Introduction
The tumor microenvironment is now recognized as a
critical participant in cancer progression (1–4). In
epithelial tumors, out of the various types of stromal cells,
cancer-associated fibroblasts (CAFs) are the predominant subset. Of
CAFs, myofibroblasts (MFs) constitute the most abundant subtype,
occupying the majority of this particular niche (5,6). In
MFs the characteristic features of smooth muscle cells and
fibroblasts are combined, as MFs acquire the ability to contract
due to the de novo expression of smooth muscle actin (α-SMA)
and by the assembly of novel stress fibers (7,8), and
also maintain the extracellular matrix (ECM) synthesizing and
secreting functions of fibroblasts. MFs are also mediators of tumor
development and metastasis. However, it is still under debate
whether their stimulating effects on cancer cell invasion are
executed directly through paracrine interactions with tumor cells
or they promote cancer cell spreading indirectly by rendering the
ECM more permissive for dissemination via remodeling (9–12). MFs
express cytokines, chemokines, growth factors, different collagens,
fibronectin, various adhesion molecules and matrix metalloproteases
(MMPs), and secrete these molecules directly or within exosomes
into the surrounding extracellular milieu (13,14).
It has been proposed that this secretome alone would not be
competent enough to trigger cancer cell invasion, but it is
definitely sufficient for intensive paracrine dialogues between
stromal cells (9,10). On the other hand, as MFs can
modulate the composition and deregulate the biomechanical features
of stromal ECM; in part by overproducing various MMPs, which will
ultimately degrade the matrix and sever cell-matrix interactions.
These MF-induced actions would undoubtedly favour cell mobilization
and contribute to cancer progression (11,15).
In fact, several studies verified that the appearance of MMPs in
the tumor microenvironment is associated with increased cancer
aggressiveness and bad prognosis (16,17).
Due to the pivotal role of stromal MFs in tumor
development and progression, targeting MFs may be a viable approach
to fight cancer invasion and metastasis. Nevertheless, prior to
establishing MF-targeting clinical trials, it is essential to
understand the major genetic, transcriptional and functional
features of MFs residing in tumor-affected as well as in normal
tissues. This knowledge is absolutely necessary in order to
identify the key mechanisms that primarily govern the contribution
of MFs in tumor invasion (10,18).
In line with this, a comparative study was performed on matched
pairs of primary MF cultures, isolated from cancerous specimens and
from the tumor-adjacent normal tissues of patients diagnosed with
different gastrointestinal cancers. Detailed knowledge on the
modulated MF behavior of the human esophagus or other
gastrointestinal regions is relevant not only in cancer progression
and invasion, but also in various injuries, inflammation, repair as
well as in gastro-esophageal reflux disease (19).
Therefore, pure MF cell cultures were first
established then genetic polymorphisms were analyzed, epigenetic
marks and gene expression profiles of normal tissue- and
tumor-derived MFs with a special emphasis on genes involved in
tumorigenesis, invasion, matrix remodeling, cell migration or
proliferation were compared. The present study's particular focus
was to reveal whether any notable genetic, epigenetic or
expressional variations could be identified that would account for
the tumor promoting effects of tumor-associated MFs. Concomitantly,
targeted genome analysis of the esophageal tumor sample was also
conducted.
Materials and methods
Chemicals and solutions
General laboratory reagents were from Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany) and Molar Chemicals Kft (Budapest,
Hungary). Cell culture media and associated reagents were from
Sigma-Aldrich (Merck KGaA), specific reagents for polymerase chain
reaction (PCR) were from Thermo Fisher Scientific, Inc., (Waltham,
MA, USA).
Ethics
The present study was approved by the Ethics
Committee of the University of Szeged, (Szeged, Hungary). All
patients gave informed consent.
Patients
Esophageal as well as cecum, sigmoid colon and
rectum resections were performed on patients diagnosed with
gastrointestinal cancer at the Department of Surgery, University of
Szeged. The esophageal cancer patient was a 71-year-old male, the
cecum cancer patient was a 69-year-old female, sigmoid colon cancer
patient was a 71-year-old male and the rectum cancer patient was a
65-year-old female. The patients were recruited between January
2007 and May 2010. The status of the patients was evaluated
regarding clinicopathological features, histopathological
parameters, the stage, grade as well as histological type of the
tumor. Lymphatic vessel or vascular invasion and positive margins
of resections were also assessed.
Tissue specimens
Tissue specimens were obtained intraoperatively
during cancer resection. Specimens were taken from the tumor
tissues and also from the adjacent normal area (at least 3–4 cm
distant from the tumors).
Tissue specimens were evaluated following routine
pathology procedures. These included passage from formalin into
paraffin, sectioning, staining the 4-µm thick sections with
hematoxylin and eosin (5 min, room temperature), Giemsa technique
(15 min, room temperature) and periodic acid-Schiff
technique/Alcian blue (30 min, room temperature). Finally,
immunostaining for α-SMA (ab: cat. no. ms-113; 1:800; Thermo Fisher
Scientific, Inc.), vimentin (ab: cat. no. rm-9120; 1:200; Thermo
Fisher Scientific, Inc.) and desmin (ab: cat. no. ms-376; 1:100;
Thermo Fisher Scientific, Inc.) was carried out using EnVision Flex
system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's protocol. Sections were
counterstained with hematoxylin at room temperature for 1 min. All
slides were stained simultaneously using a computer-controlled
autostainer (Ventana BenchMark Ultra, Ventana Medical Systems,
Inc., Tucson, AZ, USA).
The images captured for histopathological assessment
using a Zeiss Axiocam 506 color microscope and were analyzed using
ImageJ 1.4.3.67 software (National Institutes of Health, Bethesda,
MD, USA). Densitometry was performed to determine brown color
intensity. Images were converted to RGB format and color-based
thresholding was set as follows: Hue 0–136, saturation 57–255,
brightness 0–255. On 8-bit images threshold was adjusted to 0–242,
then particle analysis was performed.
Establishment of MF cultures
The isolation and culturing of normal as well as
tumor-derived MFs was performed as described previously by Czepan
et al (20). Briefly, normal
tissue specimens and esophageal tumor tissue as well as cecum,
sigmoid colon or rectum carcinoma samples were washed with 1 mM
DTT, incubated four times with 1 mM EDTA solution for 30 min, then
placed into RPMI (Merck KGA) selection medium supplemented with 10%
fetal bovine serum (FBS, Merck KGA); 1% penicillin-streptomycin, 2%
antibiotic-antimycotic solution. Following reaching confluence, the
cells were trypsinized (0.25% trypsin-EDTA) and were transferred
into growth medium [Dulbecco's modified Eagle's medium (DMEM, Merck
KGA) supplemented with 4 mM L-glutamine, 10% FBS, 1% amino acid
solution, 1% penicillin-streptomycin and 2% antibiotic-antimycotic
solution]. Cells were cultured in 5% CO2 atmosphere at
37°C. Aliquots were frozen in liquid nitrogen and used in the
following experiments with care to minimize passage numbers. To
monitor the purity of MF cultures, cells were stained for MF
markers α-SMA and vimentin as previously described (20).
Immunocytochemistry
For the detection of histone modifications by
immunostaining, 2×104 cells were seeded onto chamber
slides (BD Biosciences, Franklin Lakes, NJ, USA) and were allowed
to adhere overnight. Cells were fixed using 4% paraformaldehyde for
20 min at room temperature, permeabilized by adding PBS
supplemented with 0.3% Triton X-100. Non-specific protein binding
sites were blocked in 5% bovine serum albumin (BSA; Sigma-Aldrich;
Merck KGaA) containing PBS and finally, cells were incubated with
primary antibodies for 2 h at room temperature. The used primary
antibodies, their sources and the applied dilutions were:
Pan-acetylated histone 3 (H3)-specific antibody (Bio-Rad
Laboratories, Inc., Hercules, CA, USA); cat. no. AHP412; diluted in
1:300 with 1% BSA containing PBS), anti-lysine 9 acetylated H3
antibody (Abcam, Cambridge, UK; cat. no. Ab4441; 1:750), antibody
against H3K18ac (Abcam; cat. no. Ab1199; 1:500), anti-H3K14ac
antibody (Sigma-Aldrich; Merck KGaA) cat. no. 06-911; 1:450),
antibody specific for lysine 8 acetylated histone 4 (Abcam; cat.
no. Ab1760; 1:400), H4K12ac-specific antibody (Abcam; cat. no.
Ab1761; 1:300), antibody against H4K16ac (Abcam; cat. no. Ab1762;
1:300) and antibody to detect pan-acetylated H4 (Sigma-Aldrich;
Merck KGaA; cat. no. 06-598; 1:500). Antibodies specific for H3 di-
and trimethylated at lysine 9 were from Upstate (Merck KGaA; cat.
no. 07-521) and Abcam (cat. no. Ab8898) and used in 1:750 and 1:500
dilutions, respectively. Samples were incubated with the secondary
antibody (Alexa Fluor 555-conjugated anti-rabbit IgG; Molecular
Probes; Thermo Fisher Scientific, Inc.) at room temperature in the
dark, for 1 h. For DNA labeling, 4,6-diamino-2-phenylindole (DAPI)
staining was performed (dilution 1:3,000 in PBS solution, for 5
min). Following extensive washing in PBS, slides were covered with
fluoromount mounting medium (Sigma-Aldrich; Merck KGaA).
Quantification of fluorescence intensity was performed using ImageJ
1.4.3.67 software (National Institute of Health, Bethesda, MD,
USA). Corrected total cell fluorescence (CTCF) was calculated based
on fluorescence values from ~30 cells/sample according to the
following formula: CTCF= integrated density-(area of selected cell
× mean fluorescence of background readings).
Variant analysis
DNA variant analysis of selected tumor genes was
performed by targeted sequencing on DNA samples derived from
tumor-associated and normal MFs and from cells of the tumor mass.
DNA was prepared using Macherey-Nagel NucleoSpin Tissue columns
according to the manufacturer's protocol. Samples were quantitated
by Qubit dsDNA BR assay kit (Thermo Fisher Scientific, Inc.) with a
Qubit 2.0 fluorimeter. For each sample, 50 ng DNA was used to
prepare Illumina sequencing libraries using Illumina TruSight Rapid
Capture kit (Illumina, Inc., San Diego, CA, USA) with TruSight
Cancer sequencing panel. The TruSight Cancer panel targets 94 genes
and sites of 284 single nucleotide polymorphisms (SNPs) associated
with tumors. The gene list of the TruSight Cancer panel is
available online on the manufacturer's website (https://emea.illumina.com/content/dam/illumina-marketing/documents/products/gene_lists/gene_list_trusight_cancer.xlsx).
Sequencing libraries were prepared by the TruSight Rapid Capture
workflow and quality tested by capillary electrophoresis on an
Agilent Bioanalyzer 2100 instrument with Agilent High Sensitivity
DNA chip (Agilent Technologies). Pooled mixtures of indexed
libraries were denatured and sequenced with Illumina MiSeq using
Reagent kit V2-300. Variants were called by MiSeq Report (MSR)
enrichment workflow of the BaseSpace cloud computing environment.
Vcf files containing the identified variants were annotated using
Illumina VariantStudio 2.1.46 software (Illumina, Inc.) with the
following annotation sources: Variant Effect Predictor, v2.8; 1000
Genomes (April 2012 v3); Cosmic (v65); ClinVar (September 5, 2013);
dbSNP (v137); NHLBI Exome Variant Server (vESP6500SI-V2); USCS
(hg19).
RNA extraction
Total RNA was isolated from confluent MF cultures
with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
using the manufacturer's protocol, followed by removal of DNA
contamination with DNase I (Thermo Fisher Scientific, Inc.). Total
RNA from Hker E6SFM human keratinocytes was a gift from Dr. Vilmos
Tubak (Creative Laboratory Ltd., Szeged, Hungary). Quality of total
RNA was assessed using Agilent Bioanalyzer 2100 (Agilent
Technologies).
TaqMan low-density gene expression
array
For gene expression the mRNA levels of 190 genes
were analyzed simultaneously by reverse transcription-quantitative
(RT-q)PCR by TaqMan Low Density Array (TLDA; Applied Biosystems;
Thermo Fisher Scientific, Inc.) using 384-well microfluidic cards.
Each sample was loaded in duplicates. In the TLDA experiments 600
ng RNA was reverse transcribed with a High Capacity Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and cDNAs were mixed with TaqMan Universal
PCR Master mix (Thermo Fisher Scientific, Inc.). UD-GenoMed Medical
Genomic Technologies Company (Debrecen, Hungary) has performed PCR
amplifications using an ABI PRISM 7900HT real-time PCR system
(Thermo Fisher Scientific, Inc.). PCR reaction cycles were: 2 min
at 50°C and 10 min at 94.5°C, followed by 40 cycles of 30 sec at
97°C and 1 min at 59.7°C.
The genes included in the analysis were manually
selected from TaqMan Gene Sets (https://products.appliedbiosystems.com/ab/en/US/adirect/ab?cmd=catNavigate2&catID=604535).
The expression levels were normalized to five internal controls;
GAPDH, β-2-microglobulin, β-actin, 18S RNA and glucuronidase-β.
Increases or decreases of 2-fold in the expression levels were
considered as significant alterations.
Quantitative and conventional
RT-PCR
For quantitative and conventional RT-PCR reactions
first strand cDNA was synthesized from 1 µg total RNA obtained from
MFs and from Hker E6SFM human keratinocytes used as control, with
random hexamer primers using TaqMan Reverse Transcription Reagent
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol.
Conventional PCR was carried out using GeneAmp PCR
System 9700 (Thermo Fisher Scientific, Inc.). Thermal cycling was
performed as follows: 5 min at 95°C, 30 cycles of 95°C for 20 sec,
60°C for 40 sec and 72°C for 45 sec. PCR products (10 µl of 30 µl
total products per lane) were resolved on 2% agarose gel and
visualized using ethidium-bromide.
qPCR reactions were carried out in duplicates in an
ABI PRISM 7500 real-time thermocycler and Thermo PikoReal 96
Real-Time PCR system using SYBR-Green chemistry (Maxima SYBR Green
qPCR Master Mix 2X; K0252, Thermo Fisher Scientific, Inc.). The
thermocycling conditions were the following: 1X (95°C, 5 min); 40X
(95°C, 15 sec - 60°C, 1 min). Individual quantification cycle (Cq)
values were normalized to Cq values of an internal control gene
(18S ribosomal RNA). The tumor-associated samples were compared
with the appropriate controls following normalization. Alterations
in the expression levels were calculated by the 2−ΔΔCq
method (21). Primer sequences were
as follows: Matrix metalloproteinase (MMP)1-F:
5′-GATGTGGAGTGCCTGATGTG-3′; MMP1-R: 5′-CTGCTTGACCCTCAGAGACC-3′;
MMP2-F: 5′-ATGACAGCTGCACCACTGAG-3′, MMP2-R:
5′-ATTTGTTGCCCAGGAAAGTG-3′, MMP3-F: 5′-GGCAGTTTGCTCAGCCTATC-3′,
MMP3-R: 5′-TCACCTCCAATCCAAGGAAC-3′; MMP10-F:
5′-CATACCCTGGGTTTTCCTCCAA-3′, MMP10-R:
5′-GTCCGCTGCAAAGAAGTATGTTTTC-3′; MMP12-F:
5′-GATGCACGCACCTCGATGT-3′; MMP12-R: 5′-GGCCCCCCTGGCATT-3′, 18
SRNA-F: 5′-AAACGGCTACCACATCCAAG-3′; 18 SRNA-R:
5′CGCTCCCAAGATCCAACTAC-3′.
Protein extraction and western
blotting
For preparation of whole cell protein extracts,
confluent MF cultures were washed with PBS, cells were collected by
scraping and lysed in loading buffer [60 mM Tris (pH 6.8), 2% SDS,
10% glycerol, 5% β-mercaptoethanol, 0.002% bromophenolblue]. Total
protein concentration of the samples was assessed by bicinchoninic
acid protein assay kit (Thermo Fisher Scientific, Inc.) prior
adding bromophenolblue. A total of 15 µg protein of each sample was
separated by SDS-PAGE on 15% gels and transferred onto
nitrocellulose membrane (Amersham; GE Healthcare, Chicago, IL,
USA). Membranes were incubated first in 5% non-fat milk (in
Tris-buffered saline supplemented with 0.5% Tween-20) to block
non-specific binding sites then with primary anti-histone
antibodies specific for H4K16ac (Abcam; cat. no. Ab1762; 1:500) and
H3K9me3 (Abcam; cat. no. Ab8898; 1:750). Detection of H4 was used
to verify equal loading. For this anti-H4 antibody (Abcam; cat. no.
mab31827) was applied in 1:8,000 dilution. Incubation with
horseradish peroxidase-conjugated IgG secondary antibodies (Dako;
Agilent Technologies, Inc., anti-rabbit IgG; cat. no. P0448;
1:2,000 for modified histones and anti-mouse IgG, cat. no. P0260;
1:10,000 for H4) was performed at room temperature, for 50 min.
Blots were developed with chemiluminescent HRP reagent (EMD
Millipore, Billerica, MA, USA).
Assessment of MMP activity by gelatine
zymography
The activities of the two gelatinolytic MMPs (MMP2
and MMP9) in cell lysates and in cell media were examined with
gelatine zymography. MFs were lysed in 50 mM Tris pH 7.4, 0.2%
Triton X-100 buffer by freeze-thawing in liquid N2 and
pressing through a 23 G needle. Following centrifugation (17,000 ×
g, 10 min, 4°C), total protein concentration was measured by the
Bradford method (Bio-Rad Laboratories, Inc.). To detect gelatinase
activity in culture media, MFs were grown to 100% confluence in 75
cm2 flasks. Cells were serum starved for 24 h, then
media were collected on ice and the proteins were precipitated by
trichloroacetic acid in the presence of sodium-deoxycholate. The
pellet was collected by centrifugation (17,000 × g, 30 min, 4°C),
washed with acetone and re-suspended in water (22).
A total of 10 µg protein samples were resolved on
10% native PAGE co-polymerized with gelatin (2.5 mg/ml, type A from
porcine skin; Sigma-Aldrich; Merck KGaA). Following
electrophoresis, gels were washed 3 times for 20 min with 2.5%
Triton X-100 solution and incubated for 20 h at 37°C in buffer
containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM
CaCl2 and 0.05% NaN3. Following incubation,
gels were stained with Coomassie Brilliant Blue for 1 h and at room
temperature and destained in 30% ethanol, 10% acetic acid
solution.
Migration assay
A total of 2×105 cells were seeded onto
six-well plates and allowed to grow overnight in growth medium. On
the following day the confluent monolayer was gently scratched by a
P2 tip in the middle of the well. Wounds were measured and images
were captured under an inverted light microscope. Cultures were
incubated at 37°C for 24 h and migration of the cells was evaluated
by counting the number of cells, which migrated into the wound area
(20). The motility of
tumor-derived MFs was expressed as percentage of the control
MFs.
Statistics
Data are presented as mean ± standard error of the
mean. Experiments were repeated 3 times using 3 independent
replicates. Statistical significance was determined using SigmaPlot
Software version 12.0 (Systat Software, San Jose, CA, USA) with
Student's t-test, Mann-Whitney U test or analysis of variance
followed by Tukey's HSD post hoc multiple comparison test.
Results
Histological analysis of MFs in normal
tissue and in the esophageal tumor of a patient
The patient involved in the major part of the study
is a 71-year-old male with a 42×39×7 mm esophageal adenocarcinoma,
which developed probably on a Barrett's esophagus background. Tumor
grading was determined as pT3N1Mx, grade 2–3. Lymphatic and
vascular invasion was identified. Sections of esophageal tumor and
normal tissue, obtained from the patient, were stained for α-SMA,
desmin and vimentin to inspect the number, morphology and
localization of MFs. Representative α-SMA immunostainings on the
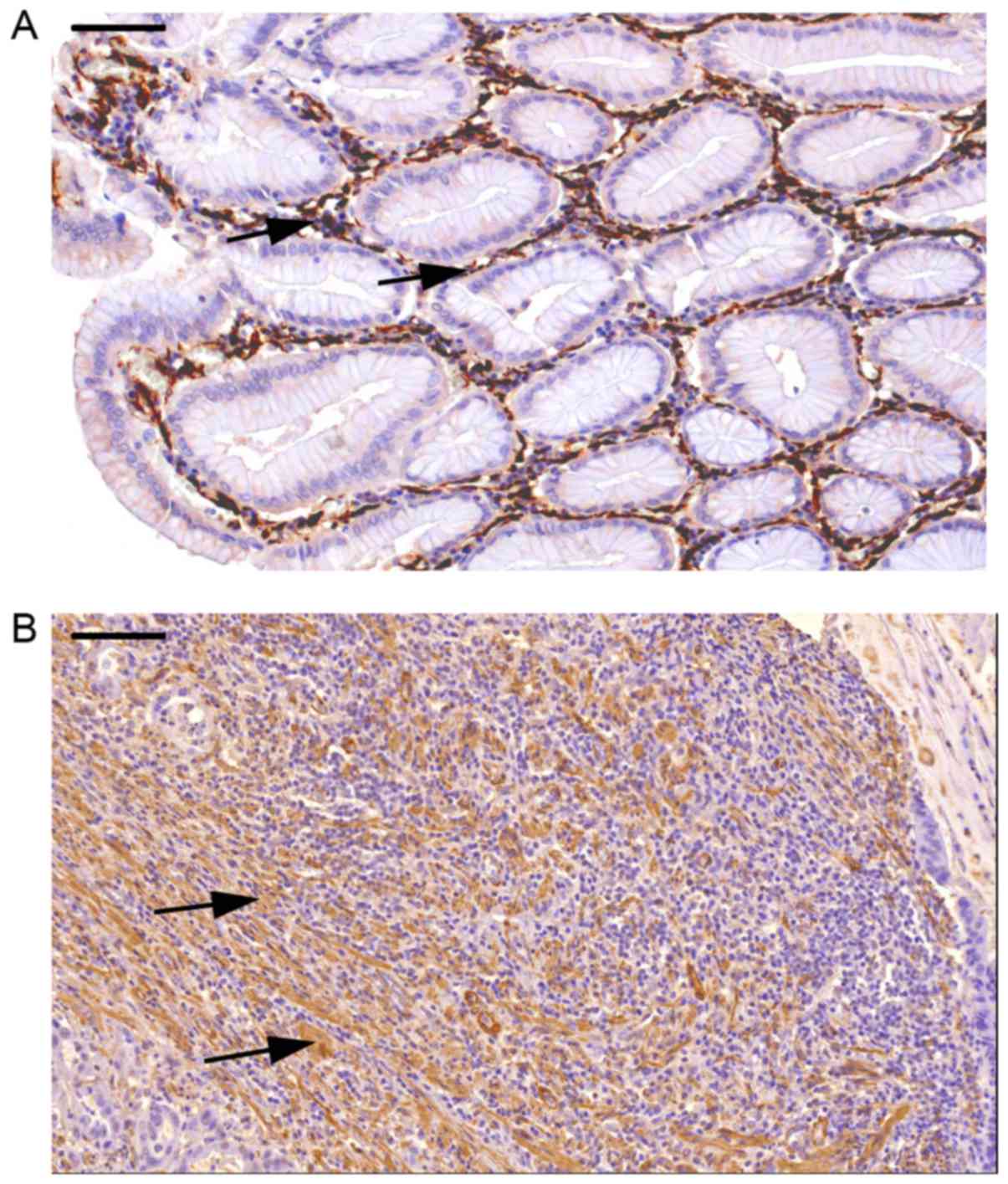
tissue sections are presented in Fig.
1 (A, normal; B, tumor-derived sample). Intensive brown
staining indicates high expression of α-SMA, a feature
characteristic to MFs. Pericytes and smooth muscle cells also
express α-SMA, however, these cells can be recognized and
distinguished from MFs based on their characteristic morphology and
on complementary immunostainings for desmin and vimentin (not
shown).
The histological analysis concluded that the number
of α-SMA- and vimentin-positive cells within the tumor was
increased (Fig. 1B).
Densitometrically quantified and normalized MF-specific brown color
staining revealed significantly elevated (230%) color density in
tumor-derived sections compared with the color intensity in
adjacent normal tissue sections (set as 100%), indicating higher
incidence of MF in the tumor-derived sections (P<0.001,
Student's t-test, data not shown). Furthermore, the spatial
arrangement and distribution of MFs were also markedly different in
the tumor environment compared with the normal tissue. MFs were
demonstrated mostly around the crypts and were confined to
pericryptal and subepithelial localization in normal tissue,
whereas in the tumor tissue, apart from the highly elevated number
and the greatly distorted shape of MFs, the arrangement of the
cells was severely transformed, composing meshwork-like structures
embedded in the tumor mass.
Genetic background, epigenetic
modifications in MFs and targeted genome analysis of the tumor
sample
To identify genetic alterations in MF cells and
possible somatic variants in esophagus tumor-derived MFs targeted
genome analysis was performed. The exonic sequences of 94
tumor-associated genes and 284 additional sites of tumor-associated
SNPs included in the Illumina TruSight Cancer Panel, were
determined by next generation deep sequencing. DNA sequences
totaling of 255 kbp of samples obtained from normal tissue-derived
and tumor-derived MF cells, and also from esophageal tumor tissue
were determined. Sequencing data were analyzed and variants of
interest were selected following annotation with Illumina
VariantStudio software.
It was demonstrated that on the sequenced 255 kbp
genomic region normal tissue-derived MFs of this patient carried
277 variants and identical number or variants were present in the
tumor-associated MFs, while 282 variants were discovered in tumor
tissue samples. Out of the 277 SNPs in MFs 22 (Table I) were predicted to be functionally
significant, based on their effect on protein structure, by their
ClinVar classification or presence in the COSMIC database, or by
their predicted effect on protein function by Sift and/or PolyPhen
analysis. These SNPs were located in the ALK receptor tyrosine
kinase, BRCA2, epidermal growth factor receptor, ERCC excision
repair 5 (ERCC5), exostosin glycosyltransferase 1, FA
complementation group I, FERM, ARH/RhoGEF and pleckstrin
domain protein 2, HNF1 homeobox A, HRas proto-oncogene, GTPase,
mutL homolog 1, mutS homolog 6 (MSH6), PMS1 homolog 2, mismatch
repair system component, patched 1, SLX4 structure-specific
endonuclease subunit (SLX4), tumor protein p53 (TP53), or
TSC complex subunit 2 (TSC2) genes and each were present in
a heterozygous state. A total of four genes ERCC5, BRCA2,
MSH6 and SLX4 were affected each by more than one SNP,
however, whether these represented mono- or diallelic state of
these variants cannot be determined.
 | Table I.Germline variants with functional
consequences in the cancer-associated myofibroblast samples. |
Table I.
Germline variants with functional
consequences in the cancer-associated myofibroblast samples.
| Gene name | Variant Freq
(%) | Type | Coordinate | Variant | Consequence | Amino acid
change | Allele Freq (%,
1000 Genomes Project) | Sift | PPhen | COSMIC ID | ClinVar
Accession | ClinVar
Significance |
|---|
| BIVM-ERCC5,
ERCC5 | G>G/C | Snv |
Chr13:103515085 | 4732 | Missense | C/S | 5 |
|
|
|
|
|
| BIVM-ERCC5,
ERCC5 | G>G/C | Snv |
Chr13:103528002 | 46.81 | Missense | D/H | 38 |
|
|
|
|
|
| BRCA2 | G>G/A | Snv | Chr13:32950896 | 49.33 | Missense | V/M | 0 | D | PrD |
|
|
|
| FARP2 | C>C/T | Snv | Chr2:242371101 | 50.26 | Missense | T/I | 8 | D | B |
|
|
|
| PTCH1 | G>G/A | Snv | Chr9:98209594 | 53.19 | Missense | P/L | 38 | D | PD |
|
|
|
| SLX4 | G>G/A | Snv | Chr16:3640784 | 46.48 | Missense | A/V | 4 | D | B |
|
|
|
| SLX4 | A>A/G | Snv | Chr16:3645607 | 51.28 | Missense | L/S | 6 | T | B |
|
|
|
| SLX4 | G>G/A | Snv | Chr16:3656625 | 37.09 | Missense | R/C | 6 | D | PrD |
|
|
|
| ALK | C>C/T | Snv | Chr2:29449819 | 48.65 | Synonymous | T | 18 |
|
| COSM148824 |
|
|
| BRCA2 | A>A/C | Snv | Chr13:32906729 | 44.92 | Missense | N/H | 24 | T | B | COSM147663 | RCV000034427.1:
RCV000009916.1 | Other:
non-pathogenic |
| EGFR | C>C/T | Snv | Chr7:55214348 | 38.01 | Synonymous | N | 45 |
|
| COSM1451542 |
|
|
| EXT1 | G>G/A | Snv | Chr8:118847782 | 49.52 | Synonymous | C | 19 |
|
| COSM150522 |
|
|
| FANCI | G>G/C | Snv | Chr15:89836228 | 51.55 | Missense | C/S | 29 | T | B | COSM1179763 |
|
|
| HNF1A | G>G/A | Snv |
Chr12:121437114 | 40 | Synonymous | T | 9 |
|
| COSM1559373 |
|
|
| HRAS | A>A/G | Snv | Chr11:534242 | 45.59 | Synonymous | H | 30 |
|
| COSM249860 |
|
|
| MLH1 | A>A/G | Snv | Chr3:37053568 | 42.39 | Missense | I/V | 17 | T | B | COSM1131469 | RCV000035355.1:
RCV000034548.1: RCV000030230.1 | Non-pathogenic:
probable-non-pathogenic |
| MSH6 | A>A/G | Snv | Chr2:48018081 | 54.02 | Synonymous | P | 10 |
|
|
| RCV000030265.1:
RCV000035321.1 | Non-pathogenic |
| MSH6 | T>T/C | Snv | Chr2:48023115 | 46.64 | Synonymous | D | 17 |
|
|
| RCV000035326.1:
RCV000030275.1 | Non-pathogenic |
| PMS2 | C>C/G | Snv | Chr7:6013049 | 26.94 | Missense | G/A | 17 | T | B |
| RCV000034628.1:
RCV000030370.1 | Non-pathogenic:
probable-non-pathogenic |
| SLX4 | G>G/A | Snv | Chr16:3639977 | 52.16 | Missense | A/V | 5 | T | B | COSM1377775 |
|
|
| TP53 | T>T/C | Snv | Chr17:7578210 | 51.63 | Synonymous | R | 1 |
|
| COSM249885 |
|
|
| TSC2 | T>T/C | Snv | Chr16:2138269 | 47.22 | Synonymous | D | 26 |
|
|
| RCV000043161.1 | Untested |
Next those variants that were present in lower
frequency and only in the esophagus tumor-derived MFs or/and in the
esophagus tumor sample but not in the adjacent normal MF samples
(Table II) were identified in
order to gain information on somatic mutation load in the tumor
tissue, and tumor-associated MFs. In the tumor-derived MF samples
variants with 5–7% allele frequency in the enhancer of zeste 2
polycomb repressive complex 2 subunit (EZH2; chr7:
148511066, C>A), TSC2 (chr16: 2120519, C>A) and
folliculin (FLCN; chr17: 17119708, T>TG) genes
were identified. The SNPs affecting EZH2 and TSC2
caused missense mutations, the insertion in FLCN causes a
frame-shift mutation and is described in the ClinVar and COSMIC
database (id: COSM1381204) as a variant associated with the large
intestine as primary tumor site. In the tumor tissue variants of
the FA complementation group D2 (FANCD2), XPC complex subunit,
BRCA1 associated protein 1 and glypican 3 were
demonstrated with 2–4% allele frequency and SNP of TP53 with
allele frequency of 14.92%. The TP53 variant (chr17: 7577539,
G>A) is present in the ClinVar and COSMIC databases (id:
COSM10656) and is associated with Li-Fraumeni syndrome and tumors
with various primary sites including the esophagus. The de
novo somatic nature of most of these variants is supported by
the fact that all of the relevant variants, except for a
FANCD2 variant in the tumor sample, have a 0% global allele
frequency in the 1,000 Genomes Project database.
 | Table II.Allelic variants present only in the
tumor or only in the tumor-associated MF samples. |
Table II.
Allelic variants present only in the
tumor or only in the tumor-associated MF samples.
| Sample | Gene | Variant | Coordinates | Freq
(%)a | GF (%)b |
Consequencec | Amino acid
change | Siftd | PPhene | ClinVar
Significancef | ClinVar
Accession | COSMIC ID |
|---|
| Tumor | EZH2 | C>C/A | chr7:
148511066 |
6.67 | 0 | Missense | Q/H | D | B |
|
|
|
| associated | TSC2 | C>C/A | chr16: 2120519 |
5.71 | 0 | Missense | H/Q | D | PrD |
|
RCV000003530.1: | COSM1381204 |
| MFs |
|
|
|
|
|
|
|
|
|
| RCV000003529.1 |
|
|
| FLCN | T>T/TG | chr17:
17119708 |
6.32 | 0 | Frameshift |
|
|
| P |
|
|
| Tumor | FANCD2 | C>C/T | chr3: 10106532 | 3.11 | 15 | Missense | P/L | T | B |
|
|
|
| tissue sample | XPC | CCCT>CCCT/C | chr3: 14219965 |
3.24 | 0 | Inframe deletion,
splice region variant | ED/D |
|
|
|
| COSM128738 |
|
| BAP1 | C>C/A | chr3: 52437502 |
3.64 | 0 | Synonymous | L |
|
|
|
|
|
|
| TP53 | G>G/A | chr17: 7577539 | 14.92 | 0 | Missense | R/W | D | PrD | P | RCV000013140.1 | COSM10656 |
|
| GPC3 | CCGG>CCGG/C | chrx:
133119383 |
2.44 | 0 | Inframe
deletion | P/- |
|
|
|
|
|
All combined, esophagus tumor-derived MFs
demonstrated 3, while the tumor sample exhibited 5 sequence
variants not present in normal MFs. The low occurrence of somatic
variants in MFs obtained from the tumor tissue and the observation
that the normal tissue-derived MFs carried nearly the same numbers
of unique somatic mutations suggest that mutation frequency is not
elevated greatly in tumor-associated MFs.
As no drastic difference in mutation frequency
between tumor-derived MFs and MFs isolated from the normal tissue
was detected, it was hypothesized that epigenetic modifications may
be involved in the development of the observed morphological
alterations, since epigenetic alterations can affect the expression
profile and ultimately the morphology and the function of
tumor-associated MFs. To substantiate this notion, a selected
number of epigenetic markers were first examined by immunostaining.
MFs were incubated with antibodies specific for pan-acetylated H3
and H4 as well as with antibodies recognizing specifically H3K9ac,
H3K14ac, H3K18ac, H4K8ac, H4K12ac, H4K16ac, and di- and
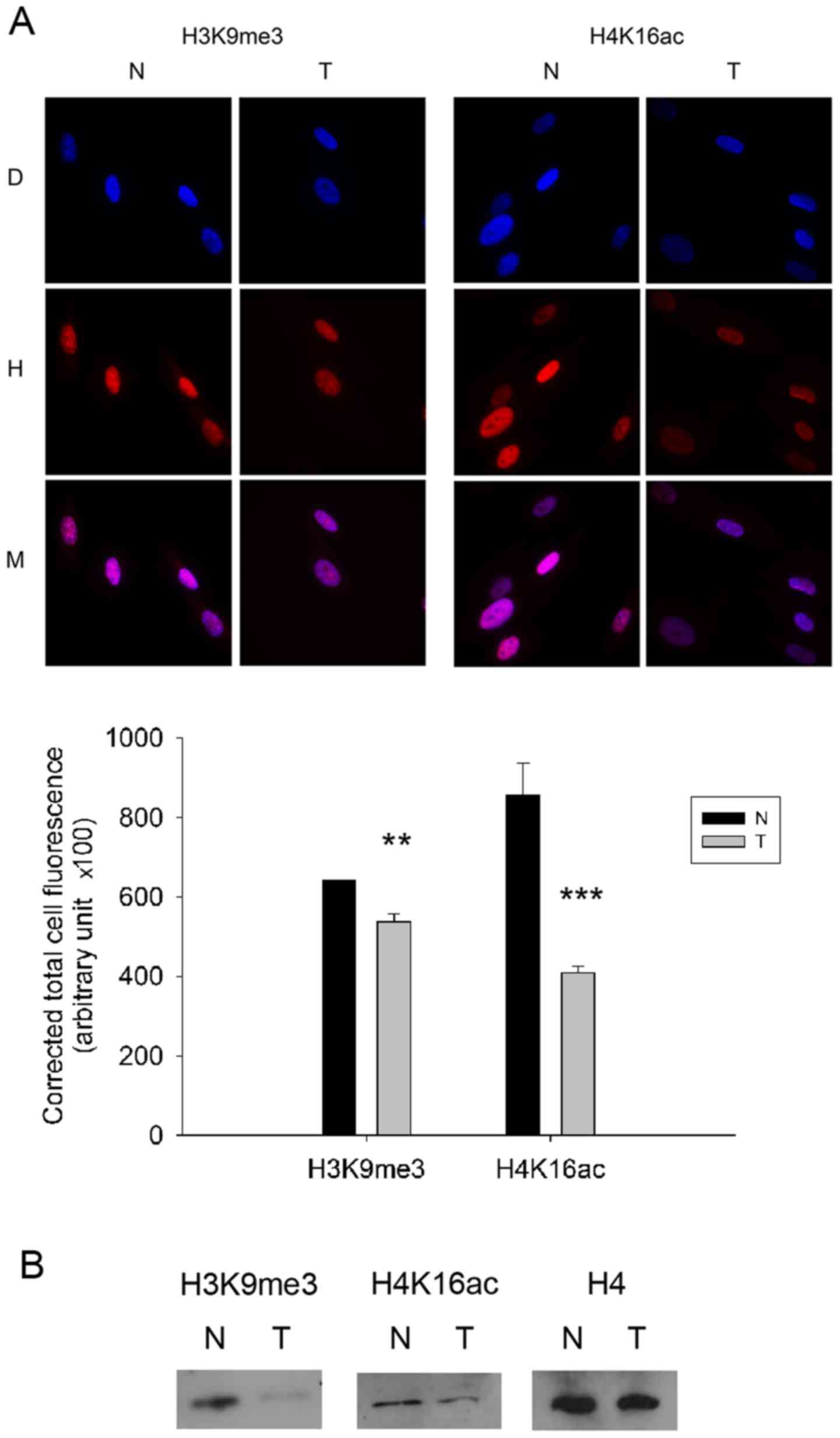
trimethylated H3K9. Immunostaining revealed significantly decreased
global levels of trimethylated H3K9 and acetylated H4K16
(P<0.01; Fig. 2A) in the
esophagus tumor-derived MFs compared to the adjacent normal MF
cells. Immunoblots verified the results obtained by
immunocytochemistry for selected histone modifications, since
western blotting also indicated lower levels of trimethylated H3K9
and acetylated H4K16 in tumor-derived MF cells (Fig. 2B). On the other hand, further
differences in global histone modifications between MFs isolated
from the tumor and from the adjacent normal tissue were not
detected (data not shown).
Transcriptome analysis of
tumor-derived and normal MFs
As a further step to compare MFs obtained from
normal and esophageal tumor tissue, whether any variations in the
expression patterns of selected genes can also be identified was
investigated. The mRNA levels of 190 chosen genes were compared on
TaqMan low-density gene expression array. In the analysis genes
associated with invasion, metastasis development, angiogenesis,
oxidative stress and apoptosis were included, as well as genes
responsible for signaling pathway components [mitogen activated
protein kinase, transforming growth factor (TGF)-β and
wingless-type MMTV integration site family (WNT)].
Furthermore, the expression level of ECM elements including
different types of collagens, basement membrane components and
matrix remodeling factors were also examined.
Results of the arrays revealed that several genes
involved in metastasis [C-X-C motif chemokine ligand 12
(CXCL12), endothelin 1 (EDN1), LY6/PLAUR domain
containing 3 (LYPD3), MMP3 and plasminogen activator, urokinase
(PLAU)], in regulation of cellular growth [cyclin D1
(CCND1), Fos proto-oncogene (FOS), mitogen-activated protein
kinase kinase kinase 8 (MAP3K8), myostatin
(GDF8), uncoupling protein 2 (UCP2) and
WNT1-inducible-signaling pathway protein 1 (WISP1)], in
apoptosis (BCL2) as well as in hypoxia-angiogenesis
[ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2),
heme oxygenase 1 (HMOX1) and pleiotrophin (PTN)]
demonstrated altered expression in matching MF cultures derived
from tumor and from the adjacent normal tissue (Table III). On the other hand, none of
the collagen genes included in the TaqMan array (collagen I α1,
collagen IV α1 and collagen XV α1) or any relevant ECM
glycoproteins and proteoglycans exhibited altered expression in
tumor-associated MFs compared with the cognate normal controls.
 | Table III.Increased or decreased expression of
specific genes in tumor-associated versus adjacent normal MFs
analyzed by TaqMan array. |
Table III.
Increased or decreased expression of
specific genes in tumor-associated versus adjacent normal MFs
analyzed by TaqMan array.
| 1. Apoptosis | Symbol | log2 change |
|---|
| B-cell CLL/lymphoma
2 | BCL2 | −1.05 |
|
| 2.
Hypoxia-angiogenesis associated | Symbol | log2
change |
|
| Ectonucleotide
pyrophosphatase/phosphodiesterase 2 (autotaxin) | ENPP2 | −3.13 |
| Heme oxygenase
(decycling) 1 | HMOX1 | −1.37 |
| Pleiotrophin | PTN | −1.28 |
|
| 3. Metastasis
associated | Symbol | log2
change |
|
| Chemokine (C-X-C
motif) ligand 12 (stromal cell-derived factor 1) | CXCL12 | −212 |
| Endothelin 1 | EDN1 |
2.55 |
| LY6/PLAUR domain
containing 3 | LYPD3 | −1.17 |
| Matrix
metallopeptidase 3 (stromelysin 1, progelatinase) | MMP3 |
2.26 |
| Plasminogen
activator, urokinase | PLAU |
1.07 |
|
| 4. Regulation of
cellular growth | Symbol | log2
change |
|
| Cyclin D1 | CCND1 |
1.03 |
| v-fos FBJ murine
osteosarcoma viral oncogene homolog | FOS | −1.6 |
| Mitogen-activated
protein kinase kinase kinase 8 | MAP3K8 | −1.2 |
| Growth
differentiation factor 8 (myostatin) | GDF8 (MTSN) | −2.72 |
| Uncoupling protein
2 (mitochondrial, proton carrier) | UCP2 |
1.4 |
| WNT1 inducible
signaling pathway protein 1 | WISP1 |
1.04 |
The detected elevated gene expression levels of
CCND1, UCP2 and Wnt signaling pathway member WISP1 in
tumor-derived MFs suggest a higher proliferation rate which can
account for the increased MF cell number within the esophageal
tumor mass. In contrast, lower levels of mRNAs corresponding to
GDF8, FOS and MAP3K8 in tumor-associated MFs
(Table III) were
demonstrated.
In tumor-derived MFs higher expression level of
metastasis-associated factor endothelin 1, a secreted peptide,
which has been demonstrated to stimulate cancer and stromal cell
proliferation and migration within the tumor microenvironment was
observed. Similarly, elevated gene expression of MMP3 and
PLAU was detected in tumor-associated MFs compared to normal
tissue-derived MFs (Table III).
On the other hand, TaqMan array data indicated lower expression of
the chemokine CXCL12 and of LYPD3.
Finally, decreased expression of
apoptosis-associated BCL2, as well as of
hypoxia-angiogenesis-linked ENPP2, HMOX1 and PTN
genes were detected in esophagus tumor-derived MFs compared with
normal counterparts (Table
III).
Migration capacity and expression of
MMPs in tumor-derived MFs
As gene expression data indicated that a number of
the genes exhibiting altered expression levels in tumor-associated
MFs are involved in the regulation of cellular growth and
metastasis, the expression analysis was extended to
matrix-modifying factors. As MFs can secrete MMP enzymes into their
surroundings, the MMP expression pattern of MFs was examined to see
whether there is a difference in this respect between the
esophageal tumor- and normal tissue-derived MFs.
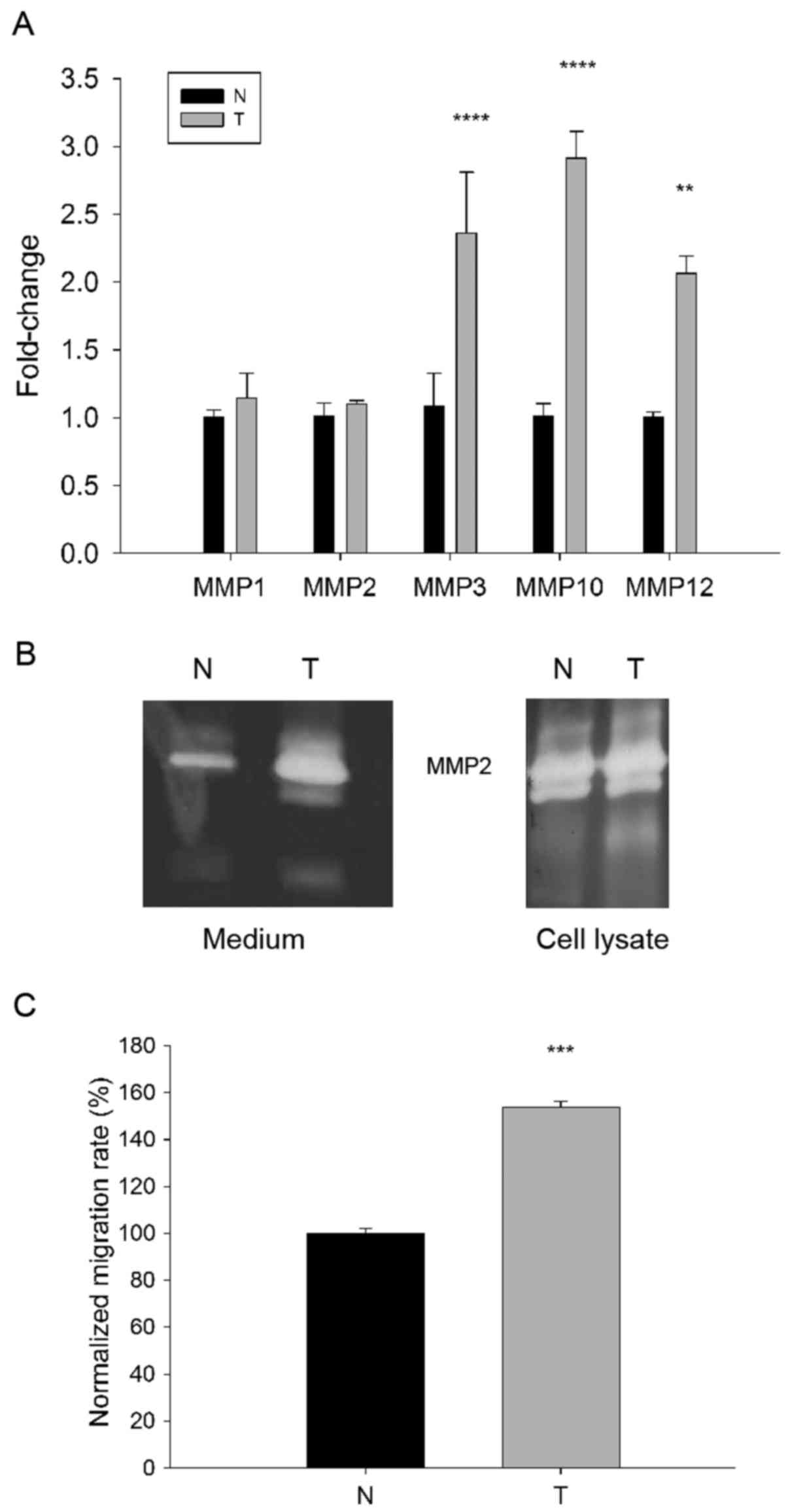
Using RT-qPCR significantly increased levels of mRNA
corresponding to MMP3, MMP10 and MMP12 were detected
in MFs isolated from the esophageal tumor compared with the normal
tissue MFs (P<0.01), however no difference was demonstrated in
MMP1 and MMP2 levels (Fig. 3A). Since the secreted gelatinase
MMP2 localizes on the surface of the migrating cells (23) and accumulates mainly on the leading
edge of invasive tumors (24),
in-gel zymography was used to test gelatinolytic activity in MF
cell lysates and conditioned cell media. Although zymograms
indicated no significant differences in MMP2 activities between
cell lysates of esophageal tumor-derived and normal tissue MFs,
conditioned media of the tumor-associated cells exhibited markedly
higher MMP2 activity compared with the medium of normal MFs
(Fig. 3B). Altogether these results
indicate that MFs in the tumor tissue are programmed to express and
secrete higher amounts of various MMP enzymes than normal tissue
residing counterparts. This feature can contribute to the
capability of tumor stroma residing MFs of degrading pericellular
ECM, as well as to promoting tumor invasion and metastasis.
In addition to the MMP expression profile and
activity, the migratory capacity of esophageal tumor- and normal
tissue-derived MFs was also investigated. For this, scratch wound
assays were performed and the migratory activity of MFs within a
24-h time frame was compared (Fig.
3C). The motility of normal MFs was defined as 100% and
expressed data obtained for tumor-derived MFs was defined as
normalized migration rate. It was demonstrated that the
tumor-derived MFs migrated significantly more (153.09%±2.12)
compared with their normal counterparts (100%±2.39; P<0.05;
Fig. 3C), suggesting that an
elevated migratory capacity is an acquired functional feature of
the tumor-residing MF phenotype.
To verify that the observed functional differences
between normal tissue and tumor-residing MFs populations of the
same individual are not limited to one patient but can be
considered widespread, and to draw a general conclusion from the
results of the present study, additional MF cultures were
established from tissue specimens of other patients bearing tumors
in diverse regions of the gastrointestinal tract, namely
individuals with cecum, sigmoid colon or rectum carcinoma. The
matching tumor-derived and normal tissue-derived MF pairs were
subjected to a scratch wound assay and RNA extraction to determine
migration activity and MMP expression levels. Pair-wise comparisons
of tumor- and normal tissue-derived MFs revealed that
tumor-originating MFs migrated significantly more compared their
normal counterparts (P<0.01; Fig. 4A
and B). Using these MF cultures, differences were demonstrated
in the expression levels of various MMPs (represented as
fold-change in Fig. 4C), proteases
that can modulate cell-matrix interactions and facilitate cell
mobilization, between stromal and normal MFs. A rather analogous
behavior of tumor-residing MFs within the whole gastrointestinal
tract is represented, therefore, it is feasible to suggest that
these traits acquired by MFs in the tumor microenvironment can in
fact develop generally in the tumor stroma and are not restricted
to one single patient. Although patient-to-patient individual
variations cannot be excluded, similar acquired features of stromal
MFs in esophagus, cecum, sigmoid colon and rectum tumor have been
demonstrated.
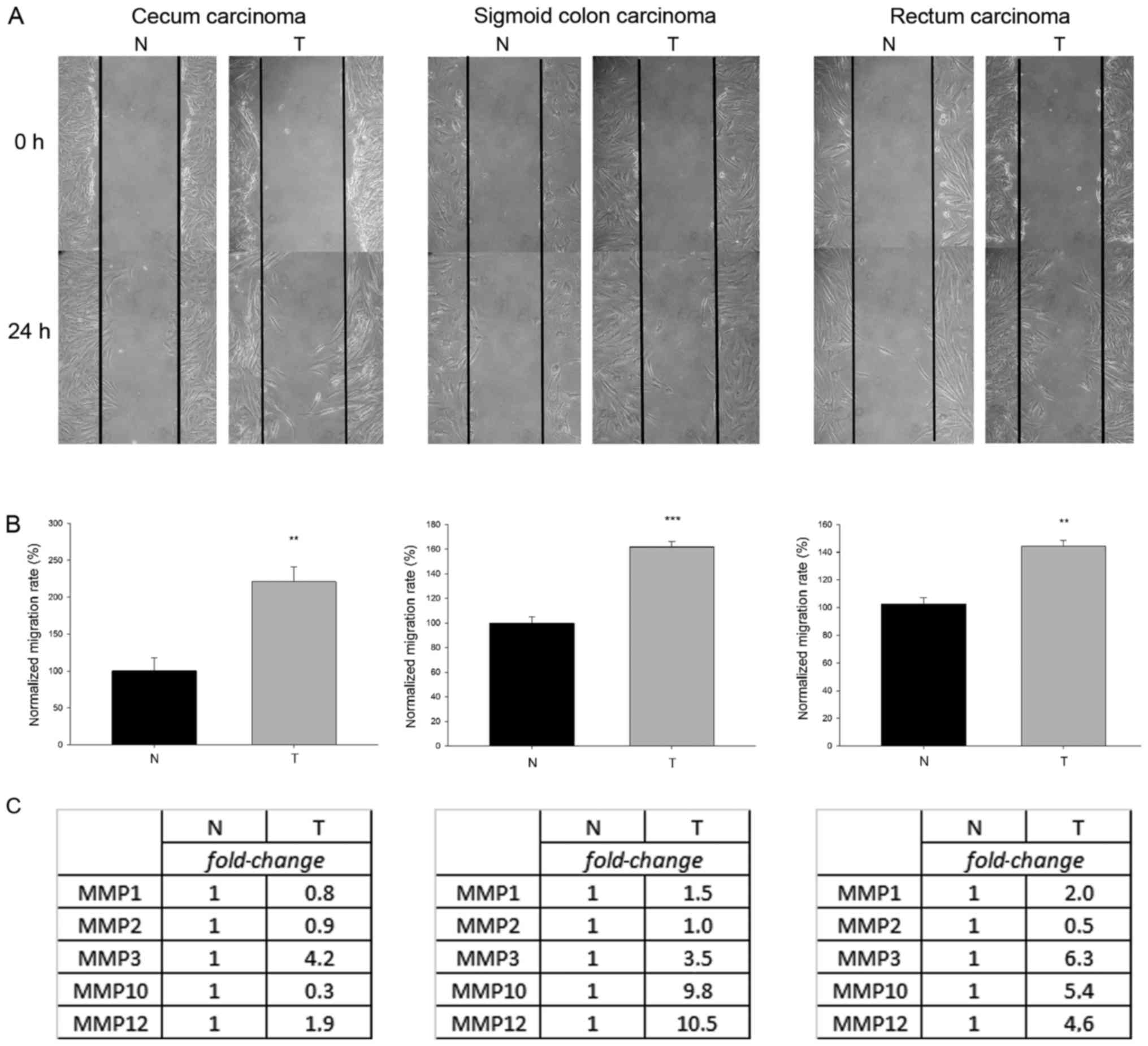 | Figure 4.Migration capacity and relative
expression levels of various MMPs of tumor-derived and normal MF
obtained from diverse regions of the gastrointestinal tract.
Representative images of scratch wound assays demonstrated
migration of MF cells into the wound area 24 h following scratching
(A). Migration rate was calculated based on the number of MF cells
in the wound zone and was normalized to the motility of the normal
tissue-derived MFs (B). Higher number of MFs could be counted
within the wound area 24 h following scratching in the
tumor-associated MF samples, therefore tumor-derived cells
exhibited a greater migratory capacity compared with the normal MFs
(**P=0.01 for cecum-derived MFs, ***P<0.001 for sigmoid
colon-derived MFs and **P=0.002 for rectum-derived MFs, t-test).
(C) Relative mRNA levels of matrix metalloproteases of
tumor-associated and normal MFs. Tables demonstrate the relative
expression of MMP1, MMP2, MMP3, MMP10 and MMP12 in the
tumor-derived MFs compared with control cells based on quantitative
polymerase chain reaction experiments. The expression levels of
MMPs were defined as 1 for the control sample and the mRNA levels
detected in tumor-derived MFs are indicated as fold-change. MMP,
matrix metalloproteinase; MF, myofibroblast; T, tumor-associated;
N, normal. |
Discussion
MFs are important stromal cells that define the
tumor tissue microenvironment through secretion of ECM components,
growth factors, cytokines, proteases and protease inhibitors
(25,26). Through direct or indirect actions
between MFs and transformed cancer cells the newly established
niche supports the active remodeling of the physical environment,
which will ultimately stimulate cancer cell proliferation and
dissemination (27). The functional
features of tumor-residing MFs are currently studied extensively,
nonetheless, knowledge on the genetic and epigenetic background, as
well as on the transcriptomic features of the evolving stromal MF
phenotypes remains limited. A comparative analysis of MFs obtained
from the tumor tissue and from tumor-adjacent normal tissue of an
esophageal cancer patient was performed, in order to identify
specific features of the exhibited MF phenotype in the tumor
microenvironment. Finally, specific acquired features on other
gastrointestinal tract tumor-derived MFs were verified. Appropriate
examination of MF-specific properties required the establishment of
pure MF cell cultures from surgically resected human samples.
To gain information on the genetic status of stromal
MFs, next generation sequencing was performed to detect the
occurrence of nucleotide variants in a set of most frequently
mutated cancer genes. As expected, over 255 kbp (~1 variant per
kbp) was demonstrated. Among these, normal tissue-derived and
tumor-associated MFs carried 22 relevant germline SNPs (Table I) predicted to be functionally
significant. A few genes (ERCC5, BRCA2, MSH6 and
SLX4) were affected by more than one SNP. A total of 4
variants were also identified that were present only in
tumor-associated MF samples and 11 variants that were present in
the tumor tissue but not in the adjacent normal MF samples
(Table II). The TP53 that were
detected to be present with 14.92% allele frequency in the tumor
tissue has already been associated with tumors in various primary
sites including the esophagus (reference in ClinVar database).
The genetic analysis in the present study indicated
low occurrence of somatic variants in tumor-associated MF samples
and that normal tissue- and tumor-derived MFs carry similar numbers
of unique somatic mutations, suggesting that mutation frequency is
not elevated drastically in tumor-associated MFs.
This lack of notable genetic alterations in
tumor-associated compared with the control MFs suggests that
genetic variations most probably cannot explain the observed
differences, including the largely elevated MF number, the severely
damaged MF architecture and the meshwork-like structures created by
tumor-residing MFs. Instead, in light of these data, epigenetic
mechanisms are responsible for the emergence of the stromal MF
phenotype. Indeed, global DNA hypomethylation that may have a
relevant role in cancer progression has already been demonstrated
in cancer-associated MFs (28).
It is well-known, that in addition to DNA
methylation, alterations in the acetylation or methylation level of
specific lysine (K) side chains of histone proteins, established by
histone modifying enzyme complexes, can lead to chromatin
modifications, thereby to major variations in the gene expression
pattern and consequently to morphological, and functional
alterations of a given cell (29–31).
Alterations in the global levels of numerous epigenetic histone
marks were examined and shifts were detected in the level of
specific acetylated and methylated histone proteins between normal
tissue- and esophageal tumor-derived MFs. The observed alterations,
including decreased levels of trimethylated H3K9 and acetylated
H4K16 in tumor-associated MFs may affect chromatin structure, which
can at least partly serve as the underlying cause of the altered
morphology of tumor-derived MFs.
In transformed cells, alterations in the expression
of hundreds of genes occur throughout tumorigenesis and tumor
invasion (32–34). Although MFs are non-transformed
cells, their expressional profile is expected to adapt to the
undergoing morphological and functional alterations within the
tumor microenvironment (35,36).
Elevated gene expression level of CCND1 were observed in
tumor-derived MFs compared with normal MFs. Significantly increased
expression of CCND1 has previously been identified in
stromal fibroblasts of patients with invasive breast cancer,
characterized by enhanced cancer growth, restrained apoptosis and
poor patient outcome (37). In the
present study it was hypothesized that the observed increased level
of the CCND1 transcript is correlated with the elevated
number of MF cells within the tumor tissue. Similarly,
WISP1, a downstream component in the WNT1 signaling pathway
[believed to be relevant to malignant transformation (38)] was also expressed at a high level in
MF cells isolated from the tumor. The encoded protein attenuates
p53-mediated apoptosis and binds to members of small leucine-rich
proteoglycans present in the ECM therefore preventing the
inhibitory activity of decorin and biglycan in tumor cell
proliferation (39,40). The modulation of apoptotic
mechanisms in tumor residing MFs is also indicated by the altered
expression of apoptosis-associated BCL2 and FOS in these
cells. The increased expression of UCP2, a critical
regulator of cellular metabolism, involved in the tumor-promoting
metabolic shift during cell transformation (41), is also detectable in MFs. These
cells must also participate in the complex metabolism of the tumor
tissue by promoting cancer cells to overcome energy depletion due
to the Wartburg effect. Finally, expression of hypoxia- and
angiogenesis-linked ENPP2, HO-1 and PTN genes were
decreased in tumor-derived MFs compared with normal counterparts
(Table III). PTN is a small
secreted cytokine and it has a potent role in tumor growth
progression, therefore, lower level of PTN mRNA in
tumor-associated compared with normal MFs was somewhat surprising,
however a study on prostate tumor CAFs reported a similar results
(42).
Although TGFβ signaling can drive tumorigenesis and
it is also the most capable factor in controlling MF
differentiation, proliferation, and the interactions with the
cellular microenvironment (43,44),
notable differences were not demonstrated between MFs of different
origins in the expression of TGFβ signaling pathway components. The
only member of the TGFβ protein family with lower levels of mRNA in
tumor-derived MFs corresponded to the secreted growth
differentiation factor GDF8/MSTN. This protein is a negative
regulator of skeletal muscle development and its high expression is
associated with cancer cachexia (45). Decreased level of MSTN was
also detectable in higher grade breast cancers and MSTN
supplementation reduced the viability, and the migratory capacity
of MCF-7 breast adenocarcinoma cell line (46). Furthermore, lower levels of mRNA
corresponding to MAP3K8 in tumor-associated MFs. This
observation is also in agreement with previous results on the
altered secretory profile of intestinal MFs of MAP3K8 mutant
mice, developing increased numbers and sizes of tumors, associated
with enhanced epithelial proliferation and decreased apoptosis
(Table III).
In tumor-derived MFs expression differences of
various genes associated with metastasis were also observed. Among
these, increased expression of EDN1 in tumor-derived MFs has
been verified. EDN1 is a metastasis-involved secreted peptide
exhibiting a strong stimulatory activity on cancer and stromal cell
proliferation and migration within the tumor microenvironment
(47). A low expression level of
HMOX1 was detected in MFs originating from the tumors. As
HMOX1 is a negative regulator of EDN 1 production, this lower
expression of HMOX1 can explain the increased transcript levels of
EDN1 (48). Like EDN1, elevated
expression of MMP3 and PLAU has been detected in
tumor-associated MFs compared with normal tissue-derived MFs
(Table III). These two genes
encode secreted zinc and serine protease enzymes, which following
activation are partly responsible for the degradation of a number
ECM proteins, including fibronectin, laminin, collagens, further
supporting the active assistance of stromal MFs in tumor
dissemination and metastasis (49).
On the other hand, TaqMan array data indicated lower transcript
levels of LYPD3 in tumor-derived MFs, which is not
surprising in the view of a recent study on the differential
diagnosis of epithelial malignancies, where the authors revealed
that LYPD3 expression is elevated only in esophageal squamous cell
carcinoma, but not in adenocarcinoma derived from esophagus tissue
(50). Also in agreement with a
previous study, lower expression of the chemokine CXCL12 was
exhibited in tumor-derived MFs, demonstrated to be a prominent
feature of clinical breast cancer lesions and together with
elevated CXCR7 levels it correlated significantly with poor
survival (51).
Tumor cells adopt the ability to migrate from the
primary tumor site by becoming motile and obtain plasticity to
mechanically navigate the tumor stroma. The invasion process
requires enzymes capable of remodeling the ECM protein scaffold to
make room for cancer cell movement. The production of ECM
remodeling proteases promotes cell invasion and motility by the
dynamical degradation of the pericellular ECM (52). When activated, these proteases,
including MMPs, cleave basement membrane components and
multiadhesion proteins. In this way, MMPs can modulate cell-matrix
interactions and facilitate cell mobilization, which ultimately
supports cell migration (53,54).
In fact, strong correlation has already been proposed between
elevated expression levels of certain MMPs in tumor tissues and
advanced tumor stages (TNM grading), increased metastasis, and poor
prognosis (16,17). Based on the mRNA expression, as well
as on the activity of various MMPs, a modified MMP profile of
tumor-derived MFs compared to normal MFs was observed. Furthermore,
strongly correlated with the altered expression and secretion of
proteases it was demonstrated that the tumor-derived MFs also
migrated more compared with their normal counterparts, revealing
another acquired functional feature of the tumor-residing MF
phenotype. These results on altered MMP expression and enhanced
migration of stromal MFs have been supported and strengthened by
tumor- and normal tissue-derived MF pairs obtained from other
regions of the gastrointestinal tract, namely from the cecum,
sigmoid colon and rectum. Therefore, it is feasible to suggest that
MFs in the tumor microenvironment assist cancer cell movement and
by acquiring migratory capacity, these stromal cells have the
potential to move collectively with cancer cells to promote the
invasiveness of cancer cells in the tumor leading edge (55).
Taken together, the results of the present study
indicate that the observed morphological, gene expressional and
concomitant functional alterations of MFs in the tumor
microenvironment are definitely not due to genetic alterations but
are more probably associated with epigenetic modifications.
Nonetheless, similarly to other attempts trying to correlate
histone modifications with altered RNA production level a careful
analysis is warranted to distinguish causes and consequences.
In conclusion, in the present study human MFs
obtained from gastrointestinal cancers were compared with those
residing in adjacent normal tissue of individual patients to
identify features of the exhibited MF phenotype that were acquired
in the tumor microenvironment. MF cells were analyzed for
differences in morphology, gene expression profile and function
with the aim to associate observed variations with underlying
genetic and/or epigenetic differences. It was demonstrated that
mutation frequency was not elevated in tumor-derived MFs. Histone
acetylation and methylation on the other hand can be more relevant
in the development of gene expressional and functional alterations
during the evolution of the stromal MF phenotype. The results of
the present study also demonstrated that though correlations
between transcriptional, functional and epigenetic results can be
drawn, general conclusions have to be considered very carefully.
Since the present study directly compared features of MFs of normal
and tumor tissues of various gastrointestinal cancer patients, it
was hypothesized that the present study's results are a valuable
contribution to the knowledge on the impact of MFs in supporting
tumor progression.
Acknowledgements
The authors would like to acknowledge the assistance
of Ms. Edina Pataki and Mrs. Zoltán Fuksz.
Funding
The present study was funded by the Hungarian
National Development Agency (TÁMOP grant no. 4.2.2–08/1/2008- 0013,
Hungarian Academy of Sciences) and by the MTA-SZTE Momentum Grant
(grant no. LP2014-10/2014) and by
GINOP-2.3.2–15-2016-00020.
Availability of data and materials
Data supporting the results of the present study are
available upon request.
Authors' contributions
IH carried out the transcriptional and epigenetic
studies, analyzed the data and performed the statistical analysis.
LB carried out next generation sequencing and analyzed the
sequencing data. MC performed the analysis of the clinical data. DK
performed the scratch assays. AS carried out experiments on matrix
metalloproteinases; LT was responsible for conducting the
histopathological analysis; GL performed patient surgery and
provided resection samples. ZR made a substantial contribution to
the interpretation of the data and revised the manuscript; PH
contributed to the conception and design of the project and
coordinated the associated clinical and research activities. IMB
designed the project, coordinated the molecular genetic studies and
revised the manuscript; MK carried out immunoblotting,
substantially contributed to the analysis and interpretation of the
data and drafted the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The patient gave written consent. The study was
approved by the Ethics Committee of the University of Szeged,
Hungary. All surgical patients gave informed consent. The scheme of
the experiments complies with the ethics of research and agrees
with the declaration of the Medical World Federation proclaimed in
Helsinki in 1964. Human Investigation Board of the University of
Szeged. Date: March 20 2006. South Sefton Research Ethics Committee
PO/JO/EC.90.03-2 NHS Trust Research Ethics Committee Ref.: EC
90.03.
Patient consent for publication
The patient gave written consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu
Y, Gong Z, Zhang S, Zhou J, Cao K, et al: Role of tumor
microenvironment in tumorigenesis. J Cancer. 8:761–773. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Polyak K, Haviv I and Campbell IG:
Co-evolution of tumor cells and their microenvironment. Trends
Genet. 25:30–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiga K, Hara M, Nagasaki T, Sato T,
Takahashi H and Takeyama H: Cancer-associated fibroblasts: their
characteristics and their roles in tumor growth. Cancers (Basel).
7:2443–2458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orimo A and Weinberg RA: Heterogeneity of
stromal fibroblasts in tumors. Cancer Biol Ther. 6:618–619. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eyden B: The myofibroblast: Phenotypic
characterization as a prerequisite to understanding its functions
in translational medicine. J Cell Mol Med. 12:22–37. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gabbiani G: The biology of the
myofibroblast. Kidney Int. 41:530–532. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hinz B, Phan SH, Thannickal VJ, Prunotto
M, Desmoulière A, Varga J, De Wever O, Mareel M and Gabbiani G:
Recent developments in myofibroblast biology: Paradigms for
connective tissue remodeling. Am J Pathol. 180:1340–1355. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hinz B, Phan SH, Thannickal VJ, Galli A,
Bochaton-Piallat ML and Gabbiani G: The myofibroblast: One
function, multiple origins. Am J Pathol. 170:1807–1816. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Desmoulière A, Guyot C and Gabbiani G: The
stroma reaction myofibroblast: A key player in the control of tumor
cell behavior. Int J Dev Biol. 48:509–517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pietras K and Ostman A: Hallmarks of
cancer: Interactions with the tumor stroma. Exp Cell Res.
316:1324–1331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grugan KD, Miller CG, Yao Y, Michaylira
CZ, Ohashi S, Klein-Szanto AJ, Diehl JA, Herlyn M, Han M, Nakagawa
H, et al: Fibroblast-secreted hepatocyte growth factor plays a
functional role in esophageal squamous cell carcinoma invasion.
Proc Natl Acad Sci USA. 107:11026–11031. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Couto N, Caja S, Maia J, Strano Moraes MC
and Costa-Silva B: Exosomes as emerging players in cancer biology.
Biochimie. S0300-9084(18)30067-1. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
De Wever O, Demetter P, Mareel M and
Bracke M: Stromal myofibroblasts are drivers of invasive cancer
growth. Int J Cancer. 123:2229–2238. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bauvois B: New facets of matrix
metalloproteinases MMP-2 and MMP-9 as cell surface transducers:
Outside-in signaling and relationship to tumor progression. Biochim
Biophys Acta. 1825:29–36. 2012.PubMed/NCBI
|
|
17
|
Shay G, Lynch CC and Fingleton B: Moving
targets: Emerging roles for MMPs in cancer progression and
metastasis. Matrix Biol. 44-46:200–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mehner C and Radisky DC: Triggering the
landslide: The tumor-promotional effects of myofibroblasts. Exp
Cell Res. 319:1657–1662. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gargus M, Niu C, Vallone JG, Binkley J,
Rubin DC and Shaker A: Human esophageal myofibroblasts secrete
proinflammatory cytokines in response to acid and Toll-like
receptor 4 ligands. Am J Physiol Gastrointest Liver Physiol.
308:G904–G923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Czepán M, Rakonczay Z Jr, Varró A, Steele
I, Dimaline R, Lertkowit N, Lonovics J, Schnúr A, Biczó G, Geisz A,
et al: NHE1 activity contributes to migration and is necessary for
proliferation of human gastric myofibroblasts. Pflugers Arch.
463:459–475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bensadoun A and Weinstein D: Assay of
proteins in the presence of interfering materials. Anal Biochem.
70:241–250. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiao Y, Feng X, Zhan Y, Wang R, Zheng S,
Liu W and Zeng X: Matrix metalloproteinase-2 promotes αvβ3
integrin-mediated adhesion and migration of human melanoma cells by
cleaving fibronectin. PLoS One. 7:e415912012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu CF, Chen FH, Lu MH, Hong JH and Chiang
CS: Dual roles of tumour cells-derived matrix metalloproteinase 2
on brain tumour growth and invasion. Br J Cancer. 117:1828–1836.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kalluri R: The biology and function of
fibroblasts in cancer. Nat Rev Cancer. 16:582–598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yazdani S, Bansal R and Prakash J: Drug
targeting to myofibroblasts: Implications for fibrosis and cancer.
Adv Drug Deliv Rev. 121:101–116. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Holmberg C, Ghesquière B, Impens F,
Gevaert K, Kumar JD, Cash N, Kandola S, Hegyi P, Wang TC, Dockray
GJ, et al: Mapping proteolytic processing in the secretome of
gastric cancer-associated myofibroblasts reveals activation of
MMP-1, MMP-2, and MMP-3. J Proteome Res. 12:3413–3422. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang L, Gonda TA, Gamble MV, Salas M,
Seshan V, Tu S, Twaddell WS, Hegyi P, Lazar G, Steele I, et al:
Global hypomethylation of genomic DNA in cancer-associated
myofibroblasts. Cancer Res. 68:9900–9908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baxter E, Windloch K, Gannon F and Lee JS:
Epigenetic regulation in cancer progression. Cell Biosci. 4:452014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang G and Pradhan S: Mammalian
epigenetic mechanisms. IUBMB Life. 66:240–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Albrengues J, Bertero T, Grasset E, Bonan
S, Maiel M, Bourget I, Philippe C, Herraiz Serrano C, Benamar S,
Croce O, et al: Epigenetic switch drives the conversion of
fibroblasts into proinvasive cancer-associated fibroblasts. Nat
Commun. 6:102042015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Andrade VP, Morrogh M, Qin LX, Olvera N,
Giri D, Muhsen S, Sakr RA, Schizas M, Ng CK, Arroyo CD, et al: Gene
expression profiling of lobular carcinoma in situ reveals candidate
precursor genes for invasion. Mol Oncol. 9:772–782. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu Y, Wang X, Wu F, Huang R, Xue F, Liang
G, Tao M, Cai P and Huang Y: Transcriptome profiling of the cancer,
adjacent non-tumor and distant normal tissues from a colorectal
cancer patient by deep sequencing. PLoS One. 7:e410012012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Coskunpinar E, Oltulu YM, Orhan KS,
Tiryakioglu NO, Kanliada D and Akbas F: Identification of a
differential expression signature associated with tumorigenesis and
metastasis of laryngeal carcinoma. Gene. 534:183–188. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Berdiel-Acer M, Sanz-Pamplona R, Calon A,
Cuadras D, Berenguer A, Sanjuan X, Paules MJ, Salazar R, Moreno V,
Batlle E, et al: Differences between CAFs and their paired NCF from
adjacent colonic mucosa reveal functional heterogeneity of CAFs,
providing prognostic information. Mol Oncol. 8:1290–1305. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Navab R, Strumpf D, Bandarchi B, Zhu CQ,
Pintilie M, Ramnarine VR, Ibrahimov E, Radulovich N, Leung L,
Barczyk M, et al: Prognostic gene-expression signature of
carcinoma-associated fibroblasts in non-small cell lung cancer.
Proc Natl Acad Sci USA. 108:7160–7165. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pestell TG, Jiao X, Kumar M, Peck AR,
Prisco M, Deng S, Li Z, Ertel A, Casimiro MC, Ju X, et al: Stromal
cyclin D1 promotes heterotypic immune signaling and breast cancer
growth. Oncotarget. 8:81754–81775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu J, Long Z, Cai H, Du C, Liu X, Yu S and
Wang Y: High expression of WISP1 in colon cancer is associated with
apoptosis, invasion and poor prognosis. Oncotarget. 7:49834–49847.
2016.PubMed/NCBI
|
|
39
|
Desnoyers L, Arnott D and Pennica D:
WISP-1 binds to decorin and biglycan. J Biol Chem. 276:47599–47607.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Su F, Overholtzer M, Besser D and Levine
AJ: WISP-1 attenuates p53-mediated apoptosis in response to DNA
damage through activation of the Akt kinase. Genes Dev. 16:46–57.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sreedhar A, Petruska P, Miriyala S,
Panchatcharam M and Zhao Y: UCP2 overexpression enhanced glycolysis
via activation of PFKFB2 during skin cell transformation.
Oncotarget. 8:95504–95515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Orr B, Vanpoucke G, Grace OC, Smith L,
Anderson RA, Riddick AC, Franco OE, Hayward SW and Thomson AA:
Expression of pleiotrophin in the prostate is androgen regulated
and it functions as an autocrine regulator of mesenchyme and cancer
associated fibroblasts and as a paracrine regulator of epithelia.
Prostate. 71:305–317. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Costanza B, Umelo IA, Bellier J,
Castronovo V and Turtoi A: Stromal Modulators of TGF-β in cancer. J
Clin Med. 6:62017. View Article : Google Scholar :
|
|
44
|
Tian M, Neil JR and Schiemann WP:
Transforming growth factor-β and the hallmarks of cancer. Cell
Signal. 23:951–962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miyamoto Y, Hanna DL, Zhang W, Baba H and
Lenz HJ: Molecular pathways: Cachexia signaling - a targeted
approach to cancer treatment. Clin Cancer Res. 22:3999–4004. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wallner C, Drysch M, Becerikli M, Jaurich
H, Wagner JM, Dittfeld S, Nagler J, Harati K, Dadras M, Philippou
S, et al: Interaction with the GDF8/11 pathway reveals treatment
options for adenocarcinoma of the breast. Breast. 37:134–141. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rosanò L and Bagnato A: Endothelin
therapeutics in cancer: Where are we? Am J Physiol Regul Integr
Comp Physiol. 310:R469–R475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bakrania BA, Spradley FT, Satchell SC,
Stec DE, Rimoldi JM, Gadepalli RSV and Granger JP: Heme oxygenase-1
is a potent inhibitor of placental ischemia-mediated endothelin-1
production in cultured human glomerular endothelial cells. Am J
Physiol Regul Integr Comp Physiol. 314:R427–R432. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Barajas-Castañeda LM, Cortés-Gutiérrez E,
García-Rodríguez FM, Campos-Rodríguez R, Lara-Padilla E,
Enríquez-Rincón F, Castro-Mussot ME and Figueroa-Arredondo P:
Overexpression of MMP-3 and uPA with diminished PAI-1 related to
metastasis in ductal breast cancer patients attending a public
hospital in Mexico City. J Immunol Res. 2016:85196482016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang W, Ding YQ, Li ZG, Han HX and Yang L:
Expression and diagnostic application of C4.4A protein in squamous
cell carcinoma and adenocarcinoma. Zhonghua Bing Li Xue Za Zhi.
35:277–280. 2006.(In Chinese). PubMed/NCBI
|
|
51
|
Yu PF, Huang Y, Xu CL, Lin LY, Han YY, Sun
WH, Hu GH, Rabson AB, Wang Y and Shi YF: Downregulation of CXCL12
in mesenchymal stromal cells by TGFβ promotes breast cancer
metastasis. Oncogene. 36:840–849. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hua H, Li M, Luo T, Yin Y and Jiang Y:
Matrix metalloproteinases in tumorigenesis: An evolving paradigm.
Cell Mol Life Sci. 68:3853–3868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Koyama S: Coordinate cell-surface
expression of matrix metalloproteinases and their inhibitors on
cancer-associated myofibroblasts from malignant ascites in patients
with gastric carcinoma. J Cancer Res Clin Oncol. 131:809–814. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sameni M, Anbalagan A, Olive MB, Moin K,
Mattingly RR and Sloane BF: MAME models for 4D live-cell imaging of
tumor: Microenvironment interactions that impact malignant
progression. J Vis Exp. 60:36612012.
|