Introduction
Myeloid-derived suppressor cells (MDSCs) form a
heterogenous population that consists of myeloid progenitor cells,
immature macrophages, immature granulocytes and immature dendritic
cells (DCs). The phenotype of MDSCs in mice is granulocyte receptor
1 (Gr-1)+CD11b+. In the steady state, MDSCs
have no inhibitory activity and are mostly present in the bone
marrow. The percentage of MDSCs is only 2–4% in the spleen, and
they are absent from the lymph nodes in mice. In tumor hosts, MDSCs
accumulate in lymphatic organs and tumor tissue, and they produce a
large amount of immune-inhibitory factors, including arginase I,
inducible nitric oxide synthase and reactive oxygen species, which
inhibit T-cell immune responses (1–3),
induce regulatory T-cell production, suppress natural killer (NK)
cell functions, and affect cytokine production and secretion by
macrophages. Furthermore, MDSCs can facilitate
epithelial-mesenchymal transition, angiogenesis and metastasis
(2,4–6).
The spleen is the largest immune organ, containing
nearly 25% of the body's immune cells. It has a vital role in the
immune response to tumors, and splenic NK cells and T cells exert
important anti-tumor functions (7–10).
However, the spleen is also a reservoir of precursor monocytes and
granulocytes, which may be mobilized to the tumor tissue and
differentiate into tumor-associated macrophages (TAM) and
neutrophils (TAN) that promote tumor progression. Splenectomy has
been indicated to markedly reduce TAM and TAN responses, thus
retarding tumor growth (11). A
previous study by our group identified an immune-suppressive status
in the spleen of a H22 orthotopic hepatoma mouse model. MDSCs were
the major inhibitory cells causing the immunosuppression in the
spleen (12). Similarly, Levy et
al (13) reported that the
spleen may be a reservoir of MDSCs and that splenectomy may reduce
the percentage of MDSCs in peripheral blood and tumor tissue.
To date, numerous studies have reported on the
mechanisms of MDSC accumulation in tumor tissue, and the factors
involved include chemokines, inflammatory factors,
colony-stimulating factor and complements (14–16).
Tumor-derived chemokines, including C-C motif chemokine ligand
(CCL)2, CCL5, CCL21 and C-X-C motif ligand (CXCL)8 were able to
recruit MDSCs to the tumor tissue, while blocking of these
chemokines or their receptors reduced the number of MDSCs at the
tumor sites and inhibited tumor growth (17–23).
The accumulation of MDSCs in the tumor may also be regulated by
inflammatory environmental factors, including interleukin (IL)-1β,
IL-6, prostaglandin E2, S100 calcium-binding protein A8/9 and tumor
necrosis factor (24–28).
By contrast, few studies have assessed MDSC
accumulation in the spleen under pathological conditions. In the
murine MCA203 fibrosarcoma model, nestin-positive splenocytes were
able to secrete CCL2 that attracted MDSCs to the spleen in a
CCR2-dependent manner (29). The
spleen is the primary site of immune responses, and its immune
status influences tumor growth. Therefore, it is important to
explore the mechanisms of MDSC localization and kinetics in the
spleen under pathological conditions. This may facilitate the
identification of novel factors and pathways involved in MDSC
accumulation in the spleen that may have a role in the contribution
of MDSCs to tumor progression. These factors and pathways may be
novel targets in immune therapy to block MDSCs in order to suppress
tumor growth.
In the present study, the mechanisms of MDSC
accumulation in the spleen of tumor-bearing (TB) mice were
explored. MDSC proliferation, apoptosis and the expression of
cytokines in the spleen were assessed. Chemokines upregulated in TB
mice were validated and their cellular origin was identified.
Materials and methods
Cell culture
The H22 murine hepatoma cell line was purchased from
the China Center for Type Culture Collection (Wuhan, China). The
cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare,
Little Chalfont, UK) supplemented with 10% fetal calf serum
(HyClone; GE Healthcare) and 1% penicillin/streptomycin (HyClone;
GE Healthcare). The cells were grown at 37°C in an atmosphere
containing 5% CO2.
Mice
A total of 96 female BALB/c mice (age, 6–8 weeks;
weight, 20–25 g) were purchased from the animal center of Xi'an
Jiaotong University (Xi'an, China). The mice were housed under
specific pathogen-free conditions at the animal facility under a
12-h light/dark cycle and free access to food and water. All animal
procedures complied with the Guide for the Care and Use of
Laboratory Animals [National Institutes of Health (NIH);
publication from 1996] and were approved by the Xi'an Jiaotong
University Animal Care and Use Committee (Xi'an, China).
Orthotopic mouse model of
hepatocellular carcinoma
Mice were intraperitoneally injected with
106 H22 cells (at the concentration of 107
cells/ml in saline). After 12 days, ascites fluid containing
floating tumor cells was collected from these mice, which was
centrifuged at 300 × g for 5 min at 4°C to retrieve the cells,
which were washed twice with saline. These cells were subsequently
injected under the capsule of the left liver lobe of each mouse (2
×105 tumor cells at the concentration of 107
cells/ml). Four days later, white tumor nodules had formed on the
liver, indicating the successful generation of the H22 hepatoma
model.
Generation of single-cell suspensions
of splenocytes
Mice were sacrificed and the spleens were collected
at one, two and three weeks after tumor inoculation. Spleens were
dissociated into single-cell suspensions with the gentleMACS
Dissociator (Miltenyi Biotech, Bergisch Gladbach, Germany) in the
buffer [0.01 mol/l PBS, 0.5% bovine serum albumin (Amresco, Solon,
OH, USA) and 2 mmol/l EDTA], using the C tube and the ‘m_spleen_01’
program according to the manufacturer's protocols. Subsequently,
the cell suspensions were centrifuged at 300 × g for 30 sec at room
temperature. Next, the splenocytes suspensions were strained
through a 70-µm mesh to remove clumps and generate single-cell
suspensions.
Flow cytometric analysis
The ammonium-chloride-potassium lysis buffer (0.15
mol/l NH4Cl, 1 mmol/l KHCO3 and 0.1 mmol/l
EDTA, pH 7.2) was added to the splenocytes to remove red blood
cells. Subsequently, the cells were washed twice with PBS. Cells
(106) were incubated with anti-CD16/CD32 (cat. no.
101302; 10 µg/ml; BioLegend, San Diego, CA, USA) for 10 min at 4°C
to prevent non-specific labeling of surface receptors. Next, cells
were incubated with monoclonal antibodies, including anti-mouse
granulocyte receptor 1 (Gr-1)-fluorescein isothiocyanate (FITC)
(cat. no. 108405), anti-mouse Ly6G-FITC (cat. no. 127605),
anti-mouse Ly6C-phycoerythrin (PE) (cat. no. 128007), anti-mouse
CD11b-allophycocyanine (APC)-(cyanine)Cy7 (cat. no. 101226),
anti-mouse C-C motif chemokine receptor 1 (CCR1)-APC (cat. no.
152503) (all from BioLegend; 2.5 µg/ml dilution), anti-mouse
CD11b-PE (cat. no. 12-0112; 0.6 µg/ml; eBioscience, San Diego, CA,
USA), anti-mouse Fas-APC (cat. no. 17-0951; 10 µg/ml; eBioscience)
and their corresponding isotype antibodies, including FITC rat
immunoglobulin (Ig)G2b, κ (cat. no. 400605); FITC rat IgG2a, κ
(cat. no. 400505); PE rat IgG2c, κ (cat. no. 400707); APC/Cy7 rat
IgG2b, κ (cat. no. 400623); APC rat IgG2b, κ (cat. no. 400611) (all
from BioLegend; 2.5 µg/ml dilution); PE rat IgG2b, κ (cat. no.
12-4031; 0.6 µg/ml); APC mouse IgG1, κ (cat. no. 17-4714; 10 µg/ml)
(both from eBioscience) as the control for 30 min at 4°C in the
dark. Following washing, the cells were analyzed using a FACSCanto
instrument with FACSDiva 7.0 software (BD Biosciences, Franklin
Lakes, NJ, USA).
To assess splenic MDSC proliferation, splenic cell
suspensions were stained with anti-mouse Gr-1-FITC and anti-mouse
CD11b-PE antibodies for 30 min at 4°C, followed by incubation with
cold 70% ethanol (precooled to −20°C; added in a drop-wise manner
to the cells while vortexing) at −20°C for 1 h. Subsequently, the
cells were washed three times, stained with anti-mouse Ki-67-APC
antibody (cat. no. 652405; 2.5 µg/ml; BioLegend) for 30 min at 4°C
in the dark, washed and analysed with a flow cytometer.
To assess splenic MDSC apoptosis, splenic cell
suspensions were stained with anti-mouse Gr-1-FITC and anti-mouse
CD11b-PE, followed by Annexin V-APC (cat. no. 640920; BioLegend)
and 7-aminoactinomycin D (7-AAD; cat. no. 420403; 2.5 µg/ml;
BioLegend). In brief, 106 cells were washed twice and
re-suspended in Annexin V binding buffer (cat. no. 422201;
BioLegend). Next, 5 µl Annexin V-APC and 5 µl 7-AAD were added to
the cells. Cells were incubated for 15 min at 4°C in the dark,
re-suspended in 400 µl binding buffer and analyzed using a
FACSCanto II flow cytometer (BD Biosciences) within 1 h.
Fluorescence-activated sorting of
splenic macrophages
To isolate splenic macrophages, single-cell
suspensions of splenocytes were first incubated with anti-mouse
CD16/32 antibody (cat. no. 101302; 10 µg/ml; BioLegend) for 10 min
at 4°C to prevent non-specific labeling of surface receptors,
followed by incubation with monoclonal antibodies, including
anti-mouse F4/80-PE and (cat. no. 123109; 10 µg/ml; BioLegend)
anti-mouse CD11b-APC-Cy7 (cat. no. 101226; 2.5 µg/ml; BioLegend),
for 30 min at 4°C in the dark. Cells were washed twice and
F4/80+CD11b+ double-positive cells were
isolated with a FACSAria II instrument (BD Biosciences). The purity
of the sorted macrophages was verified by flow cytometry (FACSAria
II; BD Biosciences).
Cytokine array
The concentrations of cytokines in the spleens of
normal and TB mice were quantified using the Mouse Cytokine Array
G3 (cat. no. AAM-CYT-G3-4; RayBiotech, Norcross, GA, USA) according
to the manufacturer's protocols. After all of the procedure steps,
the chip was scanned with an Axon GenePix scanner (GenePix 4000B;
Axon Instruments, Inc., San Jose, CA, USA). Protein array data were
annotated and processed with GenePix Pro 6.0 software (Molecular
Devices, LLC, Sunnyvale, CA, USA). The expression levels
(fluorescent signal intensities) of each cytokine were calculated
by subtracting the mean absorbance of the blank sample, with
normalization to a positive control.
ELISA
The levels of murine CCL9 and CXCL2 in the spleens
of normal and TB mice were measured using ELISA kits (cat. nos.
F111602 and F1117; Westang, Shanghai, China). Assays were performed
in duplicate according to the manufacturer's protocols.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For analysis of CCL9 gene expression, total RNA was
extracted from the isolated splenic macrophages using TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. Complementary (c)DNA
was synthesized using the PrimeScript™ RT reagent kit with a
genomic DNA eraser (cat. no. RR047A, Takara Bio, Inc., Otsu,
Japan). Real-time PCR was performed using SYBR® Premix
Ex Taq™ II (Tli RNaseH Plus) kit (cat. no. RR820A, Takara Bio,
Inc.) in a 20-µl reaction system, including 10 µl Premix Ex Taq II
(Takara Bio, Inc.), 0.4 µl 5-carboxy rhodamine X reference dye II
(Takara Bio, Inc.), 0.8 µl of 0.4 µM forward primer and 0.8 µl of
0.4 µM reverse primer, 2 µl cDNA (resembling the transcription
product of 50 ng RNA) and 6 µl distilled water. The following
primers were used: CCL9 FP, 5′-CCCTCTCCTTCCTCATTCTTACA-3′ and RP,
5′-AGTCTTGAAAGCCCATGTGAAA-3′; GAPDH FP, 5′-AGGTCGGTGTGAACGGATTTG-3′
and RP, 5′-TGTAGACCATGTAGTTGAGGTCA-3′. Amplifications of CCL9 and
GAPDH transcripts were performed during 40 PCR cycles using the ABI
7500 fast real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The initial denaturation step was at 95°C for 30
sec. Each PCR amplification cycle included a denaturation step at
95°C for 5 sec, and a primer annealing and elongation step at 60°C
for 30 sec. The expression levels were calculated as the relative
cDNA content normalized to GAPDH expression (2−ΔΔCq
method) (30). Three independent
replicates were performed.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Data were analyzed with Prism 5 software (GraphPad
Software, Inc., La Jolla, CA, USA). Student's t-test was used to
compare the difference between the two groups. For more than two
groups, one-way ANOVA followed by Dunnett's test was performed.
P<0.05 was considered to indicate a statistically significant
difference.
Results
H22 tumor induces accumulation of
MDSCs in the spleen
The murine H22 orthotopic HCC model was established
and it was determined whether the development of H22 tumors was
associated with an increased number of MDSCs in the spleen. The
percentage of splenic MDSCs increased significantly in TB mice
compared with that in normal mice at week 1 (6.82±2.92 vs.
3.44±0.60%, P<0.05), week 2 (43.20±11.03 vs. 4.90±0.90%,
P<0.001) and week 3 (13.28±4.66 vs. 3.42±0.48%, P<0.001)
post-tumor inoculation, with the greatest difference observed at
week 2 (Fig. 1). These results
suggest that a H22 tumor may induce the accumulation of
CD11b+Gr-1+ cells in the spleen in
vivo.
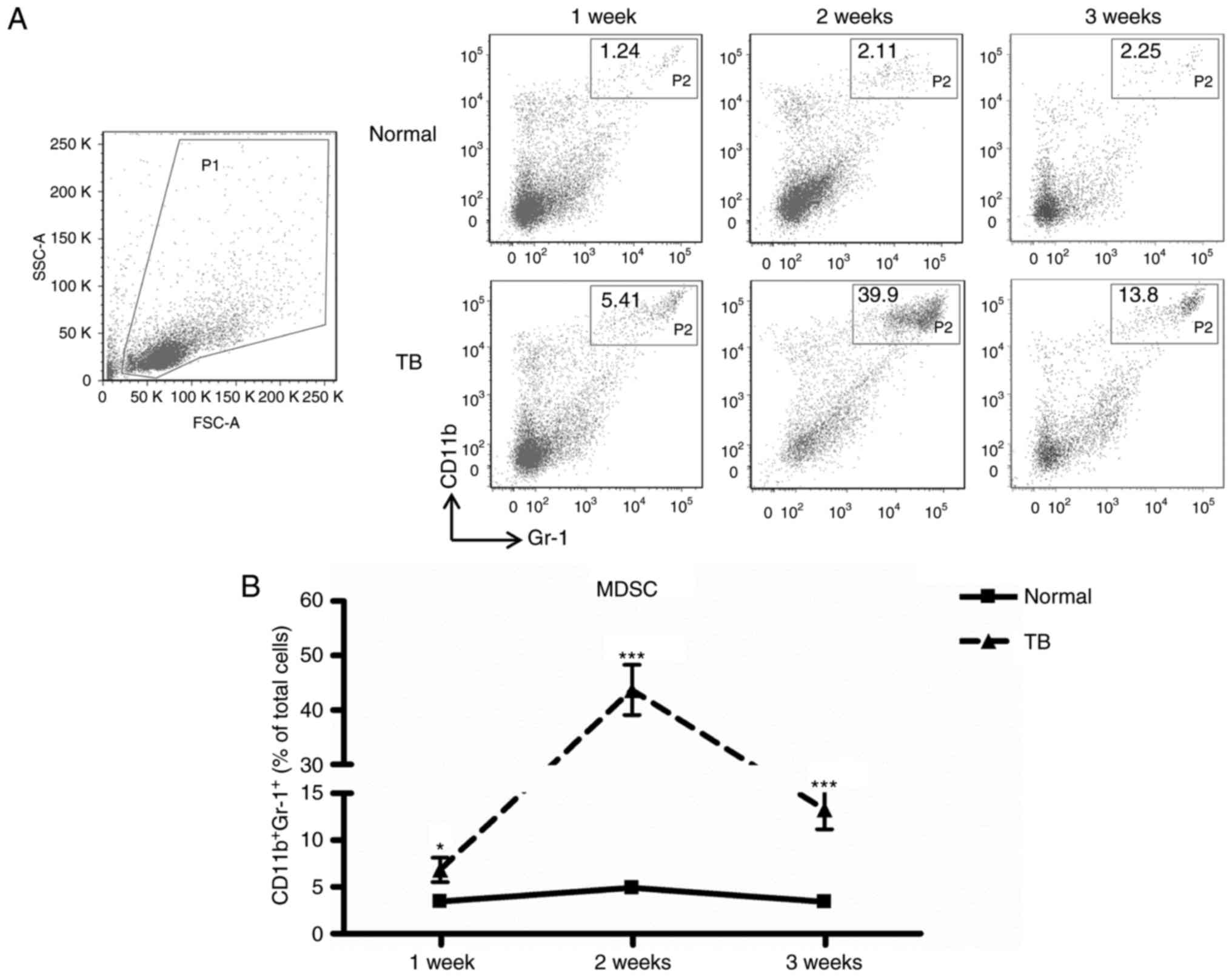 | Figure 1.MDSCs accumulate in the spleen during
tumor growth. Single-cell suspensions of splenocytes were prepared
and stained with anti-mouse-Gr-1 monoclonal antibody conjugated
with FITC and anti-mouse CD11b monoclonal antibody conjugated with
PE, and analyzed by flow cytometry. (A) Representative
fluorescence-assisted cell sorting plots of splenocyte
preparations. A live gate P1 was set in the FSC/SSC plot.
Subsequently, the populations of CD11b-PE and Gr-1-FITC cells were
displayed in dot plots. (B) The percentage of splenic MDSCs
(Gr-1+CD11b+, gate P2 within live gate) in
H22 hepatoma mice (▲) and normal mice (■) at weeks 1, 2 and 3
post-tumor inoculation (n=6); Values are expressed as the mean ±
standard deviation. *P<0.05 and ***P<0.001 compared with
normal mice. TB, tumor-bearing; MDSCs, myeloid-derived suppressor
cells; FITC, fluorescein isothiocyanate; PE, phycoerythrin; SSC,
side scatter; FSC, forward scatter; Gr-1, granulocyte receptor
1. |
Accumulation of MDSCs in the spleen is
not associated with their proliferation and apoptosis
It was investigated whether the accumulation of
splenic MDSCs in TB mice resulted from their increased
proliferation and/or reduced apoptosis. First the proliferation
status of splenic MDSCs was assessed by staining for Ki-67, a
marker of cell proliferation. The percentage of Ki-67+
splenic MDSCs was not significantly different between TB mice and
normal mice at any of the time-points. The mean fluorescent
intensity (MFI) of Ki-67 in splenic MDSCs was also not
significantly different between TB mice and normal mice (Fig. 2).
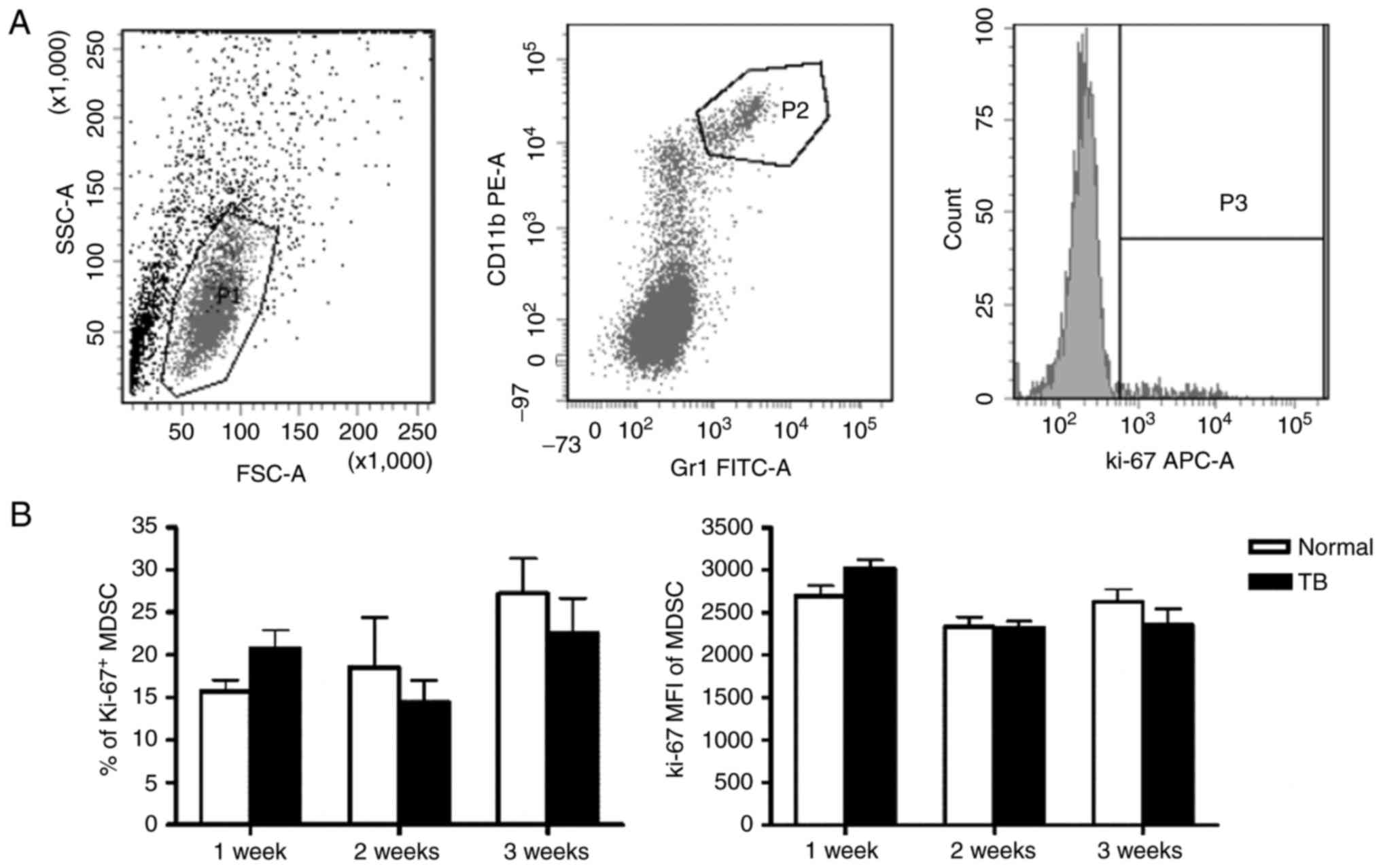 | Figure 2.Accumulation of splenic MDSCs in H22
hepatoma mice is not associated with their proliferative status.
Single-cell suspensions of splenocytes were stained with anti-mouse
Gr-1 monoclonal antibody conjugated with FITC and anti-mouse CD11b
monoclonal antibody conjugated with PE, followed by
permeabilization with 70% ethanol and staining with anti-mouse
Ki-67 antibody conjugated with APC. (A) Representative flow
cytometry plots and gating strategy. A live gate P1 was set in the
FSC/SSC plot and CD11b+/Gr-1+ were included
in the further analysis with gate P2. Subsequently, the selected
cells were displayed in a histogram for Ki-67-APC. (B) The
percentage of Ki-67+ MDSCs (left panel) and the MFI of
Ki-67 in MDSCs (right panel) in H22 hepatoma and normal mice (n=6).
Values are expressed as the mean ± standard deviation. TB,
tumor-bearing; APC, allophycocyanine; MDSCs, myeloid-derived
suppressor cells; FITC, fluorescein isothiocyanate; PE,
phycoerythrin; SSC, side scatter; FSC, forward scatter; Gr-1,
granulocyte receptor 1; MFI, mean fluorescent intensity. |
Next, the apoptosis of splenic MDSCs was assessed.
No difference in the amount of total (AnnexinV+)
apoptotic MDSCs was identified between TB and normal mice at weeks
1, 2 and 3 post-tumor inoculation (Fig.
3). Fas protein is an apoptotic protein expressed on the cell
surface, and therefore, Fas expression on the splenic MDSCs was
also assessed. Neither the percentage of Fas+ splenic
MDSCs nor the MFI of Fas in splenic MDSCs was different between TB
mice and normal mice at any of the time-points assessed (Fig. 4).
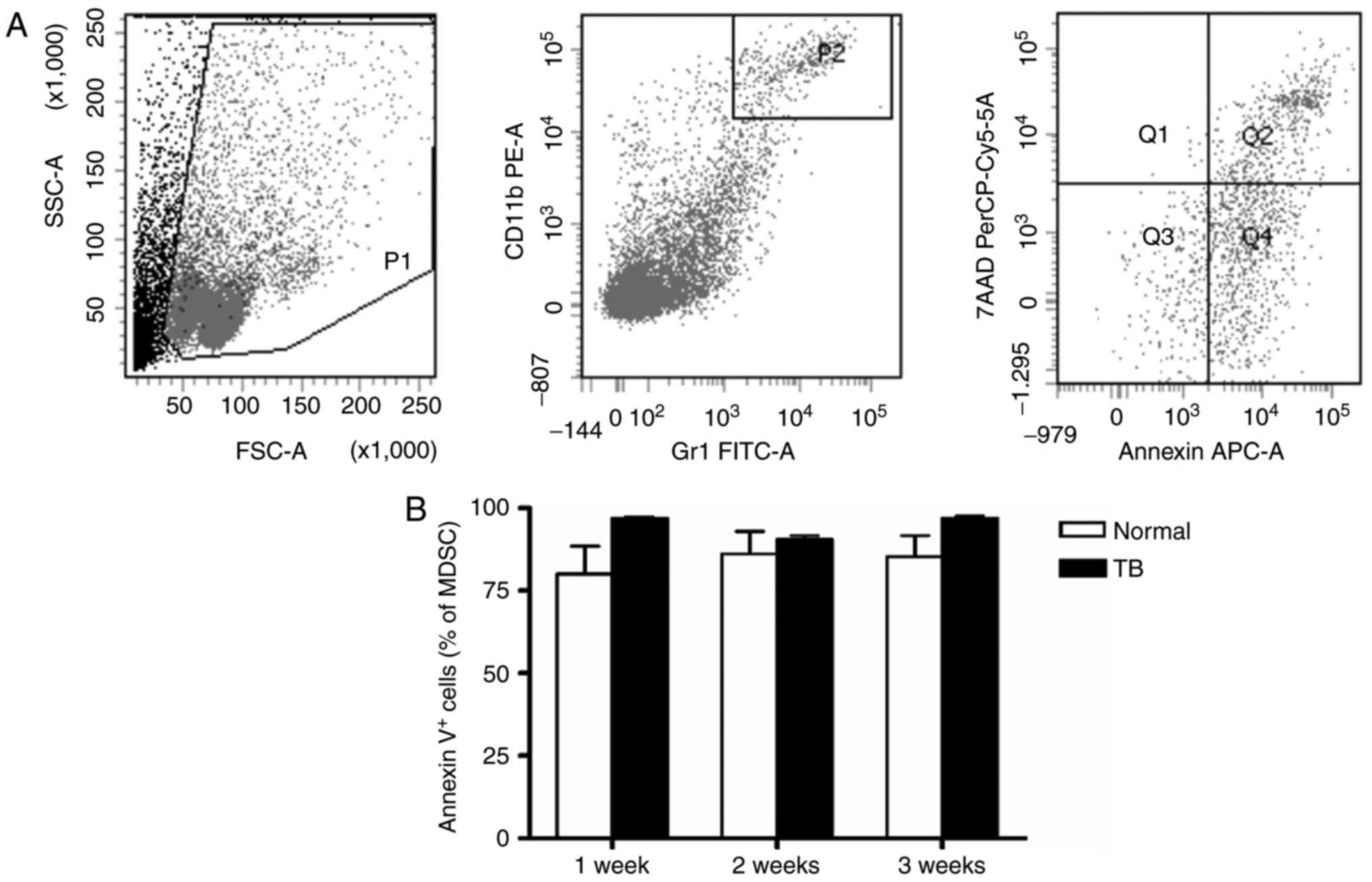 | Figure 3.Accumulation of splenic MDSCs in H22
hepatoma mice is not caused by alterations in their apoptotic rate.
Single-cell suspensions of splenocytes were stained with anti-mouse
Gr-1 monoclonal antibody conjugated with FITC and anti-mouse CD11b
monoclonal antibody conjugated with PE, followed by staining with
anti-mouse AnnexinV antibody conjugated with APC and viability dye
7-AAD. (A) Representative flow cytometry plots and gating strategy.
A live gate P1 was set in the FSC/SSC plot and
CD11b+/Gr-1+ cells were included in the
further analysis with gate P2. Subsequently, the selected cells
were displayed in histograms for Annexin-APC or 7-AAD. Annexin
V+/7-AAD− indicates cells in early apoptosis
and Annexin V+/7-AAD+ indicates cells in late
apoptosis. (B) Total apoptotic MDSC populations
(AnnexinV+) in TB and normal mice are presented. Values
are expressed as the mean ± standard deviation (n=6). TB,
tumor-bearing; APC, allophycocyanine; MDSCs, myeloid-derived
suppressor cells; FITC, fluorescein isothiocyanate; PE,
phycoerythrin; SSC, side scatter; FSC, forward scatter; Gr-1,
granulocyte receptor 1; 7-AAD, 7-amino actinomycin D; PerCP,
peridinin chlorphyll protein; Cy, cyanine. |
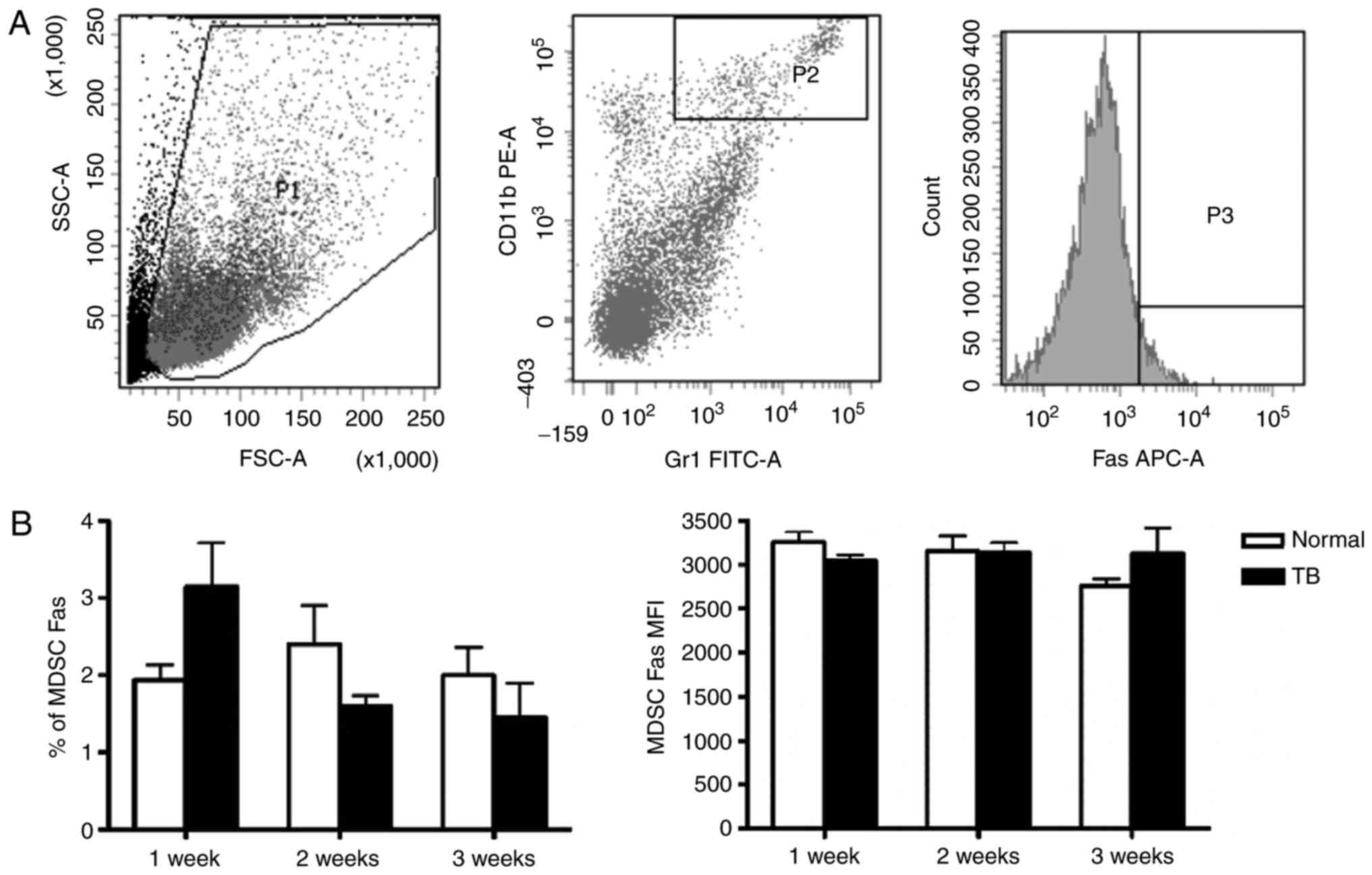 | Figure 4.Mice with hepatoma and normal mice do
not exhibit any difference in the expression of apoptotic protein
Fas in splenic MDSCs. Single-cell suspensions of splenocytes were
stained with anti-mouse Gr-1 antibody conjugated with FITC,
anti-mouse CD11b antibody conjugated with PE and anti-mouse Fas
antibody conjugated with APC. (A) Representative flow cytometry
plots and gating strategy. A live gate P1 was set in the FSC/SSC
plot and CD11b+/Gr-1+ cells were included in
a further analysis with gate P2. Subsequently, the selected cells
were displayed in a histogram for Fas-APC. (B) The percentage of
Fas+ MDSCs (left panel) and the MFI of Fas in MDSCs
(right panel) in H22 hepatoma and normal mice. Values are expressed
as the mean ± standard deviation (n=6). TB, tumor-bearing; APC,
allophycocyanine; MDSCs, myeloid-derived suppressor cells; FITC,
fluorescein isothiocyanate; PE, phycoerythrin; SSC, side scatter;
FSC, forward scatter; Gr-1, granulocyte receptor 1; MFI, mean
fluorescent intensity. |
These results indicate that the accumulation of
MDSCs in the spleen was not associated with their proliferation or
apoptosis.
Accumulation of splenic MDSCs in TB
mice is associated with increases in the level of splenic CCL9
The accumulation of splenic MDSCs in TB mice may be
caused by their chemotaxis to the spleen (29). To identify the factors involved, a
protein chip assay was then performed for the simultaneous
detection of 62 cytokines in the spleen of TB and untreated mice at
weeks 1, 2 and 3 post-tumor inoculation (Fig. 5A). At certain time-points, the
fluorescent signals of 11 cytokines were higher and those of 8
cytokines were lower in TB mice compared with those in untreated
mice. Among the upregulated cytokines, there were 7 chemokines:
Chemokine (C-X-C motif) ligand 13 (CXCL13), CXCL16, chemokine (C-C
motif) ligand 5 (CCL5), CCL12, CCL9, CXCL2 and CXCL4 (Fig. 5B). However, two of them, CXCL16 and
CCL12, were only expressed at low levels and one of them, CXCL13,
also known as B lymphocyte chemoattractant, is a chemotactic factor
for B lymphocytes. Another chemokine, CCL5, is a chemotactic factor
for T cells, eosinophils and basophils. CXCL4 has been reported to
negatively control the amount of MDSCs (31), as well as to inhibit angiogenesis
and tumor growth (32,33). Therefore, its elevated expression
was not correlated with the accumulation of MDSCs (Table I). Subsequently, the role of CCL9
and CXCL2 in MDSC accumulation in the spleen of TB mice was further
investigated.
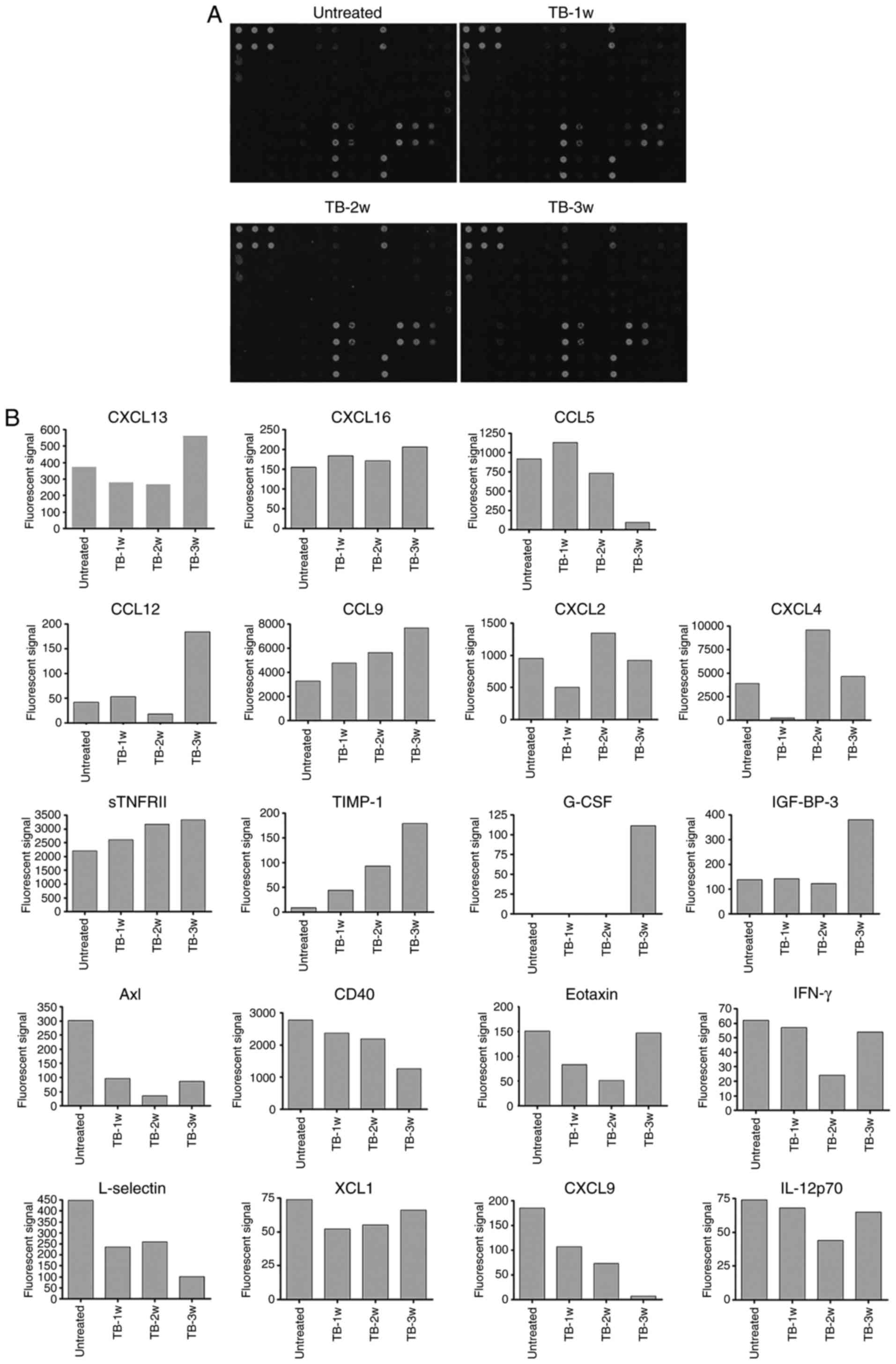 | Figure 5.Differential expression of 62
cytokines in the spleen of H22 hepatoma and normal mice. A
RayBiotech mouse cytokine antibody array was used to compare the
expression of 62 cytokines in the splenic tissues of untreated and
TB mice. (A) Mouse cytokine antibody array membranes. (B)
Quantification of A. The upregulated (upper part) and downregulated
(lower part) cytokines in the spleen of TB mice compared with
untreated mice. Among the upregulated cytokines, seven chemokines
are indicated by rectangles. TB, tumor-bearing; 1w, 1 week; G-CSF,
granulocyte-colony stimulating factor; IGFBP3, insulin-like growth
factor-binding protein 3; CCL12, chemokine (C-C motif) ligand 12;
CXCL2, chemokine (C-X-C motif) ligand 2; sTNFR, soluble tumor
necrosis factor receptors; TIMP1, tissue inhibitors of
metalloproteinase 1; IFN, interferon; IL, interleukin; XCL1,
chemokine (C motif) ligand. |
 | Table I.Upregulated chemokines in spleen of
H22 hepatoma mice. A total of 62 cytokines were detected in spleen
tissue of untreated and TB mice at weeks 1, 2 and 3 post-tumor
inoculation using a mouse cytokine antibody array. |
Table I.
Upregulated chemokines in spleen of
H22 hepatoma mice. A total of 62 cytokines were detected in spleen
tissue of untreated and TB mice at weeks 1, 2 and 3 post-tumor
inoculation using a mouse cytokine antibody array.
|
| Fluorescent signal
intensity (normalized to positive control) |
|
|---|
|
|
|
|
|---|
| Abbreviation | Untreated | TB-1w | TB-2w | TB-3w | Chemotactic
cells |
|---|
| CXCL13 | 374 | 282 | 270 | 563 | B |
| CXCL16 | 155 | 184 | 171 | 206 | T, NKT |
| CCL5 | 916 | 1132 | 729 | 91 | T, eosinophils,
basophils |
| CCL12 | 42 | 53 | 18 | 184 | Eosinophils,
monocytes, lymphocytes |
| CCL9 | 3298 | 4770 | 5644 | 7677 | DC |
| CXCL2 | 955 | 502 | 1343 | 926 | Polymorphonuclear
leukocytes |
| CXCL4 | 3907 | 256 | 9609 | 4638 | Neutrophils,
monocytes, fibroblasts |
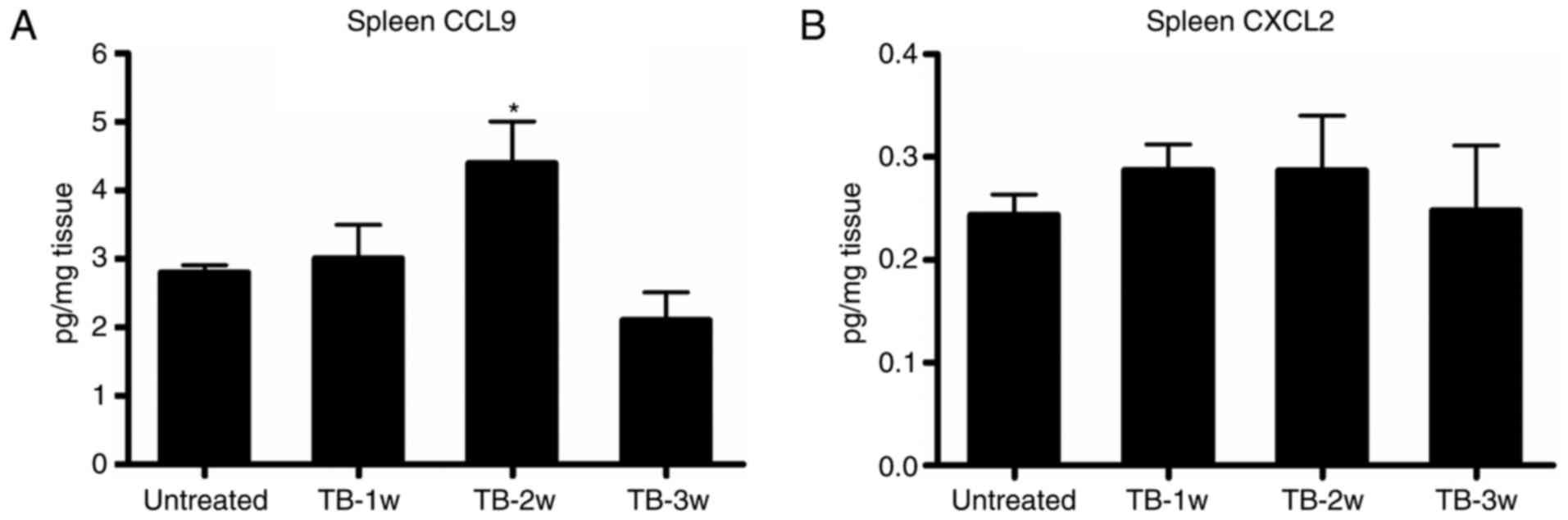
The expression of CCL9 was significantly higher in
the spleen of TB mice compared with that in untreated mice at week
2 post-tumor inoculation (Fig. 6A),
while there was no significant difference in the expression of
CXCL2 at any of the time-points assessed (Fig. 6B). Next, it was determined whether
splenic MDSCs expressed CCR1, the receptor for CCL9. It was
revealed that splenic MDSCs isolated from TB mice and normal mice
expressed CCR1. Furthermore, granulocyte-like MDSCs (G-MDSCs,
CD11b+ly6G+ly6Clow) and
monocyte-like MDSCs (MO-MDSCs,
CD11b+ly6G−ly6Chi) expressed CCR1
(Fig. 7A), suggesting that the
increased expression of CCL9 in the spleen of TB mice may attract
MDSCs in a CCR1-dependent manner. Of note, the expression of CCR1
on MO-MDSCs in TB mice was higher compared with that in normal
mice, while such a difference was not observed for G-MDSCs
(Fig. 7B).
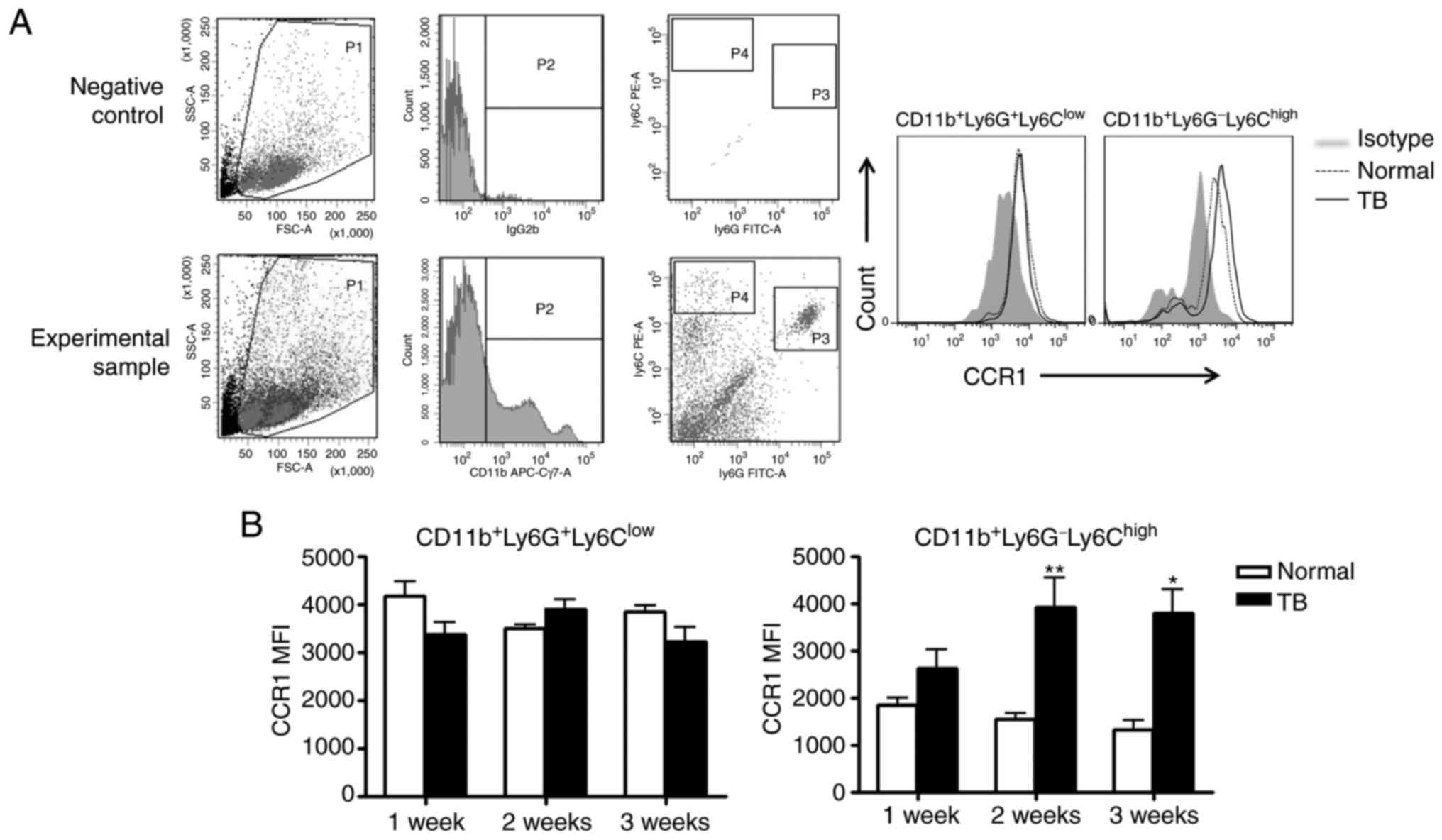 | Figure 7.Splenic MDSCs express CCR1, the
receptor for chemokine (C-C motif) ligand 9. Splenic single-cell
suspensions were stained with anti-mouse ly6G monoclonal antibody
conjugated with FITC, anti-mouse ly6C monoclonal antibody
conjugated with PE, anti-mouse CD11b monoclonal antibody conjugated
with APC-Cy7 and anti-mouse CCR1 monoclonal antibody conjugated
with APC. (A) Representative flow cytometry plots and gating
strategy. Left panel: A live gate P1 was set in the FSC/SSC plot
and CD11b+ cells were included in further analysis with
gate P2 (APC/Cy7 rat immunoglobulin G2b, κ isotype antibody was
used as the negative control for the separation of negative and
positive cell populations). Subsequently, the selected cells were
displayed in a histogram for ly6C-PE and ly6G-FITC. The population
of CD11b+ly6G+ly6Clow cells refers
to G-MDSCs, and the population of
CD11b+ly6G−ly6Chigh cells refers
to MO-MDSCs. Right panel: Representative plots for the expression
of CCR1 on G-MDSCs and MO-MDSCs from the spleen of normal and TB
mice. (B) The MFI of CCR1 on G-MDSCs (left panel) and MO-MDSCs
(right panel) from H22 hepatoma and normal mice (n=6). Values are
expressed as the mean ± standard deviation. *P<0.05 and
**P<0.01. TB, tumor-bearing; G-MDSCs, granulocytic
myeloid-derived suppressor cells; MO-MDSCs, monocytic MDSCs; TB,
tumor-bearing; APC, allophycocyanine; FITC, fluorescein
isothiocyanate; PE, phycoerythrin; SSC, side scatter; FSC, forward
scatter; MFI, mean fluorescent intensity; CCR1, C-C motif chemokine
receptor 1; Cy, cyanine. |
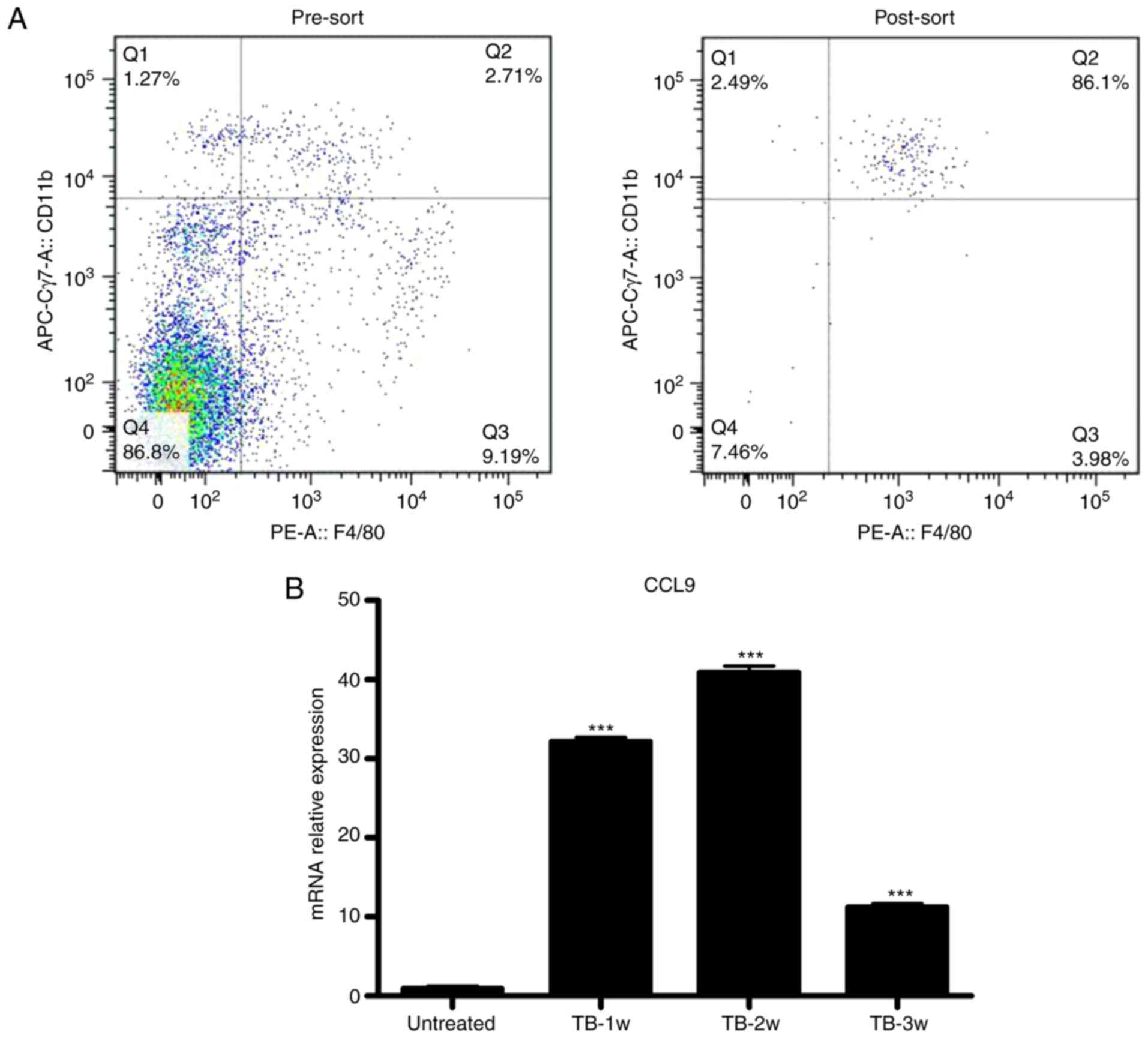
Splenic macrophages produce elevated
levels of CCL9 in tumor-bearing mice
Next, it was determined which splenic cells secrete
CCL9. CCL9 may be secreted by mononuclear phagocytic cells
(34). It was examined whether
macrophages were the source of CCL9 in spleen of TB mice. Splenic
macrophages (CD11b+/F4/80+) from untreated
and TB mice were isolated by fluorescence-assisted cell sorting
(Fig. 8A) and the expression levels
of CCL9 were detected by RT-qPCR. Splenic macrophages from TB mice
produced higher levels of CCL9 than control macrophages at weeks 1,
2 and 3 (Fig. 8B). These results
suggest that macrophages may be the source of CCL9 the in the
spleen of H22 hepatoma mice.
Discussion
Previous studies have reported that the frequencies
of MDSCs increased not only in tumor tissue but also in the spleen
of TB mice (12,24,35).
The accumulation of splenic MDSCs in TB mice may be caused by their
increased proliferation, decreased apoptosis or enhanced chemotaxis
to the spleen. In the present study, all three aspects were
investigated.
It remains elusive whether the proliferation of
MDSCs occurs mainly in the bone marrow or in the spleen. The spleen
is the major site of MDSC proliferation in the 4T1 mammary cancer
model, and the expansion of splenic MDSCs was caused by their
increased survival and decreased apoptosis (36). By contrast, in murine 3LL lung
cancer, B16 melanomas and Meth A fibrosarcomas, the proliferation
of MDSCs primarily occurs in the bone marrow and not in the
peripheral blood, the spleen or the tumor tissue (37). Therefore, in the murine H22
orthotopic hepatoma model of the present study, changes in splenic
MDSC proliferation and apoptosis were first assessed. It was
identified that the expansion of splenic MDSCs was not associated
with their proliferation or apoptosis.
Next, the mechanisms of MDSCs recruitment to the
spleen were assessed. In the murine MCA203 fibrosarcoma model,
nestin-positive splenocytes are able to secrete CCL2, which caused
an elevated level of CCL2 in the spleen and induced MDSC
accumulation in the spleen via CCR2 (29). In the present H22 hepatoma model, it
was investigated which factor is able to attract MDSCs to the
spleen by screening for 62 cytokines. The cytokine protein array
and ELISA results revealed that the CCL9 chemokine was highly
expressed in the spleen of TB mice.
The overexpressed CCL9 exerts its chemotactic
function through binding to its receptor CCR1. Therefore, it was
next determined whether MDSCs expressed CCR1. The splenic MDSCs
(including G-MDSCs and MO-MDSCs) from normal and TB mice were
identified to express CCR1. This indicates that the recruitment of
MDSCs to the spleen of TB mice was driven by increases of CCL9 as
well as its receptor CCR1, leading to the accumulation of MDSCs in
the spleen.
It was then investigated which cell type in the
spleen secretes CCL9 in TB mice. CCL9 may be secreted by
macrophages, DCs, Langerhans cells, gliocytes, osteoclasts and
certain types of tumor cell, including intestinal cancer cells.
MDSCs may also be a source of CCL9 (38–40).
As the percentage of macrophages in the spleen is higher than that
of DCs (35,41,42),
the expression of CCL9 in macrophages was examined and it was
revealed that splenic macrophages from TB mice had significantly
elevated levels of CCL9 compared with those in normal mice.
Therefore, it was concluded that the elevated CCL9 secreted by
splenic macrophages attracted MDSCs to the spleen of TB mice.
In the H22 orthotopic hepatoma model, a reduction in
spleen weight from week 2 to week 3 was previously observed
(12). The total number of
splenocytes, including macrophages, may decrease at week 3.
Macrophages viability and activity may decrease, resulting in
reduced cytokine secretion (43).
This may explain for the lower expression of CCL9 in the spleen and
reduced recruitment of MDSCs to the spleen at week 3 compared with
that at week 2.
In conclusion, the results of the present study
suggest that in the H22 hepatoma model, splenic macrophages
secreted CCL9 that induced MDSC accumulation in the spleen in a
CCR1-dependent manner. Further studies should assess which other
splenocytes, including DCs or MDSCs, secrete CCL9, whether the
mobilization of MDSCs to the spleen may be inhibited by targeting
CCL9 or CCR1, and how such an inhibition may affect tumor
growth.
Acknowledgements
The authors would like to thank Professor Malgorzata
A. Garstka (Core Research Lab of the Second Affiliated Hospital of
Xi'an Jiaotong University, Xi'an, China) for critical reading of
the manuscript.
Funding
This work was supported by the Program for
Changjiang Scholars and Innovative Research Teams at University
(grant no. 1171), the National Natural Science Foundation of China
(grant no. 81001309) and the Key Research and Development Program
of Shaanxi Province of China (grant no. 2017ZDCXL-SF-02-05,
2018SF-197).
Availability of data and materials
All data generated and analyzed during this study
are included in this manuscript.
Authors' contributions
BL performed the experiments and wrote the first
draft of the manuscript. NH, HC and PW assisted in the
establishment of the tumor model and the sample preparation of flow
cytometry. SZ assisted in the statistical analysis and data
interpretation. JY and ZL designed the study and supervised the
experimental work. All authors reviewed the manuscript prior to its
submission and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures complied with the Guide for
the Care and Use of Laboratory Animals (NIH Publication, 1996) and
were approved by the Animal Care and Use Committee of Xi'an
Jiaotong University (Xi'an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Talmadge JE and Gabrilovich DI: History of
myeloid-derived suppressor cells. Nat Rev Cancer. 13:739–752. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schrader J: The role of MDSCs in
hepatocellular carcinoma-in vivo veritas? J Hepatol. 59:921–923.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kumar V, Patel S, Tcyganov E and
Gabrilovich DI: The nature of myeloid-derived suppressor cells in
the tumor microenvironment. Trends Immunol. 37:208–220. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Arina A and Bronte V: Myeloid-derived
suppressor cell impact on endogenous and adoptively transferred T
cells. Curr Opin Immunol. 33:120–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan PY, Ma G, Weber KJ, Ozao-Choy J, Wang
G, Yin B, Divino CM and Chen SH: Immune stimulatory receptor CD40
is required for T-cell suppression and T regulatory cell activation
mediated by myeloid-derived suppressor cells in cancer. Cancer Res.
70:99–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gabrilovich DI: Myeloid-derived suppressor
cells. Cancer Immunol Res. 5:3–8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pardoll DM: Distinct mechanisms of tumor
resistance to NK killing: Of mice and men. Immunity. 42:605–606.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
You TG, Wang HS, Yang JH, Qian QJ, Fan RF
and Wu MC: Transfection of IL-2 and/or IL-12 genes into spleen in
treatment of rat liver cancer. World J Gastroenterol. 10:2190–2194.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Imai S, Nio Y, Shiraishi T, Tsubono M,
Morimoto H, Tseng CC, Kawabata K, Masai Y and Tobe T: Effects of
splenectomy on pulmonary metastasis and growth of SC42 carcinoma
transplanted into mouse liver. J Surg Oncol. 47:178–187. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zusman I, Kossoy G and Ben-Hur H: T cell
kinetics and apoptosis in immune organs and mammary tumors of rats
treated with cyclophosphamide and soluble tumor-associated
antigens. In Vivo. 16:567–576. 2002.PubMed/NCBI
|
|
11
|
Cortez-Retamozo V, Etzrodt M, Newton A,
Rauch PJ, Chudnovskiy A, Berger C, Ryan RJ, Iwamoto Y, Marinelli B,
Gorbatov R, et al: Origins of tumor-associated macrophages and
neutrophils. Proc Natl Acad Sci USA. 109:2491–2496. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li B, Zhang S, Huang N, Chen H, Wang P, Li
J, Pu Y, Yang J and Li Z: Dynamics of the spleen and its
significance in a murine H22 orthotopic hepatoma model. Exp Biol
Med. 241:863–872. 2016. View Article : Google Scholar
|
|
13
|
Levy L, Mishalian I, Bayuch R, Zolotarov
L, Michaeli J and Fridlender ZG: Splenectomy inhibits non-small
cell lung cancer growth by modulating anti-tumor adaptive and
innate immune response. Oncoimmunology. 4:e9984692015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Serafini P, Carbley R, Noonan KA, Tan G,
Bronte V and Borrello I: High-dose granulocyte-macrophage
colony-stimulating factor-producing vaccines impair the immune
response through the recruitment of myeloid suppressor cells.
Cancer Res. 64:6337–6343. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Waight JD, Hu Q, Miller A, Liu S and
Abrams SI: Tumor-derived G-CSF facilitates neoplastic growth
through a granulocytic myeloid-derived suppressor cell-dependent
mechanism. PLoS One. 6:e276902011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Markiewski MM, DeAngelis RA, Benencia F,
Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, Coukos G and
Lambris JD: Modulation of the antitumor immune response by
complement. Nat Immunol. 9:1225–1235. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shields JD, Kourtis IC, Tomei AA, Roberts
JM and Swartz MA: Induction of lymphoidlike stroma and immune
escape by tumors that express the chemokine CCL21. Science.
328:749–752. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu F, Shi Y, Wang J, Li J, Fan D and Ai W:
Deficiency of Kruppel-like factor KLF4 in mammary tumor cells
inhibits tumor growth and pulmonary metastasis and is accompanied
by compromised recruitment of myeloid-derived suppressor cells. Int
J Cancer. 133:2872–2883. 2013.PubMed/NCBI
|
|
19
|
Zhou Y and Guo F: A selective
sphingosine-1-phosphate receptor 1 agonist SEW-2871 aggravates
gastric cancer by recruiting myeloid-derived suppressor cells. J
Biochem. 163:77–83. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang AL, Miska J, Wainwright DA, Dey M,
Rivetta CV, Yu D, Kanojia D, Pituch KC, Qiao J, Pytel P, et al:
CCL2 produced by the glioma microenvironment is essential for the
recruitment of regulatory T cells and myeloid-derived suppressor
cells. Cancer Res. 76:5671–5682. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Blattner C, Fleming V, Weber R, Himmelhan
B, Altevogt P, Gebhardt C, Schulze TJ, Razon H, Hawila E, Wildbaum
G, et al: CCR5+ myeloid-derived suppressor cells are
enriched and activated in melanoma lesions. Cancer Res. 78:157–167.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamamoto T, Kawada K, Itatani Y, Inamoto
S, Okamura R, Iwamoto M, Miyamoto E, Chen-Yoshikawa TF, Hirai H,
Hasegawa S, et al: Loss of SMAD4 promotes lung metastasis of
colorectal cancer by accumulation of CCR1+
tumor-associated neutrophils through CCL15-CCR1 axis. Clin Cancer
Res. 23:833–844. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Long X, Ye Y, Zhang L, Liu P, Yu W, Wei F,
Ren X and Yu J: I L-8, a novel messenger to cross-link inflammation
and tumor EMT via autocrine and paracrine pathways (Review). Int J
Oncol. 48:5–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kapanadze T, Gamrekelashvili J, Ma C, Chan
C, Zhao F, Hewitt S, Zender L, Kapoor V, Felsher DW, Manns MP, et
al: Regulation of accumulation and function of myeloid derived
suppressor cells in different murine models of hepatocellular
carcinoma. J Hepatol. 59:1007–1013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bunt SK, Yang L, Sinha P, Clements VK,
Leips J and Ostrand-Rosenberg S: Reduced inflammation in the tumor
microenvironment delays the accumulation of myeloid-derived
suppressor cells and limits tumor progression. Cancer Res.
67:10019–10026. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sinha P, Clements VK, Fulton AM and
Ostrand-Rosenberg S: Prostaglandin E2 promotes tumor progression by
inducing myeloid-derived suppressor cells. Cancer Res.
67:4507–4513. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eisenblaetter M, Flores-Borja F, Lee JJ,
Wefers C, Smith H, Hueting R, Cooper MS, Blower PJ, Patel D,
Rodriguez-Justo M, et al: Visualization of tumor-immune
interaction-target-specific imaging of S100A8/A9 reveals
pre-metastatic niche establishment. Theranostics. 7:2392–2401.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ba H, Li B, Li X, Li C, Feng A, Zhu Y,
Wang J, Li Z and Yin B: Transmembrane tumor necrosis factor-α
promotes the recruitment of MDSCs to tumor tissue by upregulating
CXCR4 expression via TNFR2. Int Immunopharmacol. 44:143–152. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ugel S, Peranzoni E, Desantis G, Chioda M,
Walter S, Weinschenk T, Ochando JC, Cabrelle A, Mandruzzato S and
Bronte V: Immune tolerance to tumor antigens occurs in a
specialized environment of the spleen. Cell Rep. 2:628–639. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu P, He H, Gu Y, Wang Y, Sun Z, Yang L
and Miao C: Surgical trauma contributes to progression of colon
cancer by downregulating CXCL4 and recruiting MDSCs. Exp Cell Res.
370:692–698. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vandercappellen J, Van Damme J and Struyf
S: The role of the CXC chemokines platelet factor-4 (CXCL4/PF-4)
and its variant (CXCL4L1/PF-4var) in inflammation, angiogenesis and
cancer. Cytokine Growth Factor Rev. 22:1–18. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Z and Huang H: Platelet factor-4
(CXCL4/PF-4): An angiostatic chemokine for cancer therapy. Cancer
Lett. 331:147–153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Youn BS, Jang IK, Broxmeyer HE, Cooper S,
Jenkins NA, Gilbert DJ, Copeland NG, Elick TA, Fraser MJ Jr and
Kwon BS: A novel chemokine, macrophage inflammatory protein-related
protein-2, inhibits colony formation of bone marrow myeloid
progenitors. J Immunol. 155:2661–2667. 1995.PubMed/NCBI
|
|
35
|
Clark CE, Hingorani SR, Mick R, Combs C,
Tuveson DA and Vonderheide RH: Dynamics of the immune reaction to
pancreatic cancer from inception to invasion. Cancer Res.
67:9518–9527. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Younos IH, Dafferner AJ, Gulen D, Britton
HC and Talmadge JE: Tumor regulation of myeloid-derived suppressor
cell proliferation and trafficking. Int Immunopharmacol.
13:245–256. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sawanobori Y, Ueha S, Kurachi M, Shimaoka
T, Talmadge JE, Abe J, Shono Y, Kitabatake M, Kakimi K, Mukaida N,
et al: Chemokine-mediated rapid turnover of myeloid-derived
suppressor cells in tumor-bearing mice. Blood. 111:5457–5466. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lean JM, Murphy C, Fuller K and Chambers
TJ: CC L9/MIP-1gamma and its receptor CCR1 are the major chemokine
ligand/receptor species expressed by osteoclasts. J Cell Biochem.
87:386–393. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yan HH, Jiang J, Pang Y, Achyut BR,
Lizardo M, Liang X, Hunter K, Khanna C, Hollander C and Yang L:
CCL9 induced by TGFβ signaling in myeloid cells enhances tumor cell
survival in the premetastatic organ. Cancer Res. 75:5283–5298.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kitamura T, Fujishita T, Loetscher P,
Revesz L, Hashida H, Kizaka-Kondoh S, Aoki M and Taketo MM:
Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses
colon cancer liver metastasis by blocking accumulation of immature
myeloid cells in a mouse model. Proc Natl Acad Sci USA.
107:13063–13068. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fujimi S, Lapchak PH, Zang Y, MacConmara
MP, Maung AA, Delisle AJ, Mannick JA and Lederer JA: Murine
dendritic cell antigen-presenting cell function is not altered by
burn injury. J Leukoc Biol. 85:862–870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gatto D, Wood K, Caminschi I,
Murphy-Durland D, Schofield P, Christ D, Karupiah G and Brink R:
The chemotactic receptor EBI2 regulates the homeostasis,
localization and immunological function of splenic dendritic cells.
Nat Immunol. 14:446–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang S, Li ZF, Pan D, Huang C, Zhou R and
Liu ZW: Changes of splenic macrophage during the process of liver
cancer induced by diethylnitrosamine in rats. Chin Med J.
122:3043–3047. 2009.PubMed/NCBI
|