Introduction
Promyelocytic leukemia zinc finger (PLZF), also
known as BTB-containing protein 16 (ZBTB16), was originally
identified as a gene fused to RARa in acute promyelocytic leukemia
patients (1). PLZF plays an
important role in the developmental and biological processes of
carcinogenesis, functioning as a tumor suppressor or promoter
(2,3). In early studies, PLZF expression was
lost in cancer cells and the loss of PLZF expression was related to
increased proliferation, invasiveness, motility and resistance to
apoptosis (2,4). However, with further investigation the
role of PLZF appeared to be controversial in tumor progression.
Except for hematological malignancies, PLZF has also been
implicated in various solid tumors. The loss of PLZF expression in
tumors compared with normal tissue has been observed in melanoma,
hepatocellular carcinoma, pancreatic and lung cancer, thyroid
carcinoma, prostate and gallbladder cancer (5–11), and
increased PLZF expression in tumors has been observed in clear cell
renal cell carcinoma, colon cancer, glioblastoma and testicular
seminoma (3,12). However, the expression and value of
PLZF in gastric cancer (GC) have not been studied, and it is
reasonable to think that PLZF may play an important role in GC.
Furthermore, the exact reason for PLZF regulation in cancers is not
well documented.
Long non-coding RNAs (lncRNAs) are a subset of
non-coding RNAs that mediate their biological functions using
chromatin as a substrate to interact with the genetic information
encoded in the genome (13). Among
lncRNAs, ANRIL (CDKN2B antisense RNA 1) is transcribed in the
opposite direction from the INK4b-ARF-INK4a gene cluster, which has
been identified as a shared genetic susceptibility locus associated
with coronary disease, intracranial aneurysm, type 2 diabetes and
cancers (14,15). ANRIL can be induced by the ATM-E2F1
signaling pathway and is required for the silencing of p15INK4B by
recruiting polycomb repressive complex 2 (PRC2) (16,17).
EZH2, a subunit of PRC2, which plays important roles
in epigenetic gene silencing by catalyzing di- and tri-methylation
of H3K27 can recruit the DNA methyltransferases to a target
promoter, resulting in DNA methylation and subsequent gene
silencing (18,19). DNA methylation patterns in CpG-rich
promoter regions and CpG islands regulate gene expression in
mammalian cells; usually, methylation of CpG dinucleotides in the
promoter represses gene expression (20). Particularly in cancer, gene
silencing occurs through aberrant methylation in the promoter of
tumor suppressor genes (21).
It has been reported that both of PcG activity and
DNA methylation contribute to epigenetic repression and there
exists crosstalk between them. In the present study, we found that
ANRIL expression in GC was negatively associated with the level of
PLZF and with ANRIL-recruited PRC2, which then drove PLZF silencing
by collaborating between H3K27me3 and DNA methylation. Evidence
that the methylation inhibitor 5-Aza-2′-deoxycytidine (5-Aza) could
increase the expression level of PLZF confirmed the regulatory
methylation. Our results provide new insights into the expression
level and potential status of PLZF in GC as well as a possible
mechanism of alteration.
Materials and methods
Cell lines
The human GC cell lines SGC7901, BGC823, MGC803,
MKN28 and MKN45 and the normal gastric epithelium cell line (GES1)
were obtained from the Chinese Academy of Sciences Committee on
Type Culture Collection Cell Bank (Shanghai, China) and Fuheng Cell
Center (Shanghai, China). All cell lines were authenticated by STR
profiling. Cell lines were cultured in Dulbecco's modified Eagle's
medium supplemented with 10% fetal bovine serum (FBS) in a
humidified atmosphere containing 5% CO2 at 37°C.
Study subjects
We obtained 60 paired GC and adjacent non-cancer
tissues from patients who underwent surgery at Nanjing First
Hospital of Jiangsu Province in China between 2011 and 2012 and who
were diagnosed with GC based on histopathological evaluation. No
local or systemic treatment was administered to these patients
prior to surgery. All collected tissue samples were preserved in
RNA Transport (OMEGA, Norwalk, CT, USA) and immediately frozen at
−80°C until required. The clinical characteristics of all patients
are listed in Table I. The present
study was approved by the Ethics Committee of Nanjing Medical
University.
 | Table I.The relationship between PLZF
expression and clinicopathological factors of GC patients. |
Table I.
The relationship between PLZF
expression and clinicopathological factors of GC patients.
|
| PLZF levels |
|
|---|
|
|
|
|
|---|
| Clinical
parameters | High (n=10) | Low (n=50) |
P-valuea |
|---|
| Age, years |
|
| 0.540 |
|
≤60 | 2 | 18 |
|
|
>60 | 8 | 32 |
|
| Sex |
| 0.723 |
|
|
Male | 6 | 30 |
|
|
Female | 4 | 20 |
|
| Histologic
differentiation |
| 0.002c |
|
| Low or
undifferentiation | 1 | 35 |
|
| Middle
or high | 9 | 15 |
|
| Invasion depth |
| 0.006c |
|
| T1 | 8 | 14 |
|
| T2 or
above | 2 | 36 |
|
| TNM stages |
| 0.037b |
|
|
I/II | 8 | 19 |
|
|
III/IV | 2 | 31 |
|
| Lymphatic
metastasis |
| 0.037b |
|
|
Yes | 0 | 20 |
|
| No | 10 | 30 |
|
| Distant
metastasis |
| 0.278 |
|
|
Yes | 0 | 10 |
|
| No | 10 | 40 |
|
Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from cultivated cells or
tissue using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's protocol.
RNA concentrations were determined spectrophotometrically at 260
nm. Total RNA (1 µg) was reverse transcribed to cDNA in a total
volume of 20 µl. Quantitative PCR was performed with the following
thermocycling conditions: 95°C 30 sec for initial denaturation,
95°C 5 sec, 60°C 34 sec for PCR reaction, 40 cycles, using a 7300
Plus Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and SYBR Premix Ex Taq (Takara Biotechnology Co.,
Ltd., Dalian, Liaoning, China) as a DNA-specific fluorescent dye.
Notably, cDNA synthesis of coding gene mRNA used
PrimeScript™ RT reagent Kit (cat. no. RR037A; Takara
Biotechnology Co., Ltd.) and cDNA synthesis of lncRNA used
PrimeScript RT Reagent kit with gDNA Eraser (cat. no. RR047A;
Takara Biotechnology Co., Ltd.). Gene expression levels were
calculated relative to GAPDH and β-actin using the
2−ΔΔCt method (22).
GAPDH and β-actin were used as endogenous controls. The primer
sequences are listed in Table
II.
 | Table II.Primers used for qRT-PCR and
MPCR. |
Table II.
Primers used for qRT-PCR and
MPCR.
| Primers | Sequences |
|---|
| PLZF-F |
AGCGGTTCCTGGATAGTTT |
| PLZF-R |
ATGCGTTTGTGGCTCTTG |
| Methylated
PLZF-F |
TCGTTAGTATTAAAGATGGAGAGGC |
| Methylated
PLZF-R |
GAAATTAACAAATCACCACCGAC |
| Unmethylated
PLZF-F |
GGTTGTTAGTATTAAAGATGGAGAGGT |
| Unmethylated
PLZF-R |
CTACAAAATTAACAAATCACCACCA |
| ANRIL-F |
TGCTCTATCCGCCAATCAGG |
| ANRIL-R |
GCGTGCAGCGGTTTAGTTT |
| E-cadherin-F |
GAACGCATTGCCACATACAC |
| E-cadherin-R |
GAGGATGGTGTAAGCGATGG |
| N-cadherin-F |
ATGGAAGGCAATCCCACATA |
| N-cadherin-R |
CAGTAGGATCTCCGCCACTG |
| Vimentin-F |
GTACCGGAGACAGGTGCAGT |
| Vimentin-R |
CTCAATGTCAAGGGCCATCT |
| EZH2-F |
ACATCCTTTTCATGCAACACC |
| EZH2-R |
GCTCCCTCCAAATGCTGGTA |
| SUZ12-F |
ACATCAAAAGCTTGTCAGCTC |
| SUZ12-R |
GCACCTGCTTTTTACCTGTGG |
| Snail-F |
CAAGATGCACATCCGAAGCC |
| Snail-R |
CATCTGAGTGGGTCTGGAGG |
| Slug-F |
GCTACCCAATGGCCTCTCTC |
| Slug-R |
CTTCAATGGCATGGGGGTCT |
| ZEB1-F |
GATGACCTGCCAACAGACCA |
| ZEB1-R |
CCCCAGGATTTCTTGCCCTT |
| GAPDH-F |
AGCCACATCGCTCAGACAC |
| GAPDH-R |
GCCCAATACGACCAAATCC |
| β-actin-F |
CTGGGACGACATGGAGAAAA |
| β-actin-R |
AAGGAAGGCTGGAAGAGTGC |
| MEG3-F |
GTGGAAGCACGCTCACAAAG |
| MEG3-R |
GAAGACTTGGGTCCGGGATG |
| TUG1-F |
CAAGAAACAGCAACACCAGAAG |
| TUG1-R |
TAAGGTCCCCATTCAAGTCAGT |
| LINC00152-F |
TTGATGGCTTGAACATTTGG |
| LINC00152-R |
TCGTGATTTTCGGTGTCTGT |
| H19-F |
GCCTTGACGTGCTGGATCT |
| H19-R |
TCCGATGCTTTACTCAAGAAGTT |
| HOTAIR-F |
GCGCTGCAAGTGCTTACTGTGCA |
| HOTAIR-R |
CCGAGGTATTCGCACTGGATAC |
| PVT1-F |
CAGTGGGGAACTCTGACTCG |
| PVT1-R |
GTGCCTGGTGCTCTCTTACC |
| LINC00261-F |
ATCAAGAGGCAATGGTCCCA |
| LINC00261-R |
TTCAGCTCTTAGGGCAGGAC |
| SNHG1-F | AGGCTGAAGT
TACAGGTC |
| SNHG1-R |
TTGGCTCCCAGTGTCTTA |
| UCA1-F |
AGTGGCTGAAGACTGATGC |
| UCA1-R |
AGATGGACGGCAGTTGGT |
| GAS5-F |
CCCAAGGAAGGATGAG |
| GAS5-R |
ACCAGGAGCAGAACCA |
| SPRY4-IT1-F |
AGCCACATAAATTCAGCAGA |
| SPRY4-IT1-R |
CGATGTAGTAGGATTCCTTTCA |
Plasmid, siRNA and transfection
PLZF (ZBTB16) (NM_006006) Human Tagged ORF Clone
(cat. no. RG206745) and pCMV6-AC-GFP Tagged Cloning Vector (cat.
no. PS100010) were purchased from OriGene Technologies, Inc.
(Rockville, MD, USA). Plasmid vectors for transfection were
prepared using DNA Midiprep Kits (Qiagen, Duesseldorf, NRW,
Germany) and transfected into GC cells using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The siRNAs were
synthesized by Invitrogen and transfected into GC cells using
Lipofectamine 2000 according to the manufacturer's instructions.
Cells were seeded at ~2–4×105 cells/well in 6-well
plates and transfected with 100 nmol/l siRNAs in Opti-MEM medium
using Lipofectamine 2000 reagent. The cells were then allowed to
reach ~50% confluence on the day of transfection. The cells were
switched into medium containing 10% FBS after transfection for ~4–6
h and incubated for another ~24–48 h until extraction for the
assessment of interfering efficiencies and targeted genes. All
siRNA sequences are listed in Table
III.
 | Table III.siRNA sequences of targets. |
Table III.
siRNA sequences of targets.
| Targets | siRNA
sequences |
|---|
| EZH2 |
AUCAGCUCGUCUGAACCUCUU |
| MEG3 |
UCCCUCUUACCUAAAGACUUAAA |
| TUG1 |
CAGUCCUGGUGAUUUAGACAGUCUU |
| LINC00152 |
UGAUCGAAUAUGACAGACACCGAAA |
| H19 |
CCCACAACAUGAAAGAAACTT |
| HOTAIR |
GCAACCTAAACCAGCAATT |
| PVT1 |
GCUUGGAGGCUGAGGAGUUTT |
| LINC00261 |
CAGTCGCTTGGTTTGAGCTCAAATA |
| SNHG1 | CAGCAGT
TGAGGGTTTGCTGTGTAT |
| UCA1 |
GCCATATGAAGACACCCTA |
| GAS5 |
CTTGCCTGGACCAGCTTAATT |
| SPRY4-IT1 |
GCTTTCTGATTCCAAGGCCTATTAA |
| siCtrl |
GCATCAACAACCGAACATT |
Cell proliferation, migration and
invasion
The cell proliferation ability was examined using
the CellTiter 96® AQueous One Solution Cell
Proliferation Assay (MTS; Promega Corporation, Madison, WI, USA).
Absorbance values were assessed at a wavelength of 570 nm. Colony
formation assays were performed to monitor the cloning capability
of GC cells. The wound healing assay was used to evaluate the cell
migration capabilities. The Transwell assay was performed to assess
invasion. Procedural details were previously described (23).
Establishment of xenografts and in
vivo study
Four-week-old female athymic BALB/c nude mice were
maintained under specific pathogen-free conditions, using a 10-h
light/14-h dark cycle at temperature of 25±1°C and relative
humidity of 40~60% with free access to food and water, and
manipulated according to protocols approved by the Ethics Committee
of Animal Experiments of the Nanjing Medical University. The mice
were divided into two groups of the PLZF and the empty vector with
5 mice/group. PLZF and empty vector stably transfected SGC7901
cells were harvested and 107 cells were subcutaneously
injected into a single side of each mouse. Tumor sizes were
measured by calipers and recorded every 3 days. The tumor volumes
were calculated from the length (the longest diameter across the
tumor) and width (the corresponding perpendicular diameter) using
the following formula: π/6 × length × width2. Twenty
days later, the animals were sacrificed by carbon dioxide, and
tumors were harvested and preserved at −80°C and fixed in
formaldehyde for qRT-PCR and immunohistochemistry (IHC) staining,
respectively.
Western blot analysis
Western blot analysis was performed according to
standard protocols as previously described (24). The primary antibodies of H3K27me3
(cat. no. 9733), EZH2 (cat. no. 5246), PLZF (cat. no. 39784) and
GAPDH (cat. no. 5174) and the secondary antibody of HRP-linked
anti-rabbit IgG (cat. no. 7074) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). The primary antibodies were
diluted at ~1:500-1,000 and incubated at 4°C overnight and the
secondary antibody was diluted at 1:2,000 at room temperature for 2
h.
Methylation-specific PCR (MSP)
DNA is modified by sodium bisulfite treatment
converting unmethylated, but not methylated, cytosines to uracil.
Following the removal of bisulfite and completion of the chemical
conversion, this modified DNA is used as a template for PCR.
Primers were designed to discriminate between methylated and
unmethylated alleles following bisulfite treatment and to
discriminate between DNA modified (vs. unmodified) by bisulfite.
The primer sequences of methylated and unmethylated PLZF are listed
in Table II.
Bioinformatics analysis
RNA-Protein Interaction Prediction (RPISeq)
(http://pridb.gdcb.iastate.edu/RPISeq/) was used to
analyze the interaction of ANRIL and PLZF. In the RPISeq website,
the sequences of ANRIL and PLZF were copied into the designated
spaces and submitted to the system, and then the interaction
probability was revealed. Interaction probabilities generated by
RPISeq range from 0 to 1. In performance evaluation experiments,
predictions with probabilities >0.5 were considered ‘positive’,
indicating that the corresponding RNA and protein are likely to
interact.
RNA immunoprecipitation
The EZMagna RNA immunoprecipitation (RIP) Kit (EMD
Millipore, Billerica, MA, USA) was performed based on the
manufacturer's protocol. Cells were lysed in complete RIP lysis
buffer (containing cOmplete™ Protease Inhibitor Cocktail, Roche,
Branford, CT, USA). The lysis mixture was incubated with magnetic
beads conjugated with specific antibodies or control IgG overnight
at 4°C. Beads were washed and incubated with proteinase K to remove
unconjugated proteins. Finally, RNAs were purified and subjected to
qRT-PCR assay to analyze possible RNAs.
Chemical treatment
5-Aza (cat. no. S1200) and suberoylanilide
hydroxamic acid (SAHA) (cat. no. S1047) were purchased from Selleck
Chemicals (Houston, TX, USA) and dissolved in dimethylsulphoxide
(DMSO), and used at 10 and 2 µM, respectively. Subconfluent cell
cultures were treated with 5-Aza and SAHA for 48 h and then
extracted for MPCR and western blot analysis.
Statistical analysis
All results were represented as the mean ± standard
deviation (SD) from at least 3 independent experiments. Differences
were determined by the two-tailed Student's t-test (comparison for
two groups) and the one-way ANOVA (comparison for >2 groups) and
the Tukey's post hoc test for multiple comparisons. Pearson's
Chi-squared test was used to analyze the pathological features of
PLZF expression in GC. Survival curves were drawn using a
Kaplan-Meier survival plot and assessed using log-rank tests. Cox
regression analysis was used to perform univariate and multivariate
analyses of the clinicopathological factors for disease-free
survival. All statistical analyses were performed using SPSS 19.0
software (IBM Corp., Armonk, NY, USA).
Results
PLZF is downregulated in the majority
of GC cells
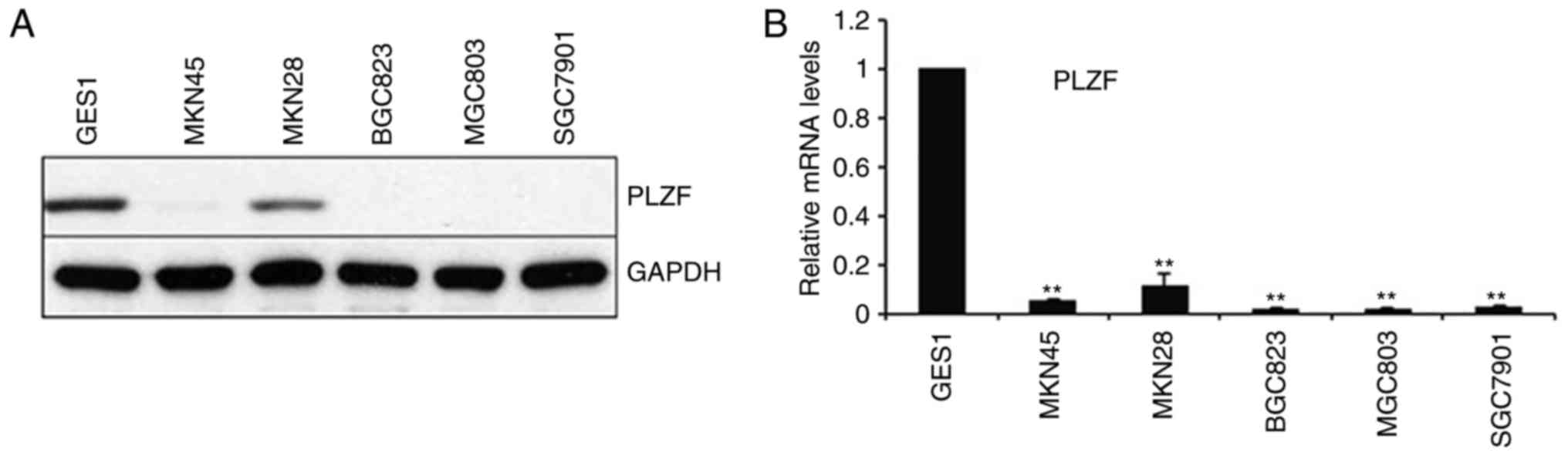
We evaluated PLZF expression in the normal human
gastric epithelial cell line GES1 and in a panel of GC cell lines,
and we found that PLZF had low expression in all the examined GC
cell lines even after 40 cycles of amplification and western blot
analysis, whereas it was expressed in the normal epithelial cell
line at both the mRNA and protein levels (Fig. 1).
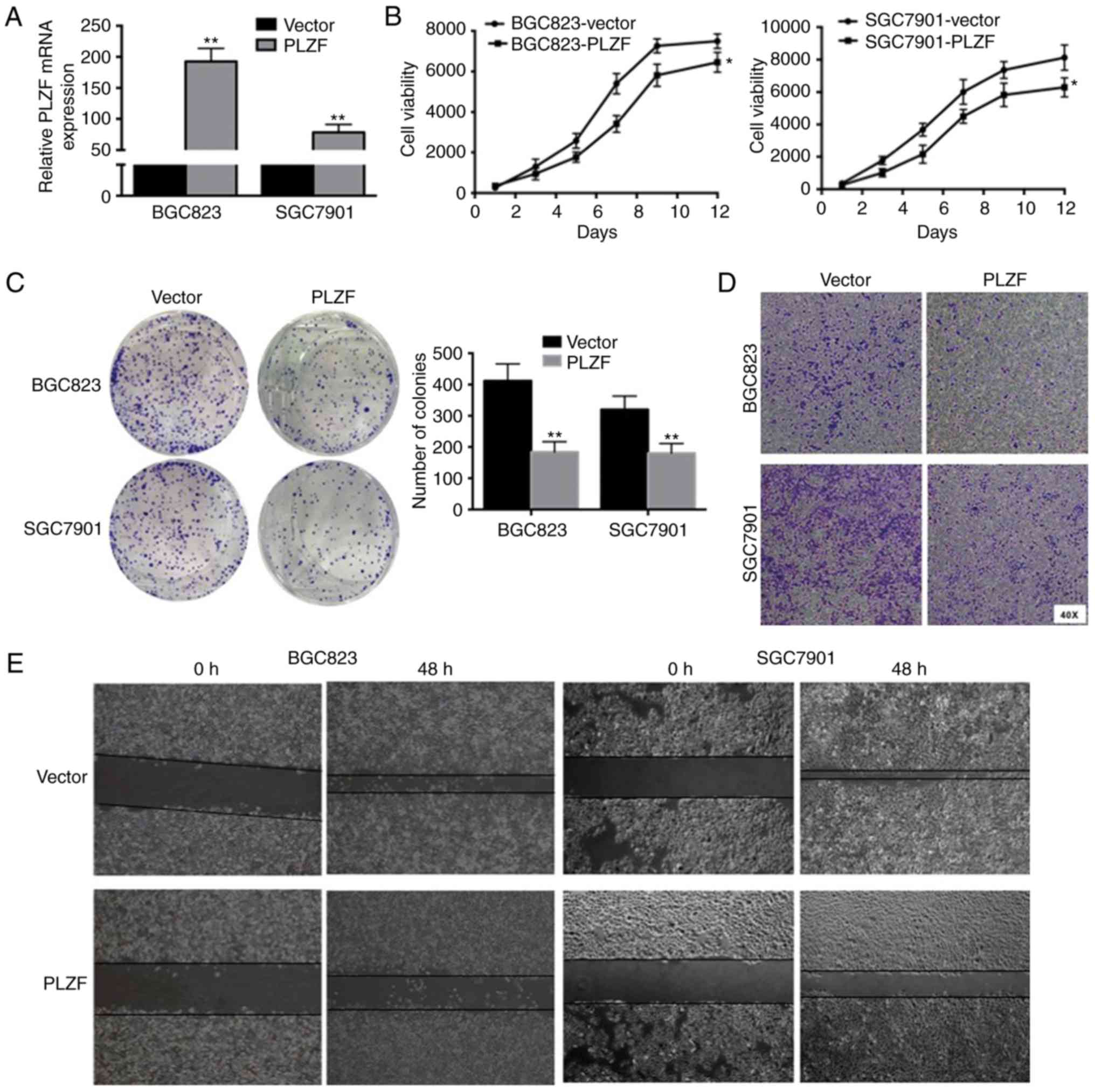
Overexpression of PLZF inhibits the proliferation,
migration and invasion of GC cells. To determine the role of PLZF
in GC, we monitored the effect of PLZF overexpression on GC cell
lines. Fig. 2A revealed the
efficiencies of PLZF overexpression. PLZF overexpression inhibited
the viability of BGC823 and SGC7901 cells (Fig. 2B). Similarly, PLZF overexpression
alleviated clone formation in BGC823 and SGC7901 cells (Fig. 2C). Transwell assay revealed that the
number of invaded cells in the control group were higher than those
in the PLZF overexpression group (Fig.
2D). In the wound healing assay, PLZF overexpression suppressed
wound healing in both BGC823 and SGC7901 cells (Fig. 2E). Collectively, these findings
indicated that increased PLZF expression in GC inhibited GC cell
proliferation, migration and invasion.
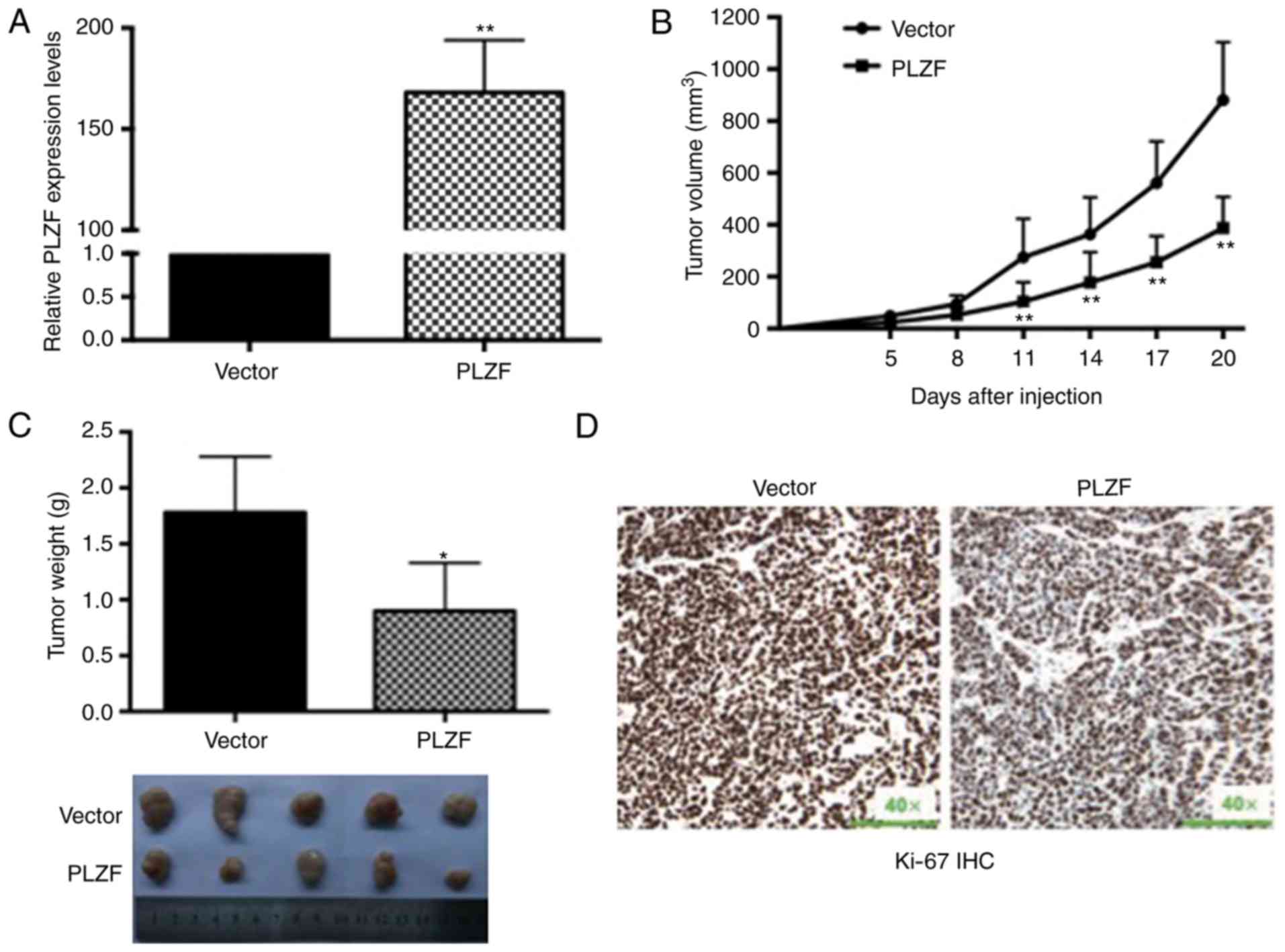
Overexpression of PLZF suppresses the
proliferation of nude mouse xenografts
In nude mouse xenografts, SGC7901 cells stably
transfected with the PLZF plasmid were inoculated into the flanks
of mice, and SGC7901 cells stably transfected with the empty vector
were also inoculated into the flanks of mice as a control. The
efficiencies of PLZF overexpression were assessed by qRT-PCR and
presented in Fig. 3A. PLZF
overexpression significantly inhibited tumorigenesis in vivo
given that the tumor weight and size were significantly decreased
compared with those of the controls (Fig. 3B and C). Furthermore, we detected
stronger Ki-67 expression in tumors derived from control nude mice
than in those derived from PLZF overexpression (Fig. 3D).
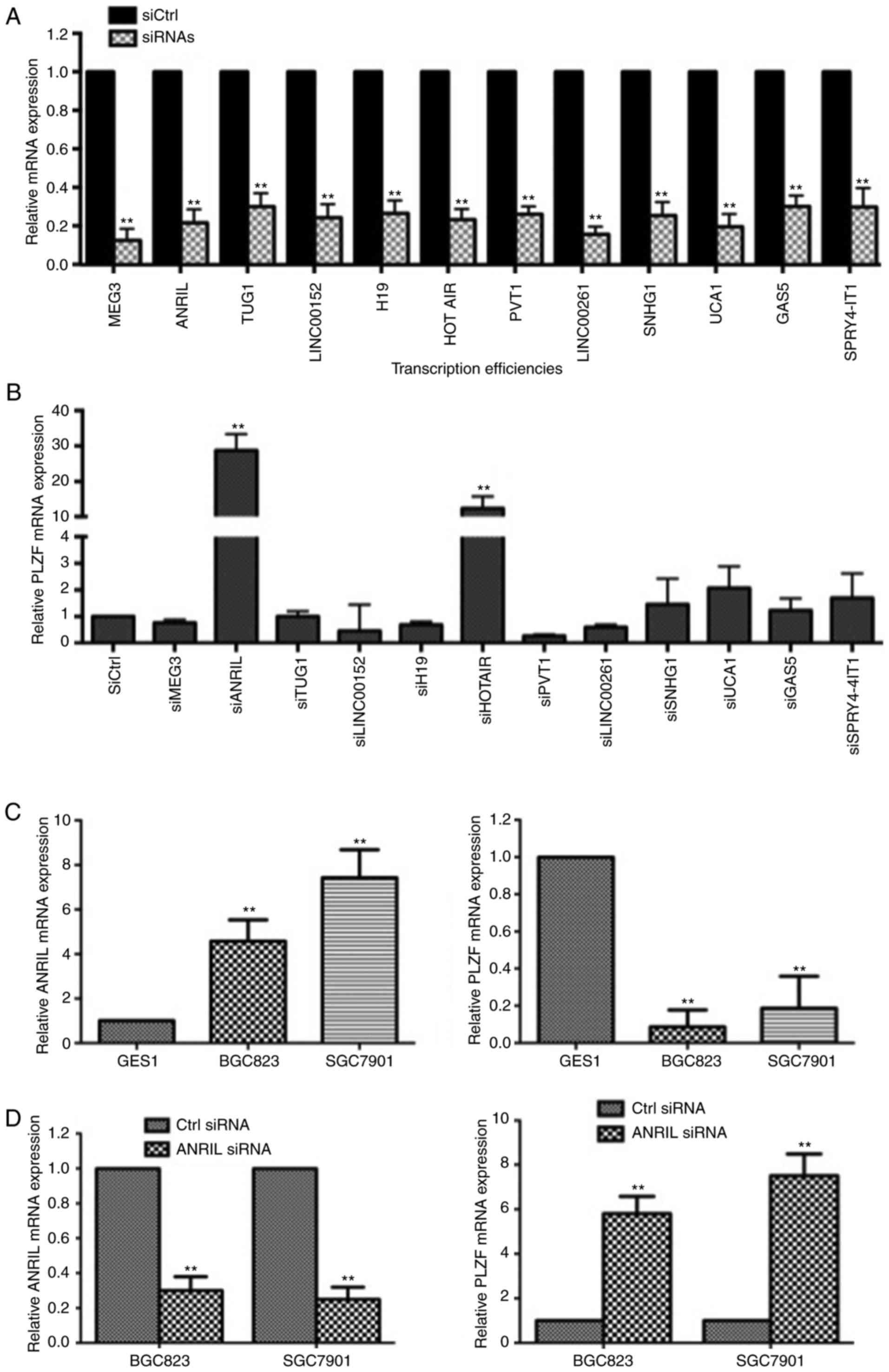
Negative association of PLZF and ANRIL
in GC cells
By acting as oncogenes or tumor suppressors, many
lncRNAs contribute to cancer occurrence and development by
epigenetically regulating gene expression (25). We screened a panel of lncRNAs to
search for the associated lncRNA of PLZF expression and found that
both ANRIL and HOTAIR knockdown recovered the expression of PLZF in
BGC823 cells (Fig. 4A and B). Since
ANRIL knockdown increased PLZF expression more than HOTAIR
knockdown, we chose ANRIL to further study PLZF regulation.
Furthermore, we confirmed the negative association between PLZF and
ANRIL expression in BGC823 and SGC7901 cells; ANRIL knockdown
increased the expression of PLZF in both BGC823 and SGC7901 cells
(Fig. 4C and D).
The interaction of ANRIL and PLZF
induces DNA methylation of PLZF
To verify crosstalk between ANRIL and PLZF, we
performed bioinformatics analysis and found that PLZF could
possibly interact with ANRIL. In our analysis, the RF classifier
was 0.7, and the SVM classifier was 0.99; both were larger than
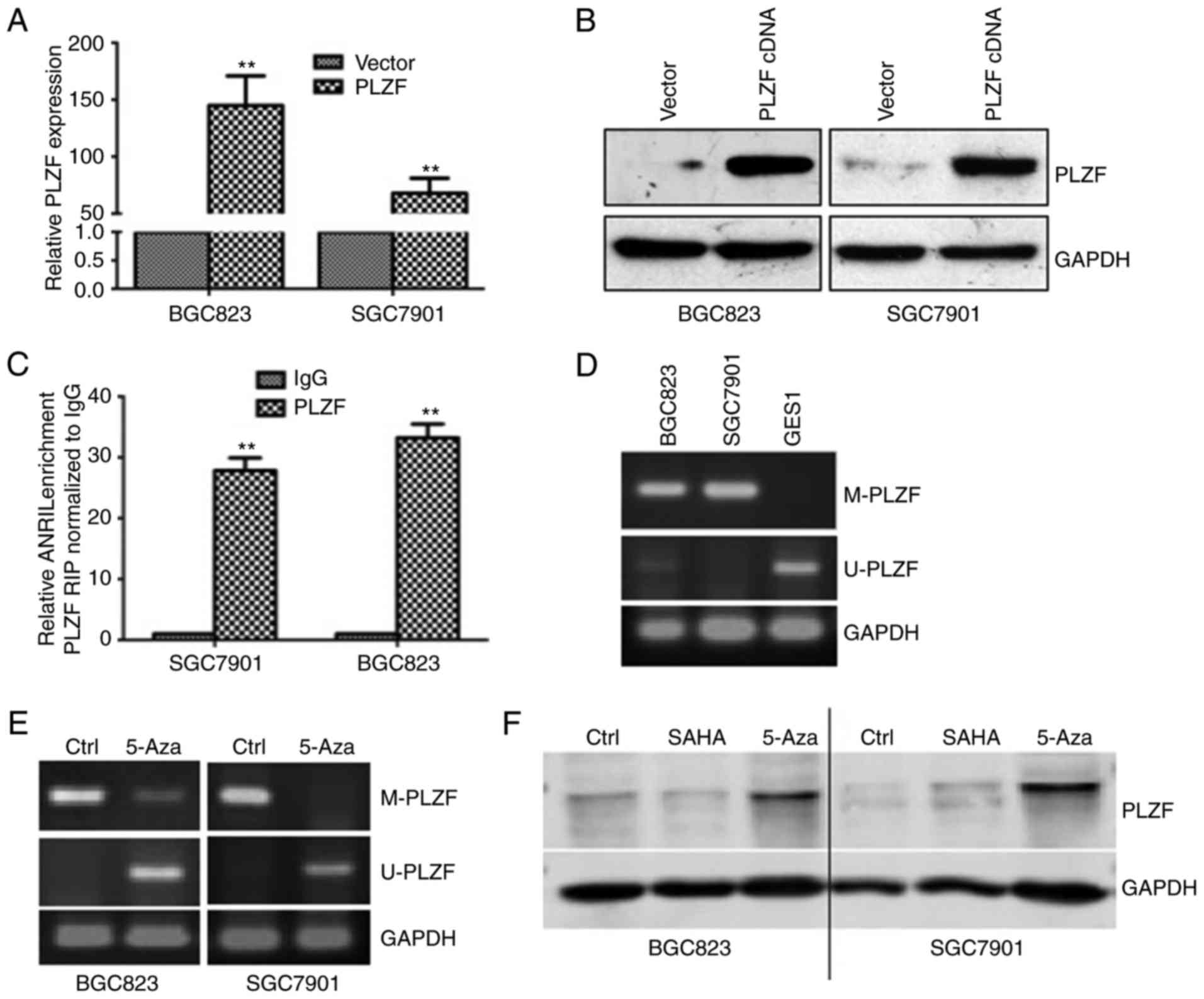
0.5. In our study, efficiencies of PLZF overexpressed cell lines
were presented in Fig. 5A and B.
RIP experiment confirmed the interaction between ANRIL and PLZF in
GC cells (Fig. 5C). Recent
discoveries have marked lncRNAs as new important players in DNA
methylation regulation (26).
Therefore, we inferred that ANRIL may regulate PLZF methylation by
binding to PRC2. MSP is a method for analyzing DNA methylation
patterns in CpG islands, and it revealed that PLZF was methylated
in BGC823 and SGC7901 cells, not in the normal cell lines GES1
(Fig. 5D). BGC823 and SGC7901 cells
were treated with the methylation inhibitor 5-Aza, DNA methylation
of PLZF was decreased (Fig. 5E),
but PLZF expression was increased (Fig.
5F). In addition, the histone deacetylase (HDAC) inhibitor SAHA
did not have the same effect, which suggested that PLZF was
methylated, not acetylated (Fig.
5F).
PLZF is negatively associated with
EZH2 and H3K27me3
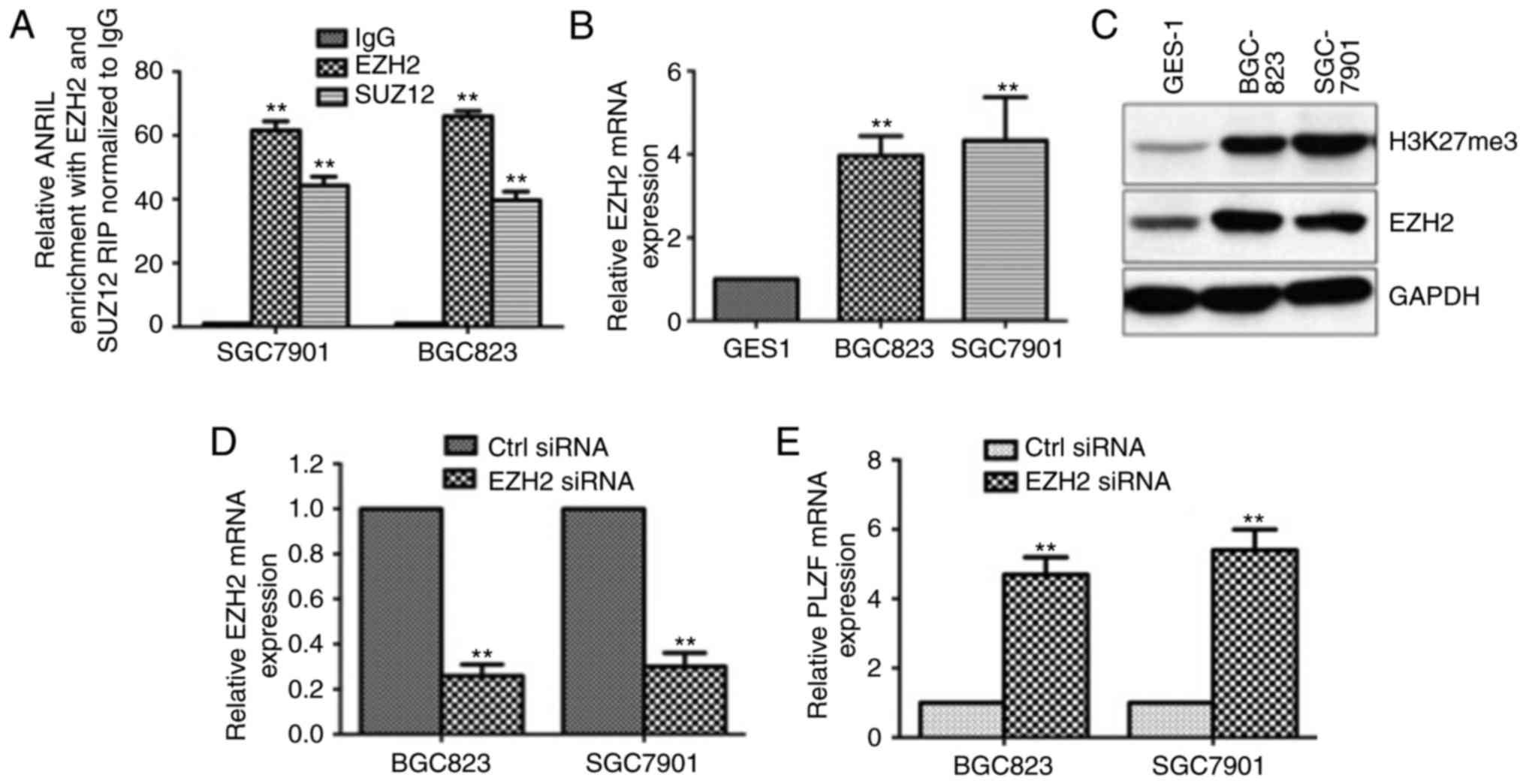
It has been reported that ANRIL strongly enriches
PRC2, which was confirmed (Fig. 6A)
(27). To further investigate the
relationship between PLZF and PRC2, we assessed the expression
levels of EZH2 and H3K27me3. We found that EZH2 and H3K27me3 were
highly expressed in GC cell lines compared to that in the normal
cell line GES1, which suggested that EZH2 and H3K27me3 were
negatively associated with PLZF (Fig.
6B and C). Moreover, upon EZH2 knockdown by siRNA transfection,
PLZF expression was markedly increased (Fig. 6D and E), confirming that PLZF was
negatively associated with EZH2 and H3K27me3. Therefore, we
concluded that ANRIL regulated PLZF level through collaboration
between DNA methylation and H3K27me3.
Absence of PLZF modulates
epithelial-mesenchymal transformation (EMT) of GC cells and is
correlated with poor prognosis
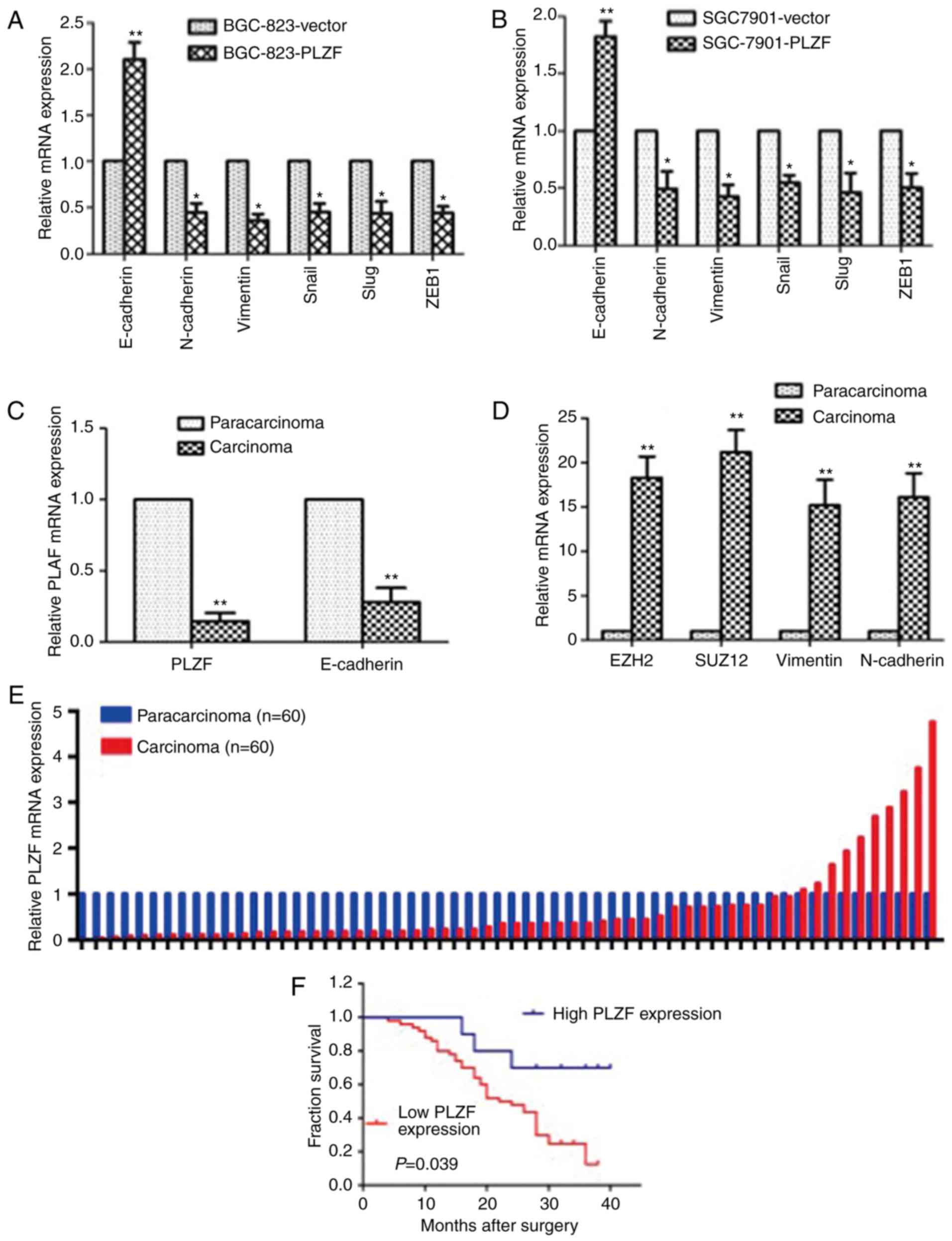
To investigate the effect of PLZF on EMT, we
identified EMT markers in the PLZF-overexpressed GC cell lines
BGC823/PLZF and SGC7901/PLZF. The results revealed that the
epithelial marker E-cadherin was enhanced and that the mesenchymal
markers N-cadherin, vimentin, Snail, Slug and ZEB1 were reduced
compared to those in GC cells transfected with the empty vector
(Fig. 7A and B). In GC patient
tissue samples, we found that PLZF and E-cadherin expression levels
in carcinoma tissues had parallel trends and were less than those
in adjacent non-cancer tissues, while EZH2, SUZ12, vimentin and
N-cadherin expression levels in carcinoma tissues were higher than
those in adjacent non-cancer tissues (Fig. 7C and D). These results indicated
that the level of PLZF expression contributed to
mesenchymal-epithelial transformation (MET).
To validate the differential expression of PLZF in
GC patients, we assessed the PLZF expression levels in tissue
samples from 60 GC patients by qRT-PCR assay. Relative PLZF mRNA
expression was assessed and normal adjacent non-cancer tissue was
presented as 1. PLZF expression of cancer tissue was normalized to
the adjacent non-cancer tissue, and a ratio <1 represented a low
PLZF level, while a ratio of >1 represented a high PLZF level.
The results revealed that PLZF expression was significantly lower
in cancer tissues than in adjacent normal tissues (Fig. 7E). By analyzing clinicopathological
factors, we found that the low PLZF expression was correlated with
poor prognosis in GC patients (Fig.
7F). The PLZF levels were also correlated with histological
differentiation, tumor invasion depth, tumor-node-metastasis (TNM)
stage and lymphatic metastasis. No relation was found between PLZF
expression and other factors, e.g., sex, age and distant metastasis
(Table I). Multivariate analysis
further revealed that PLZF expression could be regarded as a
potential diagnostic biomarker for disease-free survival in
patients with GC, as well as distant metastasis (Table IV).
 | Table IV.Univariate and multivariate Cox
regression analyses for disease-free survival of 60 patients with
GC. |
Table IV.
Univariate and multivariate Cox
regression analyses for disease-free survival of 60 patients with
GC.
|
| DFS |
|---|
|
|
|
|---|
| Risk factors | HR | 95% CI | P-value |
|---|
| Univariate
analysis |
| Age
(≤60 years, >60 years) | 2.199 | 1.124–4.302 | 0.021a |
| Sex
(male, female) | 0.661 | 0.347–1.259 | 0.208 |
| PLZF
expression | 0.049 | 0.010–0.244 | 0.000b |
|
Histological grade (low or
undifferent, middle or high) | 1.797 | 0.943–3.426 | 0.075 |
| Lymph
node metastasis (yes or no) | 2.221 | 1.152–4.281 | 0.017a |
| Tumor
invasion depth (T1, T2 or above) | 5.906 | 2.761–12.632 | 0.000b |
| TNM
stage (I/II, III/IV) | 1.925 | 0.851–4.353 | 0.116 |
| Distant
metastasis (yes or no) | 4.760 | 2.303–9.838 | 0.000b |
| Multivariate
analysis |
| PLZF
expression | 0.087 | 0.018–0.421 | 0.002b |
| Distant
metastasis (yes or no) | 2.355 | 1.089–5.094 | 0.030a |
Discussion
GC is ranked as the fifth most common malignant
neoplasm in the world, with approximately 951,600 new diagnoses and
723,100 deaths in 2012 (28).
Despite the decreased mortality rate of GC in recent years, it is
still the second leading cause of cancer-related deaths (29,30).
GC remains a major clinical challenge worldwide due to its poor
prognosis and limited therapeutic approaches; currently, there are
no effective prognostic biomarkers for GC. Accumulating studies
have demonstrated that both oncogenes and anti-oncogenes play
important roles in the tumorigenesis, metastasis, prognosis and
drug resistance of GC (31–33). In the present study, it was reported
that the absence of the tumor suppressor PLZF led to aggressive
proliferation and malignant invasion of GC by inducing EMT and
mechanistic methylation via the binding of ANRIL to PRC2.
In the present study, we observed that PLZF had low
expression levels in the majority of GC cells and tumor tissues
compared to those in the normal cell line GES1 and adjacent
non-tumor tissues, respectively (Figs.
1 and 7E). Moreover, low PLZF
expression in GC tissues was associated with a poor prognosis
(Fig. 7F) and may be an independent
prognostic factor. To determine the function of decreased PLZF
expression in GC, we investigated the effects of PLZF
overexpression in GC cell lines. PLZF overexpression by cDNA
transfection decreased cell viability, clone formation, wound
healing and cell invasion in both BGC823 and SGC7901 cells
(Fig. 2). These findings indicated
that decreased PLZF expression in GC contributes to GC cell
proliferation and migration. In addition, in a nude mice xenograft
model, SGC7901 cells with PLZF overexpression significantly
inhibited tumorigenesis and had lower tumor weights, tumor sizes
and Ki-67 expression levels compared to those in the control group
(Fig. 3). These effects were
similar to observations in lung cancer, melanoma, prostate cancer
and hepatocellular carcinoma (7,8,34,35).
The lack of PLZF expression may be due to genomic
alteration, epigenetic modification or a change in subcellular
location. Epigenetic regulation, particularly DNA methylation, is
well known as the most common contributing factor to a decrease in
tumor suppressor functionality. Wang et al demonstrated that
downregulation of PLZF in non-small cell lung cancer was partially
caused by methylation of the PLZF promoter (34). Several cellular processes involve
lncRNAs, and a significant number have been shown to function in
cooperation with chromatin-modifying enzymes to promote epigenetic
activation or silencing of gene expression (25). The first described epigenetic
mechanism involving an lncRNA was Xist (X-inactive specific
transcript), a product of the Xist gene, which is exclusively
transcribed from the inactive X (Xi) chromosome. X inactivation was
the first mechanism of gene silencing that was identified to depend
on an interaction between lncRNA and PRC2, providing a way to study
epigenetic regulation of gene expression mediated by lncRNAs
(36). In the present study, lncRNA
ANRIL recruited PRC2, which, in turn, EZH2, a core member of PRC2,
catalyzed trimethylation of H3K27 and then drove PLZF silencing by
promoting DNA methylation of PLZF. The observation that the
methylation inhibitor 5-Aza can increase PLZF expression confirms
regulatory methylation. There is corroborating evidence that DNA
methylation is often associated with specific types of histone
modifications that can cooperatively affect chromatin structure to
silence gene expression (37). The
methylation effect corresponds to the loss of PLZF in the
differentiated GC groups in our study. These findings, at least in
part, explain why PLZF expression is lost in GC.
The underlying molecular mechanism of PLZF in cancer
remains poorly understood. One study in melanoma demonstrated that
PLZF partially suppressed migration and invasion through integrin
β3 and MMP9 (35). We examined the
expression levels of EMT markers to identify a possible mechanism
and observed that PLZF overexpression decreased N-cadherin,
Vimentin, Snail, Slug and ZEB1 expression levels, while it enhanced
the expression level of E-cadherin, indicating that increased PLZF
expression could inversely affect EMT (also called MET).
Conversely, in gallbladder cancer, PLZF selectively induced the
mesenchymal marker N-cadherin, instead of vimentin and fibronectin,
to initiate the MET program (11).
EMT is regarded as a potent driver for cells to develop metastatic
features, and the loss of PLZF initiates EMT, which contributes to
the malignant development and poor prognosis of GC. These results
may explain why PLZF levels are correlated with histological
differentiation, tumor invasion depth, TNM stage and lymphatic
metastasis. Activation of EMT has been shown to be a very efficient
strategy adopted by epithelial cancer cells to promote local
invasion and dissemination in distant organs (38). In the present study, the limited
number of patient tissues including a limited sample of distant
metastasis cases, contributed to the lack of a correlation between
PLZF and distant metastasis.
Collectively, in the present study, we reported that
PLZF expression was markedly reduced in GC tissues and cell lines
and that the loss of PLZF expression was correlated with
histological differentiation, tumor invasion depth, TNM stage,
lymphatic metastasis and poor prognosis. Furthermore, we found that
ANRIL expression was negatively associated with PLZF and that ANRIL
recruited PRC2, which drove PLZF silencing by promoting
collaboration with H3K27me3 and DNA methylation. Therefore,
constitutive ANRIL activation was a possible cause of the lack of
PLZF expression in GC cells. These results indicated that PLZF and
ANRIL are functionally independent and that their coupled
deregulation may account for most of the alterations described in
GC. To the best of our knowledge, we identified the
tumor-suppressor role of PLZF in GC both in vitro and in
vivo, and PLZF may be a potential tumor biomarker and a
critical therapeutic target for GC.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (no. 31401094), the Natural
Science Research Project of Anhui Provincial Institutions of Higher
Education (no. KJ2015B118by), the Major Projects of the Natural
Science Research in Anhui Provincial Colleges and Universities
(grant no. KJ2018A0245), and the Nature Science Key Program of
Bengbu Medical College (no. BYKY1415ZD).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
JBW and YJ performed the molecular studies and in
vivo assays. PW and WJZ collected the clinical data and tissue
samples and measured genes' expression of clinical tissues. YL
performed the functional experiments and the statistical analysis.
JFC and WD directed the experiments and helped draft the
manuscript. FY designed the study and wrote the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Nanjing Medical University and written informed
consent was obtained from all patients. All animal experiments were
performed according the National Institutes of Health (Bethesda,
MD, USA) guidelines on the use of experimental animals and were
approved by the Ethics Committee of the Nanjing Medical
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PLZF
|
promyelocytic leukemia zinc finger
|
|
GC
|
gastric cancer
|
|
lncRNAs
|
long non-coding RNAs
|
|
ANRIL
|
CDKN2B antisense RNA 1
|
|
5-Aza
|
5-Aza-2′-deoxycytidine
|
|
PRC2
|
polycomb repressive complex 2
|
|
MSP
|
methylation-specific PCR
|
|
Xist
|
X-inactive specific transcript
|
|
EMT
|
epithelial-mesenchymal
transformation
|
|
MET
|
mesenchymal-epithelial
transformation
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1
|
Ball HJ, Melnick A, Shaknovich R, Kohanski
RA and Licht JD: The promyelocytic leukemia zinc finger (PLZF)
protein binds DNA in a high molecular weight complex associated
with cdc2 kinase. Nucleic Acids Res. 27:4106–4113. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Suliman BA, Xu D and Williams BR: The
promyelocytic leukemia zinc finger protein: Two decades of
molecular oncology. Front Oncol. 2:742012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choi WI, Kim MY, Jeon BN, Koh DI, Yun CO,
Li Y, Lee CE, Oh J, Kim K and Hur MW: Role of promyelocytic
leukemia zinc finger (PLZF) in cell proliferation and
cyclin-dependent kinase inhibitor 1A (p21WAF/CDKN1A) gene
repression. J Biol Chem. 289:18625–18640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Felicetti F, Errico MC, Bottero L,
Segnalini P, Stoppacciaro A, Biffoni M, Felli N, Mattia G, Petrini
M, Colombo MP, et al: The promyelocytic leukemia zinc
finger-microRNA-221/-222 pathway controls melanoma progression
through multiple oncogenic mechanisms. Cancer Res. 68:2745–2754.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vincent A, Omura N, Hong SM, Jaffe A,
Eshleman J and Goggins M: Genome-wide analysis of promoter
methylation associated with gene expression profile in pancreatic
adenocarcinoma. Clin Cancer Res. 17:4341–4354. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matsuzawa K, Izawa S, Ohkura T, Ohkura H,
Ishiguro K, Yoshida A, Takiyama Y, Haneda M, Shigemasa C, Yamamoto
K, et al: Implication of intracellular localization of
transcriptional repressor PLZF in thyroid neoplasms. BMC Endocr
Disord. 14:522014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsieh CL, Botta G, Gao S, Li T, Van Allen
EM, Treacy DJ, Cai C, He HH, Sweeney CJ, Brown M, et al: PLZF, a
tumor suppressor genetically lost in metastatic
castration-resistant prostate cancer, is a mediator of resistance
to androgen deprivation therapy. Cancer Res. 75:1944–1948. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hui AW, Lau HW, Cao CY, Zhou JW, Lai PB
and Tsui SK: Downregulation of PLZF in human hepatocellular
carcinoma and its clinical significance. Oncol Rep. 33:397–402.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao GQ, Li F, Findeis-Hosey J, Hyrien O,
Unger PD, Xiao L, Dunne R, Kim ES, Yang Q, McMahon L, et al:
Down-regulation of cytoplasmic PLZF correlates with high tumor
grade and tumor aggression in non-small cell lung carcinoma. Hum
Pathol. 46:1607–1615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin Y, Nenseth HZ and Saatcioglu F: Role
of PLZF as a tumor suppressor in prostate cancer. Oncotarget.
8:71317–71324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H, Zhan M, Zhang Y, Huang S, Xu S,
Huang X, He M, Yao Y, Man M and Wang J: PLZF inhibits proliferation
and metastasis of gallbladder cancer by regulating IFIT2. Cell
Death Dis. 9:712018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jones C, St-Jean S, Fréchette I, Bergeron
D, Rivard N and Boudreau F: Identification of a novel promyelocytic
leukemia zinc-finger isoform required for colorectal cancer cell
growth and survival. Int J Cancer. 133:58–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yap KL, Li S, Muñoz-Cabello AM, Raguz S,
Zeng L, Mujtaba S, Gil J, Walsh MJ and Zhou MM: Molecular interplay
of the noncoding RNA ANRIL and methylated histone H3 lysine
27 by polycomb CBX7 in transcriptional silencing of INK4a.
Mol Cell. 38:662–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pasmant E, Laurendeau I, Héron D, Vidaud
M, Vidaud D and Bièche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural
system tumor family: Identification of ANRIL, an antisense
noncoding RNA whose expression coclusters with ARF. Cancer
Res. 67:3963–3969. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen S, Zhang JQ, Chen JZ, Chen HX, Qiu
FN, Yan ML, Chen YL, Peng CH, Tian YF and Wang YD: The over
expression of long non-coding RNA ANRIL promotes
epithelial-mesenchymal transition by activating the ATM-E2F1
signaling pathway in pancreatic cancer: An in vivo and in vitro
study. Int J Biol Macromol. 102:718–728. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of
p15INK4B tumor suppressor gene. Oncogene. 30:1956–1962.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Hevi S, Kurash JK, Lei H, Gay F,
Bajko J, Su H, Sun W, Chang H, Xu G, et al: The lysine demethylase
LSD1 (KDM1) is required for maintenance of global DNA methylation.
Nat Genet. 41:125–129. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Li C, Zhang R, Xiao W, Niu X, Ye X,
Li Z, Guo Y, Tan J and Li Y: The EZH2- H3K27me3-DNMT1 complex
orchestrates epigenetic silencing of the wwc1 gene, a
Hippo/YAP pathway upstream effector, in breast cancer epithelial
cells. Cell Signal. 51:243–256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Long HK, King HW, Patient RK, Odom DT and
Klose RJ: Protection of CpG islands from DNA methylation is
DNA-encoded and evolutionarily conserved. Nucleic Acids Res.
44:6693–6706. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nawaz I, Qiu X, Wu H, Li Y, Fan Y, Hu LF,
Zhou Q and Ernberg I: Development of a multiplex methylation
specific PCR suitable for (early) detection of non-small cell lung
cancer. Epigenetics. 9:1138–1148. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu
DY, Tang CJ, De W and Yang F: STAT3-induced lncRNA HAGLROS
overexpression contributes to the malignant progression of gastric
cancer cells via mTOR signal-mediated inhibition of autophagy. Mol
Cancer. 17:62018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang F, Deng R, Qian XJ, Chang SH, Wu XQ,
Qin J, Feng GK, Ding K and Zhu XF: Feedback loops blockade
potentiates apoptosis induction and antitumor activity of a novel
AKT inhibitor DC120 in human liver cancer. Cell Death Dis.
5:e11142014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marchese FP and Huarte M: Long non-coding
RNAs and chromatin modifiers: Their place in the epigenetic code.
Epigenetics. 9:21–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cech TR and Steitz JA: The noncoding RNA
revolution-trashing old rules to forge new ones. Cell. 157:77–94.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol Oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Colquhoun A, Arnold M, Ferlay J, Goodman
KJ, Forman D and Soerjomataram I: Global patterns of cardia and
non-cardia gastric cancer incidence in 2012. Gut. 64:1881–1888.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chia NY, Deng N, Das K, Huang D, Hu L, Zhu
Y, Lim KH, Lee MH, Wu J, Sam XX, et al: Regulatory crosstalk
between lineage-survival oncogenes KLF5GATA4GATA6
cooperatively promotes gastric cancer development. Gut. 64:707–719.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thiem S, Eissmann MF, Elzer J, Jonas A,
Putoczki TL, Poh A, Nguyen P, Preaudet A, Flanagan D, Vincan E, et
al: Stomach-specific activation of oncogenic KRAS and
STAT3-dependent inflammation cooperatively promote gastric
tumorigenesis in a preclinical model. Cancer Res. 76:2277–2287.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang K, Liang Q, Li X, Tsoi H, Zhang J,
Wang H, Go MY, Chiu PW, Ng EK, Sung JJ and Yu J: MDGA2 is a novel
tumour suppressor cooperating with DMAP1 in gastric cancer and is
associated with disease outcome. Gut. 65:1619–1631. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang X, Wang L, Guo S, Bao Y, Ma Y, Yan F,
Xu K, Xu Z, Jin L, Lu D, et al: Hypermethylation reduces expression
of tumor-suppressor PLZF and regulates proliferation and apoptosis
in non-small-cell lung cancers. FASEB J. 27:4194–4203. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Felicetti F, Bottero L, Felli N, Mattia G,
Labbaye C, Alvino E, Peschle C, Colombo MP and Carè A: Role of PLZF
in melanoma progression. Oncogene. 23:4567–4576. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nguyen CT, Gonzales FA and Jones PA:
Altered chromatin structure associated with methylation-induced
gene silencing in cancer cells: Correlation of accessibility,
methylation, MeCP2 binding and acetylation. Nucleic Acids Res.
29:4598–4606. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ombrato L and Malanchi I: The EMT
universe: Space between cancer cell dissemination and metastasis
initiation. Crit Rev Oncog. 19:349–361. 2014. View Article : Google Scholar : PubMed/NCBI
|