Introduction
According to the survey results published by the
American Cancer Society (ACS), colorectal cancer (CRC) is one of
the most common tumors worldwide. The incidence rate is ranked
third among cancers (1). The number
of deaths attributed to colorectal cancer each year still reaches
610,000 (2), ranking it second in
malignant tumors (1).
Polymorphic adenoma-like protein 2 (PLAGL2) is a
zinc finger protein of the PLAG gene family with 7 C2H2 zinc finger
domains on the N-terminus (3–5). This
structure is highly conserved, can bind DNA and enables the
transcription factor PLAGL2 to activate the transcription of
specific genes (6). PLAGL2 is
closely related to the development of malignant tumors. PLAGL2 is
important in the development of acute myeloid leukemia (7), lung cancer (8), glioma (9), prostate cancer (10) and other tumors. In colorectal
cancer, Liu et al (11)
selected 225 pairs of colon cancer tissues and 66 normal tissues
for immunohistochemical studies and found that PLAGL2 expression
was significantly increased in colorectal cancer. The mechanism of
how PLAGL2 regulates the development of colon cancer has not been
clearly elucidated.
In our previous study, causes of high PLAGL2
expression in colorectal cancer were initially addressed (12). The aim of the present study was to
investigate the mechanism by which PLAGL2 affects the development
of colorectal cancer.
Materials and methods
Tissue specimens
All 31 CRC tissue specimens and pericarcinomatous
tissues were collected from the Third Affiliated Hospital of
Central South University between January 2017 and May 2017
(Changsha, China). There are 15 female and 16 male patients and
their age ranged from 35 to 70 years. All tissues were immediately
flash-frozen in liquid nitrogen after resection and were then
stored in liquid nitrogen. All cases were diagnosed as colorectal
cancer (CRC) by pathological sections, and the patients received no
chemo-, radio- or hormone therapy. All the patients signed consent
forms. The present study was approved by the Institute Research
Medical Ethics Committee of Central South University (Changsha,
China).
Animals
BALB/C nude mice (n=24, female, 5-weeks-old and
16–20 g) were purchased from the SJA Laboratory Animal Company
(Changsha, China) and housed under specific pathogen-free
conditions with a 12-h light/dark cycle and autoclaved food/water
were provided freely. The LV-PLAGL2-Homo-808 or LV–Vector (Shanghai
GenePharma Co., Ltd., Shanghai, China) was stably transfected in
SW480 and HCT116 cells following the manufacturer's instructions,
and a suspension of transfected-cells [5×106 in 100 µl
phosphate-buffered saline (PBS)] was injected into nude mice (n=6
for each group). Three groups were injected with HCT116 cells
(mock, LV-Vector and LV-PLAGL2-Homo-808). The tumor volume was
assessed every three days beginning 10 days after the injection and
was calculated with the following formula: Tumor volume
(mm3) = (length × width2)/2. All mice were
sacrificed 30 days later by cervical dislocation after the mice
were anaesthetized, and all animal protocols in this study were
approved by the Animal Ethics Committee of Central South University
(Changsha, China).
Cell culture
The human CRC cell lines SW480 and HCT116 were
purchased from Wuhan Boster Biological Technology Ltd. (Wuhan,
China). SW480 cells were cultured in L15 medium (Nanjing KeyGen
Biotech Co., Ltd., Nanjing, China) containing 10% fetal bovine
serum (FBS) (Biological Industries, Kibbutz Beit-Haemek, Israel).
HCT116 cells were cultured in McCoy's 5A medium (Nanjing KeyGen
Biotech Co., Ltd.) containing 10% FBS. All cells were cultured in a
humidified incubator at 37°C and 5% CO2.
Plasmid and lentivirus constructs
The human CRC cell lines SW480 and HCT116 were
transfected with three different vectors, which were transferred by
lentivirus, targeting PLAGL2 to knock down PLAGL2 protein
expression. Three short hairpin RNAs targeting human PLAGL2 and a
non-targeting RNA sequence serving as a negative control were
cloned into the pGPU6 vector (Shanghai GenePharma Co., Ltd.,
Shanghai, China). The details of the RNA sequences are presented in
Table I. Virus packaging was
performed in 293T cells. SW480 and HCT116 cells were cultured in
6-well plates with normal medium containing 10% FBS the day before
transfection (5×105 cells/plate). After 24 h, the cells
were transfected with 50 µl lentivirus with 5 µl Polybrene (5
µg/ml) and 1.5 ml medium. The cells were selected for 5 days and
then used for various assays. The lentivirus and vectors were
purchased from Nanjing KeyGen Biotech Co., Ltd.
 | Table I.Sequences of the vectors. |
Table I.
Sequences of the vectors.
| Serial number | Sequence
(5′-3′) |
|---|
| PLAGL2-Homo |
CCACCAGTGTATGTACTGTGA |
| −506 (LV1) |
|
| PLAGL2-Homo |
CTGCAGACCTTTGAGAGTACC |
| −699 (LV2) |
|
| PLAGL2-Homo |
GGCGGTTCTATACTCGTAAGG |
| −808 (LV3) |
|
| LV-NC |
TTCTCCGAACGTGTCACGT |
Full-length human WNT6 cDNA (NM_006522.3) was cloned
into the pCMV vector (Shanghai GenePharma Co., Ltd.). Virus
packaging and transduction were performed as aforementioned. WNT6
was ovexpressed in HCT116 cells as PLAGL2 was knocked down. In
addition, WNT6-overexpressed-cells were cultured in McCoy's 5A
medium (Nanjing KeyGen Biotech Co., Ltd.) without FBS.
Western blotting (WB)
Total protein was extracted by lysis in RIPA buffer
(Nanjing KeyGen Biotech Co., Ltd.) containing protease inhibitor
cocktail (Nanjing KeyGen Biotech Co., Ltd.) for 15 min at 4°C and
centrifuged at 16,666 × g for 15 min at 4°C. The proteins were
measured by BCA assay. Proteins (40 µg) were separated by 10%
SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)
membranes.
The membranes were blocked with 5% non-fat milk in
PBS for 2 h at room temperature and were then incubated with
primary antibodies at 4°C overnight. Subsequently, the membranes
were incubated with IRDye 800CW conjugated goat (polyclonal)
anti-rabbit IgG (H+L) (dilution 1:15,000; cat. no. 926-32211;
LI-COR Biosciences, Lincoln, NE, USA) for 2 h at room temperature.
The protein bands were visualized by Infrared fluorescence with
Odyssey CLx (LI-COR Biosciences) and the excitation wavelength was
778 nm. The primary antibodies used were as follows: Anti-PLAGL2
(cat. no. ab139509) and anti-Wnt6 (cat. no. EPR9244) (both from
Αbcam, Cambridge, UK), anti-β-catenin (cat. no. 8480) (Cell
Signaling Technology, Danvers, MA, USA) at a 1:1,000 dilution;
anti-GAPDH (cat. no. ab181602; Abcam) at a 1:3,000 dilution at a
1:3,000 dilution.
Quantitative real-time PCR
(RT-qPCR)
Total RNA was extracted by TRIzol reagent
(Invitrogen; Thermo Fisher, Scientific, Inc., Waltham, MA, USA).
Real-Time Quantitative PCR (RT-qPCR) was performed with the Toyobo
RT kit and KOD SYBR® qPCR kit (Toyobo Life Science,
Osaka, Japan) according to the manufacturer's protocol. The primers
for mRNA were produced by Sangon Biotech Co., Ltd. (Sangon Biotech,
China) and the primer sequences are listed in Table II. The thermocycling conditions are
listed in Table III. All mRNA
expressions were standardized to GAPDH, and the mRNA levels were
determined by the 2−ΔΔCq method. The method of
quantification employed by Livak and Schmittgen (13).
 | Table II.Primer sequences for RT-qPCR. |
Table II.
Primer sequences for RT-qPCR.
| Gene detected | Sequence
(5′-3′) |
|---|
| GAPDH | F:
GAAGGTGAAGGTCGGAGT |
|
| R:
CATGGGTGGAATCATATTGGAA |
| PLAGL2 | F:
GAGTCAAGTGAAGTGCCAATGT |
|
| R:
TGAGGGCAGCTATATGGTCTC |
| FZD6 | F:
TCTGCTGTCTTCTGGGTTGG |
|
| R:
CTGTAGCTCCTGTGCTGGTT |
| FZD7 | F:
TTCTACCACAGACTTAGCCACAG |
|
| R:
CTCACTTCCAGGTCACTTCTCA |
| Wnt3 | F:
TGACTTCGGCGTGTTAGTGT |
|
| R:
GTGCATGTGGTCCAGGATAG |
| Wnt6 | F:
GGTGCGAGAGTGCCAGTTC |
|
| R:
CGTCTCCCGAATGTCCTGTT |
| Wnt11 | F:
GGAGTCGGCCTTCGTGTATG |
|
| R:
GCCCGTAGCTGAGGTTGTC |
 | Table III.The thermocycling conditions used in
qPCR. |
Table III.
The thermocycling conditions used in
qPCR.
| Temperature
(°C) | Time |
|
|---|
| 95 | 5 min |
|
| 95 | 30 sec | -- |
| 60 | 30 sec | ⎪40 cycles |
| 72 | 30 sec | -- |
| 72 | 10 min |
|
| 4 | Hold |
|
Cell Counting Kit-8 assay
Cell proliferation was assessed by a Cell Counting
Kit-8 assay (CCK-8 Kit; Dojindo Laboratories, Kumamoto, Japan).
After cells were stably transfected with lentivirus, which included
the vectors to knock down the expression of PLAGL2, the cells
(4×103 cells/well) were incubated in normal medium
without serum at 37°C overnight the day before and incubated in
96-well plates with 100 µl normal culture medium at 37°C. At the
indicated time-points (12, 24, 48, 72 and 96 h), 10 µl of CCK-8
solution was added into each well and then cultured for 3 h at
37°C. The absorbance of each well at 450 nm (A450) was detected
with an EnVision microplate reader (PerkinElmer, Inc., Waltham, MA,
USA). All experiments were performed in triplicate.
Cell cycle analysis experiment
Cells were plated into a 6-well plate at a density
of 3×105 cells/well for 24 h and fixed with 2 ml 1%
paraformaldehyde and stored in 70% ethanol at −20°C. The cells were
stained with 0.5 ml PI/RNase (BD Biosciences, Fraklin Lakes, NJ,
USA) staining buffer for 15 min at room temperature and analyzed by
flow cytometry.
Immunofluorescence
A total of 3×105 cells were plated into
the 24-well plates for 24 h at −20°C and each well was covered by a
glass coverslip. Cells were fixed in 4% paraformaldehyde for 24 h
at room temperature and blocked in 10% BSA for 2 h at room
temperature. Cells were labeled with anti-PLAGL2 antibodies
(dilution 1:100; cat. no. 139509; Abcam) at 4°C for 12 h. The
following day, the cells were stained for 1 h with cy3-labeled goat
anti-rabbit IgG (dilution 1:100; cat. no. KGIF010; Nanjing KeyGen
Biotech Co., Ltd.) at 37°C and slides were washed three times for 5
min with PBS. Nuclei were stained with DAPI (Nanjing KeyGen Biotech
Co., Ltd.) for 5 min at 37°C. PLAGL2 immunofluorescence was imaged
using a Zeiss LSM 800 confocal microscope (Carl Zeiss AG,
Oberkochen, Germany).
Sequence analysis
The overexpression of PLAGL2 was analyzed by the
GEPIA database (14) and COSMIC
database (http://cancer.sanger.ac.uk/cosmic/). The NCBI gene
browser (https://www.ncbi.nlm.nih.gov/gene/) was used to find
the human Wnt6 locus (Chr2: 218,858,382-218,875,674) and the Wnt6
promoter region (Chr2: 218,858,821-218,860,072). Candidate
transcription factor binding sites were predicted by means of the
JASPAR database (http://jaspar.genereg.net/).
ChIP
Chromatin immunoprecipitation (ChIP) was performed
using an EZ-ChIP™ ChIP kit (Merck Millipore, Darmstadt,
Germany) following the manufacturer's protocol, including anti-RNA
polymerase II (Merck Millipore; cat. no. 05-623B), anti-IgG (mouse)
(Merck Millipore; cat. no. 12-371B) and GAPDH primers (Merck
Millipore; cat. no. 22-004). HCT116 cells (2×107) were
crosslinked with 37% formaldehyde. The chromatin was cleaved into
fragments between 200 and 600 bp by ultrasound. Anti-PLAGL2 was
purchased from Abcam and qPCR was performed with the KOD
SYBR® qPCR kit (Toyobo). Primer details are presented in
Table IV. The levels of DNA were
determined by the 2−ΔΔCq method.
 | Table IV.Primer sequences for qPCR in ChIP
assays. |
Table IV.
Primer sequences for qPCR in ChIP
assays.
| Gene detected | Sequence
(5′-3′) |
|---|
| GAPDH | F:
TACTAGCGGTTTTACGGGCG |
|
| R:
TCGAACAGGAGGAGCAGAGAGCGA |
| Wnt6-1 | F:
TCTCTACTCTTCTTCCCAGCCCTT |
|
| R:
TCGGGCTGTGGGACCCGCCCGCTT |
| Wnt6-2 | F:
CCAAGTCCGAGAGAGGCGAGGCGA |
|
| R:
GCGGGACTCTCGGGAGCGAGCCGT |
| Wnt6-3 | F:
AGGAGACACAGGCGCTGGCTGCCC |
|
| R:
TCAGTGCGAGCGCGGCGAGCGCAA |
Generation of luciferase reporter
constructs
The promoter fragments of the WNT6 gene were
generated by PCR using a forward primer with a KpnI
(15) site inserted at the 5′ end
and a reverse primer with a BglII (15) site inserted at the 3′ end. PCR
products were digested with KpnI/BglII and cloned
into a pGL3 firefly luciferase basic vector (Promega Corp.,
Madison, WI, USA) following the manufacturer's protocol.
Cell transfection and luciferase
assays
HCT116 cells were transfected using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Cells were then incubated at 37°C for
48 h before assaying for luciferase activity using the
Dual-Luciferase Reporter assay system (Promega Corp.) according to
the manufacturer's instructions. Firefly luciferase activity was
normalized relative to Renilla luciferase activity for each
transfection and calculated as the fold increase over pGL3 firefly
luciferase basic vector (pGL3Basic). At least two independent
transfections and two replicates per assay were performed for each
individual construct.
Statistical analysis
The statistical analysis was performed by using SPSS
19.0 software (SPSS, Inc., Chicago, IL, USA). Data were imaged with
GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA,
USA) and expressed as the mean ± SD. Differences between groups
were compared with Student's t-test (when 2 groups) and one-way
ANOVA (when >2 groups) was used, followed by
Student-Newman-Keuls (SNK) method for comparing each two groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
PLAGL2 is overexpressed in colorectal
cancer
Using the Catalogue of Somatic Mutations in Cancer
database (COSMIC; http://cancer.sanger.ac.uk/cosmic/), overexpressed
genes in colorectal cancer were screened and the top 20 were
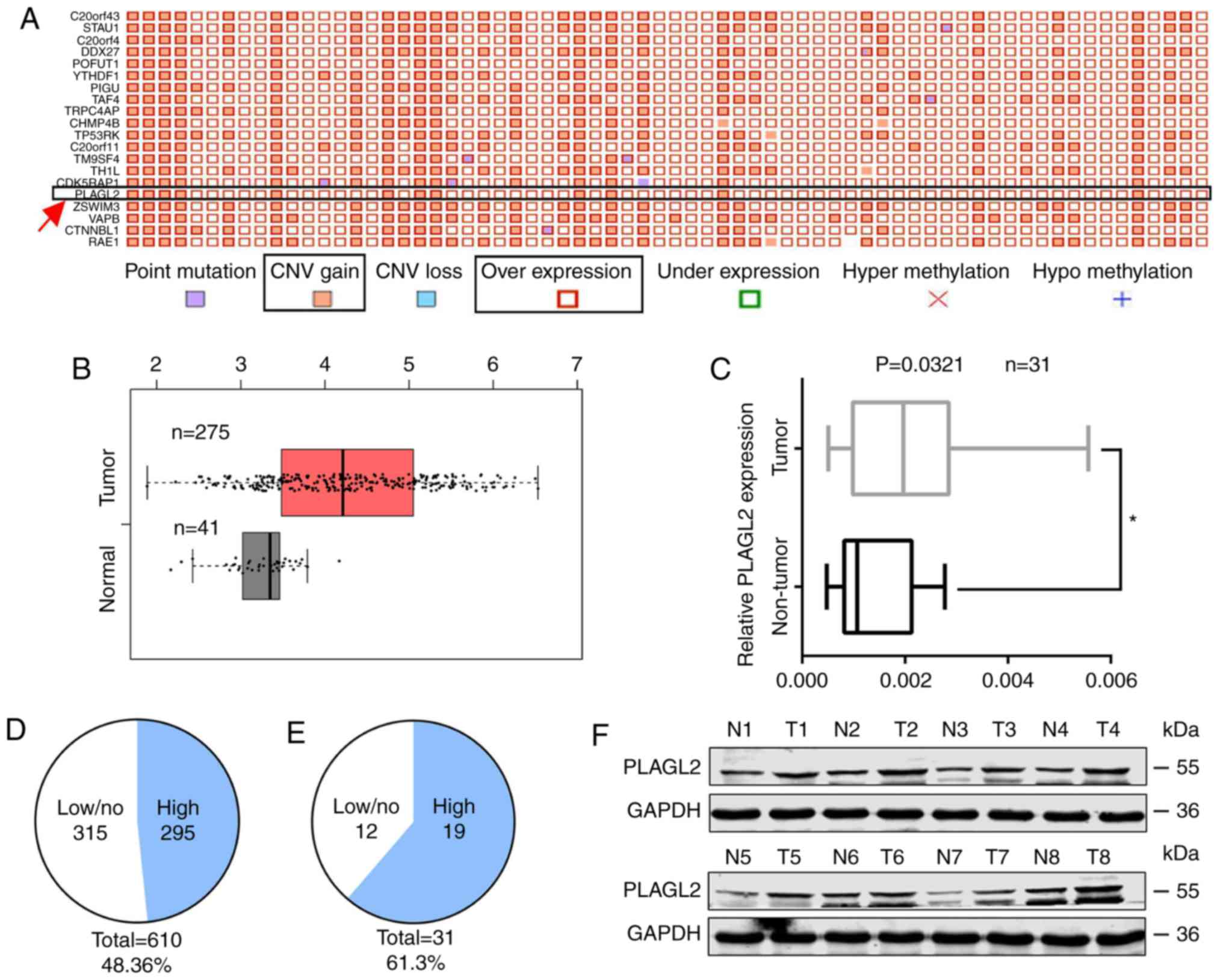
selected; PLAGL2 at position 16 was revealed (Fig. 1A). GEPIA analysis of the mRNA from
275 colon cancer specimens and 41 normal colonic tissues in TCGA
revealed that the expression of PLAGL2 in cancer tissue was
1.235-fold that of PLAGL2 in normal tissue (Fig. 1B). The mRNA of 31 colorectal cancer
tissue samples and 31 adjacent normal tissue samples was examined
by RT-qPCR. The expression of PLAGL2 in cancer tissue was
2.247-fold that of PLAGL2 in the normal tissues (Fig. 1C). In the COSMIC database, ~295 gene
fusion pairs exhibited PLAGL2 expression that was higher in cancer
tissues than in normal tissues, accounting for ~48.36% of the total
number of tissues. In the 31 pairs of tissues used in this
experiment, PLAGL2 was found in cancer tissues. Approximately 19
pairs had higher expression in cancer tissues than normal tissues,
accounting for ~61.3% of the total number of tissues (Fig. 1D and E). Eight of the 19 pairs of
tissues in which PLAGL2 expression was higher in the cancer tissues
than normal tissues were selected to detect the protein expression
of PLAGL2 by western blot analysis. The expression of PLAGL2 in
colorectal cancer tissue was significantly higher than that in the
normal tissue adjacent to the tumor (Fig. 1F). These results indicated that
PLAGL2 was overexpressed in colorectal cancer.
PLAGL2 influences the cell cycle and
promotes the proliferation of colorectal cancer
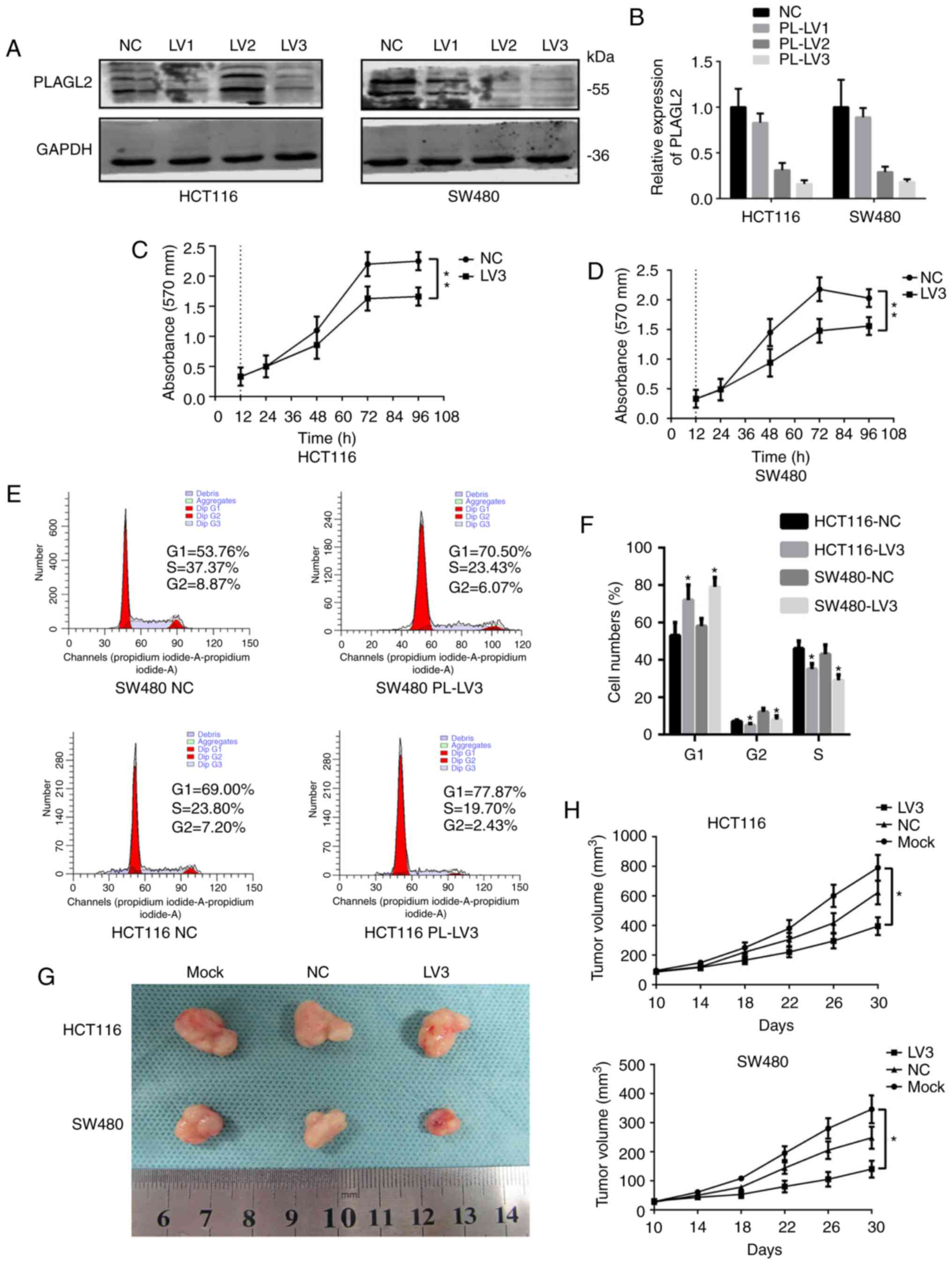
Next, we sought to reveal the function of PLAGL2 in
CRC. The CRC cell lines HCT116 and SW480 were selected and
different vectors were applied to knock down PLAGL2. It was found
that both in SW480 and HCT116 cells, the most efficient plasmid was
PLAGL2-Homo-808 (LV3) (Fig. 2A and
B). The PLAGL2-knockdown group (PL-LV3) was selected as the
experimental group and the empty plasmid group (NC) was used as the
control group for functional experiments. The results of the CCK-8
assay revealed that in SW480 cells, the growth rate of the
experimental group cells was significantly lower than that of the
control group cells after 48 h (P<0.05) and the difference among
them was more pronounced with time (P<0.01). In HCT116 cells,
from 72 h onwards, the growth rate of the experimental group of
cells was significantly lower than that of the control group cells,
with significant differences (P<0.05), and the differences among
them were more obvious with time (P<0.01) (Fig. 2C and D). Flow cytometry was used to
detect the cell cycle. The results indicated that in the HCT116 and
SW80 cells, the cells in the experimental group exhibited a
prolonged G1 phase and the S phase was shortened (Fig. 2E and F) when compared with the
control cells. Tumor formation experiments in nude mice indicated
that the tumor volume in the untreated and the control groups was
greater than that in the experimental group (Fig. 2G), and the growth curve indicated
that the growth rate of the control group was greater than that of
the experimental group (Fig. 2H).
These results indicated that PLAGL2 affected the development of
colorectal cancer in vitro and in vivo.
PLAGL2 promotes the Wnt/β-catenin
pathway
PLAGL2 plays as an important role in other tumors,
where it can always activate the Wnt/β-catenin pathway, and the
Wnt/β-catenin pathway is one of most active pathways in colorectal
cancer. Several key factors that are active in the Wnt/β-catenin
pathway were selected and primers were designed. The mRNA of SW480
and HCT116 cells was extracted and the expression was detected with
RT-qPCR. It was found that the expression of Wnt6 and Wnt11 was
significantly decreased after PLAGL2 knockdown (Fig. 3A). While Wnt11 mainly regulates the
Wnt/Ca2+ pathway, Wnt6 regulates the Wnt/β-catenin
pathway. A western blot assay was used to detect changes in the
expression of Wnt6 and β-catenin protein before and after PLAGL2
knockdown. The results revealed that the expression of Wnt6 and
β-catenin proteins decreased with the knockdown of PLAGL2 (Fig. 3B). Wnt6 was overexpressed in HCT116
cells when PLAGL2 was knocked down and the two groups were
compared. The WB assays revealed that β-catenin was increased when
WNT6 was overexpressed but PLAGL2 was knock down. It revealed that
PLAGL2 activated the Wnt/β-catenin pathway by promoting the
expression of WNT6 (Fig. 3B). The
sequence 1,000 bp upstream and 216 bp upstream of the WNT6 gene was
used to make predictions using the JASPER database, indicating that
the WNT6 upstream sequence was highly conserved among different
species and that the conserved region had 7 possible binding
domains for the PLAGL2 gene (Fig.
3C).
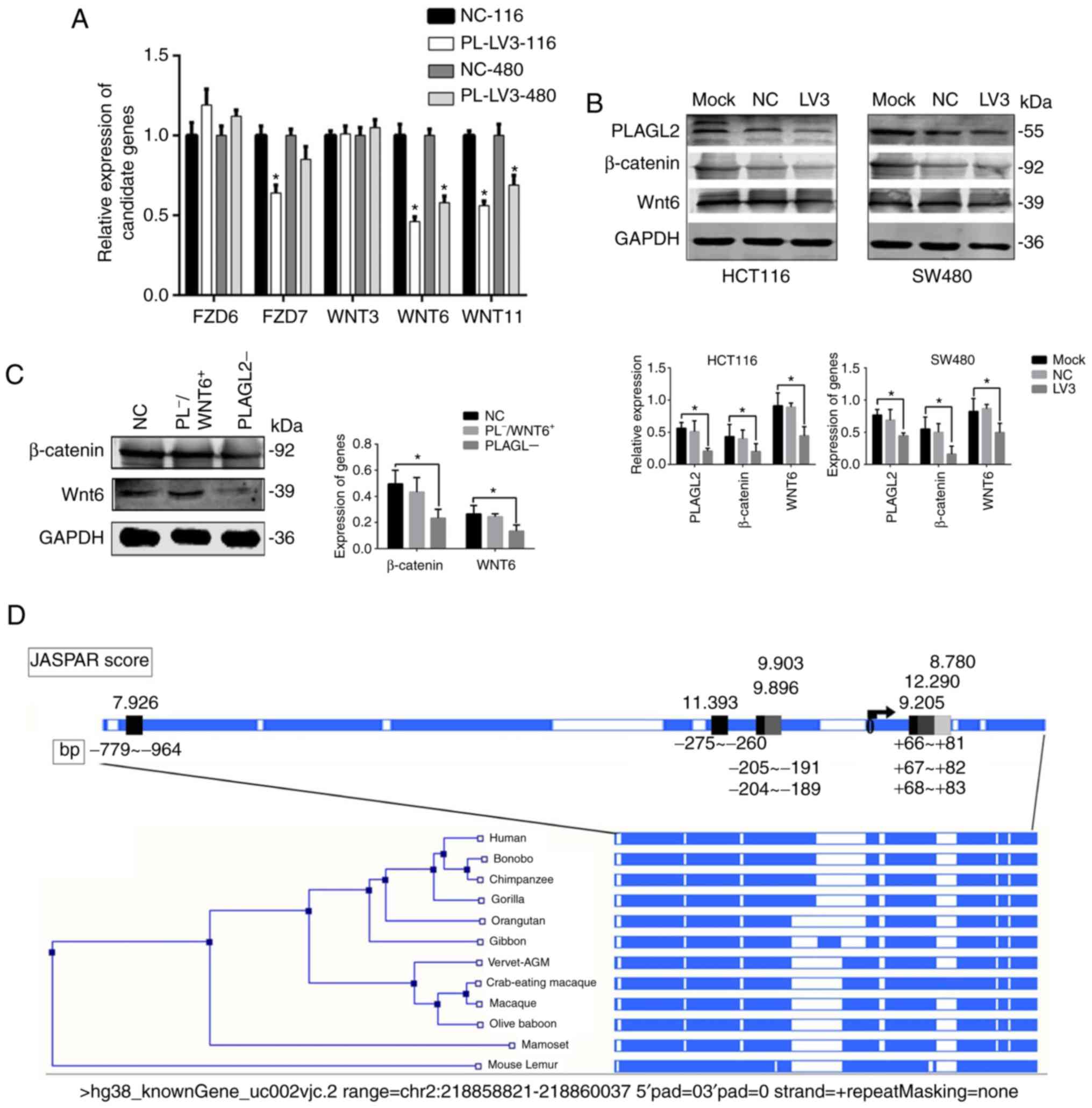 | Figure 3.Screening of signaling pathway
factors and prediction about the promoter region. (A) The factors
in the Wnt pathway were screened and the one that was related to
PLAG2 was choosen using RT-qPCR assays. (B) With western blot
assays, the expression levels of PLAGL2, Wnt6 and β-catenin were
revealed to have a positive correlation. Mock indicates the
original cancer cells, LV3 indicates PLAGL2 was knocked down, and
NC indicates the control group. (C) With western blot assays, the
expression levels of Wnt6 and β-catenin were revealed to have a
positive correlation. NC indicates the control group,
PLAGL2− indicates that PLAGL2 was knocked down, and
PL−/WNT6+ indicates PLAGL2 was knocked down
and WNT6 was overexpressed. (D) Prediction about the promoter
region that can bind with PLAGL2, which was predicted by the JASPAR
database. The data are expressed as the mean ± SD for 3
experiments, each performed in duplicate, *P<0.05. |
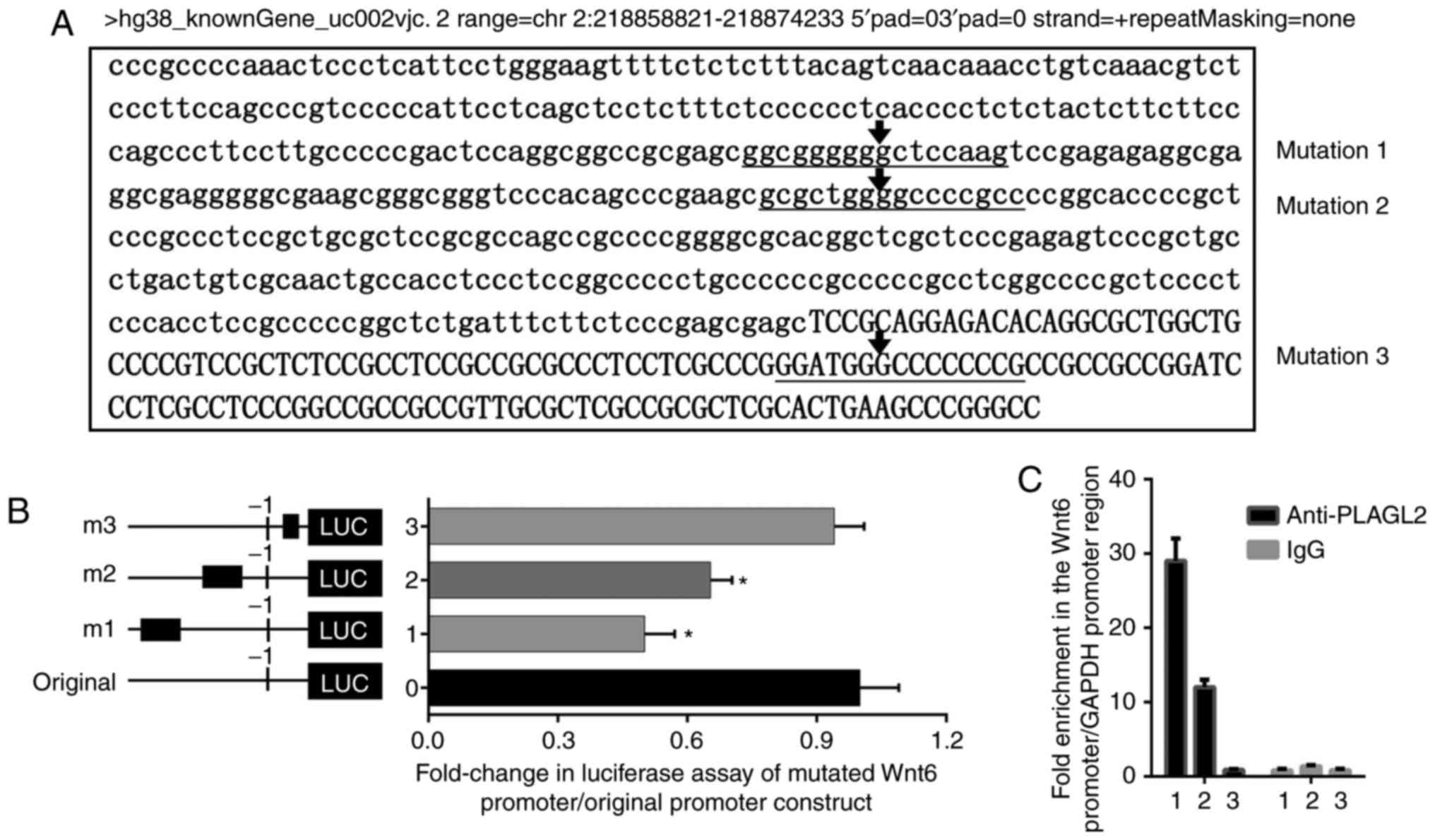
PLAGL2 binds to the promoter region of
Wnt6 and actives it
The three-ideal binding region mutation analyses
predicted by the JASPAR database were selected; the horizontal
lines represent possible binding sites and mutation regions of
PLAGL2 and Wnt6 (Fig. 4A). The
luciferase assay results revealed that after mutation of the
predicted binding regions, the promoter activity was decreased.
Compared to the activity of the original promoter region, mutation
1 resulted in a ~50% reduction in promoter activity, mutation 2
resulted in a ~35% reduction, and mutation 3 resulted in a ~10%
reduction (Fig. 4B). In HCT116
cells, we applied ChIP assays to detect the binding of PLAGL2 to
the Wnt6 promoter region. It was revealed that PALGL2 strongly
bound to the Wnt6 promoter region in HCT116 cells, with a ~28 and
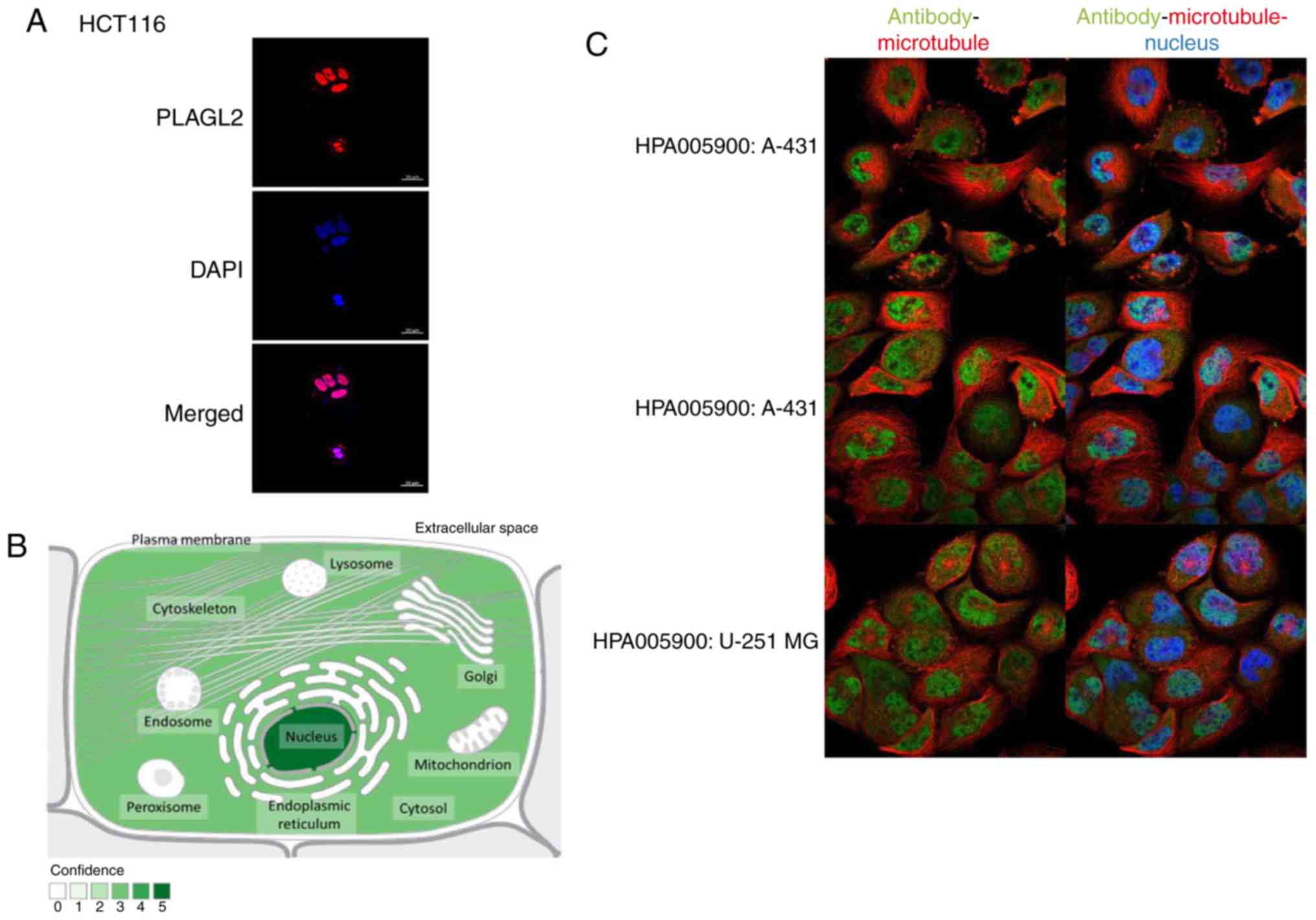
~12-fold enrichment compared to the GAPDH control (Fig. 4C). The results of immunofluorescence
also revealed that the PLAGL2 proteins were located in the cell
nucleus (Fig. 5A). These results
indicated that PLAGL2 could activate Wnt6 expression in colorectal
cancer cells, activating the Wnt/β-catenin pathway.
Discussion
The polymorphic adenoma-like protein 2 (PLAGL2),
located on chromosome 20q21.11, is a zinc finger protein derived
from the PLAG gene family. The protein can function as a
transcription factor to activate the expression of some oncogenes
and cancer-promoting signaling pathways by activating promoters,
thereby inhibiting cell differentiation and increasing the speed of
cell self-replication. The carcinogenic effects of PLAGL2 have been
found in many malignancies, including acute myeloid leukemia,
neurogenic stromal tumor, lung and breast cancer (7–9,16–18).
For the relationship between PLAGL2 and colorectal cancer, more
studies are required. In the present study, the information we
collected by screening the COSMIC database (19–21)
and GEPIA database and by RT-qPCR and WB demonstrated that PLAGL2
was overexpressed in colorectal cancer tissues.
As a proto-oncogene, PLAGL2 is overexpressed in
colorectal cancer, and it can be reasonably speculated that it
plays a role in the development of colorectal cancer. However,
PLAGL2 can act as a transcription factor in many tumors and
activate some cancer-promoting pathways relating to cancer, such as
the Wnt/β-catenin pathway.
The Wnt/β-catenin pathway is a very active signaling
pathway in colorectal cancer, and it can promote the proliferation,
cell cycle, apoptosis, invasion and migration of colorectal cancer
after activation (22). β-catenin
can be phosphorylated by GSK3-β and ultimately be hydrolyzed by the
ubiquitin-proteasome pathway. The binding of the Wnt signaling
pathway ligand to the paired frizzled-like receptors inhibits
hydrolysis of the β-catenin complex, including APC, AXIN and
GSK3-β, and a series of reactions leads to the stabilization of
β-catenin in the nucleus and eventual activation of the downstream
target gene. When the Wnt/β-catenin pathway is activated, β-catenin
is translocated into the nucleus and binds to the transcription
factor TCF/LEF in the nucleus, which leads to the activation of
expression of the downstream target genes, such as a MMP-7, CCND1,
c-Myc and survivin. The activation of these downstream genes can
promote the development of colorectal cancer (23–26).
For example, the proto-oncogene c-Myc can be activated by β-catenin
and inhibited by wild-type APC. The dependence of
β-catenin-mediated c-Myc proto-oncogene expression will maintain
the initial phenotype of colorectal cancer cells by inhibiting p21
expression and disrupting cell differentiation (27). β-catenin can also regulate the
expression of CCND1. CCND1 is an important G1 phase cyclin, and
β-catenin can increase the expression of CCND1 after it enters the
nucleus. CCND1 accumulates in the nucleus and promotes cells to
enter the S phase from the G1 phase. The cells enter the S phase
and hydrolyze, which accelerates the cell cycle (28,29).
MMP-7 which functions as a downstream target gene for the Wnt
pathway is also activated when the Wnt pathway is activated. MMP-7
is a member of the MMP family, and MMPs are
Zn2+-dependent proteolytic enzymes that can degrade a
variety of basement membrane and extracellular matrix components.
It plays an important role in tumor invasion and metastasis.
Survivin, a member of the apoptotic inhibitor protein family,
inhibits apoptosis and expression in tumors and enhances tumor cell
viability (30). High expression of
survivin was also revealed when the Wnt pathway was activated.
It was revealed that the WNT/β-catenin pathway plays
an important role in colorectal cancer. This study demonstrated
that PLAGL2 was related to the activation of the WNT/β-catenin
pathway in colorectal cancer. Now, the question that remains is how
PLAGL2 regulates the expression of the WNT/β-catenin pathway.
PLAGL2 is located on chromosome 20q11.21 and contains the
initiation codon ATG and the stop codon TAG, without the presence
of the AATAAA polyadenylation signal. Its open reading frame
encodes 496 amino acid residues. The protein consists of seven zinc
finger structure consensus sequences at the amino acid terminus and
a carboxylic end rich in proline and serine (4). The zinc finger structure is often
found in DNA-binding proteins. It consists of a ring containing
approximately 30 amino acids and a Zn2+ coordinated to 4
Cys or 2 Cys and 2 His on the ring, forming a structure like a
finger (31). The zinc finger has a
pair of cysteine residues at the N-terminus and a pair of histidine
residues at the C-terminus. The zinc finger structure can
specifically recognize the DNA sequence through the conserved
sequence at the N-terminus, and the C-terminus has a
transactivation function. The transcriptional activity of PLALG2 is
activated by binding to the bidirectional consensus sequence
GRGGC(N)(6–8)GGG (32), and its protein contains seven zinc
finger structures, of which zinc finger structure 3 is mainly
responsible for recognizing the aforementioned. In the sequence,
zinc finger structures 6 and 7 are the cores that bind to proteins
(33).
It is known from the structural features of PLAGL2
that it recognizes the promoter region of genes and promotes gene
transcription using the C-terminal activation functions. Some
researchers have claimed that PLAGL2 can be secreted to the medium
in vitro, but as a transcription factor, all the
aforementioned processes occur in the nucleus (9,34–36).
The introduction of PLAGL2 in PubMed revealed that most PLAGL2
proteins are located in the cell nucleus. The data from the
GeneCards database (https://www.genecards.org/) revealed the same results
(Fig. 5B). The immunofluorescence
results from The Human Protein Atlas (https://www.proteinatlas.org/) revealed that the
proteins of PLAGL2 are surrounded by karyotheca (Fig. 5C).
In the present study, it was demonstrated through a
ChIP assay, that PLAGL2 could be combined with the promoter region
of WNT6 in the nucleus. The luciferase assay revealed that PLAGL2
could activate the transcription of WNT6 by binding to the WNT6
promoter region.
Wnt6 protein expression activates the Wnt/β-catenin
pathway. The activation of the Wnt/β-catenin pathway leads to the
overexpression of a series of target proteins, which may play
important roles in the development of colorectal cancer.
In summary, PLAGL2 is an oncogene that is active in
a variety of malignancies. It is rarely expressed in normal adult
tissues and is active in embryos, stem cells and tumors (5). Further research on PLAGL2 may provide
a new target for the targeted therapy of colorectal cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (no. 81773130), The New Xiangya
Talent Projects of the Third Xiangya Hospital of Central South
University (no. JY201508) and The Key Projects of Postgraduate
Independent Exploration and Innovation of Central South University
(2018zzts050).
Availability of data and materials
The bioinformatics data analysis and figures used in
our study can be obtained from the following websites: GEPIA
database (http://gepia.cancer-pku.cn/); COSMIC
database (http://cancer.sanger.ac.uk/cosmic/); The NCBI gene
browser (https://www.ncbi.nlm.nih.gov/gene/); JASPAR database
(http://jaspar.genereg.net/). Genecards
database (https://www.genecards.org/); The
Human Protein Atlas (https://www.proteinatlas.org/). All the data and
figures from these websites above are open and there are no
copyright disputes. The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
Article checking and project design were carried out
by GH and XL; the experiment design, the writing and the cell
experiments were performed by NL and DL; the tissue experiments and
the animal experiments were performed by YD and CS; the figure
editing, the statistical analysis and the specimen collection was
carried out by CY and CL. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and patient consent for
participation
Signed consent forms were provided by each patient,
and the study was approved by The Institute Research Medical Ethics
Committee of Central South University (Changsha, Hunan). All animal
protocols in this study were approved by the Animal Ethics
Committee of Central South University (Changsha, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wezensky SJ, Hanks TS, Wilkison MJ, Ammons
MC, Siemsen DW and Gauss KA: Modulation of PLAGL2 transactivation
by positive cofactor 2 (PC2), a component of the ARC/mediator
complex. Gene. 452:22–34. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kas K, Voz ML, Hensen K, Meyen E and Van
de Ven WJ: Transcriptional activation capacity of the novel PLAG
family of zinc finger proteins. J Biol Chem. 273:23026–23032. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Furukawa T, Adachi Y, Fujisawa J, Kambe T,
Yamaguchi-Iwai Y, Sasaki R, Kuwahara J, Ikehara S, Tokunaga R and
Taketani S: Involvement of PLAGL2 in activation of iron deficient-
and hypoxia-induced gene expression in mouse cell lines. Oncogene.
20:4718–4727. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soto A, Arce A, M KK and Vera JH: Effect
of the cation and the anion of an electrolyte on the solubility of
DL-aminobutyric acid in aqueous solutions: Measurement and
modelling. Biophys Chem. 73:77–83. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Landrette SF, Kuo YH, Hensen K, Barjesteh
van Waalwijk van Doorn-Khosrovani S, Perrat PN, Van de Ven WJ,
Delwel R and Castilla LH: Plag1 and Plagl2 are oncogenes that
induce acute myeloid leukemia in cooperation with Cbfb-MYH11.
Blood. 105:2900–2907. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang YS, Yang MC and Weissler JC:
Pleiomorphic adenoma gene-like 2 expression is associated with the
development of lung adenocarcinoma and emphysema. Lung Cancer.
74:12–24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng H, Ying H, Wiedemeyer R, Yan H,
Quayle SN, Ivanova EV, Paik JH, Zhang H, Xiao Y, Perry SR, et al:
PLAGL2 regulates Wnt signaling to impede differentiation in neural
stem cells and gliomas. Cancer Cell. 17:497–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo J, Wang M, Wang Z and Liu X:
Overexpression of pleomorphic adenoma gene-like 2 is a novel poor
prognostic marker of prostate cancer. PLoS One. 11:e01586672016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu B, Lu C, Song YX, Gao P, Sun JX, Chen
XW, Wang MX, Dong YL, Xu HM and Wang ZN: The role of pleomorphic
adenoma gene-like 2 in gastrointestinal cancer development,
progression, and prognosis. Int J Clin Exp Pathol. 7:3089–3100.
2014.PubMed/NCBI
|
|
12
|
Su C, Li D, Li N, Du Y, Yang C, Bai Y, Lin
C, Li X and Zhang Y: Studying the mechanism of PLAGL2
overexpression and its carcinogenic characteristics based on
3′-untranslated region in colorectal cancer. Int J Oncol. 2018.
View Article : Google Scholar
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schwitalla S, Fingerle AA, Cammareri P,
Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ,
Moreaux G, et al: Intestinal tumorigenesis initiated by
dedifferentiation and acquisition of stem-cell-like properties.
Cell. 152:25–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Daughaday WH: The possible
autocrine/paracrine and endocrine roles of insulin-like growth
factors of human tumors. Endocrinology. 127:1–4. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hensen K, Van Valckenborgh IC, Kas K, Van
de Ven WJ and Voz ML: The tumorigenic diversity of the three PLAG
family members is associated with different DNA binding capacities.
Cancer Res. 62:1510–1517. 2002.PubMed/NCBI
|
|
18
|
Kas K, Voz ML, Roijer E, Aström AK, Meyen
E, Stenman G and Van de Ven WJ: Promoter swapping between the genes
for a novel zinc finger protein and beta-catenin in pleiomorphic
adenomas with t(3;8)(p21;q12) translocations. Nat Genet.
15:170–174. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Forbes SA, Beare D, Gunasekaran P, Leung
K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et
al: COSMIC: Exploring the world's knowledge of somatic mutations in
human cancer. Nucleic Acids Res. 43:D805–D811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Forbes SA, Beare D, Bindal N, Bamford S,
Ward S, Cole CG, Jia M, Kok C, Boutselakis H, De T, et al: COSMIC:
High-resolution cancer genetics using the catalogue of somatic
mutations in cancer. Curr Protoc Hum Genet. 91:1–10. 2016.
|
|
21
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45:D777–D783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Batlle E, Henderson JT, Beghtel H, van den
Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering
M, Pawson T and Clevers H: Beta-catenin and TCF mediate cell
positioning in the intestinal epithelium by controlling the
expression of EphB/ephrinB. Cell. 111:251–263. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Anderson CB, Neufeld KL and White RL:
Subcellular distribution of Wnt pathway proteins in normal and
neoplastic colon. Proc Nati Acad Sci USA. 99:8683–8688. 2002.
View Article : Google Scholar
|
|
24
|
Munemitsu S, Albert I, Souza B, Rubinfeld
B and Polakis P: Regulation of intracellular beta-catenin levels by
the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc
Nati Acad Sci USA. 92:3046–3050. 1995. View Article : Google Scholar
|
|
25
|
Rosin-Arbesfeld R, Townsley F and Bienz M:
The APC tumour suppressor has a nuclear export function. Nature.
406:1009–1012. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kolligs FT, Bommer G and Goke B:
Wnt/beta-catenin/tcf signaling: A critical pathway in
gastrointestinal tumorigenesis. Digestion. 66:131–144. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van de Wetering M, Sancho E, Verweij C, de
Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D,
Haramis AP, et al: The beta-catenin/TCF-4 complex imposes a crypt
progenitor phenotype on colorectal cancer cells. Cell. 111:241–250.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sansom OJ, Meniel VS, Muncan V, Phesse TJ,
Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H and Clarke AR:
Myc deletion rescues Apc deficiency in the small intestine. Nature.
446:676–679. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, D'Amico M, Pestell R and Ben-Ze'ev A: The cyclin D1
gene is a target of the beta-catenin/LEF-1 pathway. Proc Nati Acad
S USA. 96:5522–5527. 1999. View Article : Google Scholar
|
|
30
|
Kawasaki H, Toyoda M, Shinohara H, Okuda
J, Watanabe I, Yamamoto T, Tanaka K, Tenjo T and Tanigawa N:
Expression of survivin correlates with apoptosis, proliferation,
and angiogenesis during human colorectal tumorigenesis. Cancer.
91:2026–2032. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wolfe SA, Nekludova L and Pabo CO: DNA
recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys
Biomol Struct. 29:183–212. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Van Dyck F, Delvaux EL, Van de Ven WJ and
Chavez MV: Repression of the transactivating capacity of the
oncoprotein PLAG1 by SUMOylation. J Biol Chem. 279:36121–36131.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Voz ML, Agten NS, Van de Ven WJ and Kas K:
PLAG1, the main translocation target in pleomorphic adenoma of the
salivary glands, is a positive regulator of IGF-II. Cancer Res.
60:106–113. 2000.PubMed/NCBI
|
|
34
|
Strubberg AM, Veronese Paniagua DA, Zhao
T, Dublin L, Pritchard T, Bayguinov PO, Fitzpatrick JAJ and Madison
BB: The zinc finger transcription factor PLAGL2 enhances stem cell
fate and activates expression of ASCL2 in intestinal epithelial
cells. Stem Cell Reports. 11:410–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Van Dyck F, Declercq J, Braem CV and Van
de Ven WJ: PLAG1, the prototype of the PLAG gene family:
Versatility in tumour development (review). Int J Oncol.
30:765–774. 2007.PubMed/NCBI
|
|
36
|
Guo Y, Yang MC, Weissler JC and Yang YS:
Modulation of PLAGL2 transactivation activity by Ubc9 co-activation
not SUMOylation. Biochem Biophys Res Commun. 374:570–575. 2008.
View Article : Google Scholar : PubMed/NCBI
|