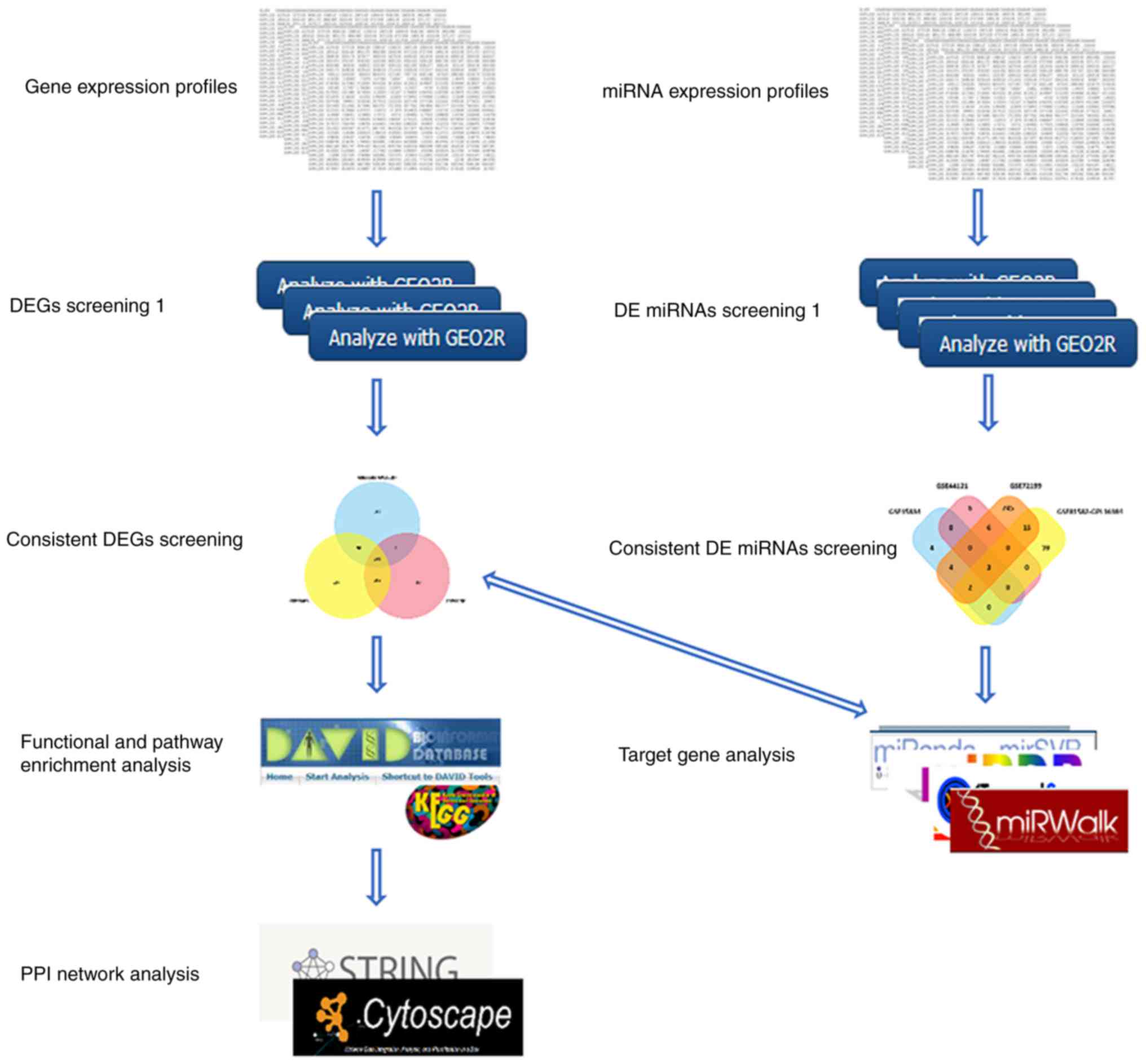
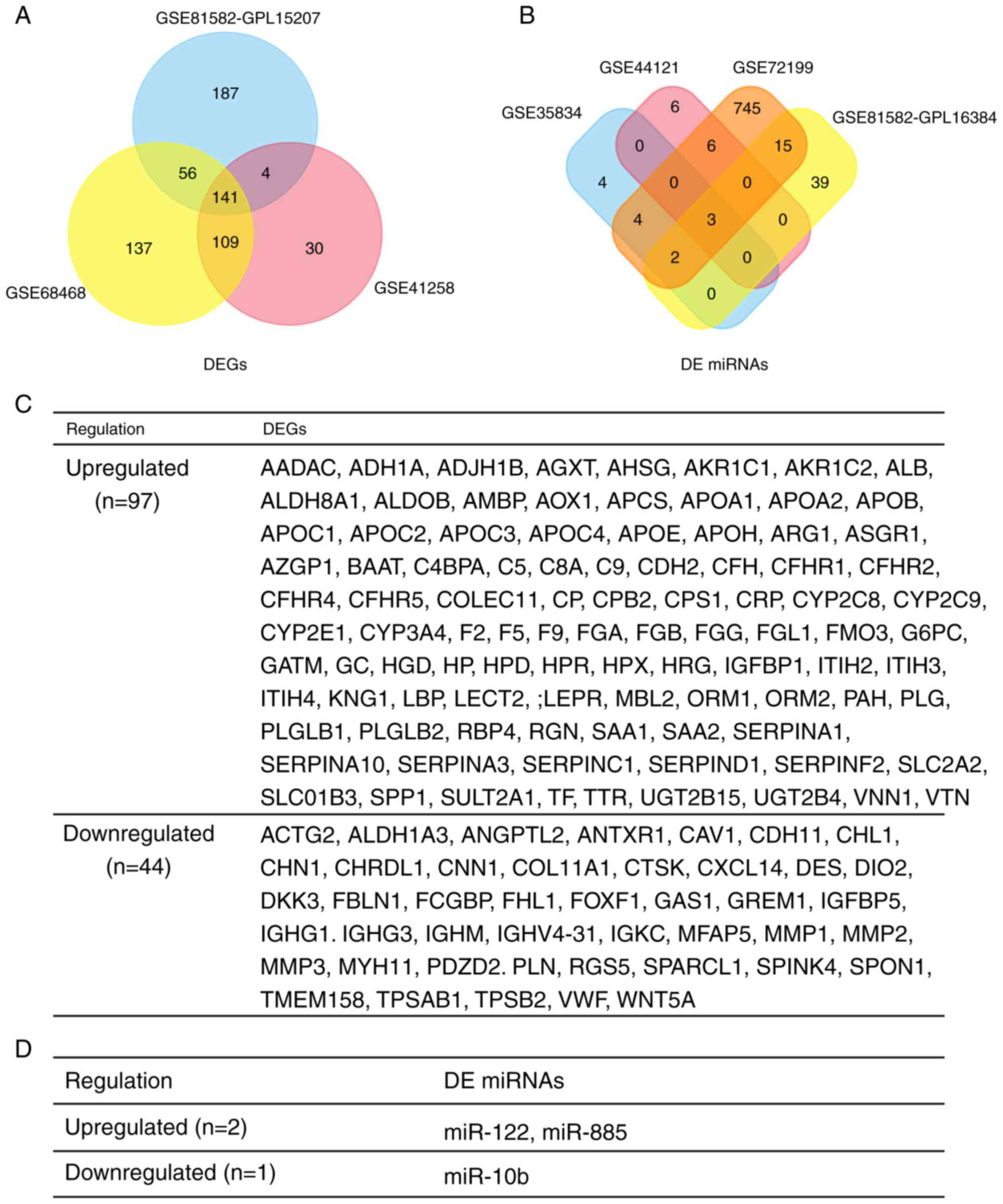
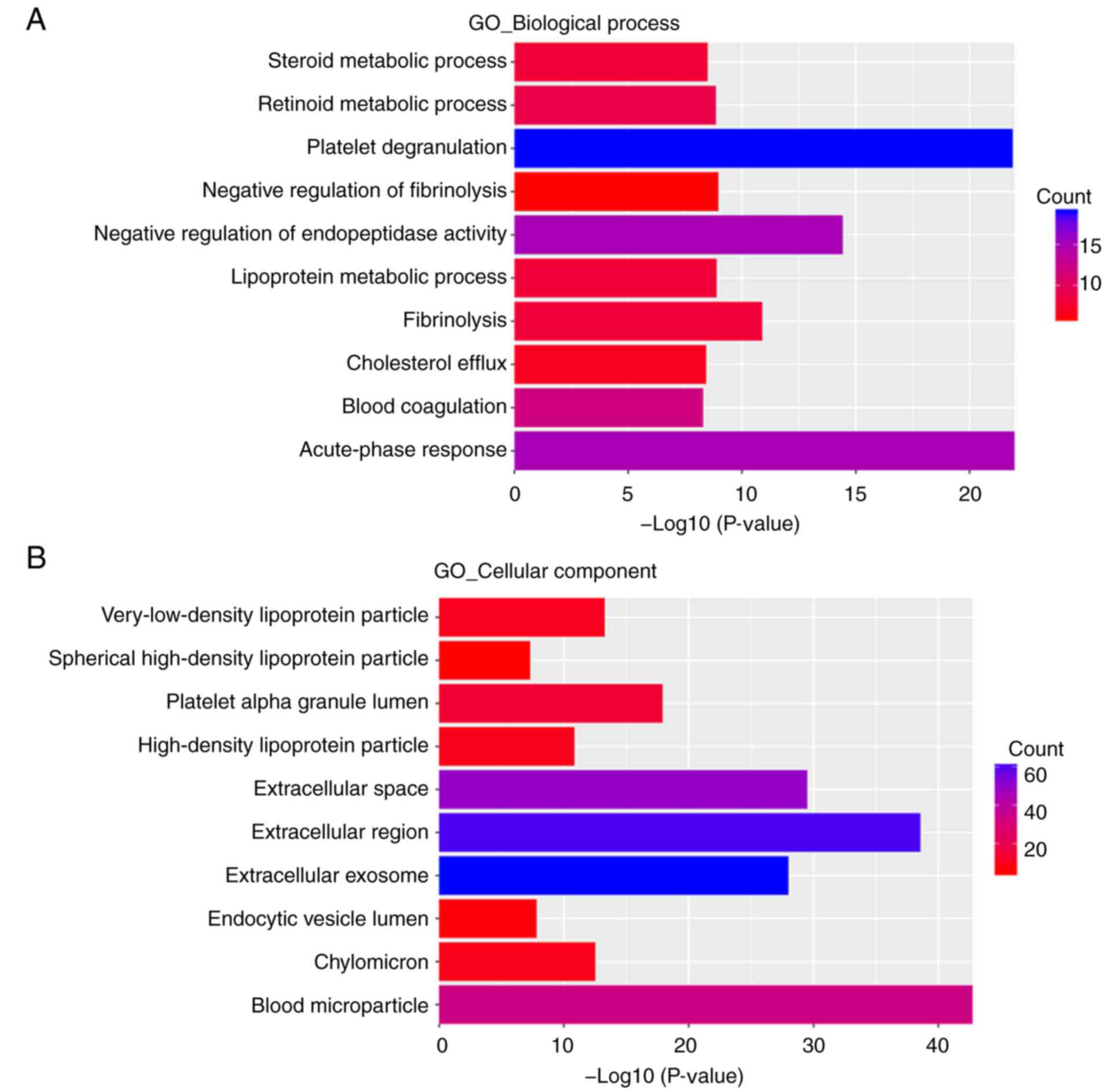
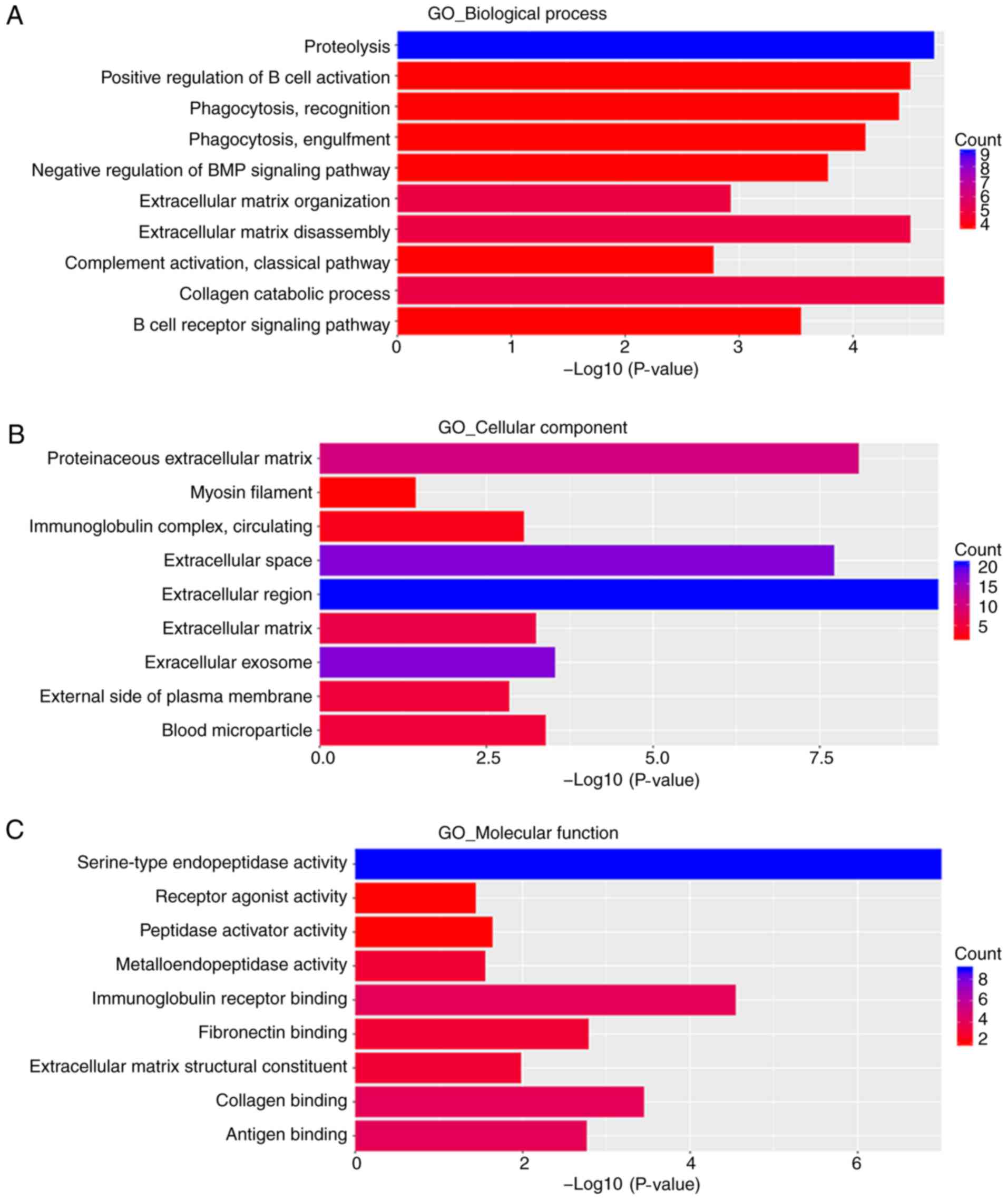
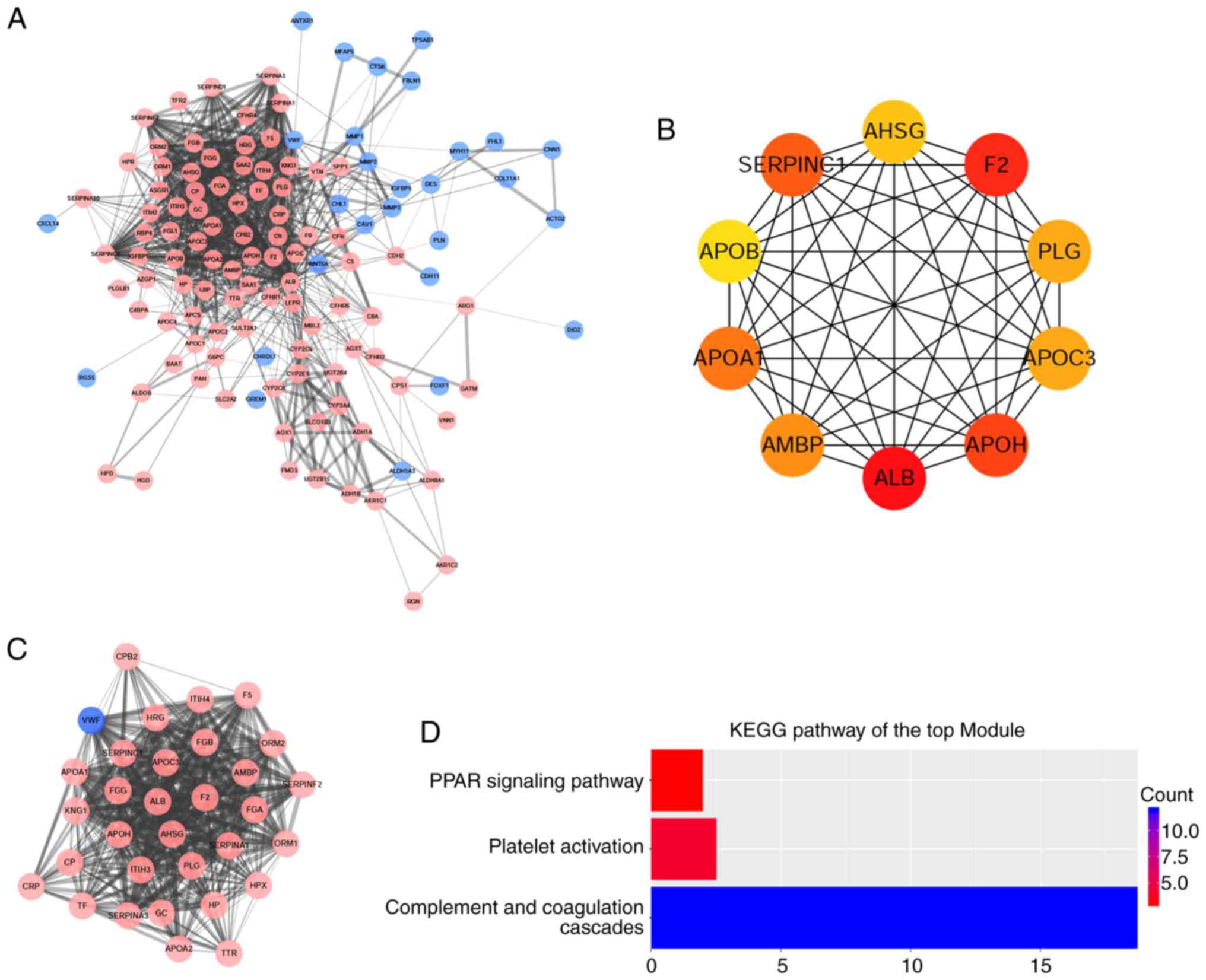
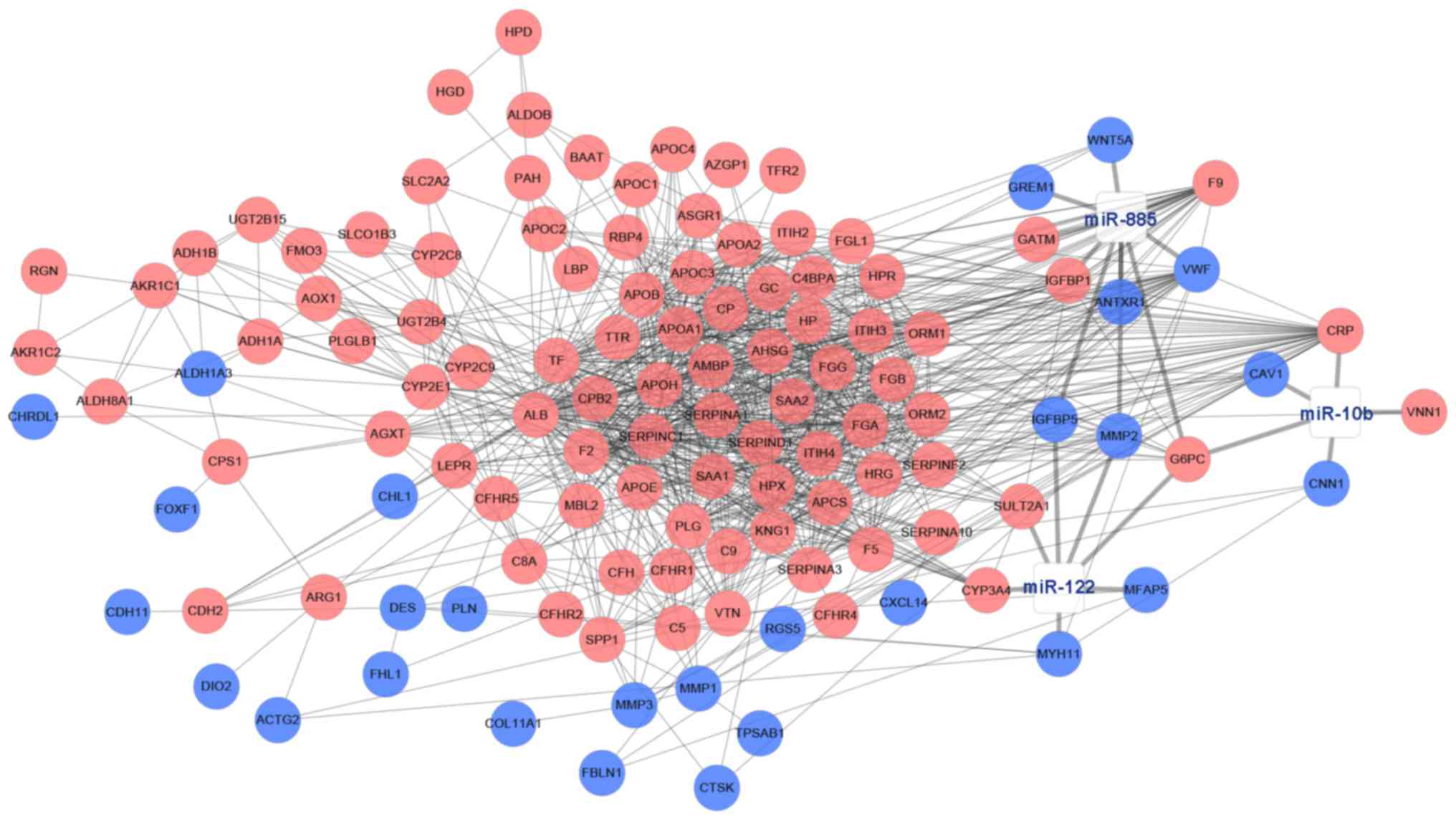
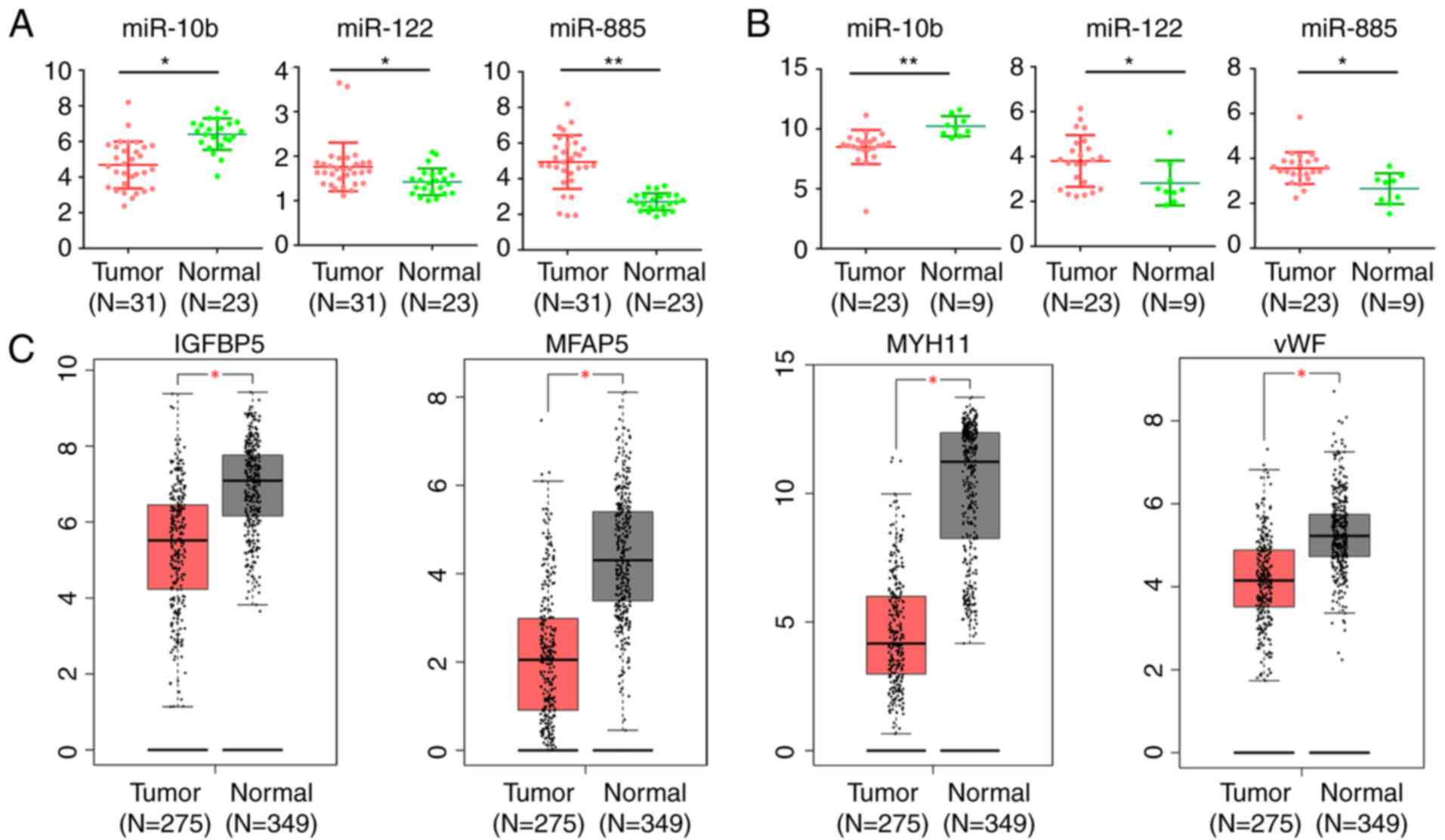
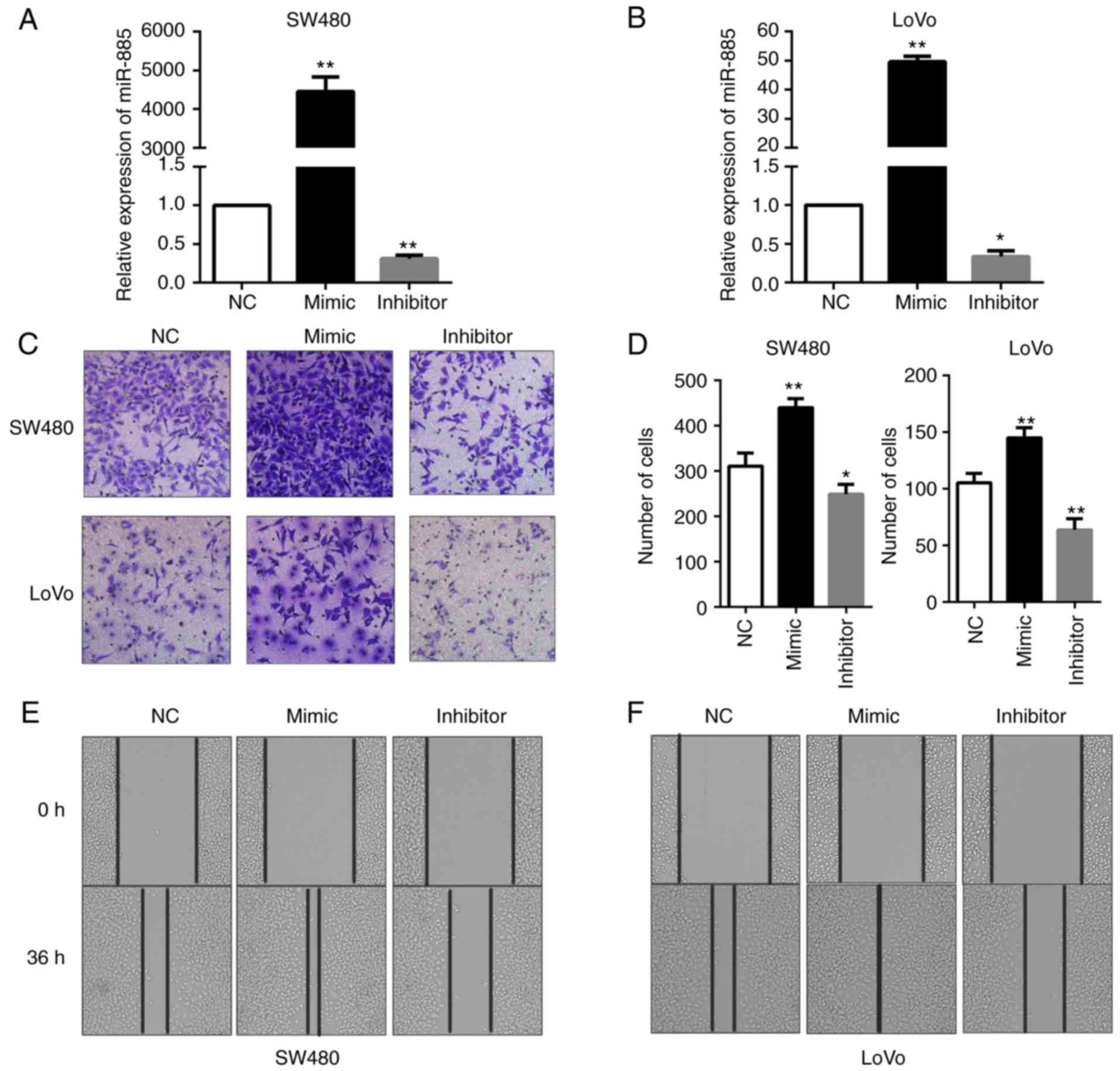
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bakalakos EA, Kim JA, Young DC and Martin
EW Jr: Determinants of survival following hepatic resection for
metastatic colorectal cancer. World J Surg. 22:399–404; discussion.
404–395. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hur K: MicroRNAs: Promising biomarkers for
diagnosis and therapeutic targets in human colorectal cancer
metastasis. BMB Rep. 48:217–222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheffer M, Bacolod MD, Zuk O, Giardina SF,
Pincas H, Barany F, Paty PB, Gerald WL, Notterman DA and Domany E:
Association of survival and disease progression with chromosomal
instability: A genomic exploration of colorectal cancer. Proc Natl
Acad Sci USA. 106:7131–7136. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsafrir D, Tsafrir I, Ein-Dor L, Zuk O,
Notterman DA and Domany E: Sorting points into neighborhoods
(SPIN): Data analysis and visualization by ordering distance
matrices. Bioinformatics. 21:2301–2308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sayagues JM, Corchete LA, Gutierrez ML,
Sarasquete ME, DelMarAbad M, Bengoechea O, Fermiñán E, Anduaga MF,
Del Carmen S, Iglesias M, et al: Genomic characterization of liver
metastases from colorectal cancer patients. Oncotarget.
7:72908–72922. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pizzini S, Bisognin A, Mandruzzato S,
Biasiolo M, Facciolli A, Perilli L, Rossi E, Esposito G, Rugge M,
Pilati P, et al: Impact of microRNAs on regulatory networks and
pathways in human colorectal carcinogenesis and development of
metastasis. BMC genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mokutani Y, Uemura M, Munakata K, Okuzaki
D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K,
Takemasa I, et al: Down-regulation of microRNA-132 is associated
with poor prognosis of colorectal cancer. Ann Surg Oncol.
23:599–608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan W, Zhang W, Sun L, Liu Y, You G, Wang
Y, Kang C, You Y and Jiang T: Identification of MMP-9 specific
microRNA expression profile as potential targets of anti-invasion
therapy in glioblastoma multiforme. Brain Res. 1411:108–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Afanasyeva EA, Mestdagh P, Kumps C,
Vandesompele J, Ehemann V, Theissen J, Fischer M, Zapatka M, Brors
B, Savelyeva L, et al: MicroRNA miR-885-5p targets CDK2 and MCM5,
activates p53 and inhibits proliferation and survival. Cell Death
Differ. 18:974–984. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Z, Yin J, Yang J, Shen W, Zhang C,
Mou W, Luo J, Yan H, Sun P, Luo Y, et al: miR-885-5p suppresses
hepatocellular carcinoma metastasis and inhibits Wnt/beta-catenin
signaling pathway. Oncotarget. 7:75038–75051. 2016.PubMed/NCBI
|
|
17
|
Lam CS, Ng L, Chow AK, Wan TM, Yau S,
Cheng NS, Wong SK, Man JH, Lo OS, Foo DC, et al: Identification of
microRNA 885-5p as a novel regulator of tumor metastasis by
targeting CPEB2 in colorectal cancer. Oncotarget. 8:26858–26870.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KE GG Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carbon S, Ireland A, Mungall CJ, Shu S,
Marshall B and Lewis S: AmiGO Hub and Web Presence Working Group:
AmiGO: Online access to ontology and annotation data.
Bioinformatics. 25:288–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu JT, Tan L and Hardy J: Apolipoprotein E
in Alzheimer's disease: An update. Annu Rev Neurosci. 37:79–100.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Borgquist S, Butt T, Almgren P, Shiffman
D, Stocks T, Orho-Melander M, Manjer J and Melander O:
Apolipoproteins, lipids and risk of cancer. Int J Cancer.
138:2648–2656. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zamanian-Daryoush M and DiDonato JA:
Apolipoprotein A-I and cancer. Front Pharmacol. 6:2652015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mangaraj M, Nanda R and Panda S:
Apolipoprotein A-I A molecule of diverse function. Indian J Clin
Biochem. 31:253–259. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peltier J, Roperch JP, Audebert S, Borg JP
and Camoin L: Quantitative proteomic analysis exploring progression
of colorectal cancer: Modulation of the serpin family. J
Proteomics. 148:139–148. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sirnio P, Vayrynen JP, Klintrup K, Makela
J, Makinen MJ, Karttunen TJ and Tuomisto A: Decreased serum
apolipoprotein A1 levels are associated with poor survival and
systemic inflammatory response in colorectal cancer. Sci Rep.
7:53742017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong Y, Zhang L, Bie P and Wang H: Roles
of ApoB-100 gene polymorphisms and the risks of gallstones and
gallbladder cancer: A meta-analysis. PLoS One. 8:e614562013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL,
Hu DF, Luo SY, Huang AC, Li LL and Li GJ: Comparative proteomic
profiles indicating genetic factors may involve in hepatocellular
carcinoma familial aggregation. Cancer Sci. 103:1833–1838. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Beecken WD, Ringel EM, Babica J, Oppermann
E, Jonas D and Blaheta RA: Plasmin-clipped
β2-glycoprotein-I inhibits endothelial cell growth by
down-regulating cyclin A, B and D1 and up-regulating p21 and p27.
Cancer Lett. 296:160–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jing X, Tian ZB, Gao PJ, Han NJ, Xu YH,
Zhang H, Ding XL, Wang XW, Man X and Zhang CP: Lipopolysaccharide
enhances beta2-glycoprotein I activation of nuclear factor κB in
liver cancer cells. Clin Lab. 61:1239–1245. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang H, Han Y, Gao J, Feng J, Zhu L, Qu
L, Shen L and Shou C: High level of serum AMBP is associated with
poor response to paclitaxel-capecitabine chemotherapy in advanced
gastric cancer patients. Med Oncol. 30:7482013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rasul S, Wagner L and Kautzky-Willer A:
Fetuin-A and angiopoietins in obesity and type 2 diabetes mellitus.
Endocrine. 42:496–505. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nangami GN, Watson K, Parker-Johnson K,
Okereke KO, Sakwe A, Thompson P, Frimpong N and Ochieng J: Fetuin-A
(alpha2HS-glycoprotein) is a serum chemo-attractant that also
promotes invasion of tumor cells through Matrigel. Biochem Biophys
Res Commun. 438:660–665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fan NJ, Kang R, Ge XY, Li M, Liu Y, Chen
HM and Gao CF: Identification alpha-2-HS-glycoprotein precursor and
tubulin beta chain as serology diagnosis biomarker of colorectal
cancer. Diagn Pathol. 9:532014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kumari S and Malla R: New insight on the
role of plasminogen receptor in cancer progression. Cancer Growth
Metastasis. 8:35–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Didiasova M, Wujak L, Wygrecka M and
Zakrzewicz D: From plasminogen to plasmin: Role of plasminogen
receptors in human cancer. Int J Mol Sci. 15:21229–21252. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Heit JA, Silverstein MD, Mohr DN,
Petterson TM, O'Fallon WM and Melton LJ III: Risk factors for deep
vein thrombosis and pulmonary embolism: A population-based
case-control study. Arch Intern Med. 160:809–815. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dicke C and Langer F: Pathophysiology of
Trousseau's syndrome. Hamostaseologie. 35:52–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guglietta S and Rescigno M:
Hypercoagulation and complement: Connected players in tumor
development and metastases. Semin Immunol. 28:578–586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Afshar-Kharghan V: The role of the
complement system in cancer. J Clin Invest. 127:780–789. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Torres S, Garcia-Palmero I, Bartolome RA,
Fernandez-Acenero MJ, Molina E, Calvino E, Segura MF and Casal JI:
Combined miRNA profiling and proteomics demonstrates that different
miRNAs target a common set of proteins to promote colorectal cancer
metastasis. J Pathol. 242:39–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hur K, Toiyama Y, Schetter AJ, Okugawa Y,
Harris CC, Boland CR and Goel A: Identification of a
metastasis-specific MicroRNA signature in human colorectal cancer.
J National Cancer Ins. 107:2015.
|
|
53
|
Vychytilova-Faltejskova P, Pesta M, Radova
L, Liska V, Daum O, Kala Z, Svoboda M, Kiss I and Slaby O:
Genome-wide microRNA expression profiling in primary tumors and
matched liver metastasis of patients with colorectal cancer. Cancer
Genomics Proteomics. 13:311–316. 2016.PubMed/NCBI
|
|
54
|
Abdelmaksoud-Dammak R, Chamtouri N, Triki
M, Saadallah-Kallel A, Ayadi W, Charfi S, Khabir A, Ayadi L,
Sallemi-Boudawara T and Mokdad-Gargouri R: Overexpression of
miR-10b in colorectal cancer patients: Correlation with TWIST-1 and
E-cadherin expression. Tumour Biol. 39:10104283176959162017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang H, Liu J, Chen Y, Ma C, Li B and Hao
T: Up-regulation of mir-10b predicate advanced clinicopathological
features and liver metastasis in colorectal cancer. Cancer Med.
5:2932–2941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gullu G, Karabulut S and Akkiprik M:
Functional roles and clinical values of insulin-like growth
factor-binding protein-5 in different types of cancers. Chin J
Cancer. 31:266–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Femia AP, Luceri C, Toti S, Giannini A,
Dolara P and Caderni G: Gene expression profile and genomic
alterations in colonic tumours induced by 1,2-dimethylhydrazine
(DMH) in rats. BMC Cancer. 10:1942010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yu L, Lu Y, Han X, Zhao W, Li J, Mao J,
Wang B, Shen J, Fan S, Wang L, et al: microRNA-140-5p inhibits
colorectal cancer invasion and metastasis by targeting ADAMTS5 and
IGFBP5. Stem Cell Res Ther. 7:1802016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Schmugge M, Rand ML and Freedman J:
Platelets and von Willebrand factor. Transfus Apher Sci.
28:269–277. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
O'Sullivan JM, Preston RJS, Robson T and
O'Donnell JS: Emerging roles for von willebrand factor in cancer
cell biology. Semin Thromb Hemost. 44:159–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang WS, Lin JK, Lin TC, Chiou TJ, Liu JH,
Yen CC and Chen PM: Plasma von Willebrand factor level as a
prognostic indicator of patients with metastatic colorectal
carcinoma. World J Gastroenterol. 11:2166–2170. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mochizuki S, Soejima K, Shimoda M, Abe H,
Sasaki A, Okano HJ, Okano H and Okada Y: Effect of ADAM28 on
carcinoma cell metastasis by cleavage of von Willebrand factor. J
NatI Cancer Inst. 104:906–922. 2012. View Article : Google Scholar
|