Introduction
Glioblastoma is the most malignant and aggressive
primary human brain tumor of the central nervous system, with a
high rate of recurrence and mortality (1). Although treatment including surgical
resection combined with radiation and chemotherapy has been
improved for glioblastoma therapy, the prognosis of glioblastoma
patients is still very poor (2).
Hypoxia is often observed in various types of solid tumors
including glioblastoma, especially in the center of rapidly growing
cancers with incomplete blood vessel networks (3). Hypoxia is considered as a main feature
of the solid tumor microenvironment, playing an important role in
tumor proliferation, metastasis and drug resistance (4). However, the molecular mechanism of how
hypoxia regulates tumor progression in glioblastoma remains unknown
and requires further research.
Hypoxia can induce the expression of
hypoxia-inducible factor 1α (HIF-1α), which is an oxygen-dependent
transcriptional activator (5).
Activation of HIF-1α during hypoxia can regulate a great number of
HIF target genes involved in cell proliferation, energy metabolism
and angiogenesis (6,7). Autophagy, which is a mechanism of
cellular degradation through lysosomes, is also involved in a
HIF-1α-mediated cell survival mechanism (8,9).
Hypoxia-induced autophagy can lead to chemoresistance and malignant
progression of cancer cells (10).
Therefore, suppression of hypoxia-induced autophagy may help
inhibit the tumorigenesis of glioblastoma.
MicroRNAs (miRNAs) are a group of small non-coding
RNAs with 17–22 nucleotides that regulate gene expression by
blocking mRNA translation and/or mediating mRNA degradation
(11). Under a hypoxic condition, a
great number of miRNAs can regulate the expression of various
autophagy-promoting genes and mediate autophagosome formation. For
example, miR-101 was reported to be a potent inhibitor of autophagy
and sensitize breast cancer cells to 4-hydroxytamoxifen
(4-OHT)-mediated cell death (12).
miR-130a was found to inhibit autophagy through targeting ATG2B and
DICER1 and triggering the killing of chronic lymphocytic leukemia
cells (13). However, it is still
unclear whether or not miRNAs modulate hypoxia-induced autophagy in
glioblastoma cells.
In the present study, we found that the relative
expression of miR-224-3p was significantly downregulated in
glioblastoma cells LN229 and astrocytoma cell line U-251MG under a
hypoxic condition. Overexpression of miR-224-3p inhibited
hypoxia-induced autophagy through the HIF-1α/miR-224-3p/ATG5
(autophagy-related gene 5) axis. Our study highlights the
relationship among hypoxia, miRNAs and autophagy in glioblastoma
and astrocytoma and aids in the identification of a novel miRNA
against glioblastoma and astrocytoma progression.
Materials and methods
Cell culture and treatment
Human glioblastoma cell line LN229 and astrocytoma
cell line U-251MG (Cell Bank, Shanghai Institutes for Biological
Sciences, Shanghai, China) were cultured in Gibco™ Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% FBS (HyClone, GE Healthcare
Life Science, Logan, UT, USA) in a humidified atmosphere of 5%
CO2 at 37°C. Hypoxia treatment was performed using a
tri-gas incubator (37°C, 5% CO2, 93% N2 and
2% O2; YCP-50S, Changsha Huaxi Electronic Technology
Co., Ltd., Hunan, China) for different periods (6, 12 and 24
h).
Temozolomide (TMZ) is an effective primary therapy
for high-grade glioma. TMZ is a novel oral chemotherapy drug that
penetrates into the brain and purportedly has a low incidence of
adverse events (14). Stock
solution of TMZ (Schering-Plough, Kenilworth, NJ, USA) was prepared
by dissolving the drug in DMSO. TMZ was used to treat cells at
different concentrations (0–80 µM). Rapamycin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), an autophagy activator, was used to
treat cells for 24 h at the concentration of 100 nM.
Western blot analysis
Cells were lysed in lysis buffer (Beyotime Institute
of Biotechnolgy, Shanghai, China) and the concentrations of
proteins were determined using a Pierce™ BCA protein assay kit
(Thermo Fisher Scientific, Inc.). A total of 20 µg of protein was
separated on 10% SDS-PAGE gel and then transferred into PVDF
membranes (EMD Millipore, Billerica, MA, USA). After blocking with
5% BSA for 1 h at room temperature, the membranes were incubated
with primary antibodies at 4°C overnight. The following antibodies
were used: anti-HIF-1α antibody (dilution 1:500; cat. no. ab51608;
Abcam, Cambridge, UK), anti-ATG5 antibody (dilution 1:2,000; cat.
no. ab108327; Abcam), anti-LC3 I/II antibody (dilution 1:1,000;
cat. no. 12741; Cell Signaling Technology, Danvers, MA, USA),
anti-p-62 antibody (dilution 1:1,000; cat. no. 88588; Cell
Signaling Technology) and anti-GAPDH antibody (1:1,000; cat. no.
5174; Cell Signaling Technology). After incubation with horseradish
peroxidase-conjugated secondary antibodies (dilution 1:2,000; cat.
no. 7056; Cell Signaling Technology), the protein bands were
detected by ImageJ software (version 1.48; National Institutes of
Health, Bethesda, MD, USA).
Real-time quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Takara Biotechnology Co., Ltd., Dalian, China). To detect
mRNA expression of miR-224-3p, extracted RNA (1 µg) was reversely
transcribed into cDNA using a miScript reverse transcription kit
(Qiagen, Dusseldorf, Germany). Primer sequences were as follows:
forward 5′-TGATGTGGGTGCTGGTGTC-3′ and reverse
5′-TTGTGTTGGGGCAGTACTG-3′; for ATG5: forward
5′-GCCGAACCCTTTGCTCAATG-3′ and reverse
5′-TGGTCACCTTAGGAAATACCCAC-3′. SYBR Green Master Mix (Life
Technologies; Thermo Fisher Scientific, Inc.) was used for gene
expression level measurement. The expression levels were calculated
using the ∆∆Cq method (15) with U6
used for the normalization of miRNA.
Cell transfection
The siRNA against HIF-1α (HIF-1α siRNA), miR-224-3p
mimic, siRNA against ATG5 (ATG5 siRNA) and the corresponding
negative control (siRNA NC, mimic NC) were synthesized by Shanghai
GenePharma Co. The sequences of HIF-1α-siRNAs were as follows:
siRNA NC, 5′-UUCUCCGAACGUGUCACGUtt-3′, HIF-1α siRNA1,
5′-UCACAGCAAUACAGAUUCAtt-3′; HIF-1α siRNA2,
5′-GCUCACCAUCAGUUAUUUAtt-3′; HIF-1α siRNA3,
5′-ACGCUCCUUGUCUUAUACCAtt-3′. The sequences of ATG5-siRNAs were as
follows: siRNA NC, 5′-TATATGAAGAAAGTTATCTGGGTAT-3′; ATG5 siRNA1,
5′-ATTATTTAAAAATCTCTCACTGTTG-3′; ATG5 siRNA2,
5′-TATAATATGAAGAAAGTTATCTGGTG-3′; ATG5 siRNA3,
5′-ATCTCACTGTTCATTATCAAAGT-3′. Cells were seeded into 6-well plates
and grown to reach 70% confluence for transfection. Transfection
was performed with these molecular productions using Invitrogen™
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions.
Bioinformatic prediction and
luciferase reporter assay
The targets of miR-224-3p were obtained from the
following target prediction programs: PicTar (https://pictar.mdc-berlin.de/), miRDB (http://mirdb.org/miRDB/custom.html) and
TargetScan (http://www.targetscan.org/vert_72/). The fragment of
ATG5 containing the target sequence of miR-224-3p was amplified by
RT-PCR and then inserted into a pmirGlO Dual-luciferase miRNA
Target Expression Vector (Promega, Madison, WI, USA) to form the
reporter vector ATG5-wild-type (ATG5-WT). Another expressing vector
was also constructed by the insertion of a mutated binding site and
was named as ATG5-mutated-type (ATG5-MUT). Cells were
co-transfected with ATG5-WT or ATG5-MUT and miR-224-3p mimic
respectively, and the Dual-Luciferase Reporter Assay system
(Promega) was used for testing the luciferase activity.
Invasion assays
Invasion assays were analyzed using Transwell
chambers coated with Matrigel (no. PIEP12R48, 8.0 µm; Millipore,
USA). Cells (1×105 in 100 µl serum-free medium) were
seeded into the top chamber and allowed to invade through the
filter into the lower chamber containing medium with serum. After
24 h, cells on the top of the filter were removed while cells on
the bottom were fixed in 4% paraformaldehyde. After that, the
chambers were stained by crystal violet at 4°C for 2 h. The cell
numbers were counted and images were captured under an inverted
microscope (Olympus Corp., Tokyo, Japan) at ×400 magnification on 5
randomly selected fields in each well.
Wound healing assay
Cells (5×105) were seeded into 6-well
plates and incubated for 24 h to reach 90–100% confluence. Then a
sterile pipette tip (1–30 µl) was used to create a straight scratch
to form a wound. After culturing for another 24 h, cells which
migrated to the wounded area were visualized under a confocal
microscope (Nikon A1; Nikon Corp., Tokyo, Japan; magnification,
×200) at 0 and 24 h. Mitomycin C (10 µg/ml) was added to the cell
culture medium to inhibit cell replication according to a previous
report (16).
Cell viability assay
MTT assay was conducted to detect cell viability.
Different groups of cells were seeded into 96-well plates at the
concentration of 5×104 cells/well. After incubating with
different concentrations of temozolomide (TMZ) for 24 h, 20 µl of 5
mg/ml MTT solution (Sigma Chemicals; Merck KGaA) was added into the
medium and incubated for 4 h at 37°C in the dark. Then, the entire
supernatant was replaced with 150 µl of dimethyl sulfoxide (DMSO;
Sigma; Merck KGaA) to dissolve the formazan crystals for an
additional 30 min at 37°C. A microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) was used to detect
absorbance at 490 nm of each well.
Cell apoptosis assay
The cells (2×105 per well) were washed
with phosphate-buffered saline (PBS) twice. Cell apoptosis was
analyzed after appropriate plasmid transfection using staining with
Annexin V and PI (BD Bioscience, San Jose, CA, USA) according to
the manufacturer's instructions. After incubation for 15 min at
room temperature in the dark, the cells were analyzed by using flow
cytometry. Annexin V-positive and PI-negative/positive staining
cells represented apoptotic cells.
In vivo animal study
All animal experiments were performed in accordance
with the NIH Guide for the Care and Use of Laboratory Animals and
were approved by the Medical Ethics Committee of Xi'an Jiaotong
University. A total of 20 BALB/c nude mice (male, 4-weeks old) were
obtained from the Animal Center of Xi'an Jiaotong University and
housed in a controlled environment at 25±3°C, humidity 60%, in a
12-h light/dark cycle with free access to food and water. LN229
cells (2×105) transfected with miR-224-3p mimic
lentivirus (LV-miR-224-3p mimic) or negative control lentivirus
(LV-mimic NC) or untreated LN229 cells were subcutaneously injected
into the flank area of mice to form tumors. The mice were divided
into 4 groups with 5 in each group: control group, mice injected
with LN229 cells without TMZ treatment; TMZ group, mice injected
with LN229 cells and received TMZ treatment by oral gavage (100 µM
daily for 5 days per week for three cycles); TMZ+LV-mimic NC group,
mice injected with LV-mimic NC transfected LN229 cells and received
TMZ treatment; TMZ+LV-miR-224-3p mimic group, mice injected with
LV-miR-224-3p mimic transfected LN229 cells and received TMZ
treatment. Tumor volume and tumor weight were measured every 5 days
post injection. Tumor volume (V) was calculated as follows: V
(mm3) = length × width2/2. After 25 days
post-injection, rats were euthanized by intraperitoneal injection
of pentobarbital sodium (200 mg/kg body weight). Tumors were
collected for the following experiments.
Immunohistochemistry
Immunohistochemical staining of nude mouse xenograft
tumor tissues was performed with antibodies against VEGF (dilution
1:1,600; cat. no. 9698; Cell Signaling Technology) as previously
described (17).
TUNEL assay
TUNEL assay was performed using Colorimetric TUNEL
Apoptosis assay kit (Beyotime Institute of Biotechnology, Jiangsu,
China) according to the manufacturer's instructions. Tumor sections
were incubated with 3% H2O2 and then the
TUNEL reaction mixture. The sections were rinsed and visualized
using DAB. Hematoxylin was used for counter-staining. The numbers
of TUNEL-positive cells from 6 random fields were counted under
light microscopy Olympus Corp.) at ×400 magnification. The cell
apoptosis rate was calculated as the percent of TUNEL-positive
cells relative to the total cells.
Statistical analysis
All experiments were performed in triplicate and the
results in our present study are presented as mean ± standard
deviation (SD). Statistical differences between experimental groups
were analyzed with the SPSS version 20.0 (IBM Corp., Armonk, NY,
USA) by utilizing one-way or two-way ANOVA followed by Bonferroni's
post hoc test. A P-value <0.05 was considered to be
statistically significant.
Results
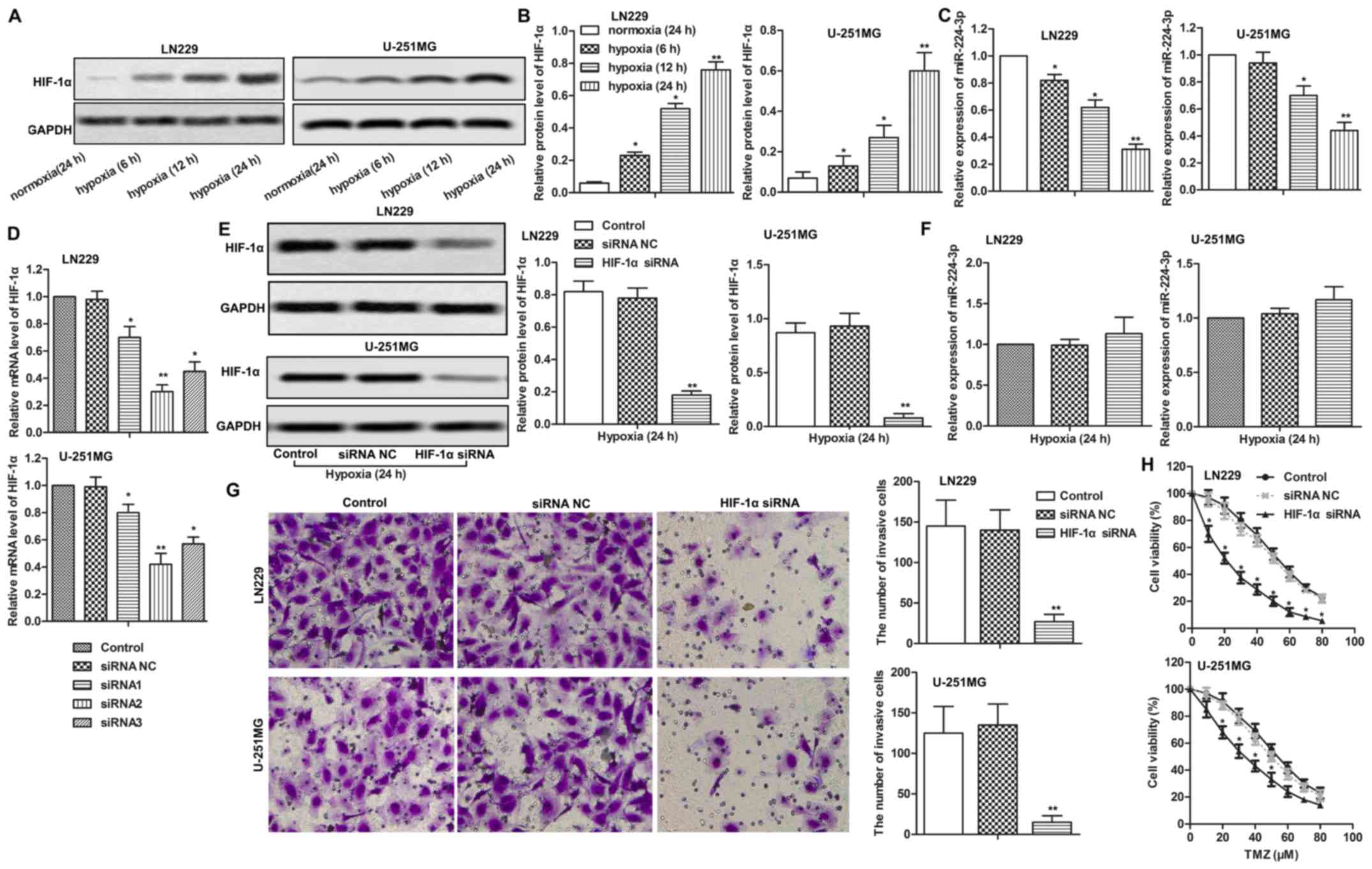
Effect of hypoxia on the expression of
HIF-1α and miR-224-3p
Firstly, we measured the expression of HIF-1α and
miR-224-3p in LN229 and U-251MG cells under a hypoxic condition.
The results showed that the expression of HIF-1α was increased in
LN229 and U-251MG cells under hypoxia. The longer the duration of
hypoxia, the higher the expression of HIF-1α (Fig. 1A and B, P<0.05 at 6 and 12 h,
P<0.01 at 24 h). On the contrary, the expression of miR-224-3p
was decreased under hypoxia condition in a time-dependent manner
(Fig. 1C, P<0.05, P<0.01 at
12 and 24 h, respectively). Then, three different siRNA sequences
were synthesized and transfected into LN229 and U-251MG cells. The
highest knockout efficiency was detected by HIF-1α siRNA2 treatment
in both LN229 and U-251MG cells (Fig.
1D, P<0.01). Thus, HIF-1α siRNA2 was selected for the
following experiments. The transfection efficiency was further
measured through western blot analysis (Fig. 1E, P<0.01). Moreover, we observed
that knockdown of HIF-1α had no significant effect on the
expression of miR-224-3p under a hypoxic condition (Fig. 1F). In addition, the number of
invasive cells was significantly decreased in the HIF-1α-knockout
cells with increased chemosensitivity (Fig. 1G and H, P<0.05, P<0.01). These
results indicated that HIF-1α influenced cell motility and
chemosensitivity by negatively regulating the expression of
miR-224-3p under a hypoxic condition.
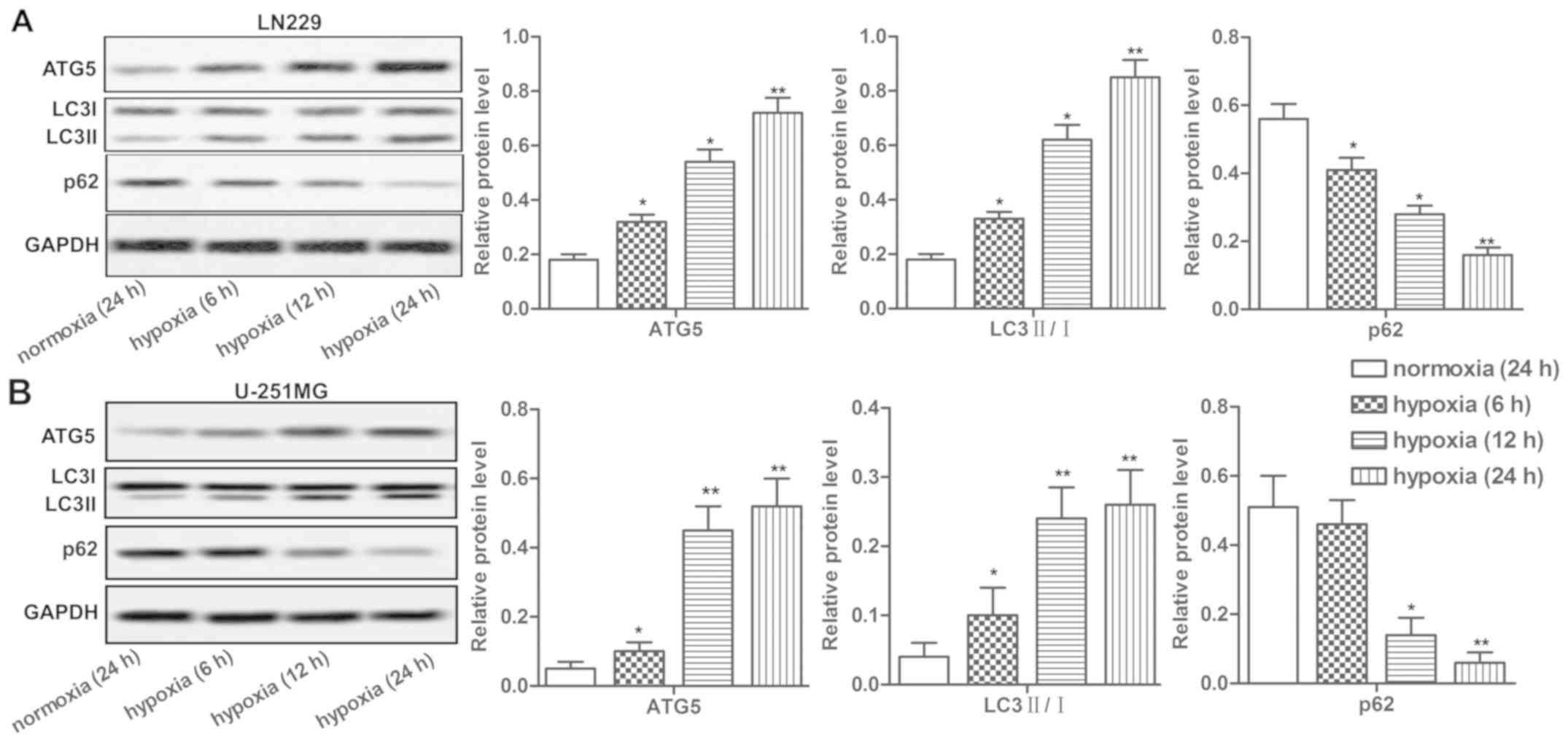
Hypoxia induces autophagy in LN229 and
U-251MG cells
Hypoxia was reported to induce autophagy of tumor
cells. Here we detected the expression of autophagy-related
proteins ATG5, LC3 I/II, p62 and found that the expression of these
proteins in LN229 and U-251MG cells was significantly upregulated
under hypoxia in a time-dependent manner. In addition, decreased
level of autophagy substrate, p62, was observed in LN229 and
U-251MG cells under hypoxia (Fig. 2A
and B, P<0.05, P<0.01). Our data suggested that hypoxia
induced autophagy in LN229 and U-251MG cells.
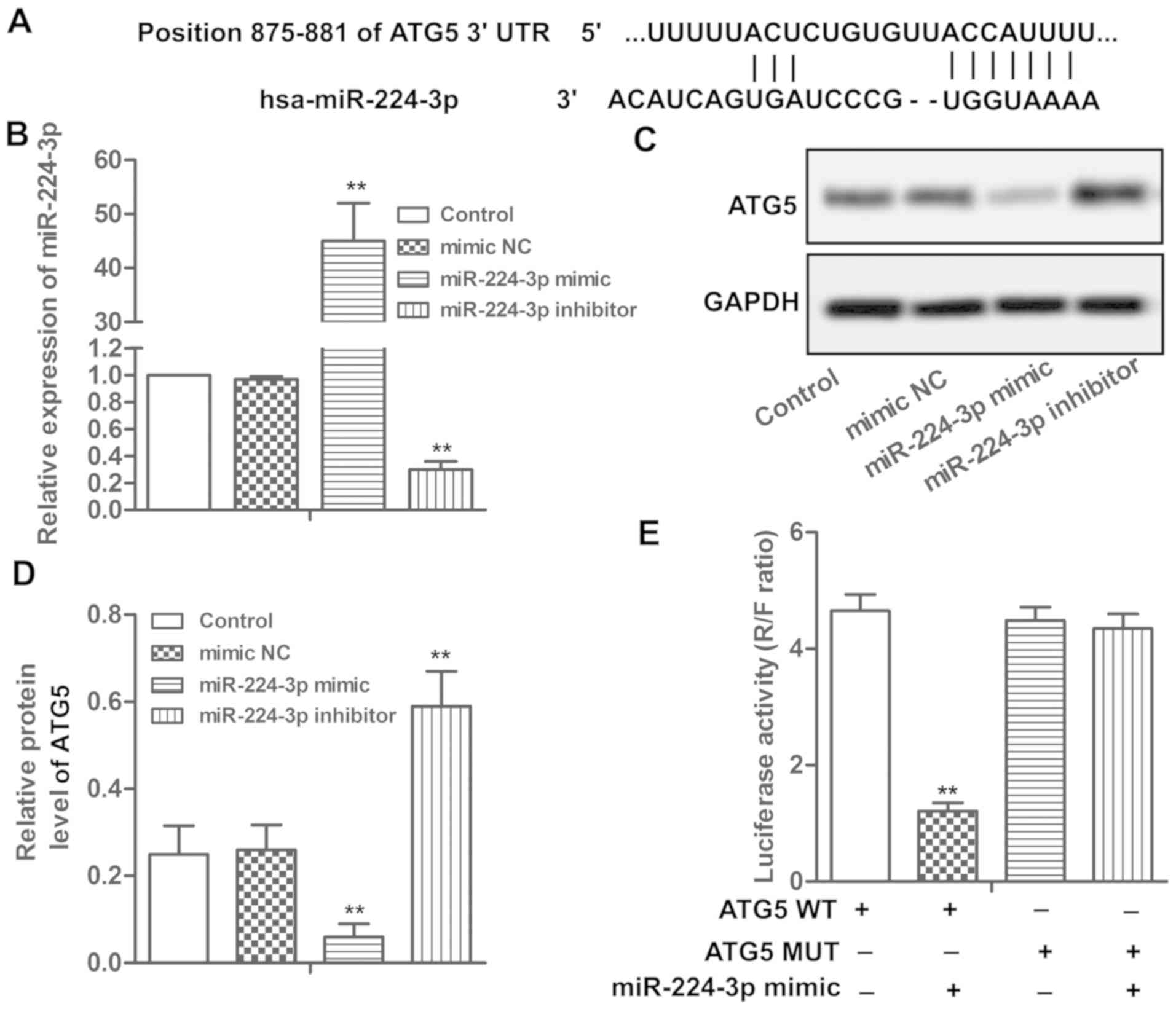
ATG5 is a target of miR-224-3p
Putative miR-224-3p targets were predicted through
bioinformatic analysis and the results indicated that ATG5 was one
of the potential targets of miR-224-3p (Fig. 3A). miR-224-3p mimic/inhibitor was
transfected into LN229 cells to increase/decrease the expression of
miR-224-3p (Fig. 3B, P<0.01).
Overexpression of miR-224-3p significantly suppressed the
expression of ATG5, while miR-224-3p inhibitor elevated the level
of ATG5 (Fig. 3C and D, P<0.01).
Results from the luciferase reporter assay showed that
overexpression of miR-224-3p significantly inhibited luciferase
activity in the wild-type ATG5 3′UTRs but not in the mutated 3′UTR
plasmids (Fig. 3E, P<0.01),
demonstrating the specificity of the miR-224-3p binding sites in
3′UTR of ATG5. These results demonstrated that ATG5 is a target of
miR-224-3p.
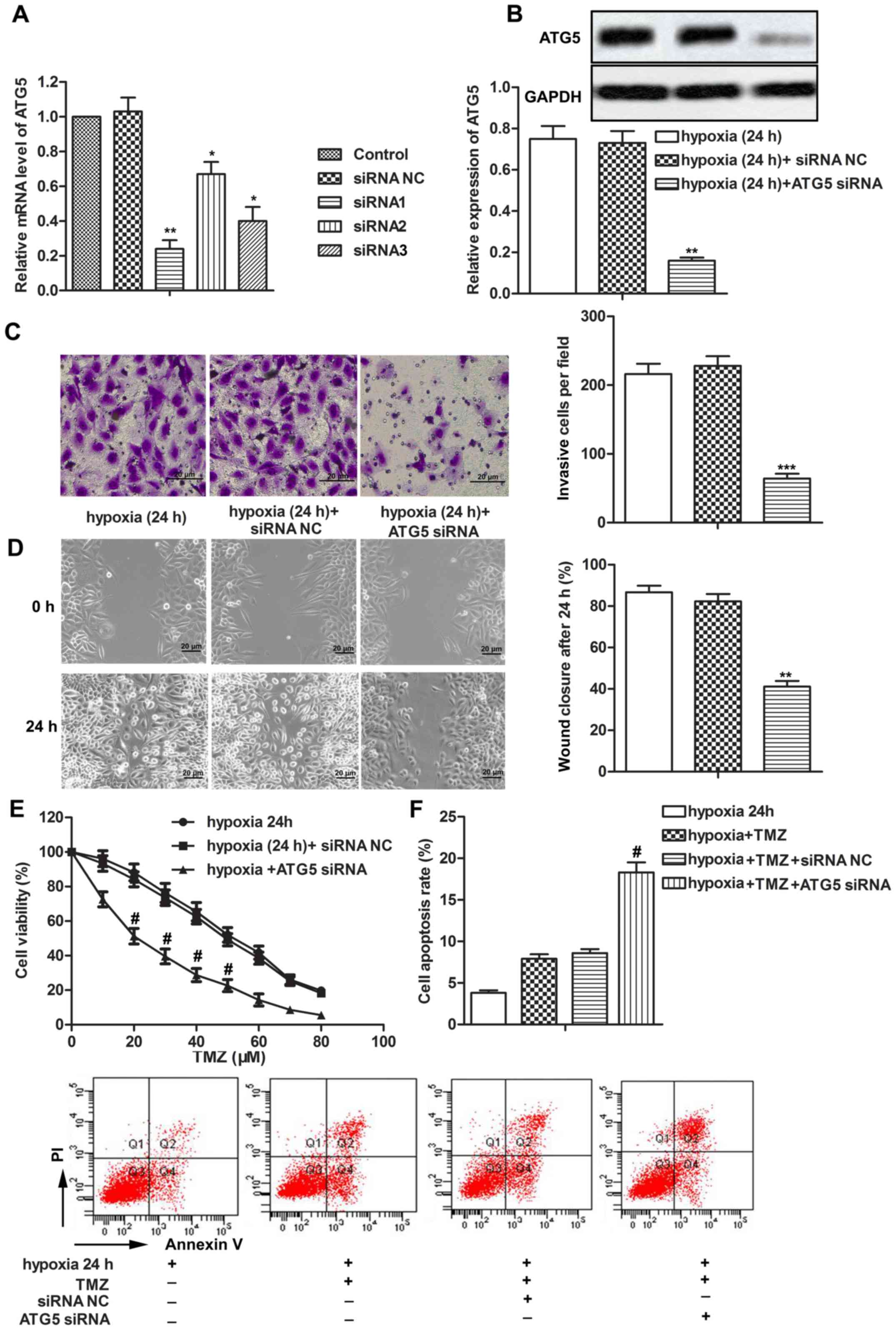
ATG5 siRNA inhibits cell metastasis
and increases chemosensitivity of LN229 cells under a hypoxic
condition
To explore the regulating role of ATG5 in tumor
progression of glioblastoma under hypoxia, three types of siRNAs
were transfected into LN229 cells. ATG5 siRNA1 with the most
effective inhibitory effect was selected to decrease the expression
of ATG5 under a hypoxic condition (Fig.
4A and B, P<0.01). Knockdown of ATG5 inhibited cell invasion
ability and migration ability of LN229 cells under hypoxia compared
with the hypoxia (24 h)+siRNA NC group (Fig. 4C and D, P<0.01, P<0.001). In
addition, the half maximal inhibitory concentration
(IC50) values of TMZ were approximately 20 and 50 µM in
the hypoxia (24 h)+ATG5 siRNA group and hypoxia (24 h)+siRNA NC
group, respectively, suggesting that ATG5 siRNA increased the
chemosensitivity of LN229 cells under a hypoxic condition (Fig. 4E, P<0.05). Moreover, ATG5 siRNA
increased cell apoptosis rates of TMZ-treated LN229 cells under a
hypoxic condition (Fig. 4F,
P<0.05). The above results indicated that ATG5 siRNA inhibited
cell metastasis and increased chemosensitivity of LN229 cells under
a hypoxic condition.
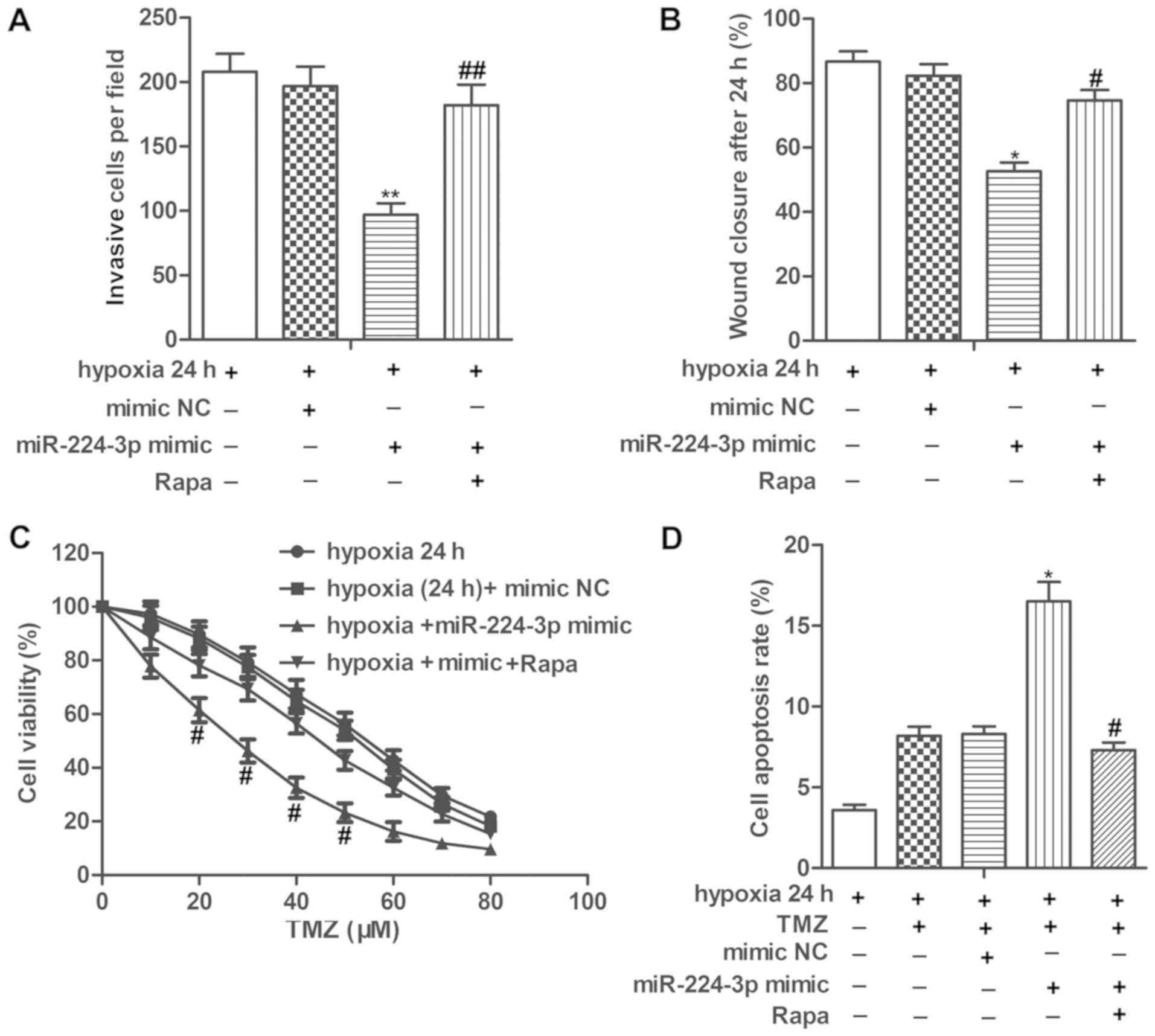
miR-224-3p mimic inhibits cell
metastasis and increases chemosensitivity of LN229 cells under a
hypoxic condition by suppressing autophagy
We then set to explore the role of miR-224-3p in
tumor progression under hypoxia. Our data showed that the
miR-224-3p mimic significantly suppressed the invasion and
migration abilities of the LN229 cells under hypoxia. However,
treatment with Rapa, an autophagy activator significantly
counteracted the inhibitory role of the miR-224-3p mimic on cell
metastasis of LN229 cells (Fig. 5A and
B, P<0.01 for invasion; P<0.05, for wound closure). In
addition, the IC50 values of TMZ were approximately 28
and 50 µM in the hypoxia (24 h)+miR-224-3p mimic group and hypoxia
(24 h)+mimic NC group, respectively. miR-224-3p mimic increased the
cell apoptosis rates of the TMZ-treated LN229 cells under hypoxia.
The above results showed that miR-224-3p increased the
chemosensitivity of LN229 cells under a hypoxic condition. However,
Rapa treatment increased the IC50 values of TMZ while
suppressing the cell apoptosis rate of the TMZ-treated LN229 cells
under hypoxia compared with the hypoxia+miR-224-3p mimic group,
indicating that activation of autophagy abolished the promoting
role of miR-224-3p mimic on the chemosensitivity of LN229 cells
under hypoxia (Fig. 5C and D,
P<0.05, for both cell viability and apoptosis). These results
elucidated that the miR-224-3p mimic inhibited cell metastasis and
increased the chemosensitivity of LN229 cells under a hypoxic
condition by suppressing autophagy.
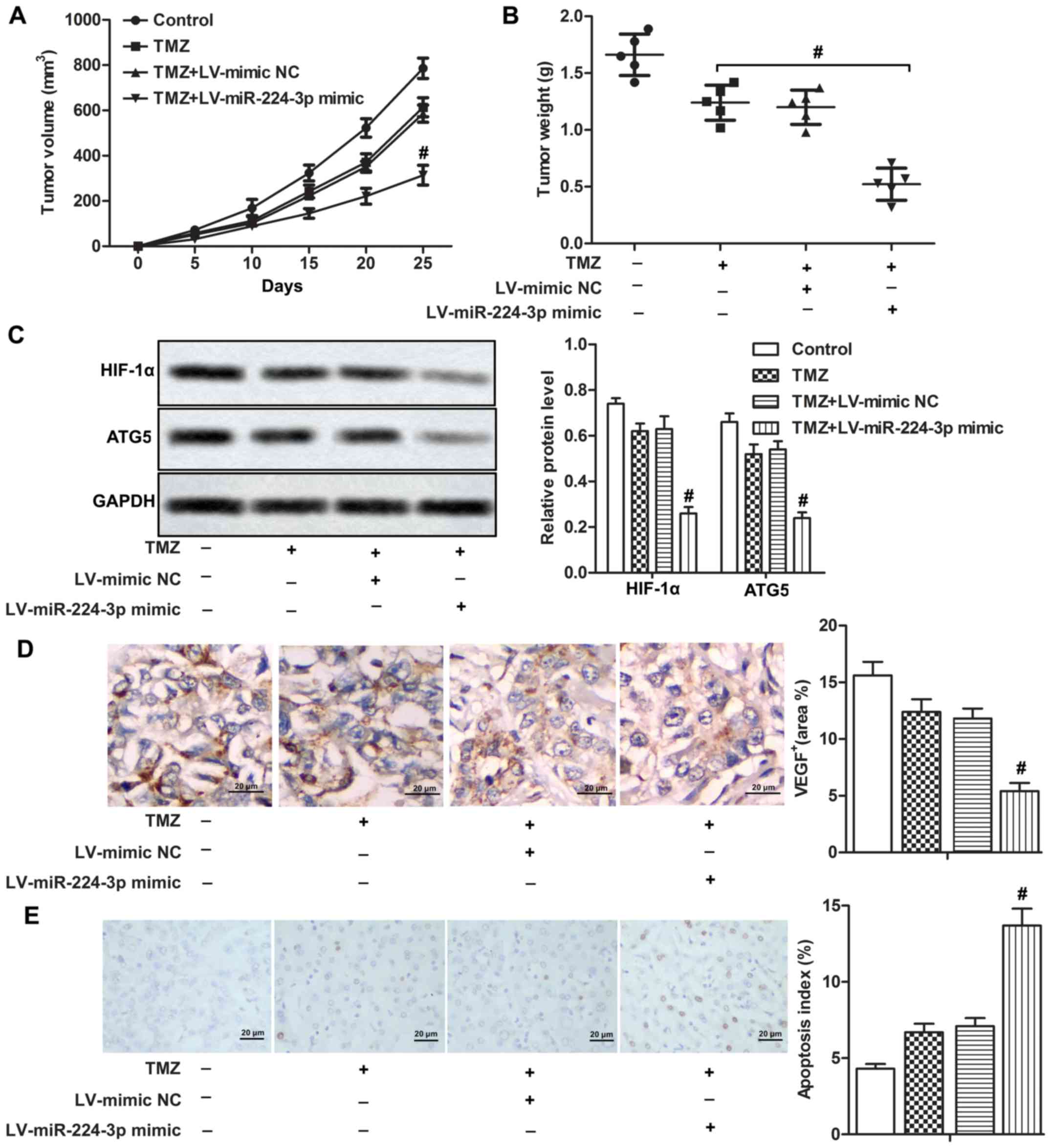
miR-224-3p mimic enhances
chemosensitivity of LN229 cells in vivo
A mouse xenograft model was constructed for our
in vivo experiments. We found that tumor volume and tumor
weight were both lower in the TMZ+LV-miR-224-3p mimic group
compared with that in the TMZ+LV-mimic NC group, suggesting that
the miR-224-3p mimic enhanced the chemosensitivity of LN229 cells
to TMZ in vivo (Fig. 6A and
B, P<0.05). Relative expression of HIF-1α and ATG5 were both
down-regulated in the TMZ+LV-miR-224-3p mimic group compared with
that noted in the TMZ+LV-mimic NC group (Fig. 6C, P<0.05). In addition, the
miR-224-3p mimic suppressed the expression of VEGF with increased
cell apoptosis rate compared with the TMZ+LV-mimic NC group
(Fig. 6D and E, P<0.05). Taken
together, our data indicated that the HIF-1α/miR-224-3p/ATG5 axis
regulated chemosensitivity of LN229 cells in vivo.
Discussion
Autophagy is a common cellular process that
eliminates intracellular damaged organelles through the lysosomal
pathway to sustain cell viability (18,19).
Previous studies have reported that autophagy can induce resistance
to radiotherapy or chemotherapy in various types of cancers,
including breast, ovarian, head and neck cancers (20–23).
Hypoxia, a well-known inducer of autophagy, is often observed in
solid tumors due to inadequate blood supply, especially in rapid
growing tumors such as glioblastoma (24,25).
Recently, the regulating roles of miRNAs in hypoxia-induced
autophagy have attracted attention. In our present study, we
identified hypoxia-mediated downregulation of miR-224-3p as a novel
inhibitor of hypoxia-induced autophagy in glioblastoma and
astrocytoma. The HIF-1α/miR-224-3p/ATG5 axis affected cell mobility
and chemosensitivity in glioblastoma and astrocytoma by regulating
hypoxia-induced autophagy both in vitro and in
vivo.
Hypoxia induced the expression of HIF-1α, which
plays a crucial role in various cellular processes during hypoxia.
HIF-1α can induce cell cycle arrest (26), increase angiogenesis (27) and influence cell metabolism
(28). Previous research reported
that HIF-1α can regulate transcription of multiple miRNAs in
hypoxia (29). The study by Silakit
et al indicated that miR-210 acted as downstream of HIF-1α
and was upregulated in various cancer cells under hypoxia condition
(30). The research by Guo et
al demonstrated that miR-224-3p was one of the downregulated
miRNAs in glioblastoma cells under hypoxia (31). Similarly, in the present study, we
found that the expression of HIF-1α was upregulated while the
expression of miR-224-3p was downregulated under hypoxia in a
time-dependent manner. Knockdown of HIF-1α significantly increased
the expression of miR-224-3p, indicating that miR-224-3p acted as a
downstream miRNA of HIF-1α under hypoxia. Moreover, we also
demonstrated that hypoxia increased the relative expression of
ATG5, LC3 II/I with decreased level of p62 which were correlated
with autophagy in a time-dependent manner, suggesting that hypoxia
induced autophagy in glioblastoma and astrocytoma cells. However,
the role of the downregulation of miR-224-3p in hypoxia-induced
autophagy warrants further investigation.
miRNAs can bind to their target genes and
participate in multiple pathophysiologic processes by regulating
their target gene expression (32).
Through bioinformatic prediction and luciferase reporter assay, we
confirmed that ATG5 was a target of miR-224-3p in our study.
Overexpression of miR-224-3p significantly reduced the protein
expression of ATG5, suggesting that the miR-224-3p mimic inhibits
hypoxia-induced autophagy by suppressing ATG5 expression in
glioblastoma.
ATG5 is a crucial molecular machinery component
involved in autophagosome formation (33). Extensive research has demonstrated
that ATG5 is a target of different miRNAs to adjust autophagy. For
instance, miR-181a was reported to inhibit autophagy of cancer
cells through targeting ATG5 (34).
miR-216b was also found to attenuate autophagy in melanoma by
targeting ATG5 (35). Moreover,
up-regulation of ATG5 was correlated with tumorigenesis of prostate
cancer (36). In agreement with
previous studies, we found that knockdown of ATG5 expression in
transfected LN229 cells with ATG5 siRNA remarkably suppressed the
invasive and migration abilities of LN229 cells under a hypoxic
condition. In addition, knockdown of ATG5 increased the
chemosensitivity of LN229 cells to TMZ and promoted cell apoptosis
under TMZ treatment. These results demonstrated that knockdown of
ATG5 suppressed cell mobility and chemoresistance of glioblastoma
cells.
According to our results, knockdown of ATG5
suppressed cell mobility and chemoresistance of glioblastoma cells.
The expression of ATG5 was inhibited by the miR-224-3p mimic; thus,
we hypothesized that the miR-224-3p mimic inhibited cell mobility
and chemoresistance of glioblastoma cells via suppressing
ATG5-mediated autophagy under hypoxia. To verify our hypothesis,
miR-224-3p was transfected into LN229 cells. Our data showed that
overexpression of miR-224-3p inhibited cell mobility while
increased chemosensitivity of glioblastoma cells under hypoxia.
However, activation of autophagy was able to counteract these
effects of miR-224-3p. In summary, our in vitro experiments
elucidated that the HIF-1α/miR-224-3p/ATG5 axis affects cell
mobility and chemosensitivity by regulating hypoxia-induced
autophagy in glioblastoma cells.
Having elucidated the regulatory role of the
HIF-1α/miR-224-3p/ATG5 axis in cell mobility and chemosensitivity
in vitro, we then explored the effects of this axis using
in vivo experiments. Previous research showed that
overexpression of miR-224-3p suppressed tumor growth in a mouse
xenograft model (31). Similarly,
we observed that tumor volume and tumor weight were both smaller in
the TMZ+LV-miR-224-3p mimic group, suggesting that the miR-224-3p
mimic enhanced chemosensitivity of the LN229 cells to TMZ in
vivo. In addition, the miR-224-3p mimic decreased HIF-1α, ATG5
and VEGF expression while promoting apoptosis of tumor cells under
TMZ treatment, suggesting that the HIF-1α/miR-224-3p/ATG5 axis
regulated chemosensitivity of LN229 cells in vivo.
Taken together, we identified miR-224-3p as a novel
inhibitor of hypoxia-induced autophagy by directly targeting ATG5
in glioblastoma and astrocytoma cells. HIF-1α influenced cell
motility and chemosensitivity by negatively regulating the
expression of miR-224-3p under hypoxia. Additionally,
overexpression of miR-224-3p inhibited cell mobility and
chemoresistance of glioblastoma cells via suppressing ATG5 mediated
autophagy under hypoxia. Therefore, miR-224-3p could be a novel
target against hypoxia-induced autophagy in glioblastoma and
astrocytoma.
Acknowledgements
The authors would like to thank the members of The
Second Affiliated Hospital, School of Medicine, Xi'an Jiaotong
University and Tangdu Hospital, Medical University of the Air
Force, for providing technical support concerning the present
study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SH interpreted the data regarding the autophagy
assay and the cell viability analysis, collected and analyzed the
data. PQ was involved in the western blot and RT-qPCR analysis. TZ
and FL were involved in the cell transfection, the animal model
establishment, immunohistochemistry and TUNEL assay. XH was
responsible for conceiving, designing, drafting and revising the
manuscript. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All animal experiments were performed in accordance
with the NIH Guide for the Care and Use of Laboratory Animals and
were approved by the Medical Ethics Committee of Xi'an Jiaotong
University. In addition, all experiments were conducted following
institutional guidelines of Xi'an Jiaotong University (Xi'an,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HIF-1α
|
hypoxia-inducible factor α
|
|
miRNAs
|
microRNAs
|
|
ATG5
|
autophagy-related gene 5
|
|
TMZ
|
temozolomide
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
SD
|
standard deviation
|
|
IC50
|
half maximal inhibitory
concentration
|
References
|
1
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pouysségur J, Dayan F and Mazure NM:
Hypoxia signalling in cancer and approaches to enforce tumour
regression. Nature. 441:437–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wilson WR and Hay MP: Targeting hypoxia in
cancer therapy. Nat Rev Cancer. 11:393–410. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Masoud GN and Li W: HIF-1α pathway: Role,
regulation and intervention for cancer therapy. Acta Pharm Sin B.
5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vallée A, Guillevin R and Vallée JN:
Vasculogenesis and angiogenesis initiation under normoxic
conditions through Wnt/β-catenin pathway in gliomas. Rev Neurosci.
29:71–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shrivastava R, Singh V, Asif M, Negi MPS
and Bhadauria S: Oncostatin M upregulates HIF-1α in breast tumor
associated macrophages independent of intracellular oxygen
concentration. Life Sci. 194:59–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodríguez ME, Catrinacio C, Ropolo A,
Rivarola VA and Vaccaro MI: A novel HIF-1α/VMP1-autophagic pathway
induces resistance to photodynamic therapy in colon cancer cells.
Photochem Photobiol Sci. 16:1631–1642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun Y, Xing X, Liu Q, Wang Z, Xin Y, Zhang
P, Hu C and Liu Y: Hypoxia-induced autophagy reduces
radiosensitivity by the HIF-1α/miR-210/Bcl-2 pathway in colon
cancer cells. Int J Oncol. 46:750–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang X, Yin H, Zhang Y, Li X, Tong H, Zeng
Y, Wang Q and He W: Hypoxia-induced autophagy promotes gemcitabine
resistance in human bladder cancer cells through hypoxia-inducible
factor 1α activation. Int J Oncol. 53:215–224. 2018.PubMed/NCBI
|
|
11
|
Wang X, Ye X, Ji J, Wang J, Xu B, Zhang Q,
Ming J and Liu X: MicroRNA155 targets myosin light chain kinase to
inhibit the migration of human bone marrowderived mesenchymal stem
cells. Int J Mol Med. 42:1585–1592. 2018.PubMed/NCBI
|
|
12
|
Frankel LB, Wen J, Lees M, Høyer-Hansen M,
Farkas T, Krogh A, Jäättelä M and Lund AH: microRNA-101 is a potent
inhibitor of autophagy. EMBO J. 30:4628–4641. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kovaleva V, Mora R, Park YJ, Plass C,
Chiramel AI, Bartenschlager R, Döhner H, Stilgenbauer S, Pscherer
A, Lichter P and Seiffert M: miRNA-130a targets ATG2B and DICER1 to
inhibit autophagy and trigger killing of chronic lymphocytic
leukemia cells. Cancer Res. 72:1763–1772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hart MG, Garside R, Rogers G, Stein K and
Grant R: Temozolomide for high grade glioma. Cochrane Database Syst
Rev. CD0074152013.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Milovic V, Teller IC, Murphy GM, Caspary
WF and Stein J: Deoxycholic acid stimulates migration in colon
cancer cells. Eur J Gastroenterol Hepatol. 13:945–949. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang YY, Sun G, Luo H, Wang XF, Lan FM,
Yue X, Fu LS, Pu PY, Kang CS, Liu N and You YP: MiR-21 modulates
hTERT through a STAT3-dependent manner on glioblastoma cell growth.
CNS Neurosci Ther. 18:722–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kroemer G, Mariño G and Levine B:
Autophagy and the integrated stress response. Mol Cell. 40:280–293.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakatogawa H, Suzuki K, Kamada Y and
Ohsumi Y: Dynamics and diversity in autophagy mechanisms: Lessons
from yeast. Nat Rev Mol Cell Biol. 10:458–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He WS, Dai XF, Jin M, Liu CW and Rent JH:
Hypoxia-induced autophagy confers resistance of breast cancer cells
to ionizing radiation. Oncol Res. 20:251–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Apel A, Herr I, Schwarz H, Rodemann HP and
Mayer A: Blocked autophagy sensitizes resistant carcinoma cells to
radiation therapy. Cancer Res. 68:1485–1494. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J and Wu GS: Role of autophagy in
cisplatin resistance in ovarian cancer cells. J Biol Chem.
289:17163–17173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sannigrahi MK, Singh V, Sharma R, Panda NK
and Khullar M: Role of autophagy in head and neck cancer and
therapeutic resistance. Oral Dis. 21:283–291. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Höckel M and Vaupel P: Tumor hypoxia:
Definitions and current clinical, biologic, and molecular aspects.
J Natl Cancer Inst. 93:266–276. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koshiji M, Kageyama Y, Pete EA, Horikawa
I, Barrett JC and Huang LE: HIF-1alpha induces cell cycle arrest by
functionally counteracting Myc. EMBO J. 23:1949–1956. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clara CA, Marie SK, de Almeida JR,
Wakamatsu A, Oba-Shinjo SM, Uno M, Neville M and Rosemberg S:
Angiogenesis and expression of PDGF-C, VEGF, CD105 and HIF-1α in
human glioblastoma. Neuropathology. 34:343–352. 2014.PubMed/NCBI
|
|
28
|
Agani F and Jiang BH: Oxygen-independent
regulation of HIF-1: Novel involvement of PI3K/AKT/mTOR pathway in
cancer. Curr Cancer Drug Targets. 13:245–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kulshreshtha R, Ferracin M, Negrini M,
Calin GA, Davuluri RV and Ivan M: Regulation of microRNA
expression: The hypoxic component. Cell Cycle. 6:1426–1431. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Silakit R, Kitirat Y, Thongchot S, Loilome
W, Techasen A, Ungarreevittaya P, Khuntikeo N, Yongvanit P, Yang
JH, Kim NH, et al: Potential role of HIF-1-responsive
microRNA210/HIF3 axis on gemcitabine resistance in
cholangiocarcinoma cells. PLoS One. 13:e01998272018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo X, Xue H, Guo X, Gao X, Xu S, Yan S,
Han X, Li T, Shen J and Li G: MiR224-3p inhibits hypoxia-induced
autophagy by targeting autophagy-related genes in human
glioblastoma cells. Oncotarget. 6:41620–41637. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jafarzadeh M, Mohammad Soltani B,
Ekhteraei Tousi S and Behmanesh M: Hsa-miR-497 as a new regulator
in TGFβ signaling pathway and cardiac differentiation process.
Gene. 675:150–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mallik A and Yammani RR: Saturated fatty
acid palmitate negatively regulates autophagy by promoting ATG5
protein degradation in meniscus cells. Biochem Biophys Res Commun.
502:370–374. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tekirdag KA, Korkmaz G, Ozturk DG, Agami R
and Gozuacik D: MIR181A regulates starvation- and rapamycin-induced
autophagy through targeting of ATG5. Autophagy. 9:374–385. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo M, Wu L, Zhang K, Wang H, Wu S,
O'Connell D, Gao T, Zhong H and Yang Y: miR-216b enhances the
efficacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in
melanoma. Cell Signal. 42:30–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li X, Li C and Zhu LH: Correlation of
autophagy-associated gene Atg5 with tumorigenesis of prostate
cancer. Zhonghua Nan Ke Xue. 21:31–34. 2015.(In Chinese).
PubMed/NCBI
|