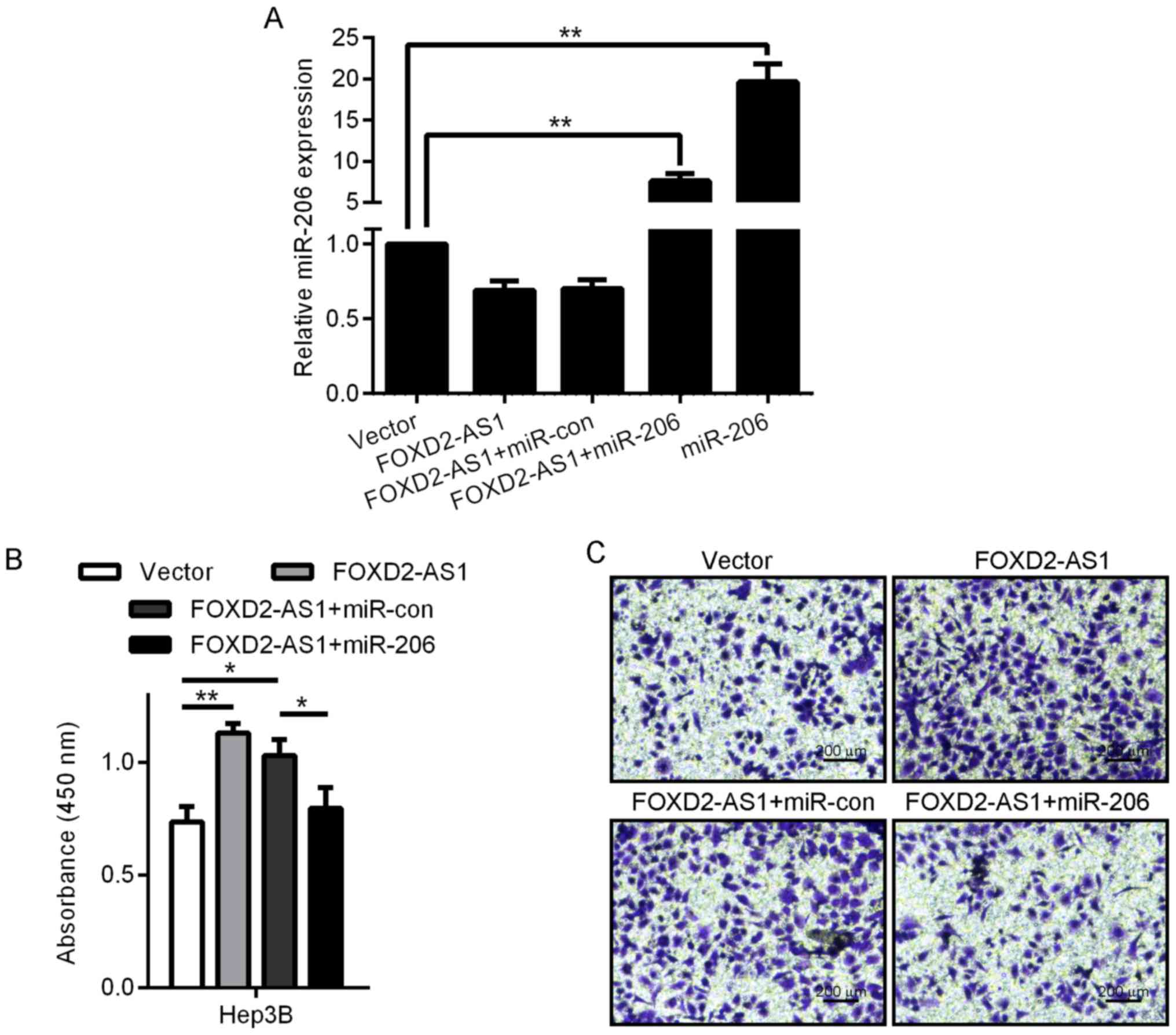
miR-206 reverses the growth-promoting effect of
FOXD2-AS1 on HCC cells. To study the significance of miR-206
binding in the FOXD2-AS1-related promotion of HCC progression, we
ectopically expressed miR-206 in stable FOXD2-AS1-overexpressing
Hep3B cells and analyzed cell viability via the MTT assay. The
expression of miR-206 was detected by real time PCR (Fig. 4A). The overexpression of miR-206
diminished the viability-promoting activity of FOXD2-AS1 (Fig. 4B); this effect was also observed in
Hep3B (Fig. 4C) cells in the
Transwell assay. These results revealed that FOXD2-AS1 promoted
tumor progression partly via competitive binding to miR-206.
FOXD2-AS1 regulates the expression of
the endogenous miR-206 target ANXA2
miR-206 operates as a tumor suppressor in humans via
the downregulation of ANXA2 (15).
To establish whether FOXD2-AS1 regulates HCC progression by
influencing miR-206 targets, we examined the impact of FOXD2-AS1 on
ANXA2 expression. After transfection with the FOXD2-AS1 expression
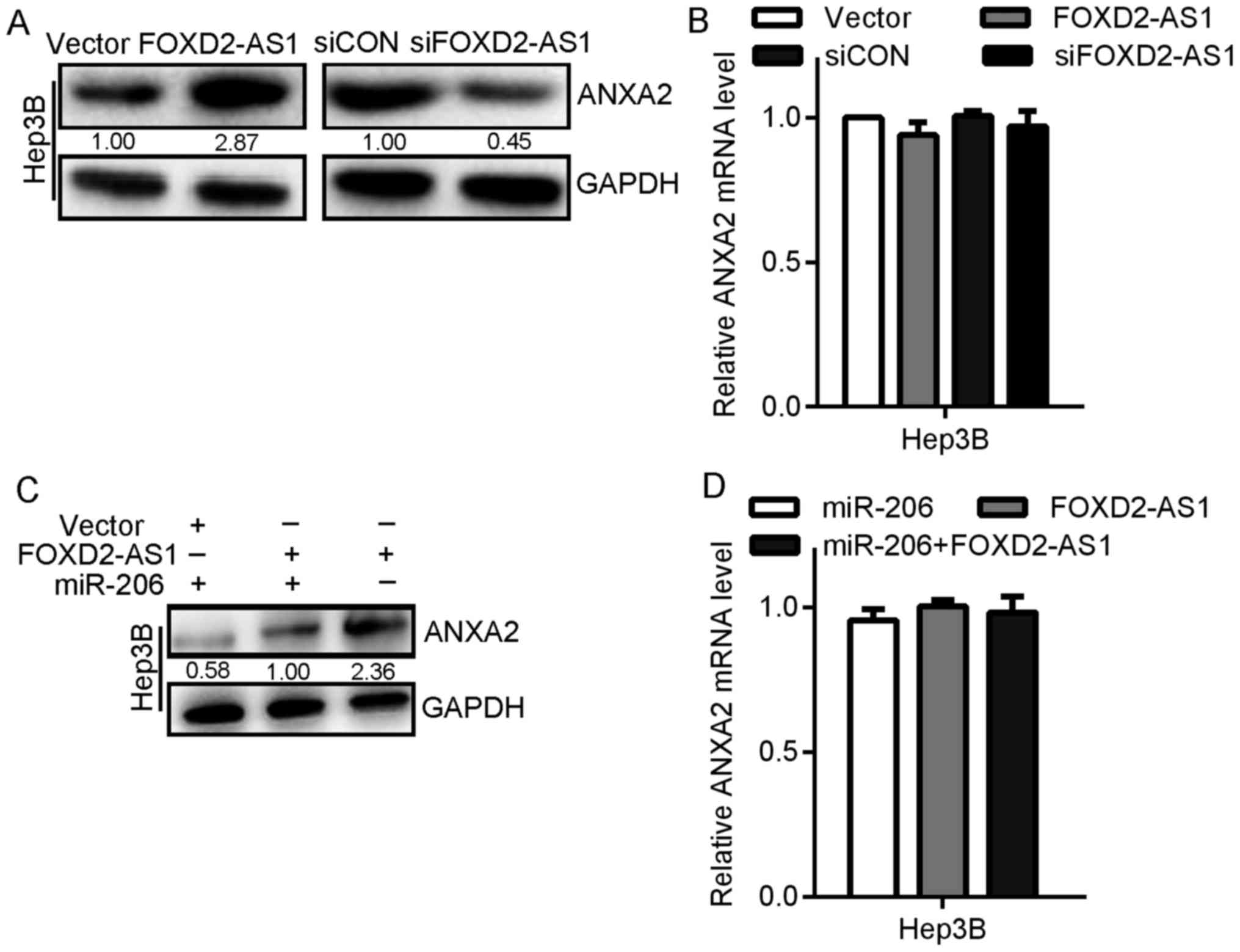
vector, the ANXA2 expression in Hep3B cells noticeably increased at
the protein level but not at the mRNA level (Fig. 5A and B). Additionally, transfection
with siFOXD2-AS1 clearly reduced ANXA2 expression in Hep3B cells at
the protein level but not at the mRNA level (Fig. 5A and B). Furthermore, similar to the
overexpression of miR-206 and FOXD2-AS1, the overexpression of
ANXA2 was detected in the Hep3B cell lines by western blotting but
not by the measurement of the mRNA levels (Fig. 5C and D). Therefore, these results
indicated that FOXD2-AS1 eliminated the suppression of ANXA2
induced by miR-206 and induced an oncogenic effect by modulating
miR-206/ANXA2.
FOXD2-AS1 upregulation promotes HCC
cell growth in vivo
FOXD2-AS1 promoted the viability and migration of
HCC cells in vitro. To elucidate the impact of FOXD2-AS1 on
tumor pathophysiology in vivo, HCC cells overexpressing
FOXD2-AS1 or the appropriate control cells were injected into
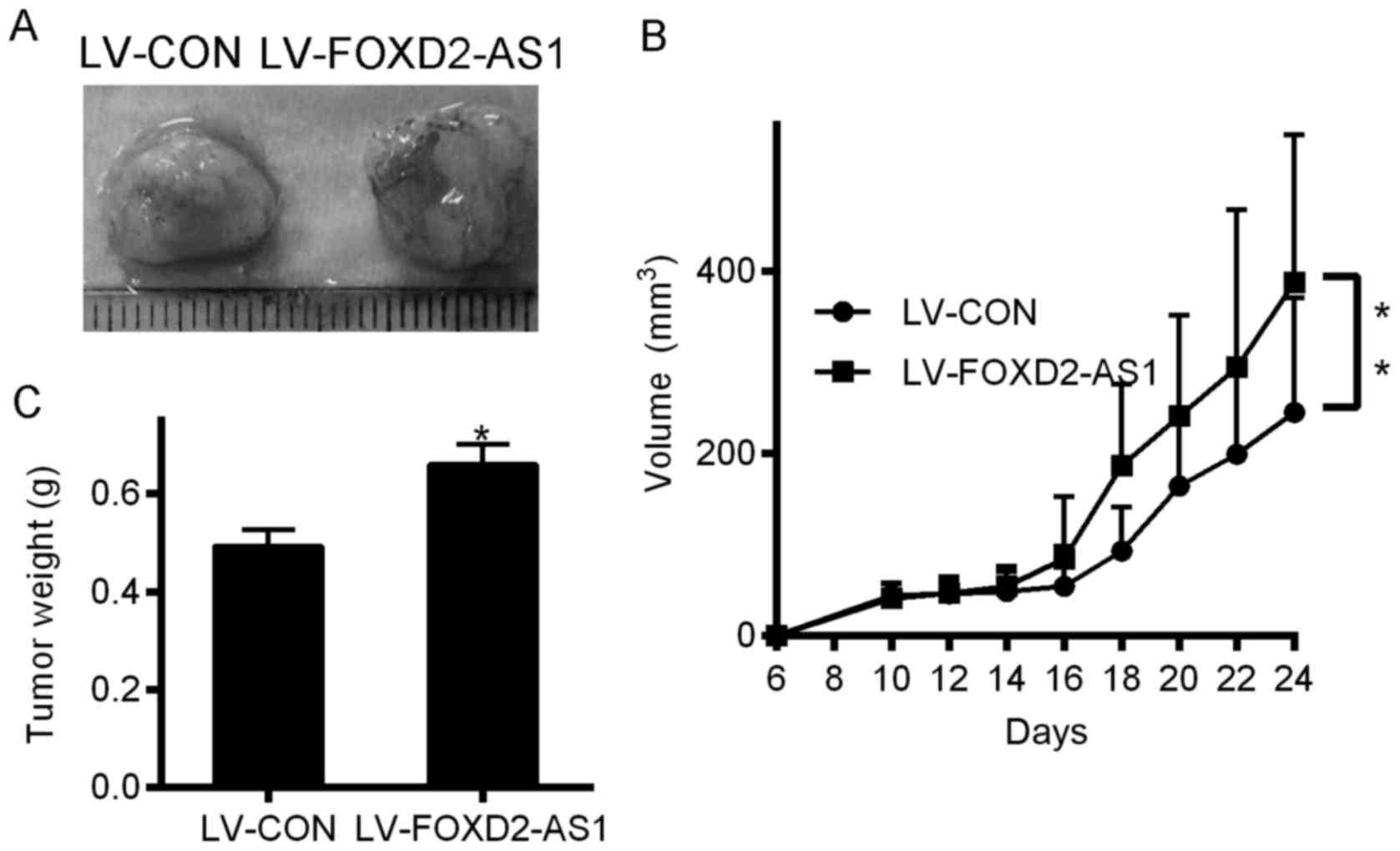
BALB/c nude mice. As displayed in Fig.
6A and B, after 24 days, the tumor growth in the FOXD2-AS1
×enograft group was considerably greater than that in the LV-CON
xenograft group; the mean tumor volumes in the two groups were
387.45±161.41 mm3 and 245.34 ± 125.64 mm3,
respectively (P<0.01). After the 24 days, the mean tumor weights
in the FOXD2-AS1 group were considerably higher than those in the
LV-CON group (Fig. 6C).
Discussion
High-throughput RNA sequencing has revealed that
approximately 70–90% of human DNA is transcribed into RNA, but over
68% of the transcripts have been classified as ncRNAs (16,17).
Tens of thousands of lncRNAs, which are functionally defined as
transcripts having a length of >200 nucleotides and no
protein-coding capacity, exist; many of these are uniquely
expressed in differentiated tissues or specific cancers (16). lncRNAs can exert either a negative
or positive effect on gene expression via several pathways
(18). Currently, the vital role of
lncRNAs in the modulation of many pathophysiological processes,
such as proliferation, development, cell cycle and apoptosis, has
been recognized. In addition, lncRNAs play a crucial role in human
tumors by functioning as either tumor suppressors or oncogenes
(19). Hence, the elucidation of
the principal lncRNA-related mechanisms underlying HCC generation
and development is critical.
Significantly diverse lncRNA profiles may act as
phenotypic signatures for the prognosis and treatment of multiple
cancers. GPC3-AS1 is overexpressed in HCC due to a probable
enhancement in histone acetylation at the promoter site; GPC3-AS1
overexpression results in the increased transcription of GPC3 via
PCAF and thus an enhanced level of cellular proliferation and
metastasis (20). SNHG12 acts as a
miR-199a/b-5p sponge; SNHG12 is upregulated in HCC, thereby
stimulating growth and hindering apoptosis. Thus, the inhibitory
effect of miR-199a/b-5p on MLK3 will be decreased, in turn
upregulating MLK3 and its targets in the NF-κB signaling pathway
(21). Via bioinformatics analysis
combined with in vitro and in vivo functional
experiments, FOXD2-AS1 was found to contribute to the progression
of colorectal cancer by activating EMT and the Notch signaling
pathway (22). In addition, lncRNA
FOXD2-AS1 is considerably upregulated in non-small cell lung cancer
(NSCLC). Loss- and gain-of-function assays revealed that FOXD2-AS1
enhanced NSCLC cell growth and tumor progression; furthermore,
FOXD2-AS1 modulated Wnt/β-catenin signaling in NSCLC cells
(8). In the present study, we
demonstrated that FOXD2-AS1 upregulation yielded an inferior
clinical outcome in HCC patients; moreover, FOXD2-AS1 could be a
potential biomarker for prognosis. Collectively, these results
implied that clinical research on lncRNAs needs to be conducted and
that additional investigations should be planned to identify
additional lncRNAs as key prognostic molecular biomarkers and vital
drug targets for HCC.
lncRNAs that act as ceRNAs or natural miRNA sponges
are major post-transcriptional modulators of gene expression that
compete with each other for binding to the same miRNAs (23,24).
For instance, lncRNA Unigene56159 enhanced metastasis by operating
as a ceRNA for miR-140-5p in HCC (25). lncRNA HOTAIR acted as a ceRNA for
miR-331-3p, thus regulating HER2 derepression and promoting tumor
progression in gastric carcinoma (26). In the present study, we identified
FOXD2-AS1 as an oncogenic player and illustrated a formerly unknown
mechanism linking FOXD2-AS1 with miRNAs in HCC biology. By in
vitro gain-and loss-of-function studies, FOXD2-AS1 was
established to be associated with the miRNA-related regulatory
network of HCC cell proliferation. Our observation that FOXD2-AS1
promoted cancer cell viability revealed a number of key aspects;
for example, FOXD2-AS1 expression was extensively upregulated in
HCC cells, thus indicating its prospective utility in HCC therapy.
Accordingly, the overexpression of FOXD2-AS1 markedly promoted cell
viability and migration in vitro. Moreover, the knockdown of
FOXD2-AS1 negatively regulated cell migration. Subsequent
mechanistic studies demonstrated that FOXD2-AS1 eliminated the
inherent inhibitory activity of miR-206 on ANXA2; furthermore, the
impact of FOXD2-AS1 on viability and migration was abrogated via
the transfection of a miR-206 mimic. Further experiments confirmed
that by competing for binding to miR-206, the ANXA2 3′-UTR acted as
a ceRNA at the mRNA level in Hep3B cells, thus affecting the
expression of ANXA2 at the protein level (Fig. 5). Therefore, targeting lncRNA-based
signaling pathways may be a novel therapeutic strategy. However,
the roles of lncRNAs in HCC carcinogenesis have not been thoroughly
explored. A thorough investigation of the molecular mechanism
underlying the initiation and progression of HCC is essential for
facilitating the exploitation of novel therapeutic targets.
To this end, a recent study established that
FOXD2-AS1 was linked to miR-363-5p and regulated the expression of
the miR-363-5p target S100A, thus revealing that FOXD2-AS1
positively modulated post-transcriptional gene expression (13). The exploration of whether FOXD2-AS1
may perform its role in human tumors through interaction with
miRNAs in addition to miR-363-5p would be of great interest.
In conclusion, this study indicated that FOXD2-AS1
expression could effectively predict the prognosis of HCC patients;
however, this result requires confirmation in future investigations
with a larger sample size. We employed mechanistic analysis to
reveal the contribution of FOXD2-AS1 to the promotion of HCC
progression by its function as a miR-206 sponge, and we illustrated
a novel FOXD2-AS1/miR-206/ANXA2 signaling pathway regulatory
network in HCC. These results indicated that FOXD2-AS1 is a key
prognostic molecular biomarker as well as a vital drug target for
HCC and that FOXD2-AS1 could contribute to the established
crosstalk among conventional pathways.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific
and Technological Project Foundation of Xi'an City
[2017113SFYX007(6)] and by the Scientific and Fundamental Research
Funds for the Central Universities of Xi'an Jiaotong
University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YC, XL and LF conceived and designed the study. YC,
JZ, CZ, GQ and GW performed the experiments. YC, LF, SW and XC
analyzed the data. YC and XL wrote the paper. LF, SW and XC
reviewed and edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China). Prior to the the study, the patients
signed written informed consent forms. All animal experiments were
performed with stringent adherence to the Guide for the Care and
Use of Laboratory Animals and were approved by the Laboratory
Animal Care Committee of the Xi'an Jiaotong University (no.
XJTULAC2018-462).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morris KV and Mattick JS: The rise of
regulatory RNA. Nat Rev Genet. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li S, Ma F, Jiang K, Shan H, Shi M and
Chen B: Long non-coding RNA metastasis-associated lung
adenocarcinoma transcript 1 promotes lung adenocarcinoma by
directly interacting with specificity protein 1. Cancer Sci.
109:1346–1356. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai T, Liu Y and Xiao J: Long noncoding
RNA MALAT1 knockdown reverses chemoresistance to temozolomide via
promoting microRNA-101 in glioblastoma. Cancer Med. 7:1404–1415.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mondal T, Subhash S, Vaid R, Enroth S,
Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al:
MEG3 long noncoding RNA regulates the TGF-β pathway genes through
formation of RNA-DNA triplex structures. Nat Commun. 6:77432015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Benetatos L, Vartholomatos G and
Hatzimichael E: MEG3 imprinted gene contribution in tumorigenesis.
Int J Cancer. 129:773–779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rong L, Zhao R and Lu J: Highly expressed
long non-coding RNA FOXD2-AS1 promotes non-small cell lung cancer
progression via Wnt/β-catenin signaling. Biochem Biophys Res
Commun. 484:586–591. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett. 589
B:1–3181. 2015.
|
|
11
|
Chen Z, Xu D and Zhang T: Inhibition of
proliferation and invasion of hepatocellular carcinoma cells by
lncRNA-ASLNC02525 silencing and the mechanism. Int J Oncol.
51:851–858. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen G, Sun W, Hua X, Zeng W and Yang L:
Long non-coding RNA FOXD2-AS1 aggravates nasopharyngeal carcinoma
carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene.
645:76–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Karginov FV, Conaco C, Xuan Z, Schmidt BH,
Parker JS, Mandel G and Hannon GJ: A biochemical approach to
identifying microRNA targets. Proc Natl Acad Sci USA.
104:19291–19296. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Keklikoglou I, Hosaka K, Bender C, Bott A,
Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H,
et al: MicroRNA-206 functions as a pleiotropic modulator of cell
proliferation, invasion and lymphangiogenesis in pancreatic
adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene.
34:4867–4878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
ENCODE Project Consortium: An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu XT, Yuan JH, Zhu TT, Li YY and Cheng
XY: Long noncoding RNA glypican 3 (GPC3) antisense transcript 1
promotes hepatocellular carcinoma progression via epigenetically
activating GPC3. FEBS J. 283:3739–3754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang X, Duan B and Zhou X: Long non-coding
RNA FOXD2-AS1 functions as a tumor promoter in colorectal cancer by
regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol
Sci. 21:3586–3591. 2017.PubMed/NCBI
|
|
23
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang
Y, Liu M and Tang H: Long non-coding RNA Unigene56159 promotes
epithelial-mesenchymal transition by acting as a ceRNA of
miR-140-5p in hepatocellular carcinoma cells. Cancer Lett.
382:166–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|