Introduction
Aberrant activation of Wnt, an outside-in signaling
pathway, is involved in the majority of malignancy types, including
gastric cancer. Secreted frizzled-related protein 1 (sFRP1) has
been reported to bind to Wnt ligands and modulate their ability to
activate signal transduction (1–3). sFRPs
are a family of secreted proteins homologous to the Frizzled (Fz)
receptors, which bind Wnt ligands (4). They possess only the cysteine-rich
domain and lack the seven trans-membrane and intracellular domains
of Fz proteins (5). The expression
of sFRP1 may vary with disease status or with the stage of
development. sFRP1 has been demonstrated to serve a critical role
in the development of the lung (6),
prostate (7) and gut (8). sFRP1 is expressed in developing
tissues but not in mature prostate epithelial cells (7).
Conflicting reports indicate that sFRP1 is able to
serve tumor-promoting and tumor-suppressing roles. Transcriptional
inactivation of sFRPs has been reported in various cancer types
(9–12), supporting the hypothesis that sFRPs
function as tumor suppressors; however, contrary results have also
been published. Loss of sFRP1 expression has been determined in
>80% of invasive breast carcinoma types, excluding the medullary
type, and is associated with the presence of hormonal receptors
(13). sFRP1 is highly expressed in
the basal-like breast cancer (14)
and brain relapses, compared with luminal tumor types and bone
relapses (15). Similarly, high
levels of sFRP1 in carcinomas are associated with the presence of
lymphoplasmacytic stroma (13). In
addition to its function of inhibiting the Wnt/canonical pathway,
sFRP1 is also reported to increase the diffusion of Wnt ligands
(16), and interact with Hedgehog
(17,18), tumor necrosis factor (19) and integrin signaling (20), which indicates that sFRP1 is a
multi-functional protein (20,21).
Gastric carcinoma is the fourth most common
malignancy globally, with an estimated 989,000 novel cases and
738,000 mortalities reported in 2008 (22). The depth of invasion and the
presence of lymph node metastases are considered to be the most
important prognostic factors in gastric cancer (23,24).
sFRP1 was overexpressed in aggressive subgroups of human gastric
cancer, and was significantly associated with lymph node metastasis
and decreased overall survival rate in patients with gastric cancer
(25), which is consistent with
another previous study that demonstrated that sFRP1 is
overexpressed only in metastatic renal carcinoma, but not in
primary tumor types (26). sFRP1
overexpression is associated with the activation of the
transforming growth factor β (TGFβ) signaling pathway and thereby
induced cell proliferation, epithelial-mesenchymal transition (EMT)
and invasion (25). Expression of
sFRP1 decreases the intracellular levels of β-catenin, indicating
the inhibition of the Wnt/canonical signaling pathway (5). Crosstalk between the Wnt and TGFβ
signaling pathways that are regulated by sFRP1 are substantially
associated with one another (25).
Despite these data indicating that sFRP1 is able to promote or
repress tumorigenesis, the mechanism by which sFRP1 governs cell
signaling remains unclear.
In the present study, the molecular mechanism
underlying sFRP1-induced signaling alterations was investigated,
based on previous data. The critical role of glycogen synthase
kinase 3β (GSK3β) and Rac family small GTPase 1 (Rac1) in mediating
the sFRP1 signaling, which regulates malignant behaviors and TGFβ
signaling in gastric cancer cells, was investigated.
Materials and methods
Cell culture and chemicals
Human gastric cancer cell lines SGC-7901 and BCG823
were obtained from the Shanghai Institute of Cell Biology, Chinese
Academy of Sciences (Shanghai, China). Cells were cultured in
RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS) (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2
incubator. GSK3β inhibitor IM-12 (10, 20 and 50 µM) and Rac1
inhibitor NSC23766 (25, 50 and 100 µM) were obtained from Merck
KGaA (Darmstadt, Germany). Cells treated with inhibitors were
cultured in a 37°C incubator at 5% CO2.
Cell proliferation assays
Cell proliferation was assessed using an MTT
(Sigma-Aldrich; Merck KGaA) assay. A total of 2×103
cells in 100 µl culture medium were plated in a 96-well plate. MTT
reagent (5 mg/ml) was added into each well for 48 h and the plate
was returned to a 37°C incubator for 3 h. The culture medium was
aspirated and 150 µl dimethyl sulfoxide (DMSO) was added into each
well. The plate was shaken in an orbital shaker for 15 min. The
absorbance at an optical density of 590 nm was measured using a
microplate reader. GSK3β inhibitor IM-12 (10, 20 and 50 µM) and
Rac1 inhibitor NSC23766 (25, 50 and 100 µM) were added 24 h after
the cells (SGC-7901/vector and SGC-7901/sFRP1) were plated. Control
groups were treated using DMSO.
Plasmids and transfection
sFRP1 vector was purchased from OriGene
Technologies, Inc. (Rockville, MD, USA). The green fluorescent
protein-fused wild-type (WT) Rac1, constitutively active (CA)
mutant Rac1 (Q61L), dominant-negative (DN) mutant Rac1 (T17N)
(27), Tag5Amyc-GSK3β WT (28), pCS2 Flag Smad3 S204A (29), pCMV5B-Flag-Smad3 (30) and pCMV5 Smad2-HA (31) were purchased from Addgene, Inc.
(Cambridge, MA, USA). Top-flash luciferase plasmid (BPS Bioscience,
San Diego, CA, USA) is a luciferase reporter plasmid that contains
two sets of 3 copies of the wild-type T-cell factor (TCF) binding
regions. If the canonical Wnt signaling is activated, β-catenin
will translocate to the nucleus to associate with TCF/lymphoid
enhancer factor transcription factors to activate transcription of
Wnt target genes. pSV-β-Galactosidase control vector was used as an
internal control for transfection and was purchased from Promega
Corporation (Madison, WI, USA). For plasmid transfection,
SGC-7901/vector cells were seeded into 6-well plate and allowed to
grow 24 h prior to transfection. Cells in each well were
transfected with 4 µg plasmid using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for 6 h at 37°C and
the medium was changed subsequent to transfection.
Transient expression reporter gene
assay
The transcriptional activity of β-catenin was
measured by co-transfection with Top-flash luciferase plasmid and
sFRP1 vector (OriGene Technologies, Inc.) or the control vector
using Lipofectamine® 2000 for 6 h at 37°C. TOPFlash
encoding the LEF/TCF binding sites (insert gene) linked to firefly
luciferase and reflecting Wnt/β-catenin signaling activity was
used. After 24 h incubation, the luciferase activity was measured
and normalized to β-galactosidase activity (Promega Corporation).
The Luciferase Reporter Gene Detection kit (Promega Corporation)
and GloMax®-Multi+ Detection system (Promega
Corporation) were used according to the manufacturer's protocol.
The data presented were the mean value of three independent
experiments.
Activity of Rac1 assay
The activation of Rac1 was measured using a Rac1
Activation Assay Biochem kit (Cytoskeleton, Inc., Denver, CO, USA)
according to the manufacturer's protocol. Briefly, cell lysates
were collected using the lysis buffer at 4°C for 30 min from the
kit. The activated forms of Rac1 were combined by Rac1 activated
kinase (PAK)-Rac/Cdc42 (p21) binding domain (PBD) affinity beads.
The beads were centrifuged at 5,000 × g at 4°C for 1 min and
activated GTPases were pulled-down into the bead pallets. Bound
GTPases were eluted by SDS buffer and analyzed by 12% SDS-PAGE and
western blotting. Rac1 levels were analyzed by the specific
antibodies.
Western blotting
Whole cell lysates (SGC-7901/vector, SGC-7901/sFRP1,
inhibitor treated cells and plasmid-transfected cells) were
harvested using radio immunoprecipitation assay cell lysis buffer
supplemented with a protease inhibitor cocktail (Sigma-Aldrich;
Merck KGaA) at 4°C for 30 min. The nuclear and cytosol extracts
were isolated using the nuclear extract kit (Active Motif,
Carlsbad, CA, USA) according to the manufacturer's protocol. The
concentration of the proteins were measured using a DC Protein
assay kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according
to the manufacturer's protocol. A total of 30 µg protein was loaded
per lane and loaded into 10–12% SDS-PAGE gels. The proteins were
then transferred to a polyvinylidene fluoride membranes followed by
blocking with 5% bovine serum albumin (BSA) for 2 h at room
temperature on a rocket shaker. Membranes were washed using
tris-buffered saline with 0.1% Tween-20 (pH 8.0) three times for 10
min each. Primary antibodies (1:1,000) against sFRP1 (cat no.
ab126613), phosphorylated (p-)Smad3L (cat no. ab63402), zinc finger
E-box binding homeobox 2 (ZEB2; cat no. ab138222) and lamin A/C
(cat no. ab108922) were purchased from Abcam (Cambridge, UK).
Antibodies against Smad3 (cat no. 9523), Smad2 (cat no. 5339),
Smad4, p-Smad3c (cat no. 9520), p-Smad2c (cat no. 3108), GSK3β (cat
no. 12456), p-GSK3β Ser9 (cat no. 9323), p21 (cat no. 2947),
β-catenin (cat no. 8480), p-Rac1/cell division cycle 42 S71 (cat
no. 2461) (all obtained from Cell Signaling Technology, Inc.,
Danvers, MA, USA) and inhibitor of DNA binding 1 (ID1; cat no.
5559-1), Vav guanine nucleotide exchange factor 2 (VAV2; cat no.
EP1067Y), plasminogen activator inhibitor 1 (PAI1; cat no.
EPR21850-82) (all obtained from Epitomics, Burlingame, CA, USA)
were used at 1:1,000 dilutions. Antibodies against GAPDH (cat no.
G8795), Lamin A/C (cat no. SAB4200236) and Lamin C (cat no.
MAB3540) (Sigma-Aldrich; Merck KGaA) were used at a 1:5,000
dilution. Primary antibodies were diluted in 5% BSA and incubated
overnight at 4°C. Horseradish peroxidase (HRP)-conjugated goat
anti-rabbit immunoglobulin G (IgG) H&L (cat no. ab6789) and
HRP-conjugated goat anti-mouse IgG H&L (cat no. ab6721)
secondary antibodies were purchased from Abcam and used at a
1:10,000 dilution in 5% BSA at room temperature for 2 h. Signals
were visualized using enhanced chemiluminescence reagent (Pierce;
Thermo Fisher Scientific, Inc.). Membranes were scanned using the
ChemiDoc Touch Imaging system (Bio-Rad Laboratories, Inc.) and the
images were captured using Image Lab Touch Software (version
1.0.0.15; Bio-Rad Laboratories, Inc.).
Immunofluorescence staining
Cells were fixed with 4% formaldehyde at room
temperature for 30 min and then permeabilized with PBS containing
0.2% Triton X-100 for 15 min at room temperature. Slides were
blocked using 5% bovine serum albumin at room temperature for 1 h.
For F-actin staining, cells were fixed with 4% formaldehyde at room
temperature for 30 min and then incubated with Alexa Fluor
555® phalloidin (Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature for 30 min. The nucleus was
counterstained using DAPI at room temperature for 5 min. Slides
were washed using PBS, mounted and observed under a microscope.
Immunofluorescence staining was visualized and captured using a
Nikon Digital Sight DS-U2 (Nikon Corporation, Tokyo, Japan) and NIS
elements F3.0 software was used (Nikon Corporation). Confocal
images were obtained using an inverted ZEISS LSM710 confocal
microscope (×40 oil lens; Carl Zeiss AG, Oberkochen, Germany). Zen
2009 Light Edition (Carl Zeiss AG) was used for measurement of the
images.
Cell migration assays
Cell migration was analyzed using a Transwell
chamber assay. A 24-well plate with 8 µm pore size inserts was
used. SGC-7901/vector and SGC-7901/sFRP1 cells treated with vehicle
(DMSO), NSC23766 (25 µM) and IM-12 (10 µM) for 24 h were used. A
total of 1×104 cells were mixed in 100 µl RPMI-1640
medium and added to the upper chamber of the Transwell insert. A
total of 600 µl RPMI 1640-medium with 10% FBS was added to the
lower chamber. The Transwell inserts were then placed into the
wells of a 24-well plate. After 12 h incubation at 37°C, the cells
were fixed using 4% formaldehyde at room temperature for 30 min and
stained by 0.01% (v/v in methanol) crystal violet at room
temperature for 10 min. Subsequent to being washed three times by
H2O, the cells on the inner side of the chamber were
removed with a cotton swab and the cells on the outer side of the
chamber were counted under a light microscope at a magnification of
×100. Cells were visualized using an Olympus BX50 microscope,
images were captured using Nikon Digital Sight DS-U2 and NIS
elements F3.0 software was used for analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured SGC7901/vector
and SGC-7901/sFRP1 cells using the RNeasy mini kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's protocols, and
cDNA was synthesized with oligo (dT) primers by using of a
SuperScript first-strand cDNA synthesis kit (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
A total of 1 µg RNA was used to synthesize cDNA. Gene expression
was assessed by RT-qPCR using an Applied Biosystems 7500 Fast
Sequence Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR reaction mixture consisted of QuantiTect
SYBR Green PCR master mix (2× QuantiTect SYBR Green kit, containing
HotStart Taq® DNA polymerase, QuantiTect SYGB Green PCR
buffer, dNTP mix, SYGB I, Rox passive reference dye and 5 mM
MgCl2; Qiagen, Inc.), 0.5 µmol/l of each primer and
cDNA. The thermocycling conditions were as follows: 95°C for 30
sec, 40 cycles at 95°C for 5 sec, 60°C for 30 sec; and the
dissociation stage at 95°C for 15 sec, 60°C for 1 min and 95°C for
15 sec. The transcript of the housekeeping gene, GAPDH was used as
an endogenous control to normalize the expression data. The
comparative Cq method was used to calculate the relative changes in
gene expression. Expression fold change was calculated using the
equation 2 − (Cq gene - Cq GAPDH) (32). The primers used were as follows:
PAI1 forward, 5′-TGGCACGGTGGCCTCCTCAT-3′ and reverse,
5′-ACTGTTCCTGTGGGGTTGTGCC-3′; ID1 forward,
5′-CGAGATCAGCGCCCTGACGG-3′ and reverse, 5′-GGCCGCCGATCGGTCTTGTT-3′;
Smad3 forward, 5′-GGAGAAATGGTGCGAGAAGG-3′ and reverse,
5′-GAAGGCGAACTCACACAGC-3′; p21 forward, 5′-GCCGAAGTCAGTTCCTTGTG-3′
and reverse, 5′-TTCTGACATGGCGCCTCCT-3′; activating transcription
factor 3 (ATF3) forward, 5′-GAGGTGGGGTTAGCTTCAGT-3′ and reverse,
5′-TTGATTTTGGGGCAAGGTGC-3′; GAPDH forward,
5′-GGGCTCTCTGCTCCTCCCTGTTCT-3′ and reverse,
5′-CAGGCGTCCGATACGGCCAAA-3′.
Statistical analysis
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analysis. Values are presented as the mean
± standard deviation of samples measured in triplicate. P<0.05
was considered to indicate a statistically significant difference.
Each experiment was repeated three times, unless otherwise
indicated. The significance of differences between experimental
groups compared to the vehicle control group was analyzed using a
paired Student's t-test and two-tailed distribution. Multiple
comparisons were analyzed using a one-way analysis of variance
(ANOVA) test. Newman-Keuls test was used following ANOVA.
Results
Overexpression of sFRP1 activates
Rac1
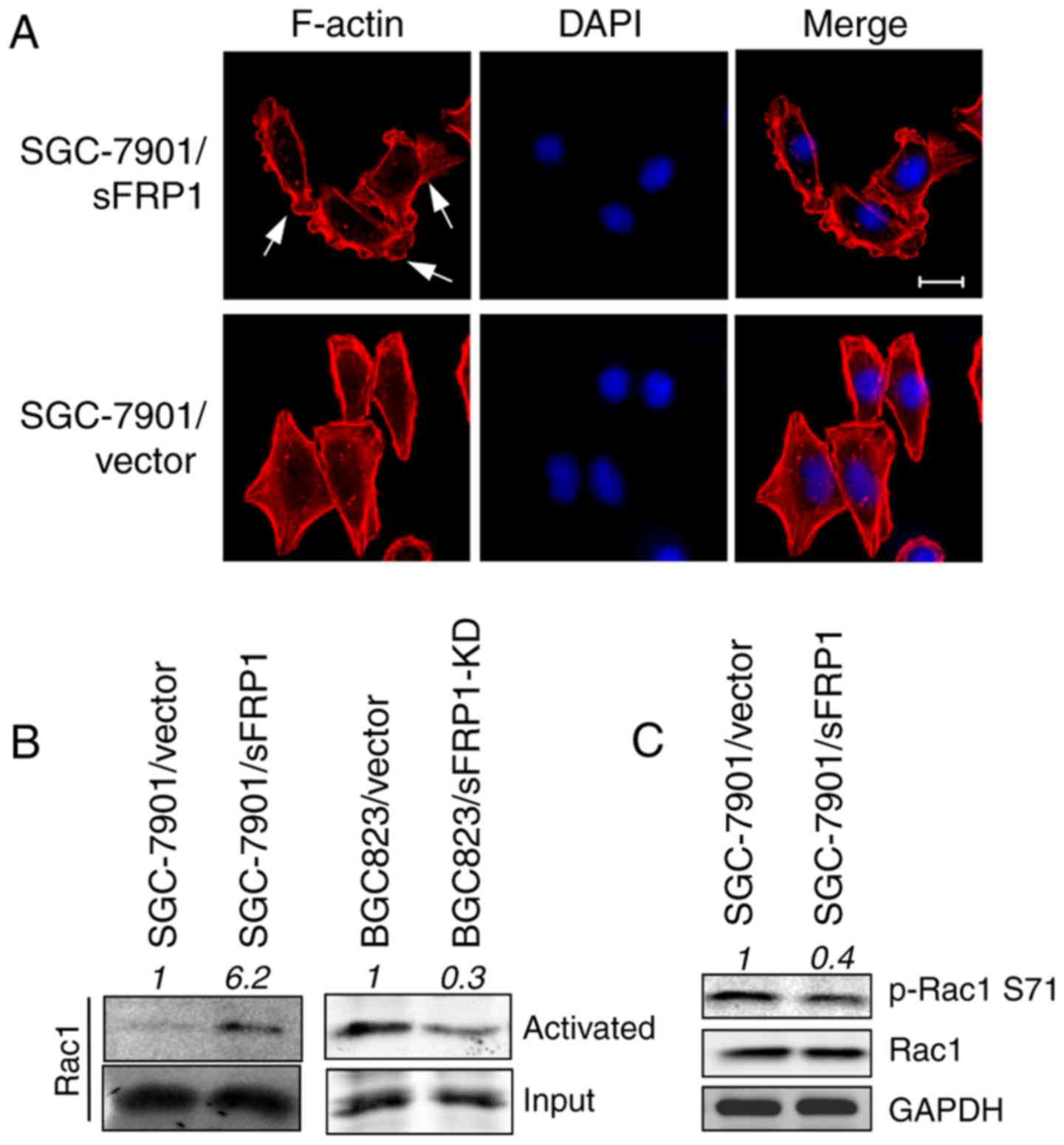
Firstly, the cell morphological changes induced by
sFRP1 overexpression were investigated. SGC-7901/sFRP1 cells
exhibited extended and protruded lamellipodia composed of F-actin
fibro stained by phalloidin (Fig.
1A, upper); however, SGC-7901/vector cells exhibited a
polygon-like shape with less lamellar extensions around the entire
cell periphery (Fig. 1A, lower).
This phenomenon indicated that Rac1, which is well known to be
involved in filopodia and lamellipodia formation and thus control
cell movement (33), was activated
by sFRP1 overexpression. Therefore, Rac1 activity was measured in
control and sFRP1-overexpressing cells by kinase activity assays.
In agreement with a previous study (33), sFRP1-overexpressing cells exhibited
increased Rac1 activity (Fig. 1B,
left); however, the loss of Rac1 activity/activation was observed
in sFRP1-knockdown BGC823 cells, compared with vector only control
cells (Fig. 1B, right).
Immunoblotting also demonstrated a lower level of its inactivated
form p-Rac1 Ser71 in sFRP1-overexpressing cells (Fig. 1C). These data indicated that sFRP1
activates Rac1 activity.
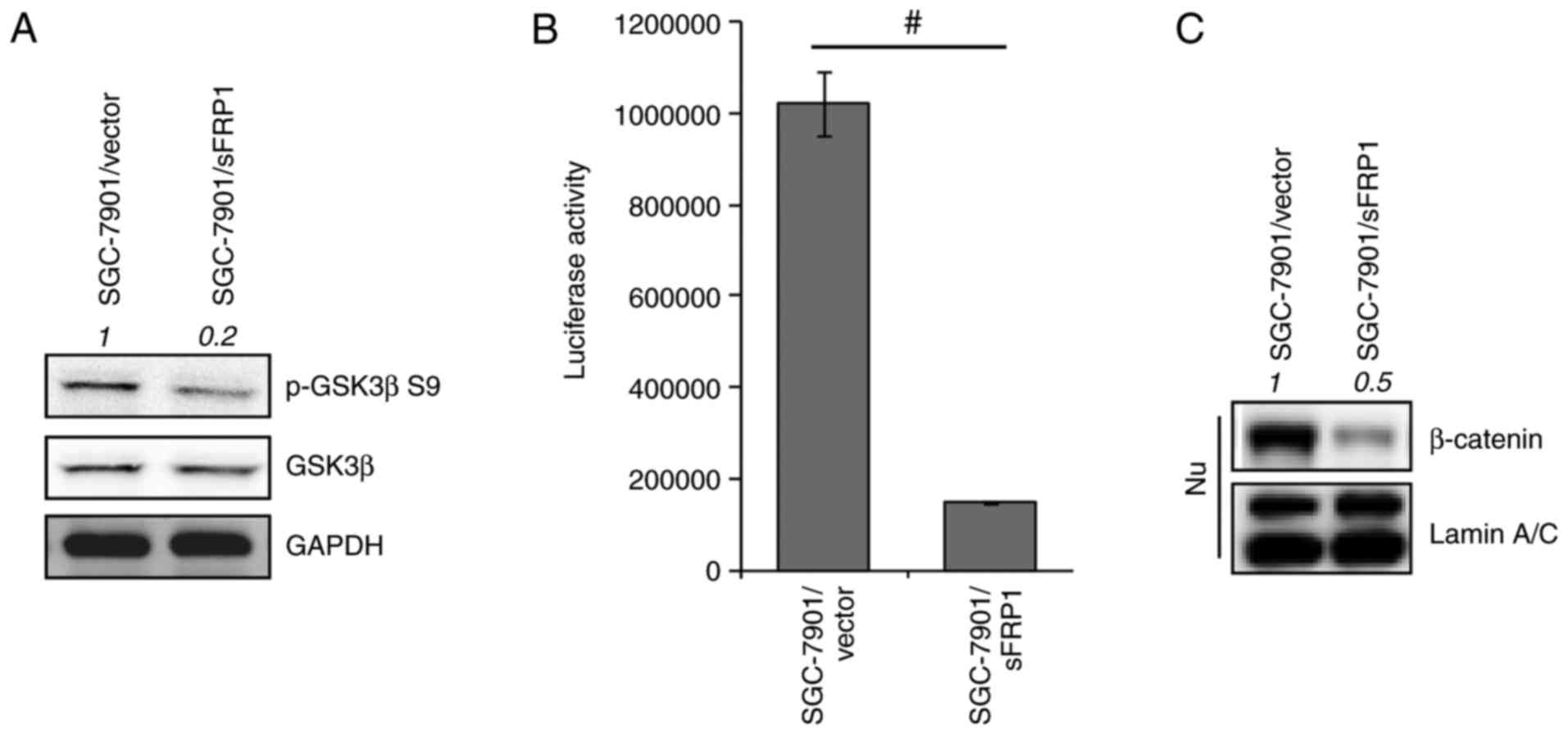
sFRP1 overexpression restores GSK3β
activity
In addition, it was reported previously that sFRP1
abrogates GSK3β inactivation by preventing its phosphorylation at
the Ser9 residue (34). The present
study also demonstrated a lower level of p-GSK3β Ser9 in
sFRP1-overexpressing cells compared with the control cells
(Fig. 2A). In agreement with the
notion that sFRP1 is an inhibitor of Wnt signaling, it was
determined that TCF-responsive luciferase activity was
significantly repressed by sFRP1 overexpression compared with the
control cells (P<0.05; Fig. 2B)
and the nuclear accumulation of β-catenin was attenuated (Fig. 2C). Consistent with other data, the
present cell model also demonstrated that sFRP1 overexpression
restored GSK3β activity and inhibited the Wnt/canonical
pathway.
sFRP1 regulates Rac1 activity through
GSK3β
Due to sFRP1 overexpression activating Rac1 and
GSK3β, and GSK3β being previously reported to modulate Rac1
activity (35), the present study
investigated whether GSK3β regulated Rac1 activity in
SGC-7901/sFRP1 cells. Decreased lamellipodia formation, a feature
of Rac1 inactivation, was observed in SGC-7901/sFRP1 cells treated
with GSK3β inhibitor IM-12 or Rac1 inhibitor NSC23766 compared with
vehicle cells (Fig. 3A). As
depicted in Fig. 3B, a reduced
amount of Rac1 bound to PAK-PBD compared with vehicle cells, which
indicated reduced Rac1 activity. Levels of VAV2, a guanine
nucleotide exchange factor (GEF) and activator of Rac1 (36), were lower in NSC23766 and IM-12
treated cells that were precipitated by PAK-PBD compared with
vehicle cells, indicating that GSK3β or Rac1 inhibition suppressed
Rac1 activity. Notably, GSK3β was also one of the components that
was precipitated by PAK-PBD beads, and its level was decreased upon
Rac1 or GSK3β inhibition compared with the vehicle control cells
(Fig. 3B, left). The total levels
of Rac1, GSK3β, and VAV2 remained consistent in cells with
different treatments (Fig. 3B,
right). Due to GSK3β being precipitated by PAK-PBD, which bound the
activated form of Rac1, this indicated that GSK3β may directly or
indirectly interact with Rac1; therefore, the levels of
precipitated GSK3β were decreased in a similar pattern to the
levels of the activated-Rac1, indicating that GSK3β may regulate
Rac1 activity. Subsequently, a GSK3β overexpression model was used
to investigate whether GSK3β was able to regulate Rac1 activity. As
expected, a low level of the inactivated form of Rac1 (p-Rac1
Ser71) was observed in GSK3β-overexpressing cells compared with the
vector cells (Fig. 3C). Due to
NSC23766 inhibiting Rac1-GEF interaction (37) and IM-12 directly suppressed GSK3β
activity (38), GSK3β activity may
be necessary for regulating Rac1 activity.
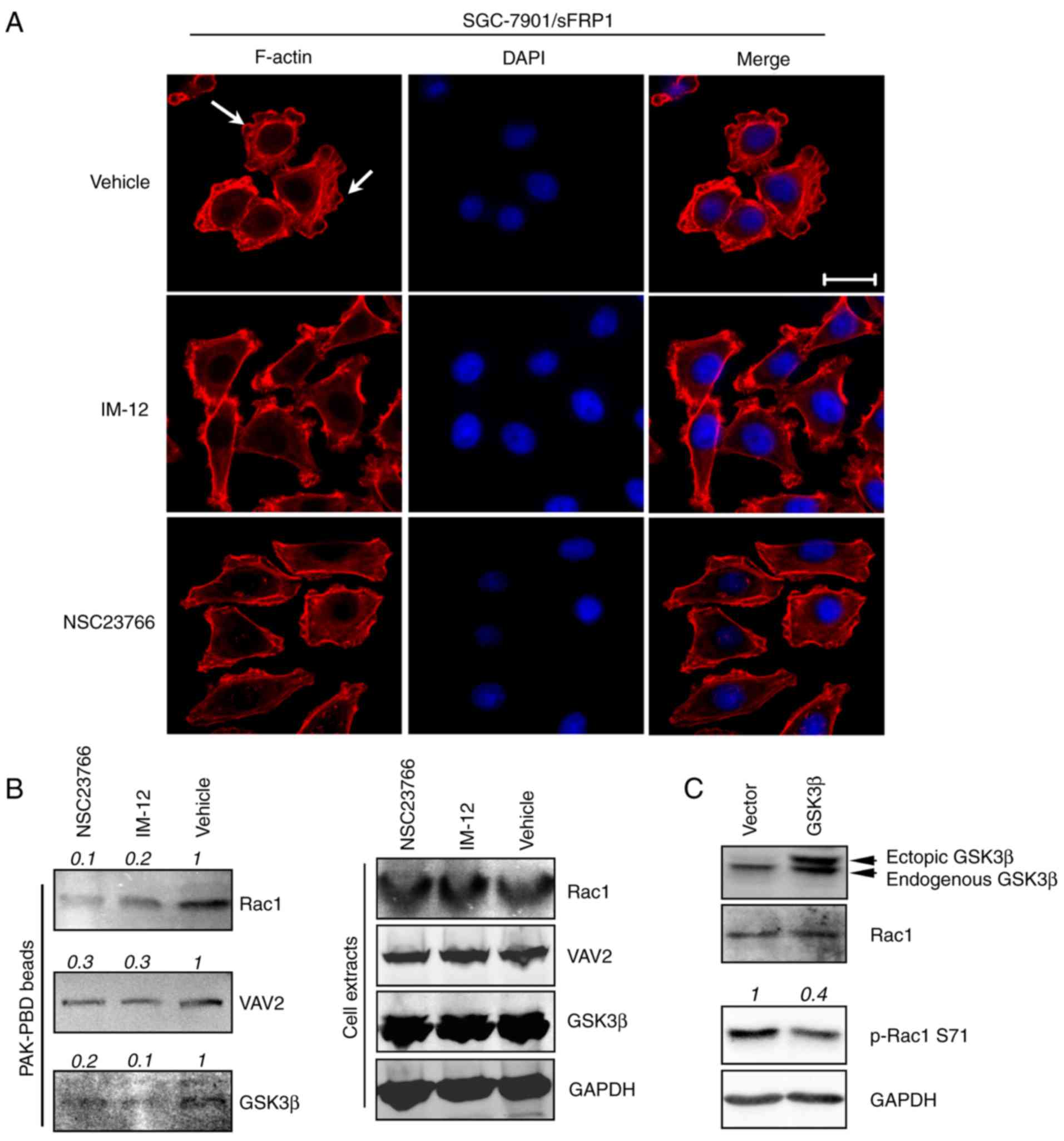 | Figure 3.GSK3β regulates Rac1 activity in
sFRP1-overexpressing SGC-7901 cells. (A) GSK3β inhibition decreased
lamellipodia formation in SGC-7901/sFRP1 cells. SGC-7901/sFRP1
cells were treated with DMSO (vehicle), GSK3β inhibitor (IM-12) or
Rac1 inhibitor (NSC23766) under normal culture medium (DMEM with
10% FBS) for 2 h. Cells were then either fixed and F-actin stained
using Alexa Fluor 555®-labeled phalloidin to depict the
cytoskeleton. Scale bar, 20 µM. (B) SGC-7901/sFRP1 cells were
treated with DMSO (vehicle), GSK3β inhibitor (IM-12) or Rac1
inhibitor (NSC23766) under normal culture medium (DMEM with 10%
FBS) for 2 h. Cells were collected for the Rac1 activity assays
(right). PAK-PBD beads were used for precipitation of activated
Rac1. Western blotting assays were performed to visualize the
activated form of Rac1 protein, in addition to GSK3β and VAV2 that
were also precipitated by PAK-PBD beads. The total levels of Rac1,
VAV2, GSK3β and GAPDH were examined using cell extracts from
different treatments. (C) Immunoblotting analysis examined the Rac1
activity in the vector only and GSK3β-overexpressing SGC-7901
cells. GAPDH was used as a loading control. Quantification of the
intensity of the bands was normalized relative to the vehicle or
vector, which are depicted on top of the bands. sFRP1, secreted
frizzled-related protein 1; Rac1, Rac family small GTPase 1; GSK3β,
glycogen synthase kinase 3β; DMSO, dimethyl sulfoxide; PAK, Rac1
activated kinase; PBD, Rac/Cdc42 (p21) binding domain; VAV2, Vav
guanine nucleotide exchange factor 2. |
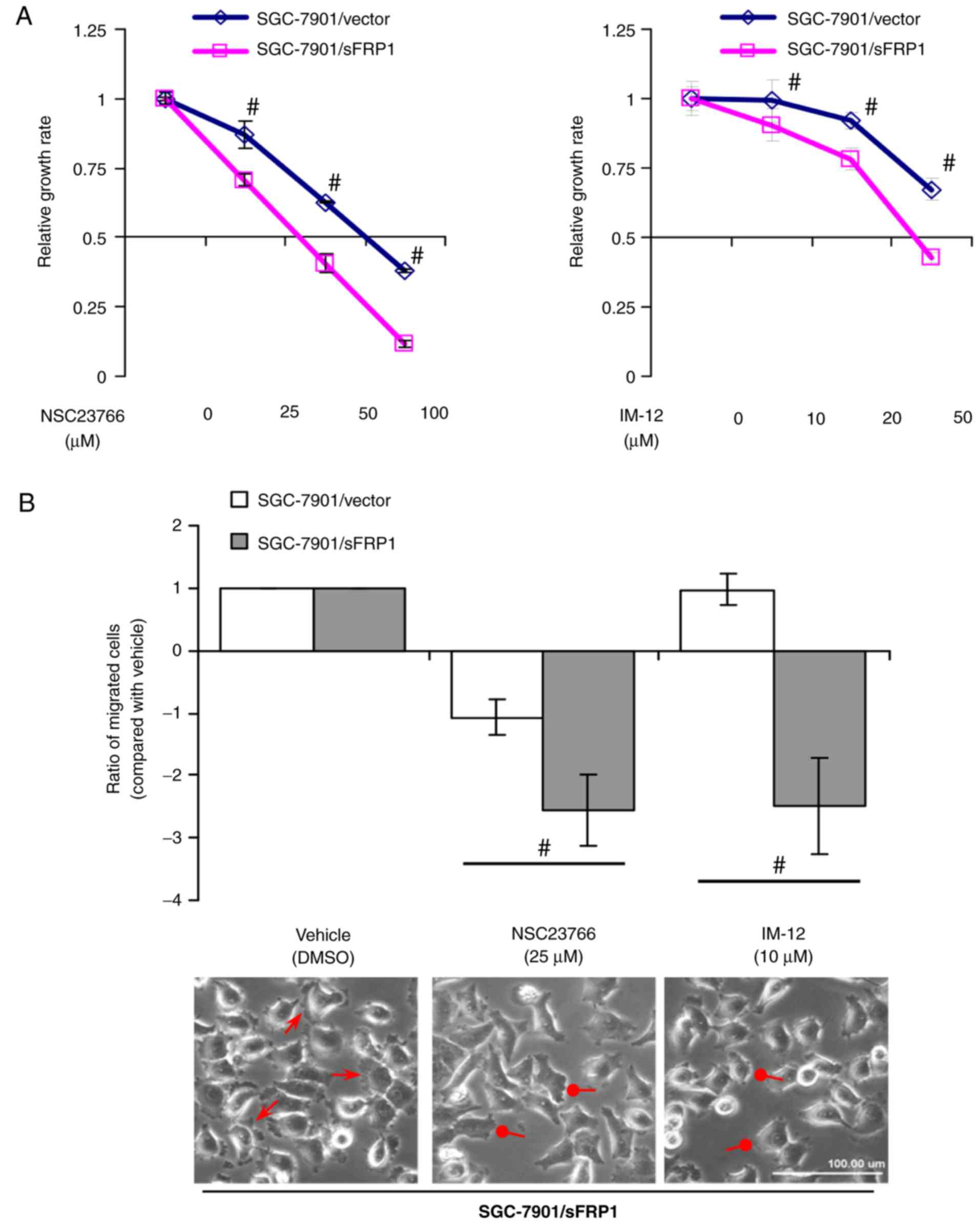
Inhibition of Rac1 or GSK3β activity
suppresses growth and metastasis in sFRP1-overexpressing cells
Activated Rac1 signaling has been determined to be
important in gastric cancer tumorigenesis (39) and induces the high mobility cell
phenotype (40). Although GSK3β is
a classic inhibitor of the Wnt/canonical pathway, it is able to
activate other signaling pathways and promote tumorigenesis
(41,42). Subsequently, whether Rac1 or GSK3β
mediated the tumor-promoting effects of sFRP1 was investigated. To
address this question, a specific Rac1 inhibitor NSC23766 (37) and a small molecule GSK3β inhibitor
IM-12 (38) were used, which were
demonstrated to inhibit Rac1-GEF interaction and GSK3β kinase
activity, respectively. Cell proliferation and migratory ability
were then investigated. The significant inhibition of
SGC-7901/sFRP1 cell growth by NSC23766 or IM-12 was depicted in
Fig. 4A (P<0.05).
sFRP1-overexpressing cells exhibited significantly inhibited
migration following NSC23766 or IM-12 treatment, compared with
control cells (P<0.05; Fig. 4B,
upper). NSC23766 or IM-12 treatment abolished the formation of
lamellipodia and membrane ruffles in SGC-7901/sFRP1 cells (Fig. 4B, lower). These data indicated that
Rac1 and GSK3β serve essential functions in regulating the growth
and migration of sFRP1-overexpressing cells.
Rac1 or GSK3β inhibition abolishes the
regulation of sFRP1 on Smad3 activity
As demonstrated previously (25), sFRP1-overexpressing cells retained
nuclear Smad2 levels but exhibited notably reduced Smad3 levels,
compared with vector control cells, indicating unbalanced Smad2 and
Smad3 activity (Fig. 5A). PAI1, ID1
and ZEB2, downstream targets of the TGFβ signaling pathway
(43,44), were also upregulated in
SGC-7901/sFRP1 cells (Fig. 5A).
Rac1 was determined to selectively antagonize TGFβ/Smad3 mediated
growth inhibition via its ability to promote Smad2 activation
(45). GSK3β was previously
reported to be responsible for the linker region of Smad3 and
inhibited its transcriptional activity on molecules that mediated
the growth inhibition activity of TGFβ signaling (46). Subsequently, whether Rac1 and GSK3β
participated in the regulation of TGFβ signaling through sFRP1 was
investigated; therefore, immunoblotting was performed using the
nuclear extracts from SGC-7901/sFRP1 cells treated with Rac1 or
GSK3β inhibitors. As depicted in Fig.
5B, nuclear Smad2 expression levels were not altered, and Smad3
and Smad4 expression levels were decreased following NSC23766 or
IM-12 treatment. Inhibition of Rac1 or GSK3β activity also
decreased ID1 and ZEB2 levels, which explained why Rac1 or GSK3β
inhibition suppressed cell growth and migration.
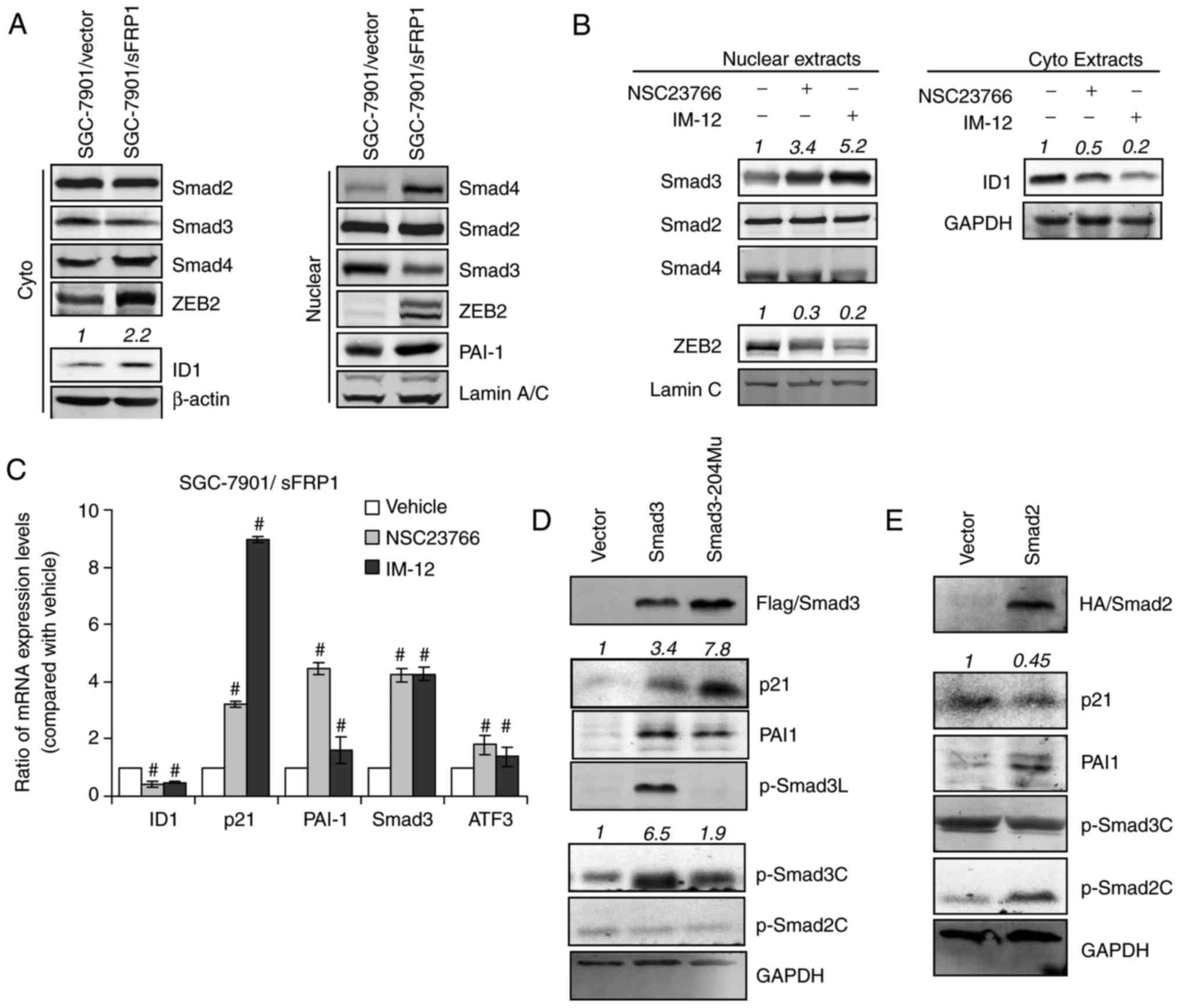 | Figure 5.Rac1 and GSK3β participate in the
regulation of the TGFβ pathway in sFRP1-overexpressing gastric
cancer cells. (A) The expression of TGFβ signaling proteins were
examined using immunoblotting. Cytoplasm and nuclear extracts from
SGC-7901/vector and SGC-7901/sFRP1 cells were collected using cells
cultured for 24 h under normal conditions. Actin and Lamin A/C were
used as loading controls for cytoplasm and nuclear proteins,
respectively. (B) Nuclear or cytoplasm extracts were collected from
SGC-7901/sFRP1 cells treated with IM-12 or NSC23766. TGFβ-signaling
downstream targets were examined by immunoblotting. (C) Reverse
transcription-quantitative polymerase chain reaction analysis was
performed to examine the expression of Smad3-responsive genes in
SGC-7901/sFRP1 cells. The cells were treated with dimethyl
sulfoxide (vehicle), Rac1 inhibitor (NSC23766) or GSK3β inhibitor
(IM-12) in normal culture conditions for 24 h. Relative expression
levels, compared with vehicle, are plotted. The data are presented
as the mean ± standard deviation of three independent experiments.
Statistical analysis was performed for each inhibitor (NSC23766 or
IM-12) compared to vehicle individually (ANOVA test).
#P<0.05 vs. vehicle. (D) Immunoblotting analysis of
protein expression levels in vector-only SGC-7901 cells transfected
with vector, WT Flag-Smad3 or Ser204 mutant Flag-Smad3 (the
phosphorylation site modulated by GSK3β). (E) Immunoblotting
analysis of protein expression levels in vector-only SGC-7901 cells
transfected with vector and WT HA-Smad2. GAPDH was used as a
loading control. Quantification of the intensity of the bands was
normalized in relative to vehicle or vector, which are depicted on
top of the bands. sFRP1, secreted frizzled-related protein 1; Rac1,
Rac family small GTPase 1; GSK3β, glycogen synthase kinase 3β;
TGFβ, transforming growth factor β; WT, wild-type. |
Elevated mRNA levels of Smad3-responsive genes
(47), including p21, ATF3, PAI1
and Smad3, were significantly elevated by Rac1 or GSK3β inhibition,
whereas the ID1 mRNA level was significantly inhibited (P<0.05;
Fig. 5C). To further observe the
different gene responses to Smad2 and Smad3 signaling, HA-Smad2 or
Flag-Smad3 constructs were transfected into SGC-7901 cells
(Fig. 5D and E). Notably, cells
overexpressing Smad3 exhibited high levels of pSmad3C, PAI1,
pSmad3L and p21. Transfection of the Smad3-S204 mutant, a mutant
form of WT Smad3 with the Ser204 mutant that cannot be
phosphorylated by GSK3β, construct into SGC-7901 cells resulted in
even higher levels of p21, without exhibiting a pSmad3L band.
Additionally, Smad2 overexpression also resulted in elevated PAI1,
which was potentially caused by increased pSmad2C levels, as
pSmad3C levels were unaltered. These observations supported the
observation that sustained Smad2 activity was able to compensate
some of the Smad3-responsive functions in sFRP1-overexpressing
cells. These data strongly indicated that the Rac1 and GSK3β were
able to suppress Smad3 function, whilst retaining the expression of
genes that were critical in mediating TGFβ-induced survival and the
EMT phenotype.
Ectopic overexpression of Rac1 or
GSK3β suppresses the Smad3 activity
To further examine the function of Rac1 or GSK3β in
suppressing Smad3 activity, Rac1 or GSK3β were ectopically
overexpressed in SGC-7901/vector cells. It is known that GSK3β
phosphorylates the linker region (Ser204) of Smad3 and inhibited
its transcriptional activity (48).
In the present study, higher levels of pSmad3L (Ser204) and
unaltered pSmad2 levels (Fig. 6A)
were also observed in GSK3β-overexpressing cells compared with the
vector cells. Subsequently, the function of Rac1 overexpression on
Smad3 activity was investigated by transfecting Rac1-WT and Rac1-CA
plasmids into SGC-7901/vector cells. Rac1-WT overexpressing cells
exhibited lower pSmad3C expression levels compared with the vector
cells (Fig. 6B, left). Rac1-CA
overexpressing cells exhibited a more notable decrease in pSmad3C
expression, compared with Rac1-WT overexpressing cells (Fig. 6B, left); however, Rac1-DN
overexpressing cells exhibited an elevated pSmad3C expression level
(Fig. 6B, right). Collectively,
these data indicated that GSK3β and Rac1 are responsible for
modulating TGFβ/Smad3 signaling in sFRP1-overexpressing cells
(Fig. 6C).
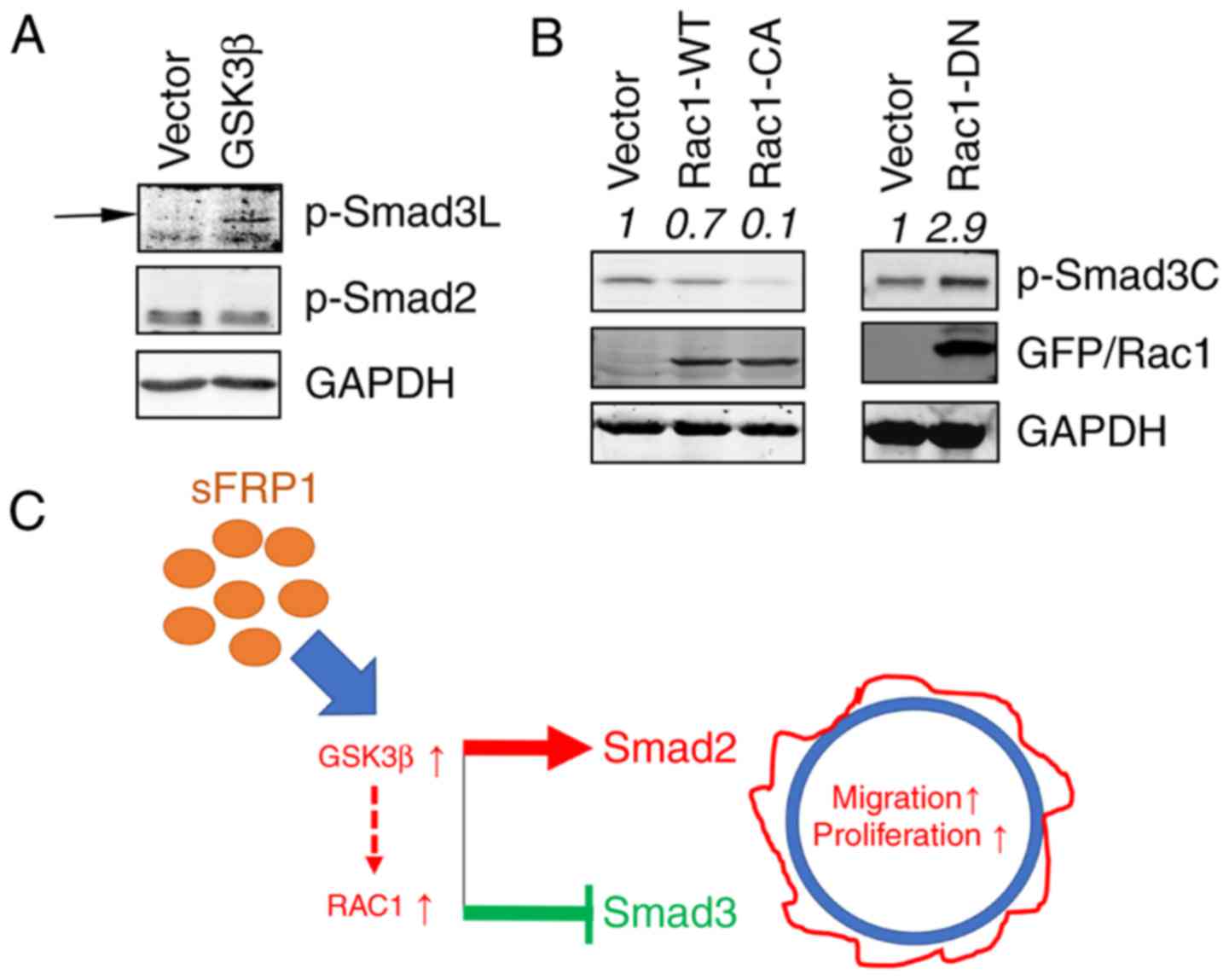 | Figure 6.sFRP1 regulates TGFβ signaling. (A)
GSK3β overexpression regulated pSmad3C and pSmad3L levels assayed
by immunoblotting. (B) Rac1 overexpression (WT, CA and DN)
regulated the pSmad3C expression level assayed by immunoblotting.
(C) A schematic diagram demonstrating the function of Rac1 and
GSK3β in sFRP1 signaling. sFRP1 activates Rac1 and GSK3β and
regulates TGFβ signaling by restraining Smad3 activity and
retaining Smad2 activity, thus enhancing epithelial-mesenchymal
transition and metastasis while suppressing the growth inhibitory
effects of TGFβ signaling. The broken line indicates currently
unknown mechanisms. Quantification of the intensity of the bands
was normalized in relative to the vehicle or vector, which are
depicted on top of the bands. sFRP1, secreted frizzled-related
protein 1; Rac1, Rac family small GTPase 1; GSK3β, glycogen
synthase kinase 3β; TGFβ, transforming growth factor β; WT,
wild-type; CA, constitutively active; DN, dominant-negative; p-,
phosphorylated; Smad, SMAD family member. |
Discussion
In the present study, it was demonstrated that there
was high GSK3β and Rac1 activity in sFRP1 overexpressing cells.
Additionally, it was observed that GSK3β and Rac1 mediated the
effect of sFRP1 overexpression on regulating cell proliferation,
migration and invasion. sFRP1-overexpression activated TGFβ and
suppressed its growth inhibitory effect through activating
GSK3β/Rac1.
Rac1 and GSK3β have been reported to be involved in
tumorigenesis. Overexpression of Rac1 occurs in a number of tumor
types, including breast (49,50),
colon (51), bladder (52) and gastric cancer (53,54).
Rac1 activation is associated with the progression of gastric
cancer (39). In vitro
studies have implicated Rac1 in cell migration (55,56),
cell-cycle progression (57,58)
and Ras-induced focus formation (59), indicating a role of Rac1 in tumor
development and progression. GSK3β was previously considered as a
tumor suppressor, due to its known inhibition of Wnt/β-catenin
activity; however, emerging evidence indicated its role in
promoting tumor formation and metastasis (41,42).
GSK3β activation is observed in gastric cancer and its signaling
pathway has been determined to be functional in gastric cancer
cells without involving Wnt signaling (60).
The first observation of the morphological changes
in sFRP1-overexpressing cells was the formation of lamellipodia,
which resulted in Rac1 activation. It was also observed that the
level of the inactivated form of GSK3β was reduced in
sFRP1-overexpressing cells, which indicated the role of sFRP1 in
restoring GSK3β activity. Inhibition of GSK3β or Rac1 suppressed
SGC-7901/sFRP1 growth and metastasis, indicating that sFRP1
overexpression may regulate cellular functions through GSK3β and
Rac1. GSK3β overexpression and GSK3β inhibition further
demonstrated a positive association between GSK3β and Rac1
activity, which is consistent with previous data indicating that
Rac1 activity may be regulated by GSK3β (35). Suppression of Wnt signaling may
result in the stabilization of Rac1 (61). High TGFβ1 expression levels observed
in sFRP1-overexpressing cells (25)
may also activate Rac1, thus promoting cell invasiveness (62); therefore, sFRP1-overexpression may
activate Rac1 and maintain its sustained activity through
multi-pathways. sFRP1 is also known to abrogate GSK3β inactivation,
by preventing its phosphorylation at the Ser9 residue (34); thus, the activation of Rac1 may be
due to the inhibition of Wnt and/or subsequent GSK3β
activation.
A previous study determined that sFRP1
overexpression was associated with the activation of the TGFβ
signaling pathway and induced cell proliferation, EMT and invasion
(25). Additionally, the
EMT-associated gene expression profile and TGFβ-induced growth
inhibitory gene expression signature, including the upregulation of
p21 and p15 and the downregulation of ID1, were not exhibited in
sFRP1-overexpressing cells. This observation indicated that the
growth inhibitory effect of TGFβ signaling was suppressed by sFRP1
overexpression. TGFβ1 may serve as a potent inhibitor of
proliferation in epithelial cells. This cytostatic activity is
dependent on the ability of TGFβ1 to increase the expression of
cyclin-dependent kinase inhibitors, including p15Ink4b and p21Cip1.
and repress the expression of the growth-promoting factors,
including ID family proteins, and is primarily controlled by a
Smad3-dependent signal (48);
however, Smad2 was not responsible for the growth inhibition and
the response of migratory induced by TGFβ1 (45). Loss of the negative regulation is
considered to contribute to tumor development (63–65).
Different mechanisms regarding how cells evade
TGFβ-meditated growth inhibition have been investigated. Among
these, GSK3β was determined to inhibit Smad3 activity as a
pro-apoptotic effector of TGFβ signaling in cancer cells (48); however, Rac1 antagonizes TGFβ/Smad3
mediated growth inhibition by promoting Smad2 activation (45). In the present study, it was
determined that Rac1 enhanced Smad2 but suppressed Smad3 signal
assayed by Rac1 overexpression and inhibition. Smad3 levels and
activity were consistently reduced in sFRP1-overexpressing cells.
Additionally, GSK3β and Rac1 were demonstrated to have increased
activation in SGC-7901/sFRP1 cells, compared with control cells;
therefore, GSK3β and Rac1 activity were conversely associated with
nuclear Smad3 levels in sFRP1-overexpressing cells.
Recent studies (46,66)
indicated that Rac1 and GSK3β can regulate TGFβ signaling. GSK3β
phosphorylates the linker region of Smad3 and inhibits its
transcriptional activity (46).
Consistent with previous data (45,48),
it was demonstrated that GSK3β and Rac1 activity were conversely
associated with nuclear Smad3 expression levels in
sFRP1-overexpressing cells. This regulation by Rac1 may be
indirectly through GSK3β, due to Rac1 not combining with Smad3
(data not shown). In the present study, apparent loss or gain of
nuclear Smad2 in sFRP1-overexpression cells was not determined.
Additionally, nuclear Smad2 expression levels were not notably
altered by GSK3β and Rac1 activity. Due to the potential of Smad2/3
activity being influenced by other proteins, including mannosidases
α class 1 (67), neural precursor
cell expressed developmentally downregulated 4-like E3 ubiquitin
protein ligase (29), sterol
carrier proteins (68) and protein
kinase B (69), the effects from
other regulators on Smad2/3 in sFRP1-overexpressing cells cannot be
excluded. It was speculated that the regulation towards TGFβ
signaling by sFRP1-overexpression is primarily through targeting
Smad3.
In conclusion, sFRP1 overexpression promotes gastric
cancer cell proliferation and metastasis by activating TGFβ
signaling; however, sFRP1 limits the growth inhibitory effect of
TGFβ signaling via Rac1 and GSK3β. The present study demonstrated
that sFRP1 has a novel role in regulating gastric cancer malignancy
and may therefore serve as a therapeutic target for gastric cancer
treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JXP conceived and designed the study. JXP, SYL and
LL performed the experiments. JXP wrote the manuscript. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Surana R, Sikka S, Cai W, Shin EM, Warrier
SR, Tan HJ, Arfuso F, Fox SA, Dharmarajan AM and Kumar AP: Secreted
frizzled related proteins: Implications in cancers. Biochim Biophys
Acta. 1845:53–65. 2014.PubMed/NCBI
|
|
2
|
Bovolenta P, Esteve P, Ruiz JM, Cisneros E
and Lopez-Rios J: Beyond Wnt inhibition: New functions of secreted
frizzled-related proteins in development and disease. J Cell Sci.
121:737–746. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kawano Y and Kypta R: Secreted antagonists
of the Wnt signalling pathway. J Cell Sci. 116:2627–2634. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rattner A, Hsieh JC, Smallwood PM, Gilbert
DJ, Copeland NG, Jenkins NA and Nathans J: A family of secreted
proteins contains homology to the cysteine-rich ligand-binding
domain of frizzled receptors. Proc Natl Acad Sci USA. 94:2859–2863.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Melkonyan HS, Chang WC, Shapiro JP,
Mahadevappa M, Fitzpatrick PA, Kiefer MC, Tomei LD and Umansky SR:
SAR Ps A family of secreted apoptosis-related proteins. Proc Natl
Acad Sci USA. 94:13636–13641. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Foronjy R, Imai K, Shiomi T, Mercer B,
Sklepkiewicz P, Thankachen J, Bodine P and D'Armiento J: The
divergent roles of secreted frizzled related protein-1 (SFRP1) in
lung morphogenesis and emphysema. Am J Pathol. 177:598–607. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Joesting MS, Cheever TR, Volzing KG,
Yamaguchi TP, Wolf V, Naf D, Rubin JS and Marker PC: Secreted
frizzled related protein 1 is a paracrine modulator of epithelial
branching morphogenesis, proliferation, and secretory gene
expression in the prostate. Dev Biol. 317:161–173. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsuyama M, Aizawa S and Shimono A: Sfrp
controls apicobasal polarity and oriented cell division in
developing gut epithelium. PLoS Genet. 5:e10004272009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki H, Gabrielson E, Chen W, Anbazhagan
R, van Engeland M, Weijenberg MP, Herman JG and Baylin SB: A
genomic screen for genes upregulated by demethylation and histone
deacetylase inhibition in human colorectal cancer. Nat Genet.
31:141–149. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chung MT, Lai HC, Sytwu HK, Yan MD, Shih
YL, Chang CC, Yu MH, Liu HS, Chu DW and Lin YW: SFR P1 and SFRP2
suppress the transformation and invasion abilities of cervical
cancer cells through Wnt signal pathway. Gynecol Oncol.
112:646–653. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Valencia A, Román-Gómez J, Cervera J, Such
E, Barragán E, Bolufer P, Moscardó F, Sanz GF and Sanz MA: Wnt
signaling pathway is epigenetically regulated by methylation of Wnt
antagonists in acute myeloid leukemia. Leukemia. 23:1658–1666.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukui T, Kondo M, Ito G, Maeda O, Sato N,
Yoshioka H, Yokoi K, Ueda Y, Shimokata K and Sekido Y:
Transcriptional silencing of secreted frizzled related protein 1
(SFRP 1) by promoter hypermethylation in non-small-cell lung
cancer. Oncogene. 24:6323–6327. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ugolini F, Charafe-Jauffret E, Bardou VJ,
Geneix J, Adélaïde J, Labat-Moleur F, Penault-Llorca F, Longy M,
Jacquemier J, Birnbaum D, et al: WNT pathway and mammary
carcinogenesis: Loss of expression of candidate tumor suppressor
gene SFRP1 in most invasive carcinomas except of the medullary
type. Oncogene. 20:5810–5817. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750–2767.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu
J, Klijn JG, Foekens JA and Martens JW: Subtypes of breast cancer
show preferential site of relapse. Cancer Res. 68:3108–3114. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mii Y and Taira M: Secreted
frizzled-related proteins enhance the diffusion of Wnt ligands and
expand their signalling range. Development. 136:4083–4088. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Katoh Y and Katoh M: Hedgehog signaling,
epithelial-to-mesenchymal transition and miRNA (Review). Int J Mol
Med. 22:271–275. 2008.PubMed/NCBI
|
|
18
|
He J, Sheng T, Stelter AA, Li C, Zhang X,
Sinha M, Luxon BA and Xie J: Suppressing Wnt signaling by the
hedgehog pathway through sFRP-1. J Biol Chem. 281:35598–35602.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Häusler KD, Horwood NJ, Chuman Y, Fisher
JL, Ellis J, Martin TJ, Rubin JS and Gillespie MT: Secreted
frizzled-related protein-1 inhibits RANKL-dependent osteoclast
formation. J Bone Miner Res. 19:1873–1881. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Esteve P and Bovolenta P: The advantages
and disadvantages of sfrp1 and sfrp2 expression in pathological
events. Tohoku J Exp Med. 221:11–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Toni F, Racaud-Sultan C, Chicanne G,
Mas VM, Cariven C, Mesange F, Salles JP, Demur C, Allouche M,
Payrastre B, et al: A crosstalk between the Wnt and the
adhesion-dependent signaling pathways governs the chemosensitivity
of acute myeloid leukemia. Oncogene. 25:3113–3122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hohenberger P and Gretschel S: Gastric
cancer. Lancet. 362:305–315. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smith DD, Schwarz RR and Schwarz RE:
Impact of total lymph node count on staging and survival after
gastrectomy for gastric cancer: Data from a large US-population
database. J Clin Oncol. 23:7114–7124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu Y, Ray PS, Li J, Cai Q, Bagaria SP,
Moran C, Sim MS, Zhang J, Turner RR, Zhu Z, et al: High levels of
secreted frizzled-related protein 1 correlate with poor prognosis
and promote tumourigenesis in gastric cancer. Eur J Cancer.
49:3718–3728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saini S, Liu J, Yamamura S, Majid S,
Kawakami K, Hirata H and Dahiya R: Functional significance of
secreted frizzled-related protein 1 in metastatic renal cell
carcinomas. Cancer Res. 69:6815–6822. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subauste MC, Von Herrath M, Benard V,
Chamberlain CE, Chuang TH, Chu K, Bokoch GM and Hahn KM: Rho family
proteins modulate rapid apoptosis induced by cytotoxic T
lymphocytes and Fas. J Biol Chem. 275:9725–9733. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou BP, Deng J, Xia W, Xu J, Li YM,
Gunduz M and Hung MC: Dual regulation of Snail by
GSK-3beta-mediated phosphorylation in control of
epithelial-mesenchymal transition. Nat Cell Biol. 6:931–940. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao S, Alarcón C, Sapkota G, Rahman S,
Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P and
Massagué J: Ubiquitin ligase Nedd4L targets activated Smad2/3 to
limit TGF-beta signaling. Mol Cell. 36:457–468. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Labbé E, Silvestri C, Hoodless PA, Wrana
JL and Attisano L: Smad2 and Smad3 positively and negatively
regulate TGF beta-dependent transcription through the forkhead
DNA-binding protein FAST2. Mol Cell. 2:109–120. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hata A, Lo RS, Wotton D, Lagna G and
Massague J: Mutations increasing autoinhibition inactivate tumour
suppressors Smad2 and Smad4. Nature. 388:82–87. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frame MC and Brunton VG: Advances in
Rho-dependent actin regulation and oncogenic transformation. Curr
Opin Genet Dev. 12:36–43. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ren J, Wang R, Song H, Huang G and Chen L:
Secreted frizzled related protein 1 modulates taxane resistance of
human lung adenocarcinoma. Mol Med. 20:164–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koivisto L, Häkkinen L, Matsumoto K,
McCulloch CA, Yamada KM and Larjava H: Glycogen synthase kinase-3
regulates cytoskeleton and translocation of Rac1 in long cellular
extensions of human keratinocytes. Exp Cell Res. 293:68–80. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Abe K, Rossman KL, Liu B, Ritola KD,
Chiang D, Campbell SL, Burridge K and Der CJ: Vav2 is an activator
of Cdc42, Rac1, and RhoA. J Biol Chem. 275:10141–10149. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao Y, Dickerson JB, Guo F, Zheng J and
Zheng Y: Rational design and characterization of a Rac
GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA.
101:7618–7623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schmöle AC, Brennführer A, Karapetyan G,
Jaster R, Pews-Davtyan A, Hübner R, Ortinau S, Beller M, Rolfs A
and Frech MJ: Novel indolylmaleimide acts as GSK-3beta inhibitor in
human neural progenitor cells. Bioorg Med Chem. 18:6785–6795. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan Y, Bi F, Liu N, Xue Y, Yao X, Zheng Y
and Fan D: Expression of seven main Rho family members in gastric
carcinoma. Biochem Biophys Res Commun. 315:686–691. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Matsuoka T, Yashiro M, Kato Y, Shinto O,
Kashiwagi S and Hirakawa K: RhoA/ROCK signaling mediates plasticity
of scirrhous gastric carcinoma motility. Clin Exp Metastasis.
28:627–636. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang QL, Xie XB, Wang J, Chen Q, Han AJ,
Zou CY, Yin JQ, Liu DW, Liang Y, Zhao ZQ, et al: Glycogen synthase
kinase-3β, NF-κB signaling, and tumorigenesis of human
osteosarcoma. J Natl Cancer Inst. 104:749–763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shakoori A, Mai W, Miyashita K, Yasumoto
K, Takahashi Y, Ooi A, Kawakami K and Minamoto T: Inhibition of
GSK-3 beta activity attenuates proliferation of human colon cancer
cells in rodents. Cancer Sci. 98:1388–1393. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu H, Wang YL, Wang GW, Wong YC, Wang XF,
Wang Y and Xu KX: A novel role of Id-1 in regulation of
epithelial-to-mesenchymal transition in bladder cancer. Urol Oncol.
31:1242–1253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sánchez-Tilló E, Siles L, de Barrios O,
Cuatrecasas M, Vaquero EC, Castells A and Postigo A: Expanding
roles of ZEB factors in tumorigenesis and tumor progression. Am J
Cancer Res. 1:897–912. 2011.PubMed/NCBI
|
|
45
|
Ungefroren H, Groth S, Sebens S, Lehnert
H, Gieseler F and Fändrich F: Differential roles of Smad2 and Smad3
in the regulation of TGF-β1-mediated growth inhibition and cell
migration in pancreatic ductal adenocarcinoma cells: Control by
Rac1. Mol Cancer. 10:672011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guo X, Ramirez A, Waddell DS, Li Z, Liu X
and Wang XF: Axin and GSK3-control Smad3 protein stability and
modulate TGF-signaling. Genes Dev. 22:106–120. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wrana JL, Attisano L, Cárcamo J, Zentella
A, Doody J, Laiho M, Wang XF and Massagué J: TGF beta signals
through a heteromeric protein kinase receptor complex. Cell.
71:1003–1014. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cohen-Solal KA, Merrigan KT, Chan JL,
Goydos JS, Chen W, Foran DJ, Liu F, Lasfar A and Reiss M:
Constitutive Smad linker phosphorylation in melanoma: A mechanism
of resistance to transforming growth factor-β-mediated growth
inhibition. Pigment Cell Melanoma Res. 24:512–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schnelzer A, Prechtel D, Knaus U, Dehne K,
Gerhard M, Graeff H, Harbeck N, Schmitt M and Lengyel E: Rac1 in
human breast cancer: Overexpression, mutation analysis, and
characterization of a new isoform, Rac1b. Oncogene. 19:3013–3020.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fritz G, Just I and Kaina B: Rho GTPases
are over-expressed in human tumors. Int J Cancer. 81:682–687. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu G, Wang Y, Huang B, Liang J, Ding Y,
Xu A and Wu W: A Rac1/PAK1 cascade controls β-catenin activation in
colon cancer cells. Oncogene. 31:1001–1012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kamai T, Shirataki H, Nakanishi K, Furuya
N, Kambara T, Abe H, Oyama T and Yoshida K: Increased Rac1 activity
and Pak1 overexpression are associated with lymphovascular invasion
and lymph node metastasis of upper urinary tract cancer. BMC
Cancer. 10:1642010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhan H, Liang H, Liu X, Deng J, Wang B and
Hao X: Expression of Rac1, HIF-1α, and VEGF in gastric carcinoma:
Correlation with angiogenesis and prognosis. Onkologie. 36:102–107.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wu YJ, Tang Y, Li ZF, Li Z, Zhao Y, Wu ZJ
and Su Q: Expression and significance of Rac1, Pak1 and Rock1 in
gastric carcinoma. Asia Pac J Clin Oncol. 10:e33–e39. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Steffen A, Ladwein M, Dimchev GA, Hein A,
Schwenkmezger L, Arens S, Ladwein KI, Margit Holleboom J, Schur F,
Victor Small J, et al: Rac function is critical for cell migration
but not required for spreading and focal adhesion formation. J Cell
Sci. 126:4572–4588. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lewis-Saravalli S, Campbell S and Claing
A: ARF1 controls Rac1 signaling to regulate migration of MDA-MB-231
invasive breast cancer cells. Cell Signal. 25:1813–1819. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang J, Rao Q, Wang M, Wei H, Xing H, Liu
H, Wang Y, Tang K, Peng L, Tian Z, et al: Overexpression of Rac1 in
leukemia patients and its role in leukemia cell migration and
growth. Biochem Biophys Res Commun. 386:769–774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fritz G and Kaina B: Rac1 GTPase, a
multifunctional player in the regulation of genotoxic stress
response. Cell Cycle. 12:2521–2522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Qiu RG, Abo A, McCormick F and Symons M:
Cdc42 regulates anchorage-independent growth and is necessary for
Ras transformation. Mol Cell Biol. 17:3449–3458. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cho YJ, Yoon J, Ko YS, Kim SY, Cho SJ, Kim
WH, Park JW, Youn HD, Kim JH and Lee BL: Glycogen synthase
kinase-3β does not correlate with the expression and activity of
β-catenin in gastric cancer. APMIS. 118:782–790. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Esufali S, Charames GS and Bapat B:
Suppression of nuclear Wnt signaling leads to stabilization of Rac1
isoforms. FEBS Lett. 581:4850–4856. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Binker MG, Binker-Cosen AA, Gaisano HY, de
Cosen RH and Cosen-Binker LI: TGF-β1 increases invasiveness of
SW1990 cells through Rac1/ROS/NF-kB/IL-6/MMP-2. Biochem Biophys Res
Commun. 405:140–145. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bachman KE and Park BH: Duel nature of
TGF-beta signaling: Tumor suppressor vs. tumor promoter. Curr Opin
Oncol. 17:49–54. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Akhurst RJ and Derynck R: TGF-beta
signaling in cancer-a double-edged sword. Trends Cell Biol.
11:S44–S51. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hubchak SC, Sparks EE, Hayashida T and
Schnaper HW: Rac1 promotes TGF-beta-stimulated mesangial cell type
I collagen expression through a PI3K/Akt-dependent mechanism. Am J
Physiol Renal Physiol. 297:F1316–F1323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kondé E, Bourgeois B, Tellier-Lebegue C,
Wu W, Pérez J, Caputo S, Attanda W, Gasparini S, Charbonnier JB,
Gilquin B, et al: Structural analysis of the Smad2-MAN1 interaction
that regulates transforming growth factor-β signaling at the inner
nuclear membrane. Biochemistry. 49:8020–8032. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wrighton KH, Willis D, Long J, Liu F, Lin
X and Feng XH: Small C-terminal domain phosphatases dephosphorylate
the regulatory linker regions of Smad2 and Smad3 to enhance
transforming growth factor-beta signaling. J Biol Chem.
281:38365–38375. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shepherd RD, Kos SM and Rinker KD:
Flow-dependent Smad2 phosphorylation and TGIF nuclear localization
in human aortic endothelial cells. Am J Physiol Heart Circ Physiol.
301:H98–H107. 2011. View Article : Google Scholar : PubMed/NCBI
|