Introduction
It is known that esophageal carcinoma is one of the
most commonly diagnosed digestive carcinomas, with ~455,800 new
cases and 400,200 cancer-related deaths worldwide in 2012 (1). However, the incidence of esophageal
carcinoma varies widely in different regions. According to
statistics, there are more esophageal cancer patients with squamous
cell carcinoma as the predominant form in Turkey, northern and
central China, Kazakhstan, and northeastern Iran, a geographical
region that is collectively known as the Asian Belt (2). In recent years, considerable
improvement has been achieved in treatment strategies, particularly
for the use of irradiation, which is an essential therapy among
therapies for patients with esophageal cancer. However, the local
recurrence rate after chemoradiotherapy remains high at ~25–40%,
and the overall 5-year survival rate is only ~20% among patients
with esophageal cancer (3). Since
tumors recur and the prognosis is still unsatisfactory after
irradiation, the study of the mechanism of radioresistance of
esophageal squamous cell carcinoma (ESCC) should be encouraged to
develop new therapeutic targets as assistance for radiotherapy
(RT).
The protein named high mobility group box 1 (HMGB1)
is a conserved and ubiquitous non-histone chromosomal protein that
functions as a DNA chaperone in the processes of transcription,
replication, recombination and repair (4,5). The
ability of HMGB1, which is located in the nucleus, to bind to small
recesses in DNA promotes the binding of p53 with DNA at its cognate
binding site in chromatin. Furthermore, HMGB1 is involved in DNA
damage repair induced by various factors, particularly physical
stimulations and chemical elements, and plays a decisive role in
that process (6,7).
According to previous studies, HMGB1 was revealed to
be strongly associated with the occurrence and progression of
tumors, and its expression level has been connected with the
radiosensitivity of various types of cancers (8–11),
however, the underlying mechanism is unclear. In the present study,
the expression level of HMGB1 was detected in both esophageal
cancer tissues and cell lines. Based on the results of the HMGB1
expression level detection, we designed the following in
vitro and in vivo studies to establish the impact of
suppressing HMGB1 expression in human esophageal squamous
carcinoma. In particular, the influence of HMGB1 on the
radiosensitivity of the ECA109 and TE13 cell lines in vitro
was thoroughly studied.
Materials and methods
Immunohistochemical (IHC)
staining
From January 2008 to December 2013, a total of 77
endoscopic biopsy specimens from patients who were diagnosed with
esophageal squamous carcinoma and received chemoradiotherapy (CRT)
were obtained at The Fourth Hospital of Hebei Medical University
(Hebei, China). We acquired the approval from patients and the
Ethics Committee of the Fourth Hospital of Hebei Medical University
for the usage of the specimens for research. We sectioned
4-µm-thick slides from paraffin blocks, deparaffinized the slides
in alcohol solutions with gradient concentrations, rehydrated the
slides with distilled water, and then soaked the slides with
citrate buffer and boiled them for 3 min for heat-induced antigen
retrieval. After blocking the endogenous peroxidases with hydrogen
peroxide solution for 15 min, normal goat serum (5%) was used to
block non-specific antibody binding for 30 min, and then the
sections were incubated with anti-HMGB1 monoclonal antibody
(dilution 1:400; cat. no. ab79823; Abcam, Cambridge, MA, USA) at
4°C overnight. Then, goat anti-rabbit polyclonal antibody (dilution
1:100; cat. no. SP-9000; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China) and horseradish
peroxidase-conjugated streptavidin working solution were each used
to cover the histological sections at 37°C for 30 min successively.
Subsequently, the DAB substrate-chromogen solution was applied to
the sample to produce a color reaction. According to the scoring
standard of previous studies (12,13), 2
pathologists who were blinded to the clinical parameters of the
patients interpreted the immunohistochemical results for the final
analysis.
Cell line culture and X-ray
irradiation
TE13, KYSE180, KYSE30, YES2 and ECA109 human
esophageal cancer cell lines were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with the
addition of 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo
Fisher Scientific, Inc.). The TE13 cell line is same with TE2, TE3,
TE7 and TE12 (14), and it has been
authenticated by STR profiling.
All cells were maintained in Thermo Fisher
Scientific, Inc., CO2-incubators with the temperature
set at 37°C and the concentration of carbon dioxide set to 5%.
Esophageal tumor cells were irradiated by a 6-MV
Siemens linear accelerator (Siemens, Buffalo Grove, IL, USA) with a
source-skin distance (SSD) of 100 cm and dose rate maintained at 5
Gy/min. Then, the cells were collected at certain time-points for
further study.
Cell transfection
Cell transfection was performed with human HMGB1
small interfering RNA (shRNA) and negative control shRNA (PPL,
Genebio Technology, Inc., Nanjing, China) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) under the guidance of
the manufacturer's instructions. The concentrated virus solution
and esophageal cancer cells were co-cultured, and the fluorescence
of the cells was observed by optical microscopy to confirm
successful transfection. The sequences were as follows: HMGB1-shRNA
sense, 5′-GGGAGGAGCAUAAGAAGAATT-3′ and antisense,
5′-UUCUUCUUAUGCUCCUCCCTT-3′; NC shRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Quantitative real-time reverse
transcription polymerase chain reaction
RNA was extracted from esophageal tumor cells with
TRIzol reagent (Takara Bio, Inc., Shiga, Japan), and then
reversed-transcribed to cDNA with a RevertAid First Strand cDNA
Synthesis Kit (Thermo Fisher Scientific, Inc.). Then, RT-PCR was
applied to analyze the expression of the HMGB1 gene using Platinum
SYBR-Green qPCR SuperMix-UDG (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. First, the
synthetic cDNA was denatured at 94°C for 30 sec, then used for the
following cycle: Denaturing cDNA at 94°C for 5 sec, annealing cDNA
at 56°C for 15 sec and extending cDNA at 72°C for 10 sec, 40 cycles
in total. The GAPDH gene was used for normalization of RT-qPCR
data. The 2−ΔΔCT method was applied to analyze HMGB1
gene expression (15).
Western blot analysis
We conducted western blot analysis following a
method previously described (16).
The relevant antibodies were anti-HMGB1 (dilution 1:10,000; cat.
no. ab79823), anti-γH2AX (dilution 1:1,000; cat. no. ab26350;
Abcam), anti-MMP-2 (dilution 1:1,000; cat. no. 10373-2-AP),
anti-MMP-9 (dilution 1:1,000; cat. no. 10375-2-AP), anti-p16 (cat.
no. 10883-1-AP), anti-caspase-9 (dilution 1:1,000; cat. no.
10380-1-AP), anti-Bcl-2 (dilution 1:2,000; cat. no. 12789-1-AP),
anti-CDK4 (dilution 1:2,000; cat. no. 11026-1-AP), anti-cyclin D1
(dilution 1:5,000; cat. no. 60186-1-lg), anti-Bax (dilution
1:5,000; cat. no. 50599-2-lg; ProteinTech Group, Inc., Chicago, IL,
USA) and anti-β-actin (dilution 1:10,000; cat. no. AP0060; Bioworld
Technology, Inc., St. Louis Park, MN, USA). The blotted protein
bands were revealed by an Odyssey system (LI-COR Biosciences,
Lincoln, NE, USA). Finally, the intensity of the protein bands was
assessed with ImageJ (National Institutes of Health, Bethesda, MD,
USA) and the ratio of the protein to corresponding β-actin was
calculated to reflect the changes in expression levels. All western
blot analyses were performed independently at least 3 times.
Cell proliferation assay
After preparing a cell suspension with a
concentration of up to 5×104 cells/ml, 100 µl/well was
seeded into 96-well plates with 5 duplicates for each sample. At
certain time-points after irradiation, the viability of the cells
under the different treatments was determined by incubation with 10
µl Cell Counting Kit-8 (CCK-8) [MedChem Express (MCE) Princeton,
NJ, USA] for 2 h. Then, the cell content was calculated by
detecting the absorbance of each well at 450 nm via a Multiskan
microplate. The experiment was repeated at least 3 times.
Clonogenic assay
Esophageal cancer cells irradiated with a set of
graded doses were cultured in 6-well culture plates for 15 days in
triplicate. After the formation of cell clones, crystal violet
(0.6%) was applied to stain the cells for 15 min. Colonies
containing 50 cells or more were taken into account. Basic data was
input into GraphPad Prism version 5.0 (GraphPad Software, Inc., La
Jolla, CA, USA), and the biological parameters of radiation and
cell survival curves were obtained.
Wound healing assay
Esophageal tumor cells were plated in 6-well plates.
When the cells grew to confluence >80%, a straight cell-free
zone was created by a 200-µl pipette tip/well. Then the scratched
areas were marked as 0 h and photographed using computer-assisted
fluorescent microscopy. Cells were cultured in 2 ml of media
without FBS, then images were captured to observe the scratched
areas at 16 and 32 h separately. The migration of cells with
various treatments was presented as the percentage of the cell-free
zone at the aforementioned time-points compared to 0 h and related
to the control group.
Transwell assay
The invasion ability of tumor cells was detected
using Transwells coated with Matrigel (both from Corning Inc.,
Corning, NY, USA) before use. Cells were diluted to
5×105/ml with serum-free medium and 200 µl was
transferred to the top chambers. Medium containing 10% FBS was
placed in the lower chamber. The cells on the upper side of the
chamber were slightly wiped with a cotton swab after penetrating
for 24 h. Concurrently, the cells passing through the filter were
fixed with formaldehyde and stained with crystal violet for 10 min.
Then, the number of stained cells was photographed and counted with
a fluorescent microscope from 5 randomly selected fields
(magnification, ×200).
Cell cycle and apoptosis
Cell cycle analysis used esophageal tumor cells that
were harvested and fixed with 70% precooled ethanol and maintained
at 4°C. The following day, the cells were stained with propidium
iodide probe solution (BD Biosciences, San Jose, CA, USA) in the
dark for 30 min. Cell apoptosis was detected with an Annexin V-FITC
apoptosis detection kit (BD Biosciences). In accordance with the
kit's instructions, cells were collected and stained with propidium
iodide (PI) and Annexin V-FITC. Flow cytometry was applied to
detect the cell cycle distribution and apoptosis rates,
encompassing both the early apoptosis (Annexin
V+/PI−) and late apoptosis/necrosis (Annexin
V+/PI+) phases.
Xenograft tumor models
Twelve 6-week-old male BALB/c nude mice (weight,
19±1 g) were obtained from Beijing Vital River Laboratory Animal
Technology Co., Ltd. (Beijing, China), and maintained in specific
pathogen-free conditions with controlled temperature (23±2°C),
humidity (55±5%) and light (12 h light/dark cycle). The mice were
provided with sterile food and water ad libitum. All
experiments with animals were carried out with the approval of the
Animal Care and Use Committee of the Fourth Hospital of Hebei
Medical University. A total of 4×106 HMGB1-shRNA or NC
tumor cells were injected into the left hind paw of the mice. Three
weeks after injection, irradiation (5 Gy) was performed each day
for 3 days with a collimator container to protect the normal
tissue. Calipers was used to assess the tumor diameter (mm) twice a
week, and the tumor volume (TV) was calculated by the following
formula: TV = AB2/2, where A represents the long
diameter and B represents the short diameter.
Statistical analysis
The data collected in the present study were
analyzed using SPSS software package version 21 (SPSS, Inc.,
Chicago, IL, USA) and recorded as the mean ± standard deviation
(SD). The endpoint of progression-free survival (PFS) was assessed
from the beginning of CRT to progression, death or the date of the
last follow-up. The survival analysis was carried out by
Kaplan-Meier method with log-rank test and the χ2 or
Fisher's exact tests was used to analyze the association between
clinical parameters and HMGB1 expression. The comparisons of data
between different groups was performed using ANOVA with least
significant difference (LSD) test. If the P-value of a two-sided
statistic test was <0.05 or <0.01, the result was considered
to be statistically significant.
Results
Overexpression of HMGB1 indicates an
adverse prognosis in esophageal cancer
The relationship between the expression level of
HMGB1 and the clinical parameters of esophageal cancer patients was
analyzed. Table I revealed that the
associations between the expression level of HMGB1 and sex, age,
lesion location, or N classification (P>0.05) had no
statistical significance. Conversely, the accumulation of HMGB1 in
esophageal cancer was significantly associated with gross tumor
volume (GTV) (P=0.014), TNM stage (7th AJCC) (P<0.001), T
classification (P=0.003), relapse (P=0.003) and distant metastasis
(P<0.001). Immunohistochemical staining revealed that the
expression of HMGB1 in ESCC samples (Fig. 1Ab, c and d) was higher in comparison
with adjacent tissues with no tumor complication (Fig. 1Aa). After the generation of survival
curves, the HMGB1 expression level in ESCC was significantly
associated with overall survival (P=0.007; Fig. 1B) and progression-free survival
(P=0.008; Fig. 1B), which were both
significantly shorter in patients with positive expression of HMGB1
compared with patients with negative expression of HMGB1 according
to the results of the log-rank test. In summary, statistical
analysis revealed the critical role that HMGB1 played in the
progression of esophageal cancer, and high levels of HMGB1
indicated poor clinical prognosis.
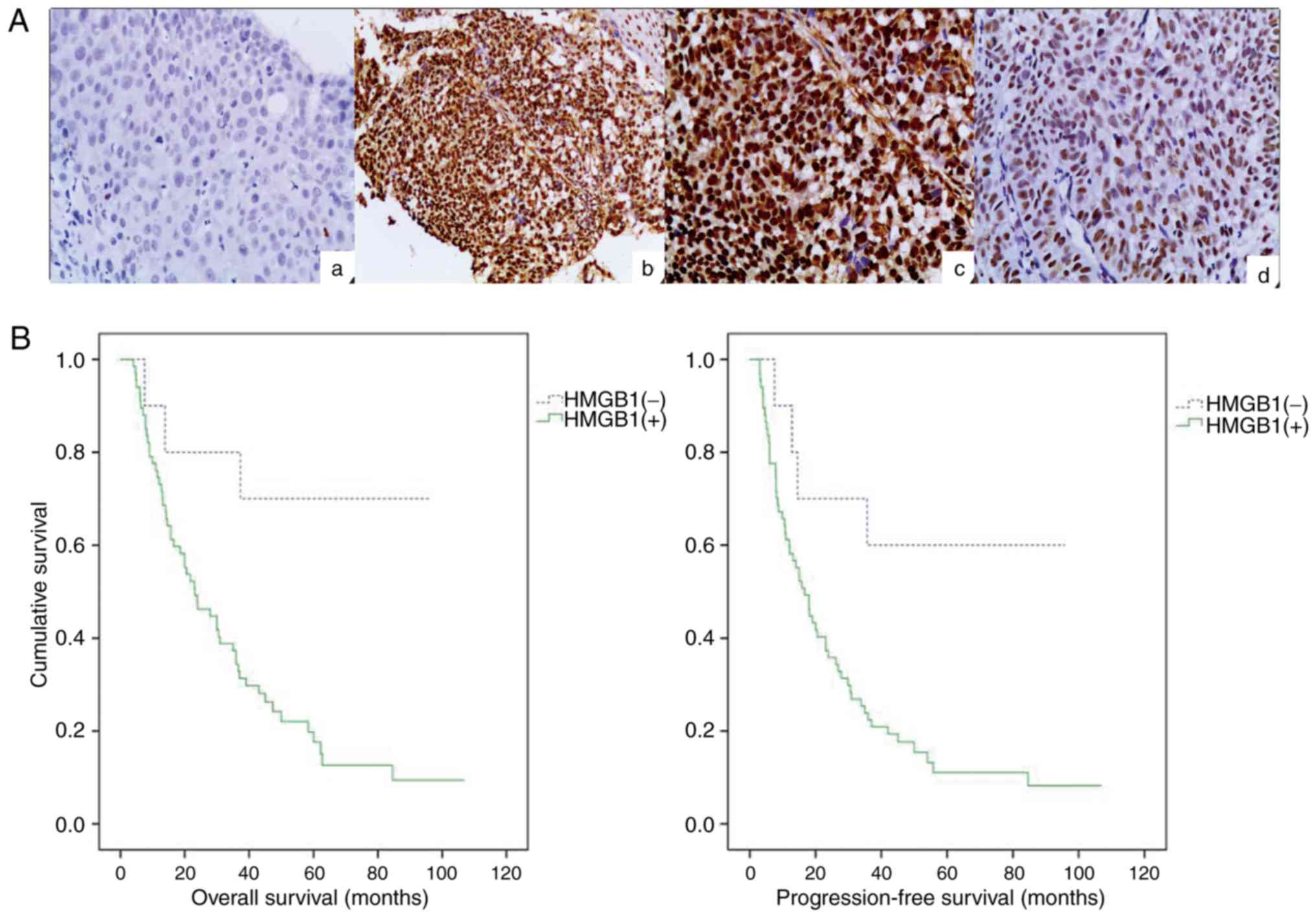 | Figure 1.High expression of HMGB1 in human
ESCC biopsy specimens and downregulation of HMGB1 by transfection
of shRNA. (A) HMGB1 staining was predominantly localized in the
nuclei. In comparison with adjacent tissues with no tumor
complication (a, negative, original magnification, ×200), the
expression of HMGB1 in ESCC samples was higher (b and c, positive,
magnification, ×100 and ×200, respectively; d, weakly positive,
magnification, ×200). (B) Overall survival and progression-free
survival among HMGB1-negative and HMGB1-positive groups revealed
statistical significance differences according to Kaplan-Meier
survival analysis (P<0.05, log-rank test). (C) The
comparison of the HMGB1 protein expression levels in 5 esophageal
cancer cell lines was performed using western blot analysis. (D)
Similarly, the suppression of HMGB1 protein expression was observed
in the HMGB1-shRNA group by western blot analysis. (E) The mRNA
expression of HMGB1 in the HMGB1-shRNA group were significantly
downregulated after transfection in both ECA109 and TE13 cells
according to the results of the RT-qPCR analysis.
**P<0.01. HMGB1, high mobility group box 1; ESCC,
esophageal squamous cell carcinoma. |
 | Table I.Association between the clinical
characteristics and expression of HMGB1 protein. |
Table I.
Association between the clinical
characteristics and expression of HMGB1 protein.
|
|
| HMGB1 |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | n | Negative | Positive | P-value |
|---|
| Age (years) |
|
<65 | 34 | 5 | 29 | 0.742 |
|
≥65 | 43 | 5 | 38 |
|
| Sex |
|
Male | 49 | 7 | 42 | 0.739 |
|
Female | 28 | 3 | 25 |
|
| Gross tumor volume
(GTV) |
|
<30 | 27 | 8 | 24 | 0.014 |
|
≥30 | 50 | 2 | 43 |
|
| Lesion
location |
|
Neck/upper | 28 | 4 | 24 | 0.798 |
|
Middle/lower | 49 | 6 | 43 |
|
| TNM
stagea |
|
I–II | 34 | 10 | 24 | <0.001 |
|
III–IV | 43 | 0 | 43 |
|
| T
classification |
|
T1-T2 | 27 | 8 | 19 | 0.003 |
|
T3-T4 | 50 | 2 | 48 |
|
| N
classification |
| N0 | 31 | 5 | 26 | 0.766 |
|
N1-N3 | 46 | 5 | 41 |
|
| Distant
metastasis |
| M0 | 16 | 7 | 9 | <0.001 |
| M1 | 61 | 3 | 58 |
|
| Relapse |
|
Negative | 15 | 6 | 9 | 0.003 |
|
Positive | 62 | 4 | 58 |
|
HMGB1 is downregulated in HMGB1-shRNA
cell lines
The expression of HMGB1 in the TE13, KYSE180,
KYSE30, YES2 and ECA109 esophageal squamous carcinoma cell lines
was determined in vitro by western blot analysis, and the
results revealed higher expression levels of HMGB1 in the ECA109
and TE13 cell lines compared with the other cell lines (Fig. 1C). Thus, ECA109 and TE13 cells were
chosen for further study, and the expression of HMGB1 after
transfection with an shRNA targeting HMGB1 (HMGB1-shRNA) was
determined through western blot and RT-qPCR analyses. Finally, no
difference was revealed in HMGB1 expression between ECA109-NC or
TE13-NC cells and their corresponding parental cells. However,
HMGB1 expression in HMGB1-shRNA cells relative to their
corresponding parental cells was significantly reduced (Fig. 1D and E). These data indicated that
HMGB1-shRNA significantly downregulated HMGB1 in esophageal cancer
ECA109 and TE13 cells.
Transfection with HMGB1-shRNA inhibits
proliferation and increases the radiosensitivity of esophageal
tumor cells in vitro and in vivo
To investigate the biological function of HMGB1 on
the cell viability of esophageal cancer cells, cells under
different conditions were collected at 24, 48, 72 and 96 h after
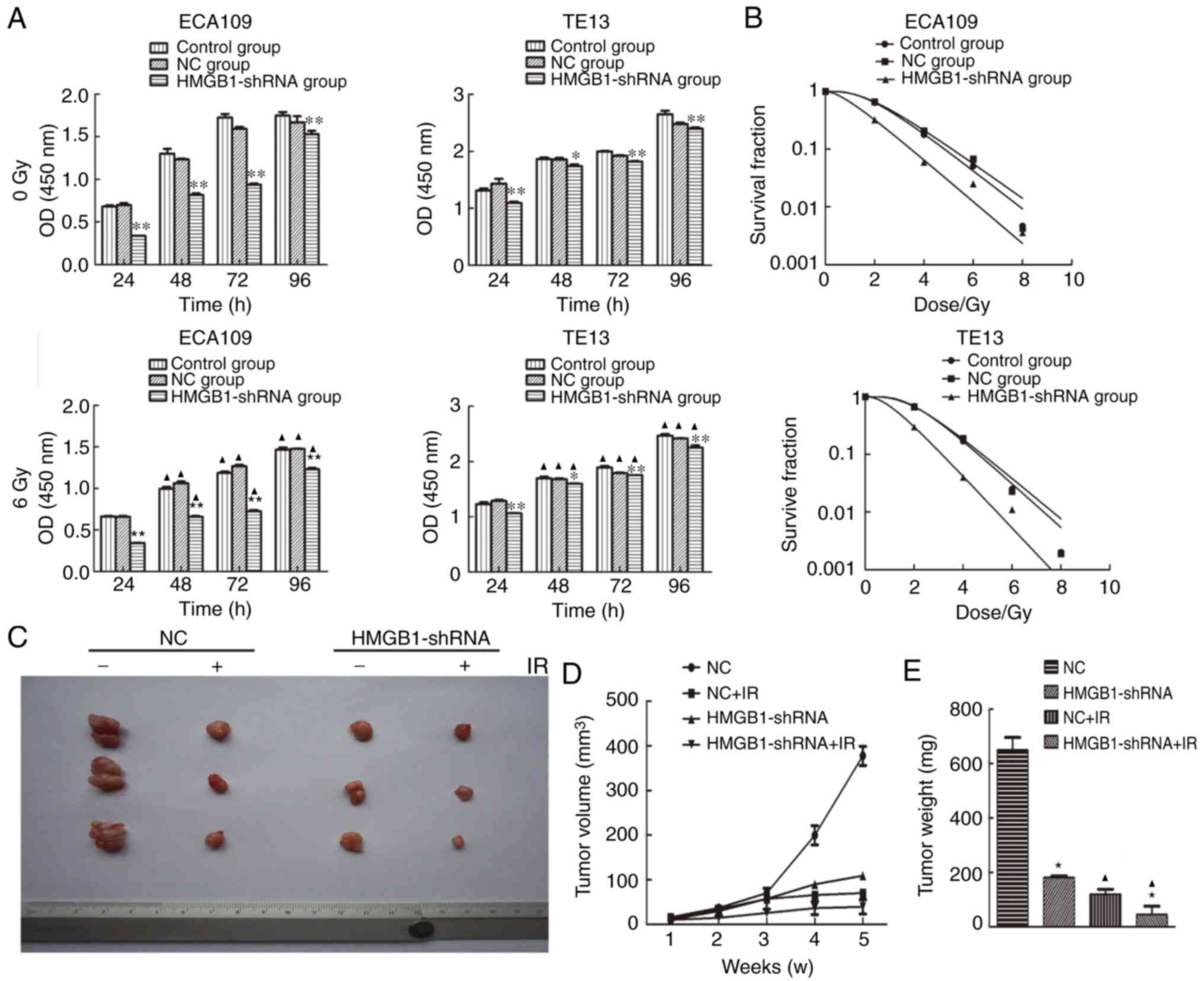
transfection and irradiation. The results of the CCK-8 assay
indicated that the proliferation rates of the HMGB1-shRNA groups
were significantly lower at each time-point than those of the
corresponding control and NC groups with or without irradiation
(Fig. 2A). In addition, a colony
formation assay was applied to detect the effect of the alteration
of HMGB1 expression on the radiosensitivity of ECA109 and TE13
cells. After the cell survival curves were plotted based on the
formed clones, and the radiobiological parameters of each group
were input into statistical analysis, it was revealed that the
radiosensitivity of the HMGB1-shRNA group was higher compared with
those of the control and NC groups (Fig. 2B). To explore the function of HMGB1
in tumor formation in vivo, the HMGB1-shRNA and NC cells of
the ECA109 cell line were implanted into nude mice. The results
revealed that the tumor volume (Fig. 2C
and D) and weight (Fig. 2E) in
the HMGB1-shRNA group were notably smaller than those of the NC
group before and after irradiation (P<0.05). These findings
indicated that transfection with HMGB1-shRNA induced proliferation
inhibition with or without irradiation and increased the
radiosensitivity of esophageal cancer cells both in vitro
and in vivo.
HMGB1 plays an essential role in the
phosphorylation of H2AX after irradiation
In the present study, whether HMGB1 was involved in
radiation-induced DNA damage repair was assessed by analyzing the
variations in HMGB1 and γH2AX expression after irradiation. The
results of the western blot analysis revealed that ionizing
radiation induced the protein expression of γH2AX and HMGB1 in a
dose-dependent manner (Fig. 2F). In
addition, the changes in expression of these 2 proteins were
consistent over the course of time after irradiation (Fig. 2G), and both of them were highest at
1 and 2 h after exposure to 6 Gy irradiation, and then their
expression gradually reduced. These findings prompted us to suppose
that HMGB1 is associated with the generation of γH2AX. Then, the
level of phosphorylated H2AX was detected to evaluate the influence
of HMGB1 deficiency with and without irradiation. The results
revealed that knockdown of HMGB1 protein in ECA109 and TE13 cells
inhibited accumulation of γH2AX following irradiation treatment,
while there were no notable changes in γH2AX concentrations in the
non-irradiated groups (Fig. 2H).
The western blot analysis demonstrated that HMGB1 deficiency
inhibited the phosphorylation of H2AX induced by irradiation.
HMGB1 silencing inhibits cell
migration and invasion
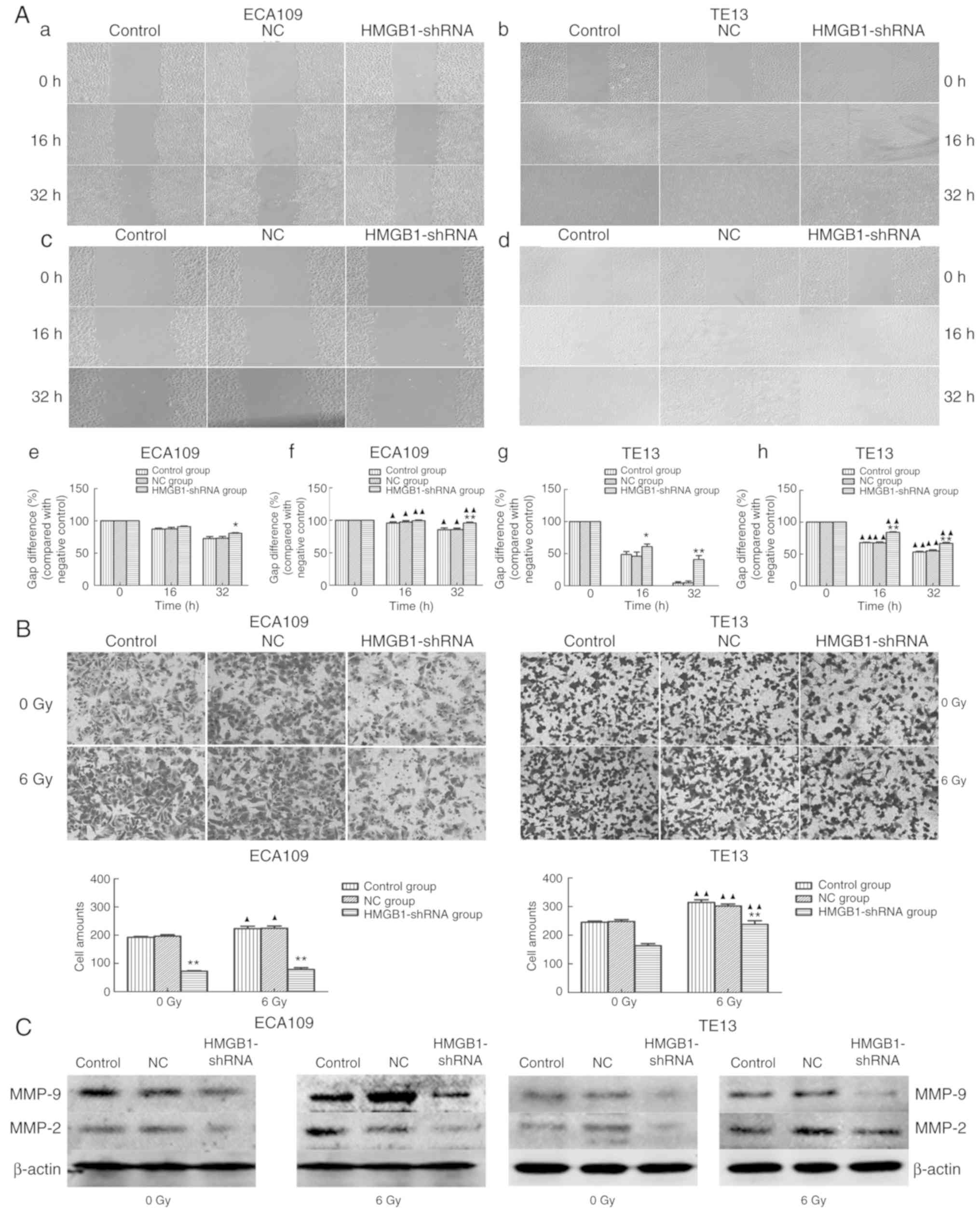
To observe the migration and invasion abilities of
esophageal carcinoma cells, a wound-healing experiment and
Transwell assay, respectively, were applied to ECA109 and TE13
cells. The results revealed that blocking the expression of HMGB1
significantly decreased the migration activity of ECA109 and TE13
cell lines both with (Fig. 3Ac, d, f
and h) and without irradiation (Fig. 3Aa, b, e and g). In addition, the
invasive ability of esophageal carcinoma cells transfected with
HMGB1-shRNA was significantly inhibited compared with those of the
control and NC groups (P<0.05) both with and without irradiation
(Fig. 3B). These results indicated
that HMGB1 deficiency decreased the migration and invasion
abilities of esophageal tumor cells. Furthermore, to analyze the
potential mechanism underlying the HMGB1-shRNA-mediated blockade of
invasion and migration, MMP-2 and MMP-9 protein expression was
examined, and the expression levels of both were reduced in the
HMGB1-shRNA group before and after irradiation in comparison to
those of the control and NC groups (Fig. 3C).
Suppression of HMGB1 increases the
apoptosis rates of esophageal carcinoma cells after irradiation in
vitro
The apoptosis rate of the HMGB1-shRNA group was
higher than that of the NC group before irradiation as evidenced by
Annexin V and PI staining (Fig.
4A). In addition, the HMGB1-shRNA group exhibited more
apoptosis after irradiation than the NC group in ECA109 and TE13
cells (Fig. 4A). To thoroughly
assess the mechanism, the expression of apoptosis-related proteins
was detected, including Bcl-2, Bax and caspase-9, before and after
irradiation by western blot analysis. The results revealed
decreased expression of Bcl-2 and increased Bax and caspase-9
levels in the HMGB1-shRNA group with and without irradiation
compared with the NC group (Fig.
4B). These results revealed that downregulation of HMGB1
increased the apoptosis of esophageal carcinoma cells by affecting
the expression levels of Bcl-2, Bax and caspase-9.
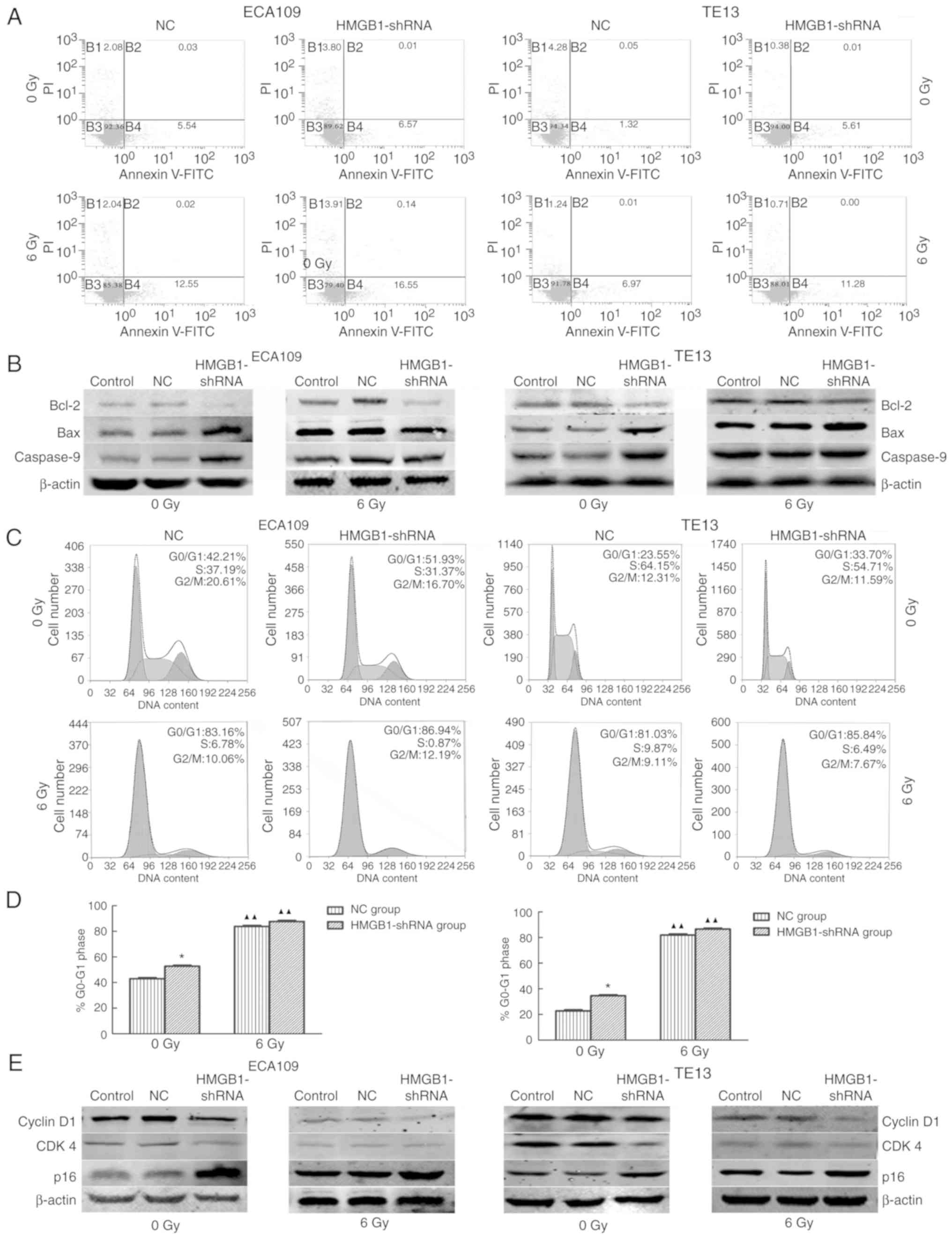 | Figure 4.Suppression of HMGB1 increases the
apoptosis rates of esophageal carcinoma cells after IR in
vitro and affects the generation of apoptosis-related proteins.
(A) The apoptotic rate was calculated as the sum of B2 and B4. As
the results revealed, compared with the NC group, silencing of
HMGB1 sensitized ECA109 and TE13 cells to apoptosis both with 6 Gy
radiation (A, lower images) and without (A, upper images). (B)
After transfection with HMGB1-shRNA, the expression of Bcl-2 was
attenuated, while the expression of Bax and caspase-9 was increased
in ECA109 and TE13 cells. (C) HMGB1-shRNA and irradiation induced
G0/G1 arrest in ESCC cells. (D) The results
of flow cytometry revealed that the percentages of
G0/G1-phase cells in the HMGB1-shRNA groups
were significantly higher compared to those of the NC groups before
IR, and the HMGB1-shRNA group percentages significantly increased
after irradiation. (E) In the HMGB1-shRNA group, the expression of
cyclin D1 and CDK4 was attenuated, while the expression of p16 was
increased. Additionally, irradiation inhibited cyclin D1 and CDK4
expression in esophageal tumor cells. A comparison with the NC
group is symbolized by an asterisk *P<0.05; and a
comparison with the corresponding non-irradiated group is
symbolized by triangle ▲▲P<0.01. HMGB1, high
mobility group box 1; IR, irradiation. |
Knockdown of HMGB1 combined with
irradiation induces G0/G1 arrest in ESCC
cells
According to flow cytometric results, silencing of
HMGB1 caused G0/G1 arrest of esophageal
carcinoma cells with and without irradiation (Fig. 4C and D). A western blot assay that
explored the expression levels of proteins related to the cell
cycle revealed downregulation of cyclin D1 and CDK4 and
upregulation of p16 in the HMGB1-shRNA groups with and without
irradiation compared with the levels in the NC group (Fig. 4E). The cyclin D1 expression was
lower in the irradiated group than the non-irradiated group.
Furthermore, cyclin D1 expression was significantly decreased in
the HMGB1-shRNA group with irradiation treatment. These data
demonstrated that suppressing the expression of HMGB1 arrested
esophageal tumor cells in the G0/G1 phase by
upregulating the expression of p16 and decreasing the expression
cyclin D1 and CDK4.
Discussion
Although combined chemoradiotherapy has improved the
prognosis for esophageal cancer, the prognosis remains poor. Less
than half of esophageal cancer patients survive 2 years without
recurrence after chemoradiation therapy with or without surgery
(17). One critical reason is that
tumors of the esophagus are found too late to perform radical and
curative surgery. In addition, esophageal cancer frequently occurs
with distal metastases, including in the lungs, bones, brain and
liver, when it is diagnosed. For advanced cases, radiotherapy
combined with chemotherapy is the mainstay treatment. Therefore,
the discovery of effective therapeutic targets to enhance
radiosensitivity may improve the survival of ESCC patients.
The overexpression of HMGB1 has been implicated in
multiple cancers, including breast (18), rectal (11), bladder (19) and gallbladder cancer (20), pleural mesothelioma (21), nasopharyngeal carcinoma (22), and esophageal cancer (23). During tumor development and cancer
treatment, diverse roles for HMGB1 have been revealed in previous
studies, including roles in inflammation (24), immune responses (25), angiogenesis (26), DNA damage repair (27), autophagy (28), proliferation, apoptosis, invasion
and metastasis (29,30). Based on the data from our
immunohistochemical staining, the positive rate of HMGB1 expression
was ~87% (67 of 77 cases) in ESCC tissue, and esophageal carcinoma
patients with positive expression of HMGB1 had poorer overall
survival (P=0.007) and progression-free survival (P=0.008) than
patients with negative expression of HMGB1. The statistical
analysis of clinical characteristics indicated that the expression
levels of HMGB1 were related to clinical cancer stage, distant
metastasis and relapse, which indicated that irradiation combined
with a new drug targeting HMGB1 may produce better results.
Therefore, the effect of downregulating HMGB1 expression on the
radiosensitivity of esophageal cancer cells was first explored;
subsequently, the effect of overexpressing the HMGB1 gene will be
determined in future investigations rendering the present study
more convincing.
Previous studies have suggested that HMGB1
participates in DNA damage repair (6,7,31). Its
direct binding sites in DNA lesions allow it to play a role in
double strand break (DSB) repair (7). The realization of the complete DNA
damage response requires the accumulation of γH2AX (32), and H2AX phosphorylation has been
found to be stimulated by HMGB1 release (33). To determine whether HMGB1
specifically regulated the DNA damage repair induced by irradiation
and the radiosensitivity of ESCC cells, the protein levels of γH2AX
were assessed and a colony formation assay was performed with cells
deficient in HMGB1 before and after irradiation. As the radiation
dose increased and time progressed, the γH2AX and HMGB1 protein
expression levels gradually changed in parallel. Furthermore, the
loss of HMGB1 inhibited the synthesis of γH2AX induced by
irradiation, as there was no significant change in the
HMGB1-deficient group without irradiation. This phenomenon
demonstrated that the existence of HMGB1 is necessary for the
generation of phosphorylated H2AX that promotes DNA damage repair.
Similar results were obtained in the colony formation assay;
HMGB1-deficient cells revealed a narrow shoulder in the survival
curves, indicating that the knockdown of HMGB1 enhanced the
radiosensitivity of ESCC cells.
Recent studies have revealed that HMGB1 has the
ability to promote tumor growth by different signaling pathways,
and the activation of the HMGB1/RAGE interaction was revealed to be
correlated with matrix metalloproteinase (MMPs) expression, tumor
proliferation, and migration (29).
In the present study, it was revealed that a deficiency in HMGB1
suppressed the proliferation of ESCC cells both before and after
irradiation in vitro and in vivo, indicating the
significant role that HMGB1 played in the growth of esophageal
cancer. In clinical studies of HMGB1 detection in nasopharyngeal
carcinoma (13), rectal cancer
(11), and our own present study of
esophageal cancer, comparative studies on metastasis in cancer
biopsies revealed that overexpression of HMGB1 may contribute to
metastasis. Then, a wound healing and Transwell assays were carried
out using 2 esophageal cancer cell lines to confirm the clinical
findings aforementioned in vitro. The results demonstrated
that silencing of HMGB1 expression with shRNA decreased both the
migration and invasion abilities of esophageal tumor cells before
and after irradiation, which coincided with the clinical
statistical analysis. Subsequently, in seeking the underlying
molecular mechanisms, the expression of MMPs, that help tumor cells
invade and metastasize by degrading extracellular matrix proteins,
were detected (34). The results
revealed decreased MMP-2 and MMP-9 expression levels after
suppression of HMGB1 expression, corresponding with the reduced
migration and invasion abilities. The aforementioned phenomena
indicated that HMGB1 facilitated the migration and invasion of
esophageal tumor cells by interacting with MMPs. The present
results were similar to previous research in which it was
discovered that HMGB1 increased cell migration through activation
of MMP-9 (35).
It has been reported that the downregulation of
cyclin D1 is a major factor during the initiation phase of G1
arrest induced by irradiation, and D cyclins primarily activate
CDK4 and CDK6, resulting in complex formation that promotes the
cells from the G1 phase into the S phase by sequestering
p21cip1 and p27kip1 away from cyclin E-CDK2
(36). In the present study, the
effects of HMGB1 expression and irradiation on the transition
process of the cell cycle were ascertained as well. Based on data
analysis, the suppression of HMGB1 induced
G0/G1 arrest before irradiation with a
decrease in cyclin D1 and CDK4 levels. Moreover, the
irradiation-induced G0/G1 arrest and decrease
in cyclin D1 and CDK4 were more notable in HMGB1-deficient cells.
The cell cycle analysis and associated-protein evaluation revealed
that the suppression of HMGB1 promoted cell cycle arrest at the
G0/G1 phase by decreasing cyclin D1 and CDK4
expression.
The present study also analyzed the effect of HMGB1
downregulation on cell apoptosis in vitro. In addition,
apoptotic cell percentages in the HMGB1-shRNA groups were
significantly greater than those in corresponding negative control
groups with irradiation, indicating that the deficiency in HMGB1
induced by shRNA was able to promote apoptosis after irradiation.
For the molecular mechanism, we explored the expression of Bcl-2,
Bax and caspase-9, all proteins related to the cell apoptosis
process. As described in a previous study, the protein Bax
increased the formation of oligomers that participate in
apoptogenic molecule releases and initiate intrinsic apoptosis;
conversely, Bcl-2 decreased apoptosis by controlling the generation
of cytochrome c and blocking the oligomerization. Moreover,
the expression of caspase-9 was associated with downstream
cytochrome c-related apoptosis (37,38).
The present study demonstrated that the expression of Bcl-2
decreased and the expression of caspase-9 and Bax increased after
HMGB1-shRNA transfection before and after irradiation. Accordingly,
HMGB1 inhibited the apoptosis of esophageal tumor cells after
irradiation by regulating pro-apoptotic Bcl-2 family members.
In conclusion, it was revealed the HMGB1 expression,
which was higher in ESCC tissue, was negatively related to survival
rates and positively associated with malignancy. Suppression of
HMGB1 significantly increased the radiosensitivity of ESCC cells by
arresting them in the G0/G1 phase and
enhancing apoptosis. Irradiation combined with treatments targeting
HMGB1 may achieve satisfactory therapeutic effects for esophageal
cancer patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (no. 81872456), the Natural
Science Foundation of China of Hebei Province (no. H2017206170),
the Medical Research Institute of Hebei Province (no. 20170154) and
a grant from the Education Department of Hebei Province (no.
CXZZBS2018071).
Availability of data and materials
All data generated or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SZ and XY conceived and designed the study. XZ and
QL performed the experiments in vitro. XZ and NZ constructed
the knock down stable cell lines. XZ, XY and NZ performed the
experiments in vivo. XZ and QL analyzed and interpreted the
data. SZ, XY and XZ drafted the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The approval from patients and the Ethics Committee
of the Fourth Hospital of Hebei Medical University was obtained for
the usage of the specimens for research. All experiments with
animals were carried out with the approval of the Animal Care and
Use Committee of the Fourth Hospital of Hebei Medical
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ishihara R, Yamamoto S, Iishi H, Takeuchi
Y, Sugimoto N, Higashino K, Uedo N, Tatsuta M, Yano M, Imai A, et
al: Factors predictive of tumor recurrence and survival after
initial complete response of esophageal squamous cell carcinoma to
definitive chemoradiotherapy. Int J Radiat Oncol Biol Phys.
76:123–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1 and cancer. Biochim Biophys Acta 1799.
131–140. 2010.
|
|
5
|
Kang R, Zhang Q, Zeh HJ III, Lotze MT and
Tang D: HMGB1 in cancer: Good, bad, or both? Clin Cancer Res.
19:4046–4057. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan F, Gu L, Guo S, Wang C and Li GM:
Evidence for involvement of HMGB1 protein in human DNA mismatch
repair. J Biol Chem. 279:20935–20940. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lange SS, Mitchell DL and Vasquez KM: High
mobility group protein B1 enhances DNA repair and chromatin
modification after DNA damage. Proc Natl Acad Sci USA.
105:10320–10325. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shrivastava S, Mansure JJ, Almajed W, Cury
F, Ferbeyre G, Popovic M, Seuntjens J and Kassouf W: The role of
HMGB1 in radioresistance of bladder cancer. Mol Cancer Ther.
15:471–479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin HJ, Liu HH, Lin CD, Kao MC, Chen YA,
Chiang-Ni C, Jiang ZP, Huang MZ, Lin CJ, Lo UG, et al: Cytolethal
distending toxin enhances radiosensitivity in prostate cancer cells
by regulating autophagy. Front Cell Infect Microbiol. 7:2232017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ke S, Zhou F, Yang H, Wei Y, Gong J, Mei
Z, Wu L, Yu H and Zhou Y: Downregulation of high mobility group box
1 modulates telomere homeostasis and increases the radiosensitivity
of human breast cancer cells. Int J Oncol. 46:1051–1058. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hongo K, Kazama S, Tsuno NH, Ishihara S,
Sunami E, Kitayama J and Watanabe T: Immunohistochemical detection
of high-mobility group box 1 correlates with resistance of
preoperative chemoradiotherapy for lower rectal cancer: A
retrospective study. World J Surg Oncol. 13:72015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Soumaoro LT, Uetake H, Higuchi T, Takagi
Y, Enomoto M and Sugihara K: Cyclooxygenase-2 expression: A
significant prognostic indicator for patients with colorectal
cancer. Clin Cancer Res. 10:8465–8471. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu D, Ding Y, Wang S, Zhang Q and Liu L:
Increased expression of high mobility group box 1 (HMGB1) is
associated with progression and poor prognosis in human
nasopharyngeal carcinoma. J Pathol. 216:167–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boonstra JJ, van der Velden AW, Beerens
EC, van Marion R, Morita-Fujimura Y, Matsui Y, Nishihira T,
Tselepis C, Hainaut P, Lowe AW, et al: Mistaken identity of widely
used esophageal adenocarcinoma cell line TE-7. Cancer Res.
67:7996–8001. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang XX, Ma M, Sang MX, Wang XX, Song H,
Liu ZK and Zhu SC: Radiosensitization of esophageal carcinoma cells
by knockdown of RNF2 expression. Int J Oncol. 48:1985–1996. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y: Epidemiology of esophageal
cancer. World J Gastroenterol. 19:5598–5606. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sohun M and Shen H: The implication and
potential applications of high-mobility group box 1 protein in
breast cancer. Ann Transl Med. 4:2172016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang H, Hu X, Zhang H and Li W:
Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder
cancer via suppressing HMGB1 expression. Radiat Oncol. 12:652017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi Z, Huang Q, Chen J, Yu P, Wang X, Qiu
H, Chen Y and Dong Y: Correlation of HMGB1 expression to
progression and poor prognosis of adenocarcinoma and squamous
cell/adenosquamous carcinoma of gallbladder. Am J Transl Res.
7:2015–2025. 2015.PubMed/NCBI
|
|
21
|
Tabata C, Shibata E, Tabata R, Kanemura S,
Mikami K, Nogi Y, Masachika E, Nishizaki T and Nakano T: Serum
HMGB1 as a prognostic marker for malignant pleural mesothelioma.
BMC Cancer. 13:2052013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng T, Hu M, Wu T, Chen Z, Zhang C, Huang
S and Zhou X: Effects of high-mobility group box 1 knockdown on
proliferation, migration and invasion of the HONE-1 human
nasopharyngeal carcinoma cell line. Mol Med Rep. 12:7531–7537.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzuki Y, Mimura K, Yoshimoto Y, Watanabe
M, Ohkubo Y, Izawa S, Murata K, Fujii H, Nakano T and Kono K:
Immunogenic tumor cell death induced by chemoradiotherapy in
patients with esophageal squamous cell carcinoma. Cancer Res.
72:3967–3976. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gebhardt C, Riehl A, Durchdewald M, Németh
J, Fürstenberger G, Müller-Decker K, Enk A, Arnold B, Bierhaus A,
Nawroth PP, et al: RAGE signaling sustains inflammation and
promotes tumor development. J Exp Med. 205:275–285. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Z, Falo LD Jr and You Z: Knockdown of
HMGB1 in tumor cells attenuates their ability to induce regulatory
T cells and uncovers naturally acquired CD8 T cell-dependent
antitumor immunity. J Immunol. 187:118–125. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van Beijnum JR, Nowak-Sliwinska P, van den
Boezem E, Hautvast P, Buurman WA and Griffioen AW: Tumor
angiogenesis is enforced by autocrine regulation of high-mobility
group box 1. Oncogene. 32:363–374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giavara S, Kosmidou E, Hande MP, Bianchi
ME, Morgan A, d'Adda di Fagagna F and Jackson SP: Yeast Nhp6A/B and
mammalian Hmgb1 facilitate the maintenance of genome stability.
Curr Biol. 15:68–72. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang D, Kang R, Livesey KM, Kroemer G,
Billiar TR, Van Houten B, Zeh HJ III and Lotze MT: High-mobility
group box 1 is essential for mitochondrial quality control. Cell
Metab. 13:701–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taguchi A, Blood DC, del Toro G, Canet A,
Lee DC, Qu W, Tanji N, Lu Y, Lalla E, Fu C, et al: Blockade of
RAGE-amphoterin signalling suppresses tumour growth and metastases.
Nature. 405:354–360. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livesey KM, Kang R, Vernon P, Buchser W,
Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ III, Li L, et
al: p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer
Res. 72:1996–2005. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krynetskaia NF, Phadke MS, Jadhav SH and
Krynetskiy EY: Chromatin-associated proteins HMGB1/2 and PDIA3
trigger cellular response to chemotherapy-induced DNA damage. Mol
Cancer Ther. 8:864–872. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Soutoglou E and Misteli T: Activation of
the cellular DNA damage response in the absence of DNA lesions.
Science. 320:1507–1510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krynetskaia N, Xie H, Vucetic S, Obradovic
Z and Krynetskiy E: High mobility group protein B1 is an activator
of apoptotic response to antimetabolite drugs. Mol Pharmacol.
73:260–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kessenbrock K, Wang CY and Werb Z: Matrix
metalloproteinases in stem cell regulation and cancer. Matrix Biol.
44-46:184–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee CC, Wang CN, Lee YL, Tsai YR and Liu
JJ: High mobility group box 1 induced human lung myofibroblasts
differentiation and enhanced migration by activation of MMP-9. PLoS
One. 10:e01163932015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Agami R and Bernards R: Distinct
initiation and maintenance mechanisms cooperate to induce G1 cell
cycle arrest in response to DNA damage. Cell. 102:55–66. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheng P, Dai W, Wang F, Lu J, Shen M, Chen
K, Li J, Zhang Y, Wang C, Yang J, et al: Ethyl pyruvate inhibits
proliferation and induces apoptosis of hepatocellular carcinoma via
regulation of the HMGB1-RAGE and AKT pathways. Biochem Biophys Res
Commun. 443:1162–1168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Allan LA and Clarke PR: Apoptosis and
autophagy: Regulation of caspase-9 by phosphorylation. FEBS J.
276:6063–6073. 2009. View Article : Google Scholar : PubMed/NCBI
|