Introduction
Tumors of the central nervous system (CNS) represent
a relatively rare but serious health burden in terms of morbidity
and mortality (1). Approximately 2%
of all adult malignancies are brain tumors, and 80% of them are
glioma (2). Despite important
progress in diagnostic and therapeutic techniques over the past
decades, the prognosis for patients with high-grade gliomas (WHO
grade III and IV tumors) remains discouraging (3). One of the most important reasons for
this, is that the molecular mechanism of glioma progression is
still unclear. Thus, it is essential to understand the mechanism of
glioma invasion and metastasis and develop new therapeutic
strategies.
It is already known that the main differences
between tumor cells and normal cells lie in the different cell
properties. Hanahan and Weinberg have concluded these properties as
the ten hallmarks of cancer, such as sustaining proliferative
signaling, activating invasion and metastasis (4). However, the precise mechanisms of
regulating these hallmarks still need to be clarified. Existing
researches have demonstrated that signaling pathways such as MYC,
TGF-β/Smad and Notch are involved in the occurrence and development
of cancer (5,6). Thereby, the malignant biological
behaviors of cancer cells which differentiate from normal cells are
mainly a result of the aberrancy of these signaling pathways.
Bromodomain PHD finger transcription factor (BPTF)
is located at chromosome 17q24.3 and is involved in transcriptional
regulation and chromatin remodeling in many biological processes
(7). The completed sequence of
human BPTF encodes a predicted protein of 2,781 amino acids, which
contain typical features of bromodomain, two PHD fingers and an
extensive glutamine-rich acidic domain (8). Previous research indicated that BPTF
mainly participated in embryonic development (9). The multiple signaling mechanisms in
embryonic development are always associated with tumor development
and progression. Another study revealed that BPTF could regulate
chromatin remodeling and control neuroectodermal differentiation,
which was also correlated with cancer development (10). In addition, BPTF was also revealed
to be involved in a variety of tumor-related signaling pathways
such as MYC and Smad (10,11). Furthermore, previous studies
confirmed the cancer-promoting role of BPTF in colorectal and lung
cancer (12,13). These findings indicated that BPTF
may play an important oncogenic role in cancer progression.
However, there are no studies on the functional role and molecular
mechanisms of BPTF in glioma.
In the present study, we first examined the
expression of BPTF in glioma and assessed the relationship of BPTF
expression with the prognosis of glioma patients. We further
identified the functional role of BPTF in the U251 cell line and
explored the potential molecular mechanism. Therefore, our findings
provided a new understanding of BPTF in glioma progression and may
reveal a novel potential therapeutic target for glioma.
Materials and methods
Patients and tissue specimens
Ten pairs of fresh-frozen glioma tissue and their
corresponding normal brain tissue were randomly selected. Another
113 cases of paraffin-embedded glioma tissue samples were also
randomly obtained from Haikou People's Hospital from January 2010
to December 2014. All the gliomas were pathologically confirmed by
2 independent pathologists according to the 2007 WHO
classification. The fresh-frozen glioma tissue and normal brain
tissue were analyzed by real-time quantitative
reverse-transcription polymerase chain reaction (real-time PCR) and
western blotting (WB). The paraffin-embedded glioma tissue samples
were detected by immunohistochemical staining (IHC). All patients
eligible for the present study had undergone surgery, and were
routinely followed-up and detailed clinicopathological and survival
data was collected. Among them, 52 were female and 61 were male and
the median age was 47 years (range, 21–82 years). Magnetic
resonance imaging (MRI) or contrast-enhanced MRI was performed
every 6 months to detect tumor relapse or metastasis after surgery.
Overall survival (OS) was defined as the time from the surgery to
the death of the patient or the last follow-up visit.
Progression-free survival (PFS) was defined as the time from the
surgery to the first evidence of recurrence, progression, or death.
All patients provided signed written informed consent. The study
was approved by the Ethics Committee of Haikou People's Hospital in
accordance with the Declaration of Helsinki.
Real-time PCR
Total RNA was extracted from the fresh-frozen
specimens using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the specifications
of the instructions. Real-time PCR was performed using the
SYBR-Green Real-Time PCR Master Mix (Toyobo Co., Ltd., Osaka,
Japan) as the manufacturer's instructions described. An ABI 7100
Real-Time Quantitative PCR System (Applied Biosystems, Foster City,
CA, USA) was used, in which each reaction (25 µl) contained 10 µl
PCR Master Mix (Ambion; Thermo Fisher Scientific, Inc.) and 1.3 µl
RT product, and each sample was analyzed in triplicate. The PCR
reaction was conducted at 95°C for 10 min, followed by 40 cycles at
95°C for 15 sec and 60°C for 1 min. The reactions were performed in
two independent assays. The primers of BPTF were as follows:
Forward, 5′-GGAGAGATGTTGGTCCTTATGGC-3′ and reverse,
5′-CTTTCCTCTGAGGTGTAGGCGT-3′; β-actin was used as a control using
the following primers: Forward, 5′-CACCATTGGCAATGAGCGGTTC-3′ and
reverse, 5′-AGGTCTTTGCGGATGTCCACGT-3′. The results were analyzed
using the 2−ΔΔCq method as previously described
(14).
Western blot (WB) analysis
Total proteins of fresh-frozen glioma and normal
brain tissues were extracted using RIPA lysis buffer (Thermo Fisher
Scientific, Inc.) and separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) then
transferred onto polyvinylidene fluoride (PVDF) membranes (EMD
Millipore, Billerica, MA, USA) as previously described (15). The protein concentration was
detected by bicinchoninic acid (BCA) protein assay (Pierce
Chemical, Rockford, IL, USA) according to the manufacturer's
instructions. The blotted membranes were incubated with antihuman
BPTF antibody (band size, 324 kDa; dilution 1:1,000; cat. no.
ab72036; Abcam, Cambridge, MA, USA), or C-MYC antibody (band size,
67 kDa; dilution 1:1,000; cat. no. sc-47694; Santa Cruz
Biotechnology, Santa Cruz CA, USA) or β-actin antibody (band size,
42 kDa; dilution 1:2,000; cat. no. ab173838; Abcam) at 4°C
overnight, then incubated with an appropriate secondary antibody
(dilution 1:2,000; cat. no. sc-2004; dilution 1:3,000; cat. no.
sc-2005; Santa Cruz Biotechnology) for 30 min at room temperature.
The western blotting band was detected by enhanced
chemiluminescence reagents (Thermo Fisher Scientific, Inc.) and the
band density was assessed by ImageJ software (version k1.45 for
windows; National Institutes of Health, Bethesda, MD, USA) and the
density detection was repeated three times.
Immunohistochemistry (IHC)
The tissue specimens were fixed with 10% formalin
and then embedded in paraffin; 4-mm sections were cut and placed on
silane-coated slides for immunohistochemical analysis. Each slide
of the specimens was stained with hematoxylin and eosin (H&E)
and microscopically examined to confirm the pathological diagnosis.
The paraffin-embedded sections were dewaxed and pretreated with
0.01 M sodium citrate buffer (pH 6.0) for 15 min at 95°C for tissue
antigen retrieval. Then, these sections were incubated with 3%
hydrogen peroxide for 20 min at room temperature to block
endogenous peroxidase. After rinsing three times for 2 min with
phosphate-buffered saline (PBS), 10% goat serum was added as a
blocking liquid, and incubated for 15 min at room temperature.
Next, the serum was removed, and the appropriate diluted primary
antibody (BPTF; dilution 1:250; cat. no. ab72036; Abcam) was added
to each section and incubated overnight at 4°C. After rinsing the
primary antibody using PBS, the sections were handled according to
the manufacturer's recommendations (PV-9000; Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd., Beijing, China) and
counterstained with hematoxylin; then dehydration was conducted by
graded ethanol followed by the addition of xylene to render the
sections transparent. Finally, the slides were covered with neutral
balsam, and then observed and mounted with a Leica DM2000 optical
microscope (Leica Microsystems, Wetzlar, Germany) and ImageJ k1.45
software (National Institutes of Health). The staining intensity of
BPTF was scored according to a previous study: - (negative), +
(weak), ++ (moderate) and +++ (strong). Stained tissues with ‘−’
and ‘+’ were considered as the BPTF low-expression group, and ‘++’
and ‘+++’ were considered as the BPTF high-expression group
(13). The staining was evaluated
by two separated senior pathologists on a multi-head microscope
with anonymous patient information.
Lentiviral vector construction and
cell transduction
Full-length human BPTF overexpression clone plasmid
and targeting shRNA (shRNA) lentivirus and their control vectors
were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China).
The transfection process was conducted according to the
manufacturer's instructions. Puromycin (2 µg/ml) (Thermo Fisher
Scientific, Inc.) was used to select stable clones if necessary.
The transfection efficiency was assessed by fluorescence
microscopy, real-time PCR and western blotting.
Proliferation assay
The proliferation capacity of cancer cells was
assessed by colony formation assay. Cells were seeded into 6-well
plates with a density of 5×102 cells/well, and cultured
for ~2 weeks with fresh medium replaced every week. Then colonies
were stained with crystal violet and counted by ImageJ software
k1.45 (National Institutes of Health) and only the colonies
containing >50 cells were counted and plotted. All the
experiments were replicated in triplicate.
Migration and invasion assays
Migration ability of cancer cells was assessed by
wound-healing and Transwell assays. In the wound-healing assay, the
cells were cultured into 6-well plates at a density of
2×105 cells/well. When the cells grew to 90% confluence,
they were incubated with mitomycin-C (10 µg/ml) for 1 h to suppress
proliferation and then starved in serum-free medium for 24 h. An
artificial wound was created by scraping the cell confluent
monolayer with a 10-µl pipette tip. The migration gap of the wound
was assessed after 48 h. The migration and invasion potential was
also evaluated by Transwell assay. The cells were treated with
mitomycin-C (10 µg/ml) for 1 h at 37°C in serum-free medium and
then plated into the upper chamber. For the migration assay the
cells were seeded with a density of 2×104 cells/insert,
and for the invasion assay cells are seeded into the top chamber
coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) at a
density of 4×104 cells/insert. Then DMEM with 10% fetal
bovine serum (FBS) was added into the bottom chamber. After
incubation for 24–48 h, the cells adhering to the lower membrane of
the inserts were stained with 0.1% crystal violet and observed by a
Leica DM2000 optical microscope (Leica Microsystems) then counted
by ImageJ k1.45 software (National Institutes of Health). All the
experiments were replicated in triplicate.
Public database analysis
The public database Oncomine (https://www.oncomine.org) was used for analyzing the
expression of BPTF in glioma. ‘BPTF’ was used as a keyword in the
Oncomine search, ‘Cancer vs. Normal Analysis’ was chosen as the
primary filter, and ‘brain and CNS cancer’ was chosen as the cancer
type, and ‘mRNA’ was chosen as the data type. The expression of
BPTF was presented in multiple data sets including Rickman's and
Murat's. The BPTF expression level was log-transformed and
median-centered per array for analysis. The GCBI (http://www.gcbi.com.cn) and COXPRESdb (http://coxpresdb.jp) databases were used to analyze
the interaction and co-expressed genes with BPTF.
Statistical analysis
All data were analyzed using the SPSS statistical
software, version 18.0, for Windows (SPSS, Inc., Chicago, IL, USA).
A Student's t-test (two-tailed) was used for statistical analysis
of continuous data between two groups such as the relative
expression level of BPTF in tumor and normal brain tissue. Multiple
comparisons of continuous data among the control,
BPTF-overexpression and BPTF-knockdown groups was analyzed by
one-way ANOVA test followed by Tukey's post hoc test. Pearson
Chi-square test was used for all of the categorical data. Survival
curves were constructed using the Kaplan-Meier method and evaluated
using the log-rank test. The univariate and multivariate Cox
proportional hazards regression models were established to identify
factors that were related or independently associated with the
survival of glioma patients. A P-value of <0.05 was considered
to indicate a statistically significant difference.
Results
BPTF expression is significantly
elevated in gliomas tissue
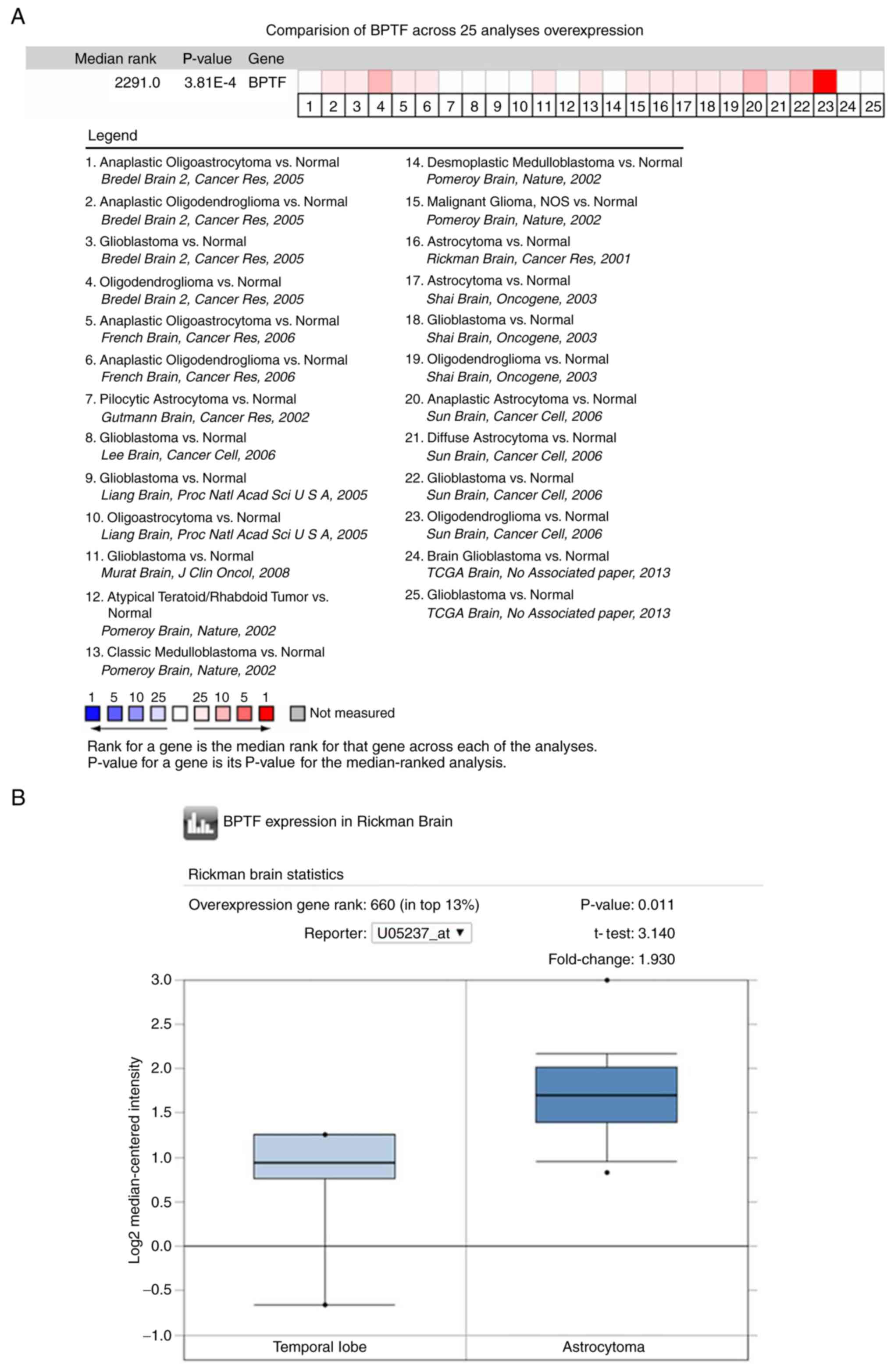
We first analyzed BPTF expression by utilizing the
public database Oncomine. The results revealed that the expression
level of BPTF in brain tumor tissue was significantly higher than
that in normal brain tissue in most of the public data (Fig. 1A). Since astrocytoma and
glioblastoma account for >70% of total glioma cases, we then
chose Rickman's (astrocytoma) and Murat's (glioblastoma) data for
further analysis. The results revealed that BPTF expression in
astrocytoma and glioblastoma were both higher than that in normal
brain tissue (Fig. 1B and C). Then,
the BPTF mRNA expression of 8 pairs of fresh-frozen glioma tumor
tissue and corresponding normal brain tissue from our hospital were
also detected by real-time PCR. The results revealed that glioma
tissues had significantly higher BPTF mRNA expression levels than
the corresponding normal brain tissues (Fig. 1D). Consistent with the mRNA
expression, the western blotting results also revealed that the
expression of BPTF protein in glioma tissue was significantly
higher than that in the corresponding normal brain tissue (Fig. 1E). These data clearly demonstrated
that BPTF expression was significantly elevated in gliomas
tissues.
High BPTF expression predicts poor
prognosis in glioma patients
To further explore the BPTF expression pattern in
glioma clinical samples and the correlation with
clinicopathological characteristics and survival, we detected the
BPTF protein expression in 113 cases of paraffin-embedded glioma
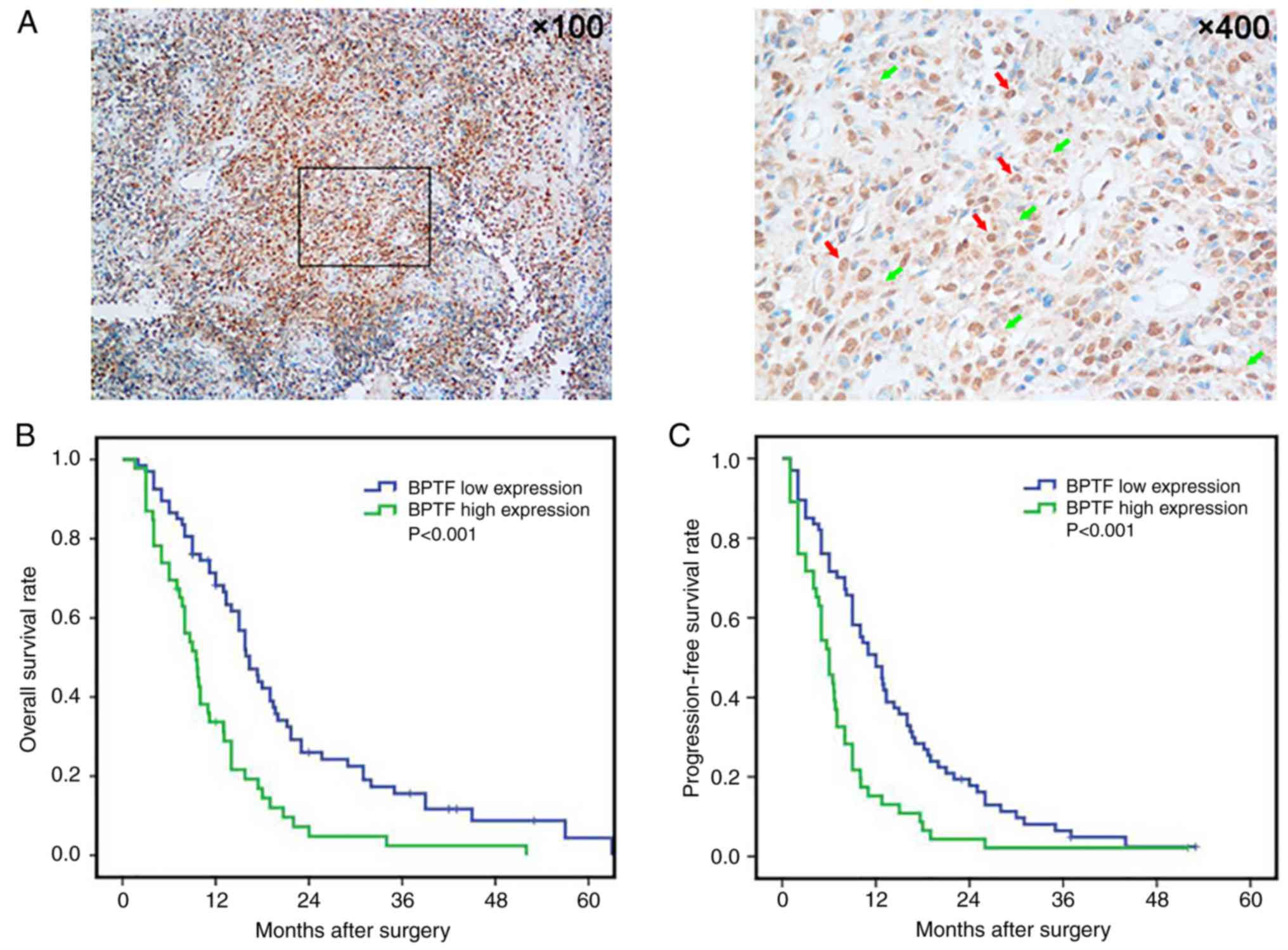
tissues by immunohistochemistry. Immunohistochemical staining
revealed BPTF positive staining mainly expressed in the cytoplasm
and/or the nuclei of the tumor cells (Fig. 2A). There were 69 (69/113, 61.1%)
cases that had BPTF positive staining, including 46 (46/113, 40.7%)
cases of high BPTF expression. Then, we analyzed the expression of
BPTF with clinicopathological features, and found that high BPTF
expression was significantly correlated with tumor size
(P<0.001) and WHO grade (P=0.001), but had no statistical
significance with sex, age, tumor location, tumor cystic change,
tumor necrosis and KPS score (Table
I). Subsequently, the survival curve was constructed and the
survival rate difference between the groups was analyzed by
log-rank test. The results revealed that the BPTF low-expression
group had favorable overall survival (OS) (P<0.001; Fig. 2B) and progression-free survival
(PFS) (P<0.001; Fig. 2C)
compared with the BPTF high-expression group. Furthermore,
univariate and multivariate Cox regression analyses both indicated
that BPTF expression was an independent prognostic factor for the
OS and PFS of glioma patients (Tables
II and III). These results
indicated that BPTF may be a useful prognostic biomarker for glioma
patients and may be involved in the progression of glioma.
 | Table I.BPTF expression and
clinicopathological features of 113 glioma cases. |
Table I.
BPTF expression and
clinicopathological features of 113 glioma cases.
|
|
| BPTF expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Total N (113) | Low (67) | High (46) | P-value |
|---|
| Sex |
|
|
| 0.482 |
|
Female | 52 | 29 | 23 |
|
| Male | 61 | 38 | 23 |
|
| Age (years) |
|
|
| 0.407 |
| ≤50 | 66 | 37 | 29 |
|
|
>50 | 47 | 30 | 17 |
|
| Location |
|
|
| 0.478 |
|
Frontal | 25 | 13 | 12 |
|
|
Temporal | 34 | 23 | 11 |
|
|
Parietal | 18 | 8 | 10 |
|
|
Occipital | 17 | 11 | 6 |
|
|
Others | 19 | 12 | 7 |
|
| Tumor size
(cm) |
|
|
|
<0.001 |
| ≤5 | 64 | 48 | 16 |
|
|
>5 | 49 | 19 | 30 |
|
| Cystic change |
|
|
| 0.335 |
|
Absence | 77 | 48 | 29 |
|
|
Presence | 36 | 19 | 17 |
|
| Necrosis |
|
|
| 0.554 |
|
Absence | 81 | 50 | 32 |
|
|
Presence | 32 | 17 | 14 |
|
| WHO grade |
|
|
| 0.001 |
| I and
II | 66 | 48 | 18 |
|
| III and
IV | 47 | 19 | 28 |
|
| KPS score |
|
|
| 0.288 |
|
≤90 | 51 | 33 | 18 |
|
|
>90 | 62 | 34 | 28 |
|
 | Table II.Univariate and multivariate analyses
of factors affecting overall survival (OS) in glioma patients. |
Table II.
Univariate and multivariate analyses
of factors affecting overall survival (OS) in glioma patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex | 0.897
(0.605–1.331) | 0.591 |
|
|
| Age (years) | 1.111
(0.749–1.648) | 0.603 |
|
|
| Location | 1.062
(0.926–1.217) | 0.389 |
|
|
| Tumor size | 2.270
(1.514–3.405) |
<0.001 | 1.683
(1.070–2.648) | 0.024 |
| Cystic change | 0.885
(0.574–1.364) | 0.579 |
|
|
| Necrosis | 1.276
(0.823–1.978) | 0.277 |
|
|
| WHO grade | 3.261
(2.121–5.014) |
<0.001 | 2.824
(1.776–4.489) |
<0.001 |
| KPS score | 0.848
(0.572–1.257) | 0.411 |
|
|
| BPTF
expression | 2.259
(1.506–3.387) |
<0.001 | 1.789
(1.125–2.846) | 0.014 |
 | Table III.Univariate and multivariate analyses
of factors affecting progression-free survival (PFS) in glioma
patients. |
Table III.
Univariate and multivariate analyses
of factors affecting progression-free survival (PFS) in glioma
patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex | 0.927
(0.635–1.355) | 0.696 |
|
|
| Age (years) | 1.122
(0.766–1.644) | 0.554 |
|
|
| Location | 1.089
(0.956–1.240) | 0.200 |
|
|
| Tumor size | 2.127
(1.441–3.140) |
<0.001 | 1.658
(1.076–2.554) | 0.022 |
| Cystic change | 0.928
(0.618–1.395) | 0.720 |
|
|
| Necrosis | 1.226
(0.800–1.877) | 0.349 |
|
|
| WHO grade | 2.611
(1.725–3.952) |
<0.001 | 2.118
(1.354–3.312) | 0.001 |
| KPS score | 0.922
(0.630–1.348) | 0.674 |
|
|
| BPTF
expression | 2.023
(1.366–2.995) |
<0.001 | 1.603
(1.020–2.521) | 0.041 |
BPTF enhances proliferation and
invasion of human glioma cells in vitro
To study the biological function of BPTF in glioma,
we constructed BPTF overexpressed (named U251BPTF-OE)
and knocked down (named U251BPTF-KD) plasmids and
transfected them into glioma U251 cells. We first examined the
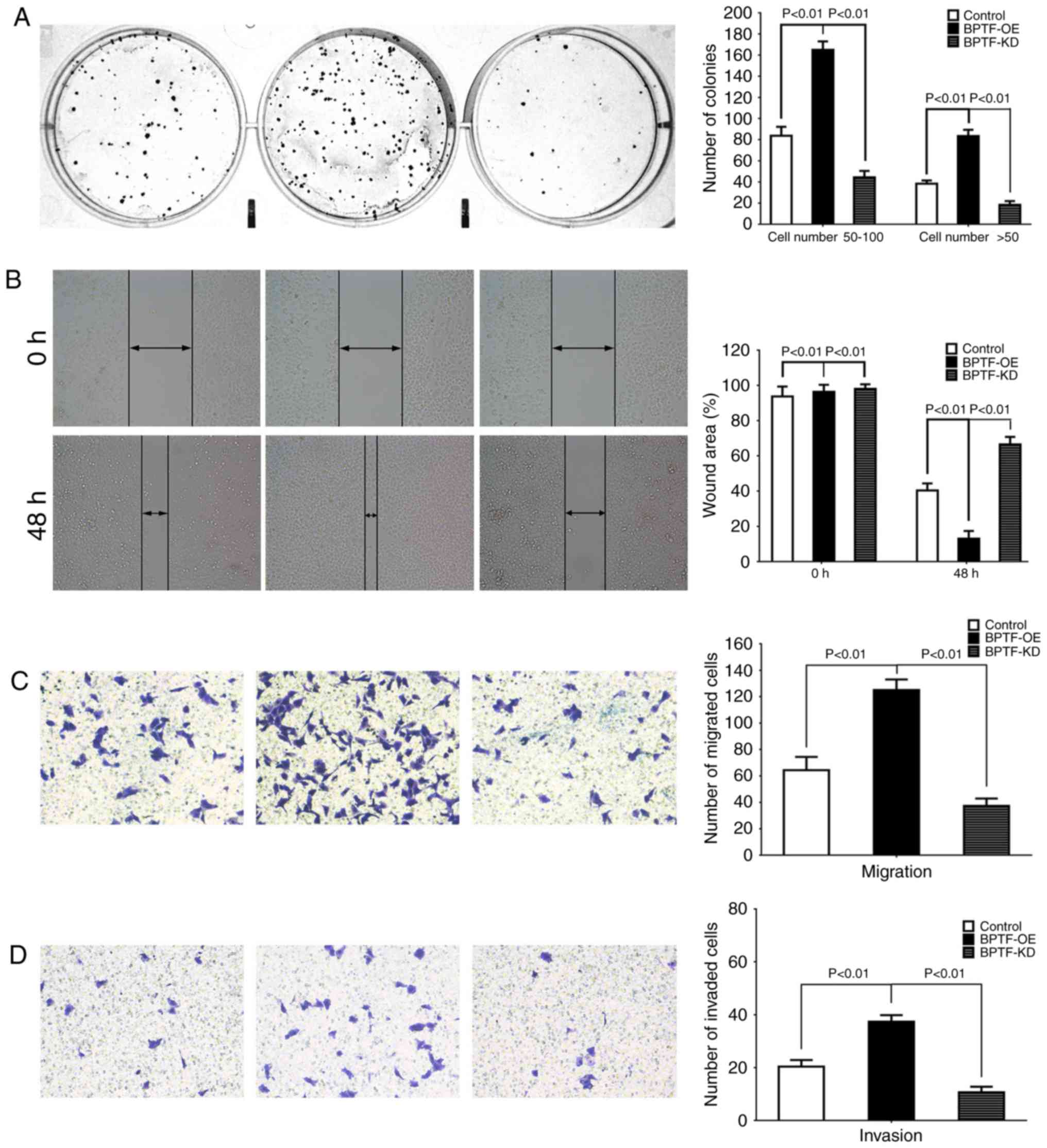
proliferation ability by colony-forming assay. The results revealed
that BPTF overexpression increased the size and number of U251 cell
colonies, while BPTF knockdown decreased the size and number of
colonies compared to their control (Fig. 3A). Then, we detected the effects of
BPTF on motility and migration by wound healing and Transwell
assays. In comparison with the control cells, wound healing and
Transwell assays revealed that BPTF overexpression significantly
enhanced the motility and migration of U251 cells (Fig. 3B and C). Then, the Transwell
Matrigel invasion assay revealed that BPTF overexpression increased
the number of cells that invaded through the Matrigel membrane
which indicated that BPTF enhanced the invasiveness ability of U251
cells (Fig. 3D). Conversely,
knockdown of BPTF in U251 cells significantly reduced the motility
and migration (Fig. 3B and C) and
invasive capacity of cells (Fig.
3D). These results indicated that BPTF could enhance malignant
characteristics of U251 cells in vitro.
BPTF promotes glioma growth and
invasion via Myc signaling
Next, we wanted to explore the potential molecular
mechanism of BPTF in the promotion of glioma proliferation and
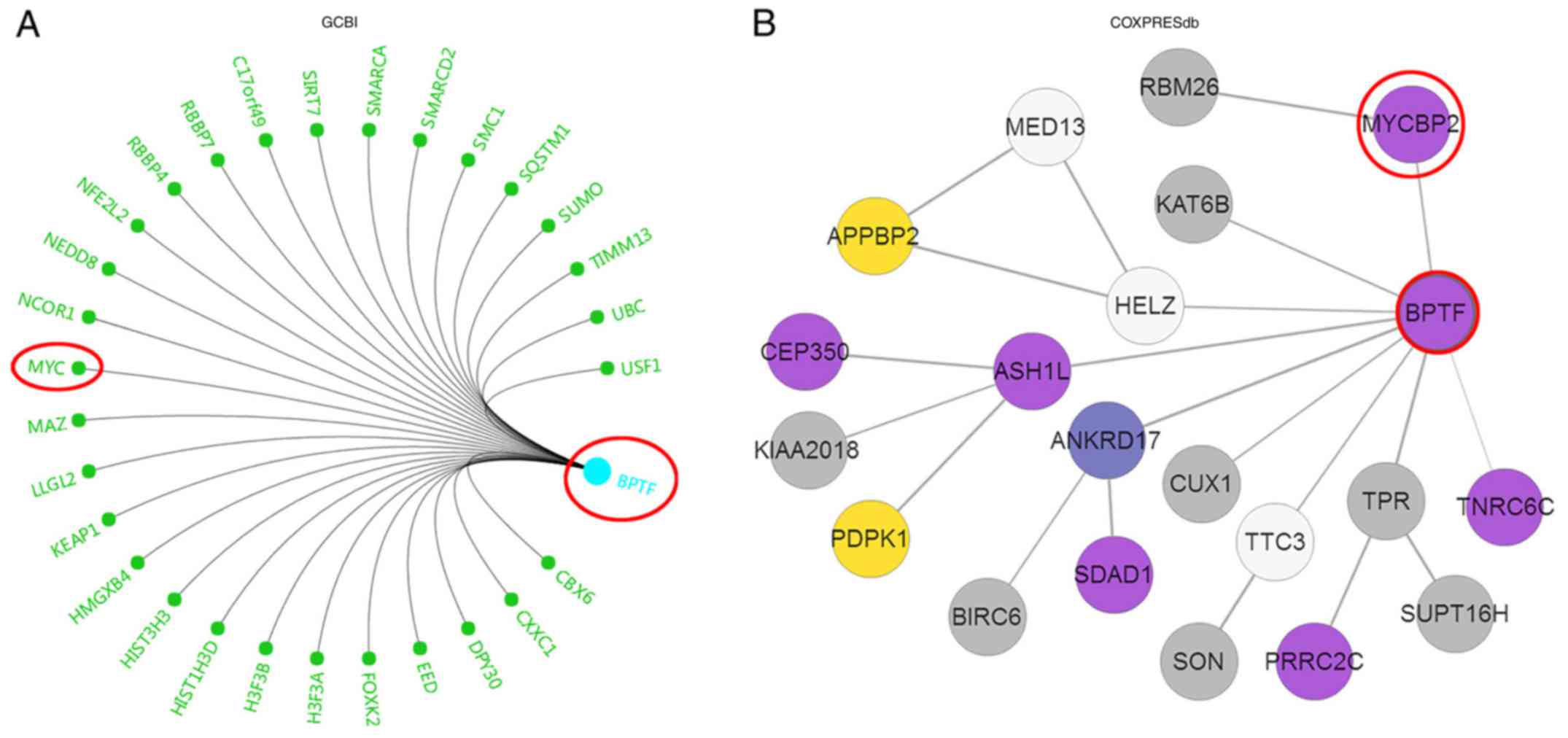
invasion. We first utilized the Gene-Cloud of Biotechnology (GCBI;
http://www.gcbi.com.cn/gclib/html/index) database to
search for potential interacting genes and found that Myc was
evidently a potential candidate (Fig.
4A). Since Myc signaling is vital for proliferation and
invasion in various tumors, we further searched for the association
of BPTF and Myc in COXPRESdb and Oncomine databases. The results
revealed that BPTF was co-expressed with MYCBP2 (Fig. 4B and C) and MYCBPAP (Fig. 4D), both of which are Myc-binding
association proteins. According to these bioinformatics results, we
further detected c-Myc expression, which is the core member of Myc
signaling, in BPTF overexpressed or knocked down U251 cells. The
results revealed that c-Myc mRNA and protein expression level were
significantly increased in U251BPTF-OE cells than the
control cells but decreased in U251BPTF-KD cells
(Fig. 4E and F). These data
indicated that BPTF promoted glioma proliferation and invasion via
Myc signaling.
Discussion
Glioma can occur at any age, regardless of sex or
ethnicity. On the basis of their histopathology, gliomas are
divided into grades I–IV according to the World Health Organisation
(WHO) grading system (3,16,17).
Despite advances in surgery, radiotherapy and chemotherapy that
have been achieved, current therapies against gliomas are still not
effective enough and with poor long-term survival. Grades III and
IV gliomas are more aggressive and difficult to treat owing to
frequent dysfunction of tumor suppressors and oncogenes (18). Therefore, the present study explored
the role of BPTF as an oncogene in glioma and provided a potential
option for prognostic prediction and targeted therapy of
glioma.
The present study by combining an online
bioinformatics database and clinical specimens, first revealed that
BPTF expression was highly expressed in gliomas and correlated with
poor survival. These findings were consistent with previous
research on other tumors, such as hepatocellular carcinoma (HCC),
and lung and colorectal cancer (CRC) (13,19).
We also explored the clinical significance of BPTF in glioma
patients and found that the expression of BPTF in glioma tissues
was correlated with histopathological grade and tumor size,
indicating that BPTF expression may be related with glioma
progression. The survival curves and univariate and multivariate
analyses for survival also revealed that high BPTF expression was
an independent risk factor affecting the prognosis of glioma
patients, indicating that the elevated expression of BPTF could be
a useful predictor for predicting prognosis of glioma patients. The
clinicopathological and prognostic findings were also consistent
with the researches in melanoma, HCC and CRC (12,19,20).
All this evidence indicated that BPTF may be a pan-oncogene in
tumors, and extended the knowledge of the biological role of BPTF,
especially in the field of tumor research.
Notably, we also investigated the functional role of
BPTF in glioma. Our in vitro experiments indicated that BPTF
could increase the proliferation and invasiveness of glioma cells.
BPTF was first identified and characterized in 2000 and was
considered to play a role in hormonally-regulated,
chromatin-mediated regulation of transcription during proliferation
(8). Then, further studies found
that BPTF played an important role in embryo development, stemness
maintenance and self-renewal capacity of stem/progenitor cells
(21–23). These known biological functions were
revealed to be correlated with tumor progression. Research also
found that BPTF was overexpressed in many cancer cell lines and had
the ability to promote cancer cell growth (24). In addition, further studies revealed
the correlation of BPTF aberrant expression with cancer and found
that BPTF could promote tumor cell proliferation, invasion and
metastasis, thus resulting in poor prognosis (13,25–27).
The present study also provided first-hand data of the promoting
effect of BPTF on proliferation and invasion in glioma cells, which
was consistent with studies in other types of cancers (24–27)
and not reported in glioma before.
The present study also explored the potential
molecular mechanism of BPTF in glioma. The bioinformatics and
online public databases provided a preliminary correlation of BPTF
with Myc signaling. Since the study conducted in 2000 first found
that BPTF could interact with the Myc-associated zinc finger
protein, studies confirmed that BPTF was a crucial c-Myc co-factor
and played a key role in c-Myc-mediated tumor progression. The
present study also revealed the correlation and potential
interaction of BPTF and c-Myc in glioma (11,26,28).
These data further indicated that BPTF may be a useful therapeutic
target for c-Myc aberrantly-expressed tumors. The main shortcomings
of our study are as follows: i) we did not conduct gain- and
loss-of-function analyses for c-Myc with BPTF interference; ii)
this study did not provide detailed molecular mechanism analysis of
how BPTF regulated c-Myc in glioma; iii) we did not provide in
vivo intervention experiments examining the therapeutic effect
of BPTF inhibition. Therefore, our future research directions will
be aimed at these shortcomings.
In summary, our results revealed that BPTF was
overexpressed in glioma and could promote proliferation and
invasion of glioma cells via Myc signaling. Moreover, BPTF may be a
valuable prognostic marker and therapeutic target for glioma
patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YP, FY, ZL and LC conceived the study and wrote the
manuscript; YP, FY, YL, GW and ZL conducted the experiments and
contributed to the analysis of data. YP, FY, YL and GW collected
the clinical samples and the corresponding clinical data. YP, FY
and LC revised the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Haikou People's Hospital in accordance with the Declaration of
Helsinki. All patients provided signed written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kohler BA, Ward E, McCarthy BJ, Schymura
MJ, Ries LA, Eheman C, Jemal A, Anderson RN, Ajani UA and Edwards
BK: Annual report to the nation on the status of cancer, 1975–2007,
featuring tumors of the brain and other nervous system. J Natl
Cancer Inst. 103:714–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Popova SN, Bergqvist M, Dimberg A, Edqvist
PH, Ekman S, Hesselager G, Ponten F, Smits A, Sooman L and
Alafuzoff I: Subtyping of gliomas of various WHO grades by the
application of immunohistochemistry. Histopathology. 64:365–379.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ho VK, Reijneveld JC, Enting RH, Bienfait
HP, Robe P, Baumert BG and Visser O: Dutch Society for
Neuro-Oncology (LWNO): Changing incidence and improved survival of
gliomas. Eur J Cancer. 50:2309–2318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hanahan D and Robert W: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC: metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Principe DR, Doll JA, Bauer J, Jung B,
Munshi HG, Bartholin L, Pasche B, Lee C and Grippo PJ: TG F-β
Duality of function between tumor prevention and carcinogenesis. J
Natil Cancer Inst. 106:djt3692014. View Article : Google Scholar
|
|
7
|
Wysocka J, Swigut T, Xiao H, Milne TA,
Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et
al: A PHD finger of NURF couples histone H3 lysine 4 trimethylation
with chromatin remodelling. Nature. 442:86–90. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones MH, Hamana N and Shimane M:
Identification and characterization of BPTF, a novel bromodomain
transcription factor. Genomics. 63:35–39. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goller T, Vauti F, Ramasamy S and Arnold
HH: Transcriptional regulator BPTF/FAC1 is essential for
trophoblast differentiation during early mouse development. Mol
Cell Biol. 28:6819–6827. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma Y, Liu X, Liu Z, Wei S, Shang H, Xue Y,
Cao Y, Meng A and Wang Q: The chromatin remodeling protein Bptf
promotes posterior neuroectodermal fate by enhancing
Smad2-activated wnt8a expression. J Neurosci. 35:8493–8506. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Richart L, Real FX and Sanchez-Arevalo
Lobo VJ: c-MYC partners with BPTF in human cancer. Mol Cell Oncol.
3:e11523462016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao S, Liu L, Lu X, Long J, Zhou X and
Fang M: The prognostic significance of bromodomain PHD-finger
transcription factor in colorectal carcinoma and association with
vimentin and E-cadherin. J Cancer Res Clin Oncol. 141:1465–1474.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dai M, Lu JJ, Guo W, Yu W, Wang Q, Tang R,
Tang Z, Xiao Y, Li Z, Sun W, et al: BPTF promotes tumor growth and
predicts poor prognosis in lung adenocarcinomas. Oncotarget.
6:33878–33892. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mayes K, Elsayed Z, Alhazmi A, Waters M,
Alkhatib SG, Roberts M, Song C, Peterson K, Chan V, Ailaney N, et
al: BPTF inhibits NK cell activity and the abundance of natural
cytotoxicity receptor co-ligands. Oncotarget. 8:64344–64357. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boruah D, Deb P, Srinivas V and Mani NS:
Morphometric study of nuclei and microvessels in gliomas and its
correlation with grades. Microvasc Res. 93:52–61. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Minniti G, Muni R, Lanzetta G, Marchetti P
and Enrici RM: Chemotherapy for glioblastoma: Current treatment and
future perspectives for cytotoxic and targeted agents. Anticancer
Res. 29:5171–5184. 2009.PubMed/NCBI
|
|
19
|
Xiao S, Liu L, Fang M, Zhou X, Peng X,
Long J and Lu X: BPTF associated with EMT indicates negative
prognosis in patients with hepatocellular carcinoma. Dig Dis Sci.
60:910–918. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dar AA, Majid S, Bezrookove V, Phan B,
Ursu S, Nosrati M, De Semir D, Sagebiel RW, Miller JR III, Debs R,
et al: BPTF transduces MITF-driven prosurvival signals in melanoma
cells. Proc Natl Acad Sci USA. 113:6254–6258. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Landry J, Sharov AA, Piao Y, Sharova LV,
Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, et al:
Essential role of chromatin remodeling protein Bptf in early mouse
embryos and embryonic stem cells. PLoS Genet. 4:e10002412008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu B, Cai L, Butler JM, Chen D, Lu X,
Allison DF, Lu R, Rafii S, Parker JS, Zheng D and Wang GG: The
chromatin remodeler BPTF activates a stemness gene-expression
program essential for the maintenance of adult hematopoietic stem
cells. Stem Cell Reports. 10:pp. 675–683. 2018, View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frey WD, Chaudhry A, Slepicka PF,
Ouellette AM, Kirberger SE, Pomerantz WCK, Hannon GJ and Dos Santos
CO: BPTF maintains chromatin accessibility and the self-renewal
capacity of mammary gland stem cells. Stem Cell Reports. 9:pp.
23–31. 2017, View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Buganim Y, Goldstein I, Lipson D,
Milyavsky M, Polak-Charcon S, Mardoukh C, Solomon H, Kalo E, Madar
S, Brosh R, et al: A novel translocation breakpoint within the BPTF
gene is associated with a pre-malignant phenotype. PLoS One.
5:e96572010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dar AA, Nosrati M, Bezrookove V, de Semir
D, Majid S, Thummala S, Sun V, Tong S, Leong SP, Minor D, et al:
The role of BPTF in melanoma progression and in response to
BRAF-targeted therapy. J Natl Cancer Inst. 107:djv0342015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Richart L, Carrillo-de Santa Pau E,
Río-Machín A, de Andrés MP, Cigudosa JC, Lobo VJ and Real FX: BPTF
is required for c-MYC transcriptional activity and in vivo
tumorigenesis. Nat Commun. 7:101532016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Li J, Luo M, Zhou C, Shi X, Yang W,
Lu Z, Chen Z, Sun N and He J: Novel long noncoding RNA NMR promotes
tumor progression via NSUN2 and BPTF in esophageal squamous cell
carcinoma. Cancer Lett. 430:57–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jordan-Sciutto KL, Dragich JM, Caltagarone
J, Hall DJ and Bowser R: Fetal Alz-50 clone 1 (FAC1) protein
interacts with the Myc-associated zinc finger protein (ZF87/MAZ)
and alters its transcriptional activity. Biochemistry.
39:3206–3215. 2000. View Article : Google Scholar : PubMed/NCBI
|