B-precursor acute lymphoblastic leukemia (B-ALL) is
the most common cancer diagnosed in children and adolescents
(1,2). One of the major causes of mortality is
relapse despite intensive multi-agent chemotherapy (3). For the past two decades, several
studies have reported that molecular abnormalities including TP53
mutations (4), deletion of
INK4A/ARF (5) and TEL deletion
(6) contribute to B-ALL relapse.
However, the pathogenesis and biological mechanisms underlying
relapsed ALL remain largely unknown. Thus, we sought to provide
novel insights by identifying prognostic biomarkers from
genome-wide expression profiling data generated by DNA
microarrays.
Microarray technology has been developed more than a
decade ago and is widely used in biomedical and clinical research.
This high-throughput strategy enables profiling genome-wide
expression simultaneously. Previously, based on committee neural
networks, the leukemia gene expression data can be subcategorized
into B-cell acute lymphoblastic leukemia, T-cell acute
lymphoblastic leukemia and acute myeloid leukemia (7). Unsupervised hierarchal clustering of
ALL gene expression could be used to reveal unique clusters with
distinct cytogenetic, genomic and transcriptomic characterizations
(8). By comparing gene expression
profiles of specimens at the time of diagnosis vs. at relapse, or
early- vs. late-relapse, several biological pathways such as cell
cycle regulation, WNT and mitogen-activated protein kinase pathways
(9,10) were identified to contribute to ALL
relapse.
However, these findings lack connections to clinical
practice, as the prediction of prognosis plays a crucial role in
facilitating clinical decision-making. The purpose of this study
was to develop a prognostic biomarker from gene expression
profiles. Unlike the previously published study (11), we integrated data sets from multiple
cohorts and implemented a comprehensive computational pipeline to
identify a 59-gene biomarker that could serve as a B-ALL prognostic
biomarker in practical applications.
For the discovery of prognostic biomarkers, we used
the gene expression data set from one previously published study
(14), where 80 samples collected
at the time of diagnosis were considered in this study. The RNA was
hybridized to Affymetrix HG-U133A oligonucleotide microarrays. The
raw intensity *CEL files were retrieved from EBI ArrayExpress
(https://www.ebi.ac.uk/arrayexpress/)
under the accession number E-MTAB-1216.
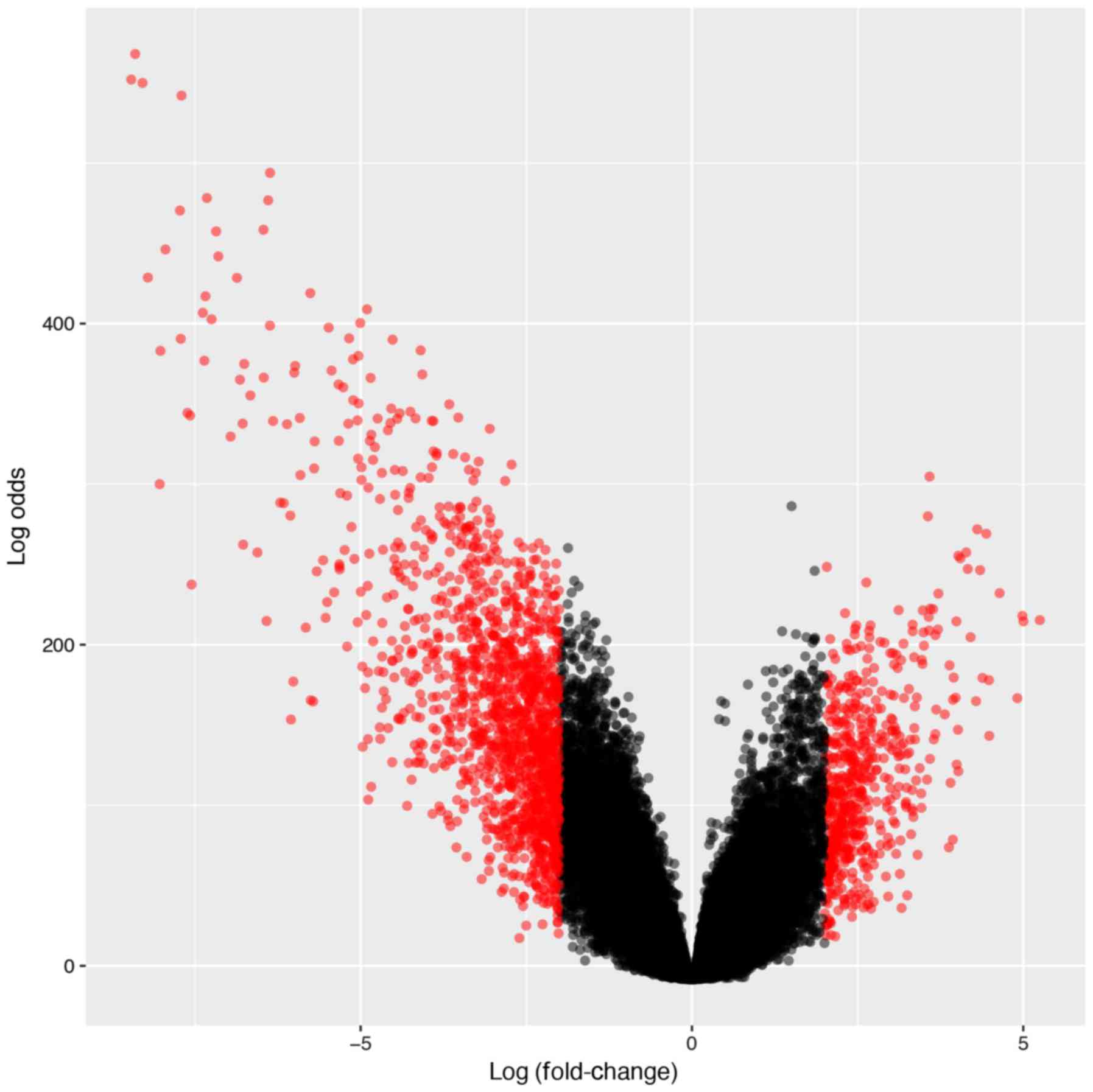
We sought to identify B-ALL-associated genes by
performing differential gene expression analysis between B-ALL
(n=207) and healthy samples (n=74). The R package limma
(16) package was used to conduct
the differential gene expression analysis. The expression data was
first Log2 transformed and then fitted into linear models with
Empirical Bayesian methods for the analysis. We filtered the
results at Log2 fold-change ≥2, or ≤-2 and B-statistics ≥4.6, which
indicates that the probabilities of the genes were differentially
expressed were >99%. After filtering, 1,273 genes were
identified. The gene set enrichment analysis was then conducted
using the R package clusterProfiler (17). The enrichment P-value was Benjamini
& Hochberg adjusted.
For the development of prognostic gene signature,
the 1,273 B-ALL-associated genes were first fitted with the Cox
proportional hazards model in 80 samples. The prognostic
significance of each gene test was assessed by log-rank test. We
selected 59 at the cut-off P-value ≤0.05. The hierarchal clustering
algorithm was then applied to the 59-gene expression profiles and
the patients were divided into high- and low-risk groups. The
Kaplan Meier-plot and log-rank test were used to test the
prognostic significance of the two groups.
In the present study, we sought to identify B-ALL
RFS biomarkers using transcriptome data. We hypothesized that the
biomarkers may be involved in B-ALL pathogenesis, thus the gene
differential expression analysis was conducted by comparing 207
B-ALL samples with 74 healthy normal blood, or bone marrow samples.
After applying the linear models and stringent cut-off [Log
fold-change ≥2, ≤-2, and false discovery rate (FDR) ≤0.01], a total
of 1,273 genes were identified as ALL-associated genes as they were
significantly upregulated or downregulated in the ALL samples
(Fig. 1).
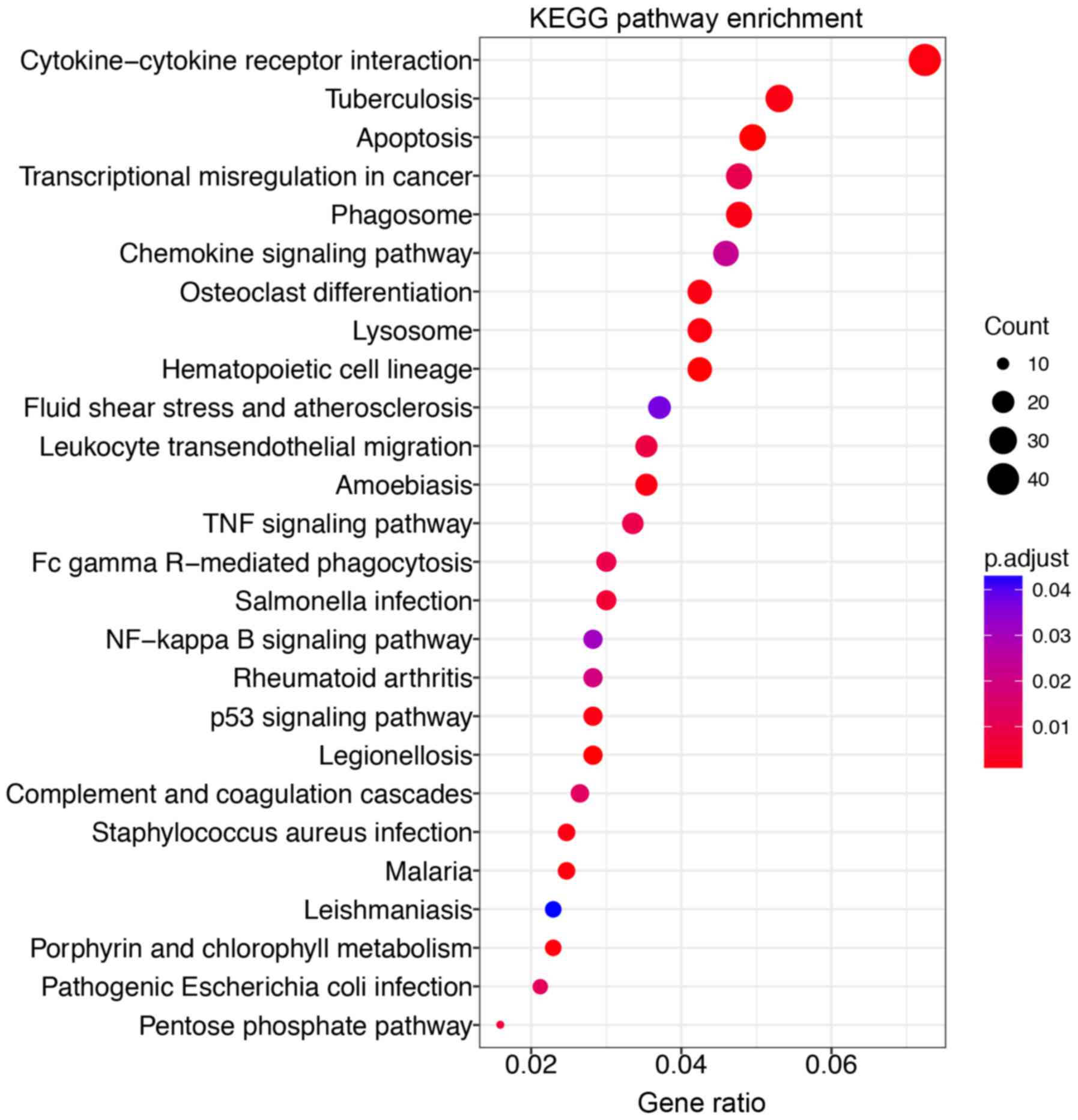
The Gene Ontology (GO) enrichment analysis revealed
that the B-ALL-associated genes were involved in various biological
processes (Table I). Surprisingly,
we found that the B-ALL-associated genes were most significantly
enriched in the biological processes that respond to bacterium
(GO:0009617). We further categorized these genes and found that
they were involved in cytokine-cytokine receptor interaction
pathway (Fig. 2). These genes
include chemokine ligands (CCL5, CXCL1, CXCL16, CXCL2, CXCL3,
CXCL8), tumor necrosis factor receptor superfamily (FAS, TNFRSF10C,
TNFRSF1B, TNFSF8), interleukins (IL-18, CXCL8) and interleukin
receptors (IL-10RA, IL-6R). These findings agreed with previous
studies on B-ALL pathogenesis. Chemokines and their receptors are
vital in many cellular activities such as migrations, targeting
developing and mature leukocytes (18). It was previously reported that the
CXCR5-CXCL13 axis plays a vital role in chronic lymphocytic
leukemia (CLL) (19). There are
also other chemokines such as CCR7, CXCR4 and CXCR5 (20), CXCR3 (21), CCL25 (22) and the CXCR4-CXCL12 axis (23) that have been identified as potential
targets in leukemia treatment.
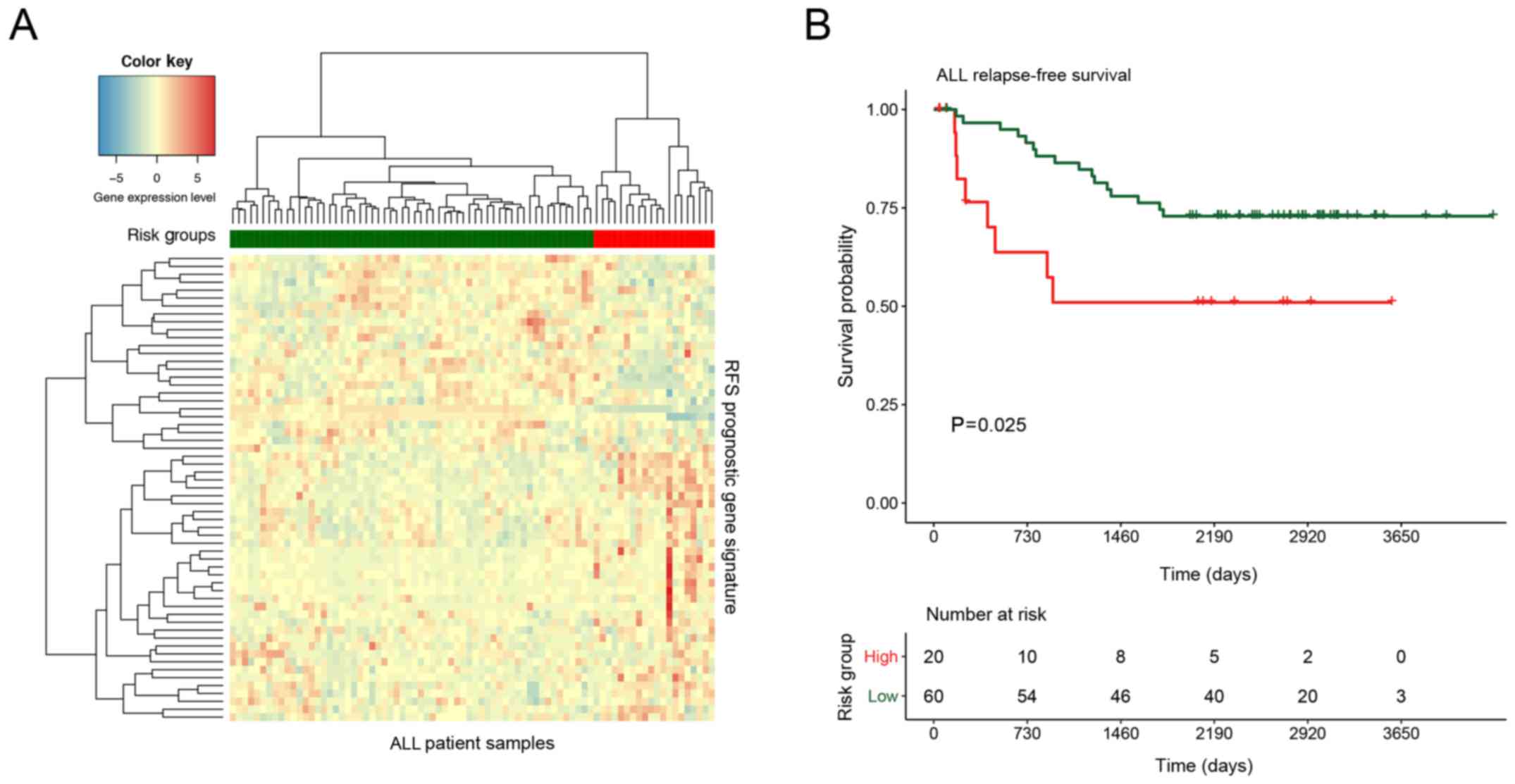
Then, we applied Cox proportional hazards modelling
to the B-ALL-associated genes and selected 59 top ranked genes
based on log-rank test P-values as B-ALL RFS genes (Table II). After applying the hierarchal
clustering to this 59-gene profile, the high- (n=20), and low-risk
(n=60) samples were identified from the B-ALL cohort with
significant (log-rank test P=0.025) different relapse-free survival
outcome (Fig. 3). The B-ALL RFS
biomarkers included genes from various families, such as
glycosyltransferases (C1GALT1 and B4GALT5), RAS oncogene GTPases
(RAB27B and RAB7A), nuclear hormone receptors (NR3C1 and RORA), RNA
binding proteins (RBM6 and U2SURP) and Zinc finger proteins
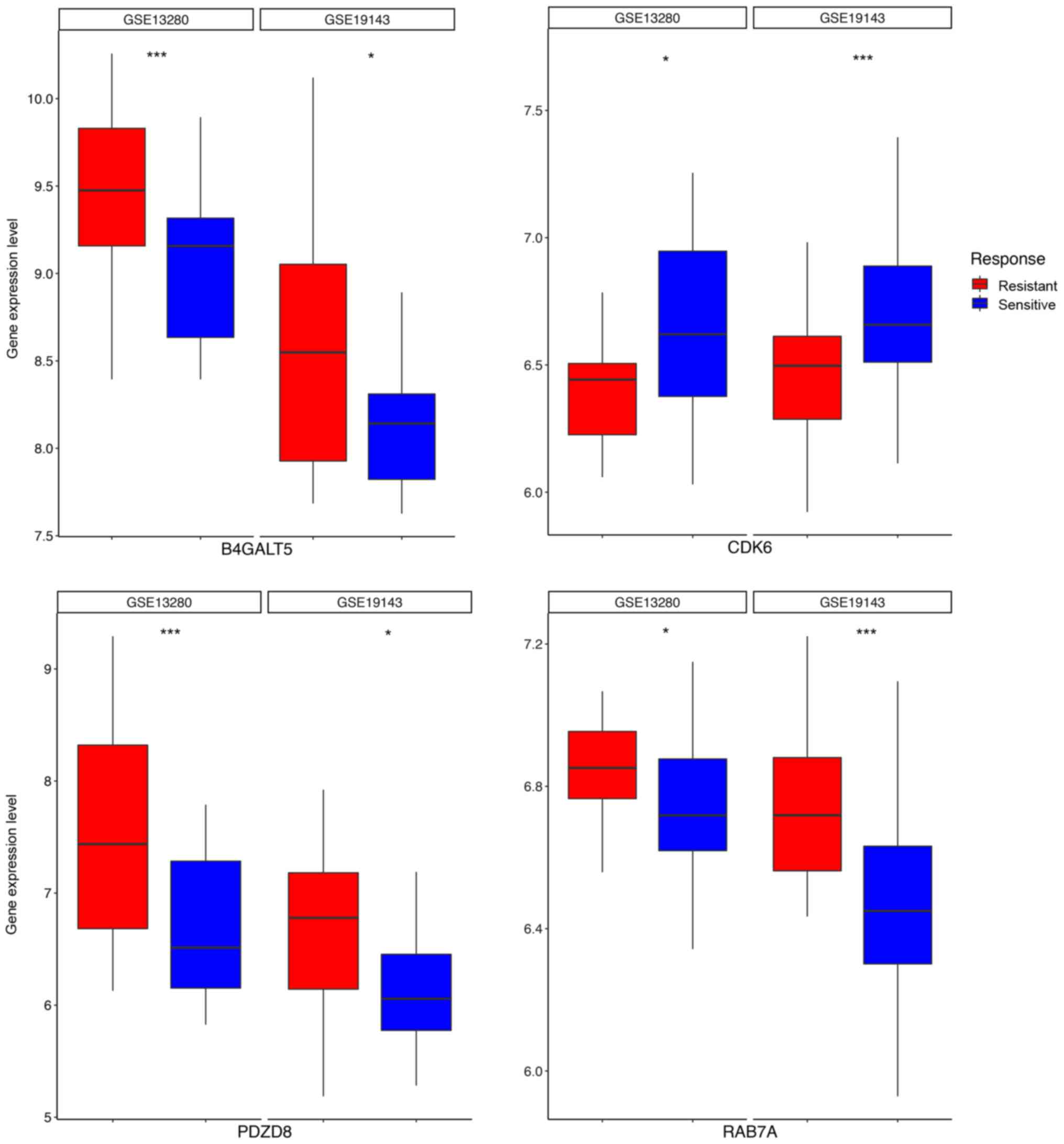
(PLAGL2, SP1 and ZNF91). We also found that among these biomarkers,
4 genes, B4GALT5, CDK6, PDZD8 and RAB7A, were candidates that were
associated with chemotherapy response (Fig. 4) in two independent ALL cohorts,
GSE19143 (n=52) (24) and GSE13280
(n=44) (25) where patients were
treated with prednisolone. Indeed, the B4GALT family is involved in
mediating drug resistance in human leukemia cells by regulating the
Hedgehog pathway (26). This
suggest that the 59-gene biomarker could also indicate drug
sensitivity in ALL treatment.
The major challenge in clinical treatments of
B-precursor acute lymphoblastic leukemia (B-ALL) is relapse after
chemotherapy; thus or the development of alternative treatments is
critical. Here, we demonstrated the ability of gene expression
profiling to reveal not only biological mechanisms but also
clinical diagnostic markers.
Among the 59 genes, several genes have been
characterized as being involved in the progression of leukemia and
solid tumors. For instance, MME, which is also known as CD10, or
CALLA, encodes a type II transmembrane glycoprotein and a common
acute lymphocytic leukemia antigen. It is an important cell surface
marker in the diagnosis of human acute lymphocytic leukemia
(27) has been well-documented in
many leukemia-related studies (28–32).
SOCS3 is a member of the suppressor of cytokine signaling (SOCS)
family and negatively regulates JAK2 kinase. Its altered expression
is associated with leukemia (33,34)
and solid tumors including melanoma (35–38),
cervical cancer (39–41), renal cell carcinoma (42–45),
prostate cancer (46,47) and gastric cancer (48). CDK6 is a serine/threonine protein
that is important for cell cycle G1 phase progression
and G1/S transition. CDK6 is dysregulated or disrupted in many
types of cancer (49–54), and it is previously reported to be
in a three-way rearrangement including other two elements: MLL and
AF-4 in a case of infant ALL (55).
This indicates the biological relevance of the 59-gene biomarker to
B-ALL and could propose new directions for investigations.
However, we do realize that there is merely a one
gene (JCHAIN known as IGJ) overlap of our 59-gene biomarker with a
previously published 38-gene classifier (11). We reasoned that this discordance is
due to differences in the analysis strategy. While in the previous
study, the biomarker selection was implemented directly on all of
the microarray probe sets, we pre-filtered the candidate list to
B-ALL-associated genes by differential gene expression analysis. We
also optimized the process of preprocessing the microarray data by
using up-to-date algorithms and software, which could lead to
higher confidence in producing final results.
In conclusion, our systematic approach provided an
intriguing guideline for the identification of B-ALL prognostic
biomarkers and revealed their potential roles in chemosensitivity.
Further investigations are expected to validate the performances of
these biomarkers before being applied to clinical management. As
the RNA-seq technologies are trending in transcriptome profiling,
we will also collect RNA-seq data to re-train and optimize our
model. We will also try to reduce the number of biomarkers while
maintaining the predictive power, so that the application in
clinical management is more feasible.
Not applicable.
No funding was received.
The datasets used during the present study are
available from NCBI GEO Database with corresponding accession
numbers.
WJ and JL conceived and designed the study. WJ
collected the data and performed the data analysis. WJ and JL wrote
and edited the manuscript. Both authors read and approved the
manuscript and agreed to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Not applicable.
Not applicable.
The authors do not have any competing interests.
|
1
|
Locatelli F, Schrappe M, Bernardo ME and
Rutella S: How I treat relapsed childhood acute lymphoblastic
leukemia. Blood. 120:2807–2816. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhojwani D and Pui CH: Relapsed childhood
acute lymphoblastic leukaemia. Lancet Oncol. 14:e205–e217. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hunger SP, Lu X, Devidas M, Camitta BM,
Gaynon PS, Winick NJ, Reaman GH and Carroll WL: Improved survival
for children and adolescents with acute lymphoblastic leukemia
between 1990 and 2005: A report from the children's oncology group.
J Clin Oncol. 30:1663–1669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu YM, Foroni L, McQuaker IG, Papaioannou
M, Haynes A and Russell HH: Mechanisms of relapse in acute
leukaemia: Involvement of p53 mutated subclones in disease
progression in acute lymphoblastic leukaemia. Br J Cancer.
79:1151–1157. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maloney KW, McGavran L, Odom LF and Hunger
SP: Acquisition of p16INK4A and p15INK4B gene
abnormalities between initial diagnosis and relapse in children
with acute lymphoblastic leukemia. Blood. 93:2380–2385.
1999.PubMed/NCBI
|
|
6
|
Zuna J: TEL deletion analysis supports a
novel view of relapse in childhood acute lymphoblastic leukemia.
Clin Cancer Res. 10:5355–5360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sewak MS, Reddy NP and Duan ZH: Gene
expression based leukemia sub-classification using committee neural
networks. Bioinform Biol Insights. 3:89–98. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harvey RC, Mullighan CG, Wang X, Dobbin
KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et
al: Identification of novel cluster groups in pediatric high-risk
B-precursor acute lymphoblastic leukemia with gene expression
profiling: Correlation with genome-wide DNA copy number
alterations, clinical characteristics, and outcome. Blood.
116:4874–4884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hogan LE, Meyer JA, Yang J, Wang J, Wong
N, Yang W, Condos G, Hunger SP, Raetz E, Saffery R, et al:
Integrated genomic analysis of relapsed childhood acute
lymphoblastic leukemia reveals therapeutic strategies. Blood.
118:5218–5226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Staal FJ, van der Burg M, Wessels LF,
Barendregt BH, Baert MR, van den Burg CM, van Huffel C, Langerak
AW, van der Velden VH, Reinders MJ, et al: DNA microarrays for
comparison of gene expression profiles between diagnosis and
relapse in precursor-B acute lymphoblastic leukemia: Choice of
technique and purification influence the identification of
potential diagnostic markers. Leukemia. 17:1324–1332. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang H, Chen IM, Wilson CS, Bedrick EJ,
Harvey RC, Atlas SR, Devidas M, Mullighan CG, Wang X, Murphy M, et
al: Gene expression classifiers for relapse-free survival and
minimal residual disease improve risk classification and outcome
prediction in pediatric B-precursor acute lymphoblastic leukemia.
Blood. 115:1394–1405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kohlmann A, Kipps TJ, Rassenti LZ, Downing
JR, Shurtleff SA, Mills KI, Gilkes AF, Hofmann WK, Basso G,
Dell'orto MC, et al: An international standardization programme
towards the application of gene expression profiling in routine
leukaemia diagnostics: The microarray innovations in LEukemia study
prephase. Br J Haematol. 142:802–807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Haferlach T, Kohlmann A, Wieczorek L,
Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK,
Mills KI, et al: Clinical utility of microarray-based gene
expression profiling in the diagnosis and subclassification of
leukemia: Report from the international microarray innovations in
leukemia Study Group. J Clin Oncol. 28:pp. 2529–2537. 2010,
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Beesley AH, Cummings AJ, Freitas JR,
Hoffmann K, Firth MJ, Ford J, de Klerk NH and Kees UR: The gene
expression signature of relapse in paediatric acute lymphoblastic
leukaemia: Implications for mechanisms of therapy failure. Br J
Haematol. 131:447–456. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCall MN, Jaffee HA and Irizarry RA: fRMA
ST Frozen robust multiarray analysis for Affymetrix Exon and Gene
ST arrays. Bioinformatics. 28:3153–3154. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. Omi A J Integr Biol. 16:284–287. 2012. View Article : Google Scholar
|
|
18
|
Campbell DJ, Kim CH and Butcher EC:
Chemokines in the systemic organization of immunity. Immunol Rev.
195:58–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Burkle A, Niedermeier M, Schmitt-Graff A,
Wierda WG, Keating MJ and Burger JA: Overexpression of the CXCR5
chemokine receptor, and its ligand, CXCL13 in B-cell chronic
lymphocytic leukemia. Blood. 110:3316–3325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
López-Giral S, Quintana NE, Cabrerizo M,
Alfonso-Pérez M, Sala-Valdés M, De Soria VG, Fernández-Rañada JM,
Fernández-Ruiz E and Muñoz C: Chemokine receptors that mediate B
cell homing to secondary lymphoid tissues are highly expressed in B
cell chronic lymphocytic leukemia and non-Hodgkin lymphomas with
widespread nodular dissemination. J Leukoc Biol. 76:462–471. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones D, Benjamin RJ, Shahsafaei A and
Dorfman DM: The chemokine receptor CXCR3 is expressed in a subset
of B-cell lymphomas and is a marker of B-cell chronic lymphocytic
leukemia. Blood. 95:627–32. 2000.PubMed/NCBI
|
|
22
|
Qiuping Z, Jei X, Youxin J, Wei J, Chun L,
Jin W, Qun W, Yan L, Chunsong H, Mingzhen Y, et al: CC chemokine
ligand 25 enhances resistance to apoptosis in CD4+ T
cells from patients with T-cell lineage acute and chronic
lymphocytic leukemia by means of livin activation. Cancer Res.
64:7579–7587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Burger JA and Peled A: CXCR4 antagonists:
Targeting the microenvironment in leukemia and other cancers.
Leukemia. 23:43–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Stam RW, Den Boer ML, Schneider P, de Boer
J, Hagelstein J, Valsecchi MG, de Lorenzo P, Sallan SE, Brady HJ,
Armstrong SA, et al: Association of high-level MCL-1 expression
with in vitro and in vivo prednisone resistance in MLL-rearranged
infant acute lymphoblastic leukemia. Blood. 115:1018–1125. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spijkers-Hagelstein JA, Schneider P,
Hulleman E, de Boer J, Williams O, Pieters R and Stam RW: Elevated
S100A8/S100A9 expression causes glucocorticoid resistance in
MLL-rearranged infant acute lymphoblastic leukemia. Leukemia.
26:1255–1165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou H, Ma H, Wei W, Ji D, Song X, Sun J,
Zhang J and Jia L: B4GALT family mediates the multidrug resistance
of human leukemia cells by regulating the hedgehog pathway and the
expression of p-glycoprotein and multidrug resistance-associated
protein 1. Cell Death Dis. 4:e6542013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Epstein J, Xiao HQ and He XY: Markers of
multiple hematopoietic-cell lineages in multiple myeloma. N Engl J
Med. 322:664–668. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dunphy CH, Gardner LJ, Evans HL and Javadi
N: CD15+ acute lymphoblastic leukemia and subsequent
monoblastic leukemia: Persistence of t(4;11) abnormality and B-cell
gene rearrangement. Arch Pathol Lab Med. 125:1227–1230.
2001.PubMed/NCBI
|
|
29
|
Rozovskaia T, Feinstein E, Mor O, Foa R,
Blechman J, Nakamura T, Croce CM, Cimino G and Canaani E:
Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with
the t(4:11) abnormality. Oncogene. 20:874–878. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Burmeister T, Meyer C, Schwartz S, Hofmann
J, Molkentin M, Kowarz E, Schneider B, Raff T, Reinhardt R,
Gökbuget N, et al: The MLL recombinome of adult CD10-negative
B-cell precursor acute lymphoblastic leukemia: Results from the
GMALL study group. Blood. 113:4011–4015. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gleissner B, Goekbuget N, Rieder H, Arnold
R, Schwartz S, Diedrich H, Schoch C, Heinze B, Fonatsch C, Bartram
CR, et al: CD10− pre-B acute lymphoblastic leukemia
(ALL) is a distinct high-risk subgroup of adult ALL associated with
a high frequency of MLL aberrations: Results of the german
multicenter trials for adult ALL (GMALL). Blood. 106:4054–4056.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ha K, Hozumi N, Hrincu A and Gelfand EW:
Lineage specific classification of leukaemia: Results of the
analysis of sixty cases of childhood leukaemia. Br J Haematol.
61:237–249. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Suzuki R, Sakamoto H, Yasukawa H, Masuhara
M, Wakioka T, Sasaki A, Yuge K, Komiya S, Inoue A and Yoshimura A:
CI S3 and JAB have different regulatory roles in interleukin-6
mediated differentiation and STAT3 activation in M1 leukemia cells.
Oncogene. 17:2271–2278. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen C, Yu K, Yan QX, Xing CY, Chen Y, Yan
Z, Shi YF, Zhao KW and Gao SM: Pure curcumin increases the
expression of SOCS1 and SOCS3 in myeloproliferative neoplasms
through suppressing class I histone deacetylases. Carcinogenesis.
34:1442–1449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Komyod W, Böhm M, Metze D, Heinrich PC and
Behrmann I: Constitutive suppressor of cytokine signaling 3
expression confers a growth advantage to a human melanoma cell
line. Mol Cancer Res. 5:271–281. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lesinski GB, Zimmerer JM, Kreiner M,
Trefry J, Bill MA, Young GS, Becknell B and Carson WE III:
Modulation of SOCS protein expression influences the interferon
responsiveness of human melanoma cells. BMC Cancer. 10:1422010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tokita T, Maesawa C, Kimura T, Kotani K,
Takahashi K, Akasaka T and Masuda T: Methylation status of the
SOCS3 gene in human malignant melanomas. Int J Oncol. 30:689–694.
2007.PubMed/NCBI
|
|
38
|
Fojtova M, Boudny V, Kovarik A, Lauerova
L, Adamkova L, Souckova K, Jarkovsky J and Kovarik J: Development
of IFN-gamma resistance is associated with attenuation of SOCS
genes induction and constitutive expression of SOCS 3 in melanoma
cells. Br J Cancer. 97:231–237. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shivapurkar N, Sherman ME, Stastny V,
Echebiri C, Rader JS, Nayar R, Bonfiglio TA, Gazdar AF and Wang SS:
Evaluation of candidate methylation markers to detect cervical
neoplasia. Gynecol Oncol. 107:549–553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang P, Yang B, Yao YY, Zhong LX, Chen
XY, Kong QY, Wu ML, Li C, Li H and Liu J: PIA S3SH P2 and SOCS3
Expression patterns in Cervical Cancers: Relevance with activation
and resveratrol-caused inactivation of STAT3 signaling. Gynecol
Oncol. 139:529–535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim MH, Kim MS, Kim W, Kang MA, Cacalano
NA, Kang SB, Shin YJ and Jeong JH: Suppressor of cytokine signaling
(SOCS) genes are silenced by DNA hypermethylation and histone
deacetylation and regulate response to radiotherapy in cervical
cancer cells. PLoS One. 10:e01231332015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Urbschat A, Stumpf S, Hänze J, Paulus P,
Maier TJ, Weipert C, Hofmann R and Hegele A: Expression of the
anti-inflammatory suppressor of cytokine signaling 3 (SOCS3) in
human clear cell renal cell carcinoma. Tumour Biol. 37:9649–9656.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stofas A, Levidou G, Piperi C, Adamopoulos
C, Dalagiorgou G, Bamias A, Karadimou A, Lainakis GA, Papadoukakis
S, Stravodimos K, et al: The role of CXC-chemokine receptor CXCR2
and suppressor of cytokine signaling-3 (SOCS-3) in renal cell
carcinoma. BMC Cancer. 14:1492014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tomita S, Ishibashi K, Hashimoto K, Sugino
T, Yanagida T, Kushida N, Shishido K, Aikawa K, Sato Y, Suzutani T,
et al: Suppression of SOCS3 increases susceptibility of renal cell
carcinoma to interferon-α. Cancer Sci. 102:57–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Oguro T, Ishibashi K, Sugino T, Hashimoto
K, Tomita S, Takahashi N, Yanagida T, Haga N, Aikawa K, Suzutani T,
et al: Humanised antihuman IL-6R antibody with interferon inhibits
renal cell carcinoma cell growth in vitro and in vivo through
suppressed SOCS3 expression. Eur J Cancer. 49:1715–1724. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Neuwirt H, Puhr M, Cavarretta IT,
Mitterberger M, Hobisch A and Culig Z: Suppressor of cytokine
signalling-3 is up-regulated by androgen in prostate cancer cell
lines and inhibits androgen-mediated proliferation and secretion.
Endocr Relat Cancer. 14:1007–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kneitz B, Krebs M, Kalogirou C, Schubert
M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Ströbel P,
et al: Survival in patients with high-risk prostate cancer is
predicted by miR-221, which regulates proliferation, apoptosis, and
invasion of prostate cancer cells by inhibiting IRF2 and SOCS3.
Cancer Res. 74:2591–2603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li G, Xu J, Wang Z, Yuan Y, Li Y, Cai S
and He Y: Low expression of SOCS-1 and SOCS-3 is a poor prognostic
indicator for gastric cancer patients. J Cancer Res Clin Oncol.
141:443–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lien HC, Lin CW, Huang PH, Chang ML and
Hsu SM: Expression of cyclin-dependent kinase 6 (cdk6) and frequent
loss of CD44 in nasal-nasopharyngeal NK/T-cell lymphomas:
Comparison with CD56-negative peripheral T-cell lymphomas. Lab
Invest. 80:893–900. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lee YH, Judge AD, Seo D, Kitade M,
Gómez-Quiroz LE, Ishikawa T, Andersen JB, Kim BK, Marquardt JU,
Raggi C, et al: Molecular targeting of CSN5 in human hepatocellular
carcinoma: A mechanism of therapeutic response. Oncogene.
30:4175–4184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ouyang Q, Chen G, Zhou J, Li L, Dong Z,
Yang R, Xu L, Cui H, Xu M and Yi L: Neurotensin signaling
stimulates glioblastoma cell proliferation by upregulating c-Myc
and inhibiting miR-29b-1 and miR-129-3p. Neuro Oncol. 18:216–226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Easton J, Wei T, Lahti JM and Kidd VJ:
Disruption of the cyclin D/cyclin-dependent
kinase/INK4/retinoblastoma protein regulatory pathway in human
neuroblastoma. Cancer Res. 58:2624–2632. 1998.PubMed/NCBI
|
|
53
|
Brandi G, Paiardini M, Cervasi B, Fiorucci
C, Filippone P, De Marco C, Zaffaroni N and Magnani M: A new
indole-3- carbinol tetrameric derivative inhibits cyclin-dependent
kinase 6 expression, and induces G1 cell cycle arrest in both
estrogen-dependent and estrogen-independent breast cancer cell
lines. Cancer Res. 63:4028–4036. 2003.PubMed/NCBI
|
|
54
|
Ye Y, Yang H, Grossman HB, Dinney C, Wu X
and Gu J: Genetic variants in cell cycle control pathway confer
susceptibility to bladder cancer. Cancer. 112:2467–2474. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Raffini LJ, Slater DJ, Rappaport EF, Lo
Nigro L, Cheung NK, Biegel JA, Nowell PC, Lange BJ and Felix CA:
Panhandle and reverse-panhandle PCR enable cloning of der(11) and
der(other) genomic breakpoint junctions of MLL translocations and
identify complex translocation of MLL, AF-4, and CDK6. Proc Natl
Acad Sci USA. 99:4568–4573. 2002. View Article : Google Scholar : PubMed/NCBI
|