Introduction
The Hippo signaling pathway was originally
discovered using genetic screens of Drosophila melanogaster
(1) and is highly conserved from
D. melanogaster to mammals (2). It has been a focus of research in the
past due to its fundamental role in modulating cell proliferation
and differentiation, apoptosis, cell survival and migration
(3,4). The Hippo pathway is considered to be a
tissue growth regulator and tumor suppressor (5). A number of studies have demonstrated
that dysregulation of the Hippo pathway closely correlates with
tumor initiation, progression and the acquisition of drug
resistance (6–12). Therefore, there is considerable
interest in and speculation about targeting the Hippo pathway to
treat human cancer. However, the activity and stability of the
pathway components contribute to internal environmental homeostasis
and are regulated in various ways, including through
ubiquitination.
Ubiquitination-deubiquitination is a common and
vital post-translational modification that serves critical roles in
protein degradation and localization, autophagy, antigen
presentation, signal transduction and other cellular processes
(13,14). Ubiquitination-deubiquitination is
primarily regulated by the E3 ubiquitin ligase and deubiquitinases
(DUBs), which target the pathway components, mediate their turnover
and activity, and lead to different biological effects (15). The balance between ubiquitination
and deubiquitination is essential for maintaining normal functional
output of the Hippo pathway. Impairment of this balance may result
in severe pathological states, including cancer. Here, the
modulatory mechanism of ubiquitination-deubiquitination in the
Hippo pathway, the recent progress in identifying therapeutic
targets, and strategies and future directions in the field are
reviewed.
Overview of the Hippo pathway
The Hippo signaling pathway, also termed the
Salvador/Warts/Hippo (SWH) pathway, was initially identified
through genetic mosaic screens of D. melanogaster by its
suppression of tissue overgrowth and was named after one of its key
signaling components, the protein kinase Hippo (Hpo) (1). The pathway comprises a large network
of proteins that have coordinating functions. At the heart of the
Hippo pathway in D. melanogaster is a kinase cassette
consisting of two serine/threonine kinases known as the Hippo and
Warts kinases, along with their regulatory proteins Salvador and
Mats (3). In mammals, the Hippo
kinases are known as mammalian STE20-like protein kinase 1 (MST1;
also known as STK4) and MST2 (also known as STK3), whereas the
Warts kinases are known as large tumor suppressor 1 (LATS1) and
LATS2 (4). In addition, Salvador
homolog 1 (SAV1) and MOB kinase activator 1A/B (MOB1A/B) are
homologs of Salvador and Mats, respectively (6). Yes-associated protein (YAP) and the
transcriptional co-activator with PDZ-binding motif (TAZ; also
known as WWTR1) are two transcriptional co-activators (Yorkie
homologs) that are the principal downstream effectors and are
negatively regulated by the Hippo pathway (16).
A variety of upstream stimuli, including apicobasal
polarity, mechanotransduction, cell-cell adhesion, cellular stress,
contact inhibition and extracellular signaling, are able to
activate the Hippo pathway (17).
Canonically, in mammals, the Hippo pathway is switched on when
activated MST1/2 phosphorylates SAV1 and they subsequently
phosphorylate and activate MOB1A, MOB1B, LATS1 and LATS2, resulting
in the phosphorylation of YAP and TAZ. Phosphorylated YAP and TAZ
bind to the 14-3-3 protein, resulting in their cytoplasmic
retention and proteasomal degradation in a β-transducin
repeat-containing E3 ubiquitin protein ligase (β-Trcp)-dependent
manner (4). YAP and TAZ are
transcriptional co-activators that lack the ability to directly
bind DNA, although they form complexes with TEA domain-containing
sequence-specific transcription factors (TEADs) as their primary
partners (16,18). YAP/TAZ also bind to other
transcription factors, including tumor protein p73 (19), mothers against decapentaplegic
homologs (SMADs) (20–22), forkhead box protein M1 (23), T-box transcription factor 5
(24,25), RUNT-related transcription factor 1
(RUNX1), and RUNX2 (26), along
with other factors, to regulate target gene expression. The
transcription cofactor vestigial-like family member 4 (VGLL4)
competes with YAP and TAZ for binding with TEADs to suppress the
co-activators of YAP and its target gene expression (27). By contrast, when the Hippo pathway
is switched off, MST1, MST2, LATS1 and LATS2 are inactivated,
resulting in hypophosphorylated YAP and TAZ and their accumulation
in the nucleus, where they associate with TEADs instead of VGLL4 to
promote the expression of target genes, including connective tissue
growth factor (CTGF), amphiregulin, cysteine-rich angiogenic
inducer 61, and surviving (28–30). A
similar mechanism of action has been identified in D.
melanogaster (31); however,
for simplicity, the present review focused primarily on the
mammalian Hippo pathway (Fig.
1).
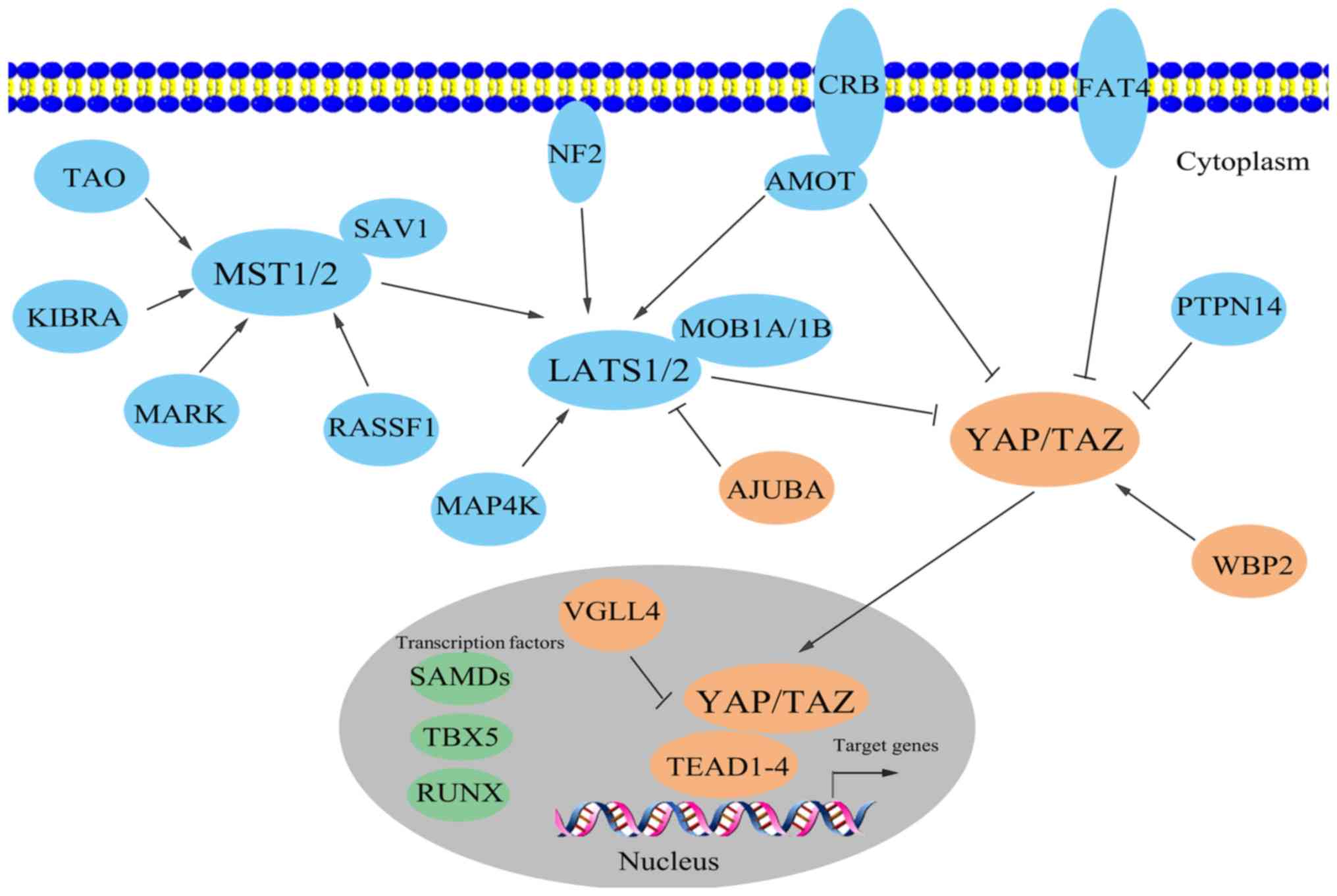 | Figure 1.Brief graphical presentation of the
mammalian Hippo pathway. The Hippo pathway is composed of a large
network of proteins that function in a coordinated manner. Putative
oncoproteins (blue) and putative tumor suppressors (orange) are
illustrated, along with a number of other transcription factors
(green) involved in the Hippo pathway, apart from TEAD1-TEAD4 and
VGLL4. AMOT, angiomotin; CRB, Crumbs; KIBRA, kidney and brain
protein; LATS, large tumor suppressor; MAP4K, mitogen-activated
protein kinase kinase kinase kinase; MARK, microtubule
affinity-regulating kinase; MOB1, MOB kinase activator 1; MST,
mammalian STE20-like protein kinase; NF2, neurofibromin 2; PTPN14,
protein tyrosine phosphatase non-receptor type 14; RASSF1, Ras
association domain family 1; RUNX, RUNT-related transcription
factor; SAV1, Salvador homologue 1; TAO, thousand and one amino
acid protein; TAZ, transcriptional co-activator with PDZ-binding
motif; TBX5, T-box transcription factor 5; TEAD, TEA
domain-containing sequence-specific transcription factor; VGLL4,
vestigial-like family member 4; WBP2, WW domain-binding protein 2;
YAP, Yes-associated protein. |
In addition to the aforementioned key components,
there are a number of other vital branches of the Hippo pathway
involved in regulating functional outputs. For example, MST kinases
may be phosphorylated and activated by thousand and one amino acid
protein (TAO) kinase (32),
microtubule affinity-regulating kinase (MARK) (33), and kidney and brain protein (KIBRA;
also known as WWC1) (34). In
addition to MST, the mammalian misshapen homolog mitogen-activated
protein kinase kinase kinase kinase (35), the apical membrane-associated
FERM-domain protein neurofibromatosis 2 (NF2) (36), angiomotin (AMOT) (37), and even heat-shock proteins
including HSP90 (38), are all able
to activate LATS kinases, whereas members of the
junction-associated Ajuba protein family directly repress LATS
activity (39). LATS, WW
domain-binding protein 2 (WBP2) (40), and protein tyrosine phosphatase
non-receptor type 14 (PTPN14) (41)
each contribute to the regulation of YAP and TAZ in different ways.
Notably, the regulatory mechanisms controlling the activity of the
Hippo pathway appear to vary depending on the specific cells and
tissues.
Further examination of the Hippo pathway in murine
embryonic stem cells and induced pluripotent cells has demonstrated
that YAP is capable of inhibiting cell differentiation, expanding
stem cell populations, and reprogramming differentiated cells to
more primitive states (42). In
human embryonic stem cells, TAZ seems to act in a similar manner
(21). Additional examples,
including stem and progenitor cells of the liver, intestine, heart
and skin, support the function of YAP and TAZ in cell stemness,
pluripotency and differentiation (43–48),
which implies their potential roles in repair and regeneration.
Furthermore, a growing body of evidence reveals pivotal roles for
the Hippo pathway in growth control in cells. In D.
melanogaster, inhibition of the Hippo or Warts kinases, or
overexpression of Yorkie, results in marked overgrowth phenotypes
leading to oversized tissues (49,50).
In mammals, deregulation of the Hippo pathway, including through
the inactivation of LATS or the overexpression of YAP, induces
liver and heart hypertrophy in mice (51,52).
It is hypothesized that there are two mechanisms leading to the
overgrowth phenotype. First, normal cell proliferation may be
disrupted, resulting in rapid cell proliferation, even when the
tissue reaches its proper size. Second, ectopic cells may become
insensitive or resistant to apoptotic signals (53). Notably, deregulation of the Hippo
pathway does not cause overgrowth of every tissue and organ, for
example the skin (47), indicating
other functions of the pathway.
Uncontrolled overgrowth tends to be a primary step
in neoplasia. Indeed, a large number of studies have reported that
dysregulation of the Hippo pathway is observed at a relatively high
frequency in various human carcinomas. For instance, overexpression
of YAP1 in mice leads to the development of hepatocellular
carcinoma (HCC) (43) and a
deficiency in MST1/2 kinases causes YAP hyperactivation, marked
liver overgrowth and the development of HCC (54). TAZ is upregulated and stabilized by
SKI like proto-oncogene, which enhances its oncogenic activity and
epithelial-mesenchymal transition (EMT) (55). Elevated YAP expression resulting
from the downregulation of LATS contributes to cell proliferation
and invasion in gastric cancer (56). However, the exact mechanisms
associated with tumorigenesis and progression are not completely
understood.
The Hippo pathway is modulated in various ways, and
diverse modulations exert different effects on physiological and
pathological processes. First, in common human cancer types,
somatic and germline mutations in Hippo pathway genes are
exceptionally rare (6). However,
evidence of mutations in Hippo pathway genes is beginning to
emerge, owing to the rapid advancements in large genomic sequencing
projects. Tumor-associated mutations have been identified in LATS2
in esophageal cancer and non-small-cell lung cancer (57,58),
and mutations occur in LATS1 and LATS2 in basal cell carcinoma of
the skin (59). NF2 (also known as
Merlin) is considered to be a tumor suppressor, and mutations in
the NF2 gene correlate with neurofibromatosis type II (53). In addition to mutations, loss of
heterozygosity, genomic deletions and promoter hypermethylation all
contribute to the ectopic activity of the Hippo pathway at the DNA
level. For example, loss of LATS1 heterozygosity has been reported
in ovarian and breast cancer (60–62).
In renal cell carcinoma, downregulation of LATS1, at least in part
by promoter hypermethylation, leads to YAP overexpression,
resulting in tumor initiation and progression (63). FAT atypical cadherin 4 (FAT4), an
important member of the FAT cadherin protein family, is a critical
regulator of the Hippo pathway and probably acts as a tumor
suppressor (64). Genomic deletion
and promoter hypermethylation of FAT4 have been detected in gastric
cancer and breast cancer (65,66),
respectively.
Second, microRNAs (miRNAs), small noncoding RNAs of
21–25 nucleotides in length that negatively modulate gene
expression at the post-transcriptional or translational level
(67), are closely associated with
the regulation of the Hippo pathway at the RNA level and with
carcinogenesis. Mature forms of miRNAs bind to the 3′-untranslated
region (UTR) of target mRNAs and cleave and/or hinder their
translation (68). Studies have
demonstrated that in endometrial cancer, miRNA 31 is able to bind
the 3′-UTR of LATS2 mRNA, causing the downregulation of LATS2, and
promoting YAP accumulation in the nucleus and the transcription of
cyclin D1 (69). Another miRNA,
miRNA (miR)-129-5p, is also able to directly inhibit YAP and TAZ
expression in a similar manner, leading to the inactivation of TEAD
transcription and a reduction in CTGF and cyclin A, offering a
possible explanation for its tumor suppressive ability in ovarian
cancer (70). Additionally, it is
noteworthy that there exists a YAP-miR-130a-VGLL4 positive feedback
loop in the control of organ size and tumorigenesis (71). miR-130a is induced by YAP and
represses VGLL4, promoting the formation of the YAP-TEAD complex
and enhancing YAP activity, which has been confirmed in liver
cancer and glioblastoma (71,72).
In addition to miRNAs, long noncoding RNAs (lncRNAs) are also
involved in the regulation of the Hippo pathway. These are
noncoding transcripts of >200 nucleotides in length that
interact with DNA, RNA and proteins, and are associated with
numerous human diseases, including tumors (73,74).
In gastric cancer, lncRNA p21 (lincRNA-p21) is capable of
negatively regulating YAP expression. Knockdown of lincRNA-p21
results in elevated expression levels of YAP mRNA and protein in a
Hippo-independent manner, with the opposite effects observed with
increased expression of lincRNA-p21 (75). However, the precise underlying
mechanism remains to be determined. Another type of noncoding RNA,
circular RNA, usually has miRNA sponge functionality and interacts
with RNA-binding proteins associated with tumorigenesis (76,77). A
recent report revealed that by sponging miR-424-5p and modulating
LATS1 expression, circular RNA_LARP4 suppresses cell proliferation
and invasion in gastric cancer (78), indicating an indirect regulatory
mechanism of the Hippo pathway.
Third, post-translational modifications (PTMs) have
attracted considerable attention in recent years due to their
crucial roles in mediating the activation and subcellular
localization of signaling components, which results in the
induction, strengthening or repression of the corresponding
signaling pathways. An increasing number of reports have revealed
that the Hippo pathway is regulated by a variety of
post-translational modifications, including phosphorylation,
acetylation, methylation and ubiquitination, whose imbalance and
malfunction are involved in tumor formation and progression
(79). Among the various types of
post-translational modifications, phosphorylation may be regarded
as the most common. In the canonical Hippo pathway, phosphorylation
of LATS1/2 promotes the phosphorylation of YAP/TAZ, leading to
YAP/TAZ cytoplasmic retention, degradation via ubiquitination, and
non-expression of relevant target genes (4). There are multiple phosphorylation
sites on proteins responsible for diverse biological effects, which
are activated by various enzymes. For instance, YAP has five
currently identified sites (T119, S138, T154, S317 and T362) that
may be phosphorylated by JUN N-terminal kinases (JNK1 and JNK2),
triggering YAP to serve a dual role under different circumstances
(25,80). Furthermore, phosphorylation of MST1
at T120 and T387 by PI3K/Akt prevents caspase-mediated cleavage of
MST1, thereby inhibiting its activation (81,82).
JNK1 facilitates MST1-mediated pro-apoptotic signaling by
phosphorylating MST1 at S82 (83).
Additionally, acetylation has been demonstrated to
specifically modify YAP at highly conserved C-terminal lysine
residues catalyzed by the nuclear acetyltransferases CREB binding
protein and p300 in response to S(N)2 alkylating agents, which may
be reversed by the nuclear deacetylase NAD-dependent protein
deacetylase sirtuin-1 (SIRT1) (84,85).
The regulation of YAP by SIRT1-mediated deacetylation may be
associated with HCC tumorigenesis and drug resistance (85).
Methylation of non-histone proteins is another
essential regulatory mechanism that controls the functions of
proteins. It has been reported that Set7 forms a complex with YAP
and directly monomethylates YAP at lysine 494, which is beneficial
for its cytoplasmic sequestration (86). This indicates a
methylation-dependent checkpoint in the Hippo pathway.
O-GlcNAcylation, catalyzed by O-GlcNAc transferase (87), is a notable type of
post-translational modification and has been associated with
tumorigenesis by mediating signal transduction, transcription,
metabolism and other cellular functions (88–90).
In a recent study, researchers reported that O-GlcNAcylation of YAP
at Thr241 within the WW domain is responsible for
high-glucose-induced liver oncogenesis (91). This kind of modification antagonizes
the phosphorylation of YAP at Ser127, partly by preventing the
binding of LATS to YAP, and enhances its stability and
pro-tumorigenic capacity (91).
Apart from the PTMs described above,
ubiquitination-deubiquitination has emerged as an important type
post-translational modification. Due to this, an introduction to
ubiquitination-deubiquitination and its modulation of the Hippo
pathway is provided below.
Overview of ubiquitination and
deubiquitination
Ubiquitin (Ub) is a highly conserved small protein
consisting of 76 amino acids with a molecular mass of ~8.5 kDa that
is widely expressed in eukaryotes (92). The process by which one or more
molecules of ubiquitin bind target proteins via enzyme catalysis is
called ubiquitination, with its fundamental function being protein
degradation (93). The
ubiquitin-proteasome system (UPS) modulates 80–85% of all protein
degradation in eukaryotic cells. It comprises ubiquitin, ubiquitin
activating enzymes (E1), ubiquitin conjugating enzymes (E2),
ubiquitin protein ligases (E3), the proteasome and the substrate
(target protein) (94). In general,
ubiquitination involves two stages. The first stage includes
ubiquitin activation by an E1 enzyme using ATP as an energy
resource, followed by the transfer of the activated ubiquitin to an
E2 enzyme via a trans(thio)esterification reaction. Finally, an
isopeptide bond is created between a lysine of the target protein
and the C-terminal glycine of ubiquitin. This step requires an E3
enzyme to transfer ubiquitin to the target protein. Additional
ubiquitin molecules may be added to the substrate by binding to the
initial ubiquitin, yielding a polyubiquitin chain. The second stage
involves the recognition of the activated target protein by the 26S
proteasome, which subsequently degrades the protein into small
peptides, typically of 3–24 amino acids in length, and releases the
ubiquitin for cyclic utilization (95).
The process in which numerous lysine residues of the
target protein are ubiquitinated is termed multiubiquitination.
Ubiquitin has seven lysine residues, K48, K63, K6, K11, K27, K29
and K33, which may serve as polyubiquitination points.
Polyubiquitination brings about different functional consequences
depending on whether the polyubiquitin chains are linked through
K48, K63 or other K residues of ubiquitin (96). For instance, K48-linked
polyubiquitination targets substrates in the 26S proteasome for
destruction, whereas K63-linked polyubiquitination is able to
stabilize proteins, direct their translocalization and transmit
signals associated with intracellular trafficking, cell signaling,
ribosomal biogenesis and DNA repair (97–99).
Ubiquitination is an important post-translational
modification. Besides regulating protein degradation (93), it also serves important roles in
modulating the activity and localization of proteins,
receptor-mediated endocytosis and the transportation of lysosomes
(100), insulin levels (101) and the transforming growth factor
(TGF)-β pathway (102). Thus,
various cellular biological processes, including gene expression,
cell proliferation and differentiation, cell senescence and
apoptosis, autophagy, antigen presentation, signal transduction,
DNA repair and immune responses are all associated with
ubiquitination to varying degrees (103).
It has been demonstrated that ubiquitination is a
reversible process that is counteracted by deubiquitinating enzymes
(DUBs). DUBs recognize specific sequences of ubiquitin, its
substrates or the proteasome, and primarily remove ubiquitin from
the target proteins to recycle the ubiquitin, to inhibit and
proofread ubiquitination, or to alter the activity state of
proteins (104). Based on their
Ub-protease domains, DUBs are divided into five subclasses
consisting of ubiquitin C-terminal hydrolases, ubiquitin-specific
proteases (USPs), ovarian tumor-related proteases,
JAB1/PAB1/MPN-domain-containing metallo-enzymes, and Machado-Joseph
disease protein domain proteases (105). Likewise, deubiquitination also
functions in metabolism, stress responses, inflammation, immunity
and other cellular activities.
An increasing number of studies have revealed that
ubiquitination and deubiquitination are associated with the
pathogenesis of numerous diseases, including cancer (106–113). These processes serve as promoters
and/or suppressors of cancer initiation and progression by
regulating cell proliferation and differentiation, cell migration,
the cell cycle, signal transduction and DNA repair (114,115). It is generally considered that the
ectopic expression of oncogenes and anti-cancer genes contributes
to tumorigenesis. Specifically, if tumor suppressors and
oncoproteins are not properly sequestered or eliminated, it may
lead to neoplasia. For instance, p53 is an anticancer protein known
as the ‘guardian of the genome’ (116). Murine double minute chromosome 2
(MDM2) acts as an E3 enzyme and marks p53 for proteasomal
degradation (117). MDM2
expression is increased in numerous tumors and is a crucial reason
for the decreased expression of wild-type p53 in malignancies
(118). The ubiquitin-specific
proteases USP7 (also known as HAUSP) and USP42 can also cleave
ubiquitin from p53, thereby protecting it from proteasome-dependent
degradation via the ubiquitin ligase pathway (119,120). Another classical example is the
nuclear factor-κB (NF-κB) signaling pathway, which is ubiquitous in
eukaryotic cells and is regarded as a facilitator of cancer
initiation (121). NF-κB inhibitor
(IκB) functions as a tumor suppressor by preventing the entry of
NF-κB into the nucleus, and thus preventing the activation of the
downstream signaling pathways (122). β-Trcp is a member of the E3
protein family that targets IκB for degradation, and activates
NF-κB signaling to drive cell malignant transformation (123). Conversely, USP15 inhibits IκB
turnover (124) and
cylindromatosis downregulates TNF receptor associated factor 6 to
repress this pathway (125).
Additionally, USP10, USP11, USP22 and USP48 are overexpressed in
malignant lymphoma, and USP17 expression is elevated in esophageal
and cervical carcinoma (126).
Notably, enzymes involved in ubiquitination and
deubiquitination may serve as either oncoproteins or tumor
suppressors, depending on the functional output of their substrate.
For example, USP2 stabilizes fatty acid synthase (FAS), an
apoptotic protein, and MDM2 via deubiquitination, leading to p53
degradation, escape from apoptosis and the development of drug
resistance in prostate cancer cells (127). On the other hand, USP2 has also
been reported to be downregulated in breast cancer (128), implying that it may be a tumor
suppressor. Taken together, it may be considered that
ubiquitination and deubiquitination are closely associated with
cancer and serve roles in oncogenesis and tumor progression via
different mechanisms. Considering the importance of the Hippo
pathway in cancer, it is of interest to investigate the precise
association between ubiquitination-deubiquitination and the Hippo
pathway.
Ubiquitination-deubiquitination in the Hippo
pathway
There are multiple components of the Hippo pathway,
including LATS1/2, YAP/TAZ, AMOT and VGLL4 which are crucial and
have been extensively studied. Each of these is described below, in
addition to a number of other components (Figs. 2 and 3; Tables I
and II).
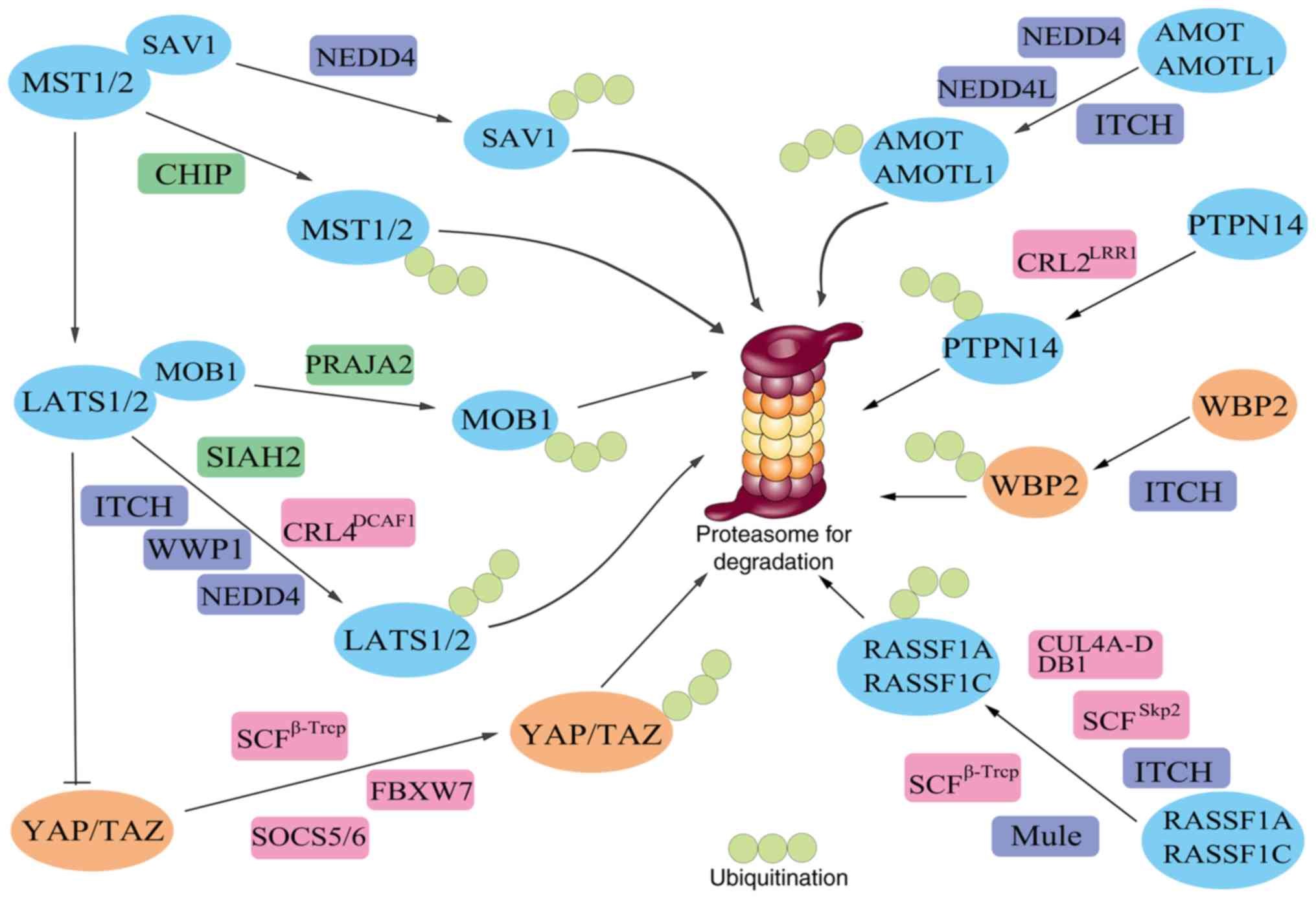 | Figure 2.Ubiquitination and proteasomal
degradation of components of the Hippo pathway. E3 ubiquitin
ligases (dark blue, green and pink) transfer ubiquitin (light
green) to the Hippo pathway components (blue and orange), which are
subsequently recognized by the 26S proteasome and targeted for
proteolysis. AIP4, atrophin-1 interacting protein 4; AMOT-p130,
angiomotin-p130; AMOTL, angiomotin-like; β-Trcp, β-transducin
repeat-containing E3 ubiquitin protein ligase; CHIP, C terminus of
Hsp70 interacting protein; CRL2, cullin 2-RING ubiquitin ligase;
CRL4, cullin 4-RING ubiquitin ligase; DCAF1, DDB1 and CUL4
associated factor 1; DDB1, DNA damage-binding protein 1; LATS,
large tumor suppressor; LRR1, leucine-rich repeat protein 1; MOB1,
MOB kinase activator 1; MST, mammalian STE20-like protein kinase;
NEDD4, neural-precursor-cell-expressed developmentally
downregulated 4; NEDD4L (NEDD4-2), (neural-precursor-cell-expressed
developmentally downregulated 4)-like; NEDL2, NEDD4-like ubiquitin
protein ligase 2; PC2, polycystin 2; PTPN14, protein tyrosine
phosphatase non-receptor type 14; RASSF1, Ras association domain
family 1; SAV1, Salvador homologue 1; SCF, Skp1-Cullin1-F-box;
SIAH2, seven in absentia homolog 2; TAZ, transcriptional
co-activator with PDZ-binding motif; WBP2, WW domain-binding
protein 2; WWP1, WW domain containing E3 ubiquitin protein ligase
1; YAP, Yes-associated protein. |
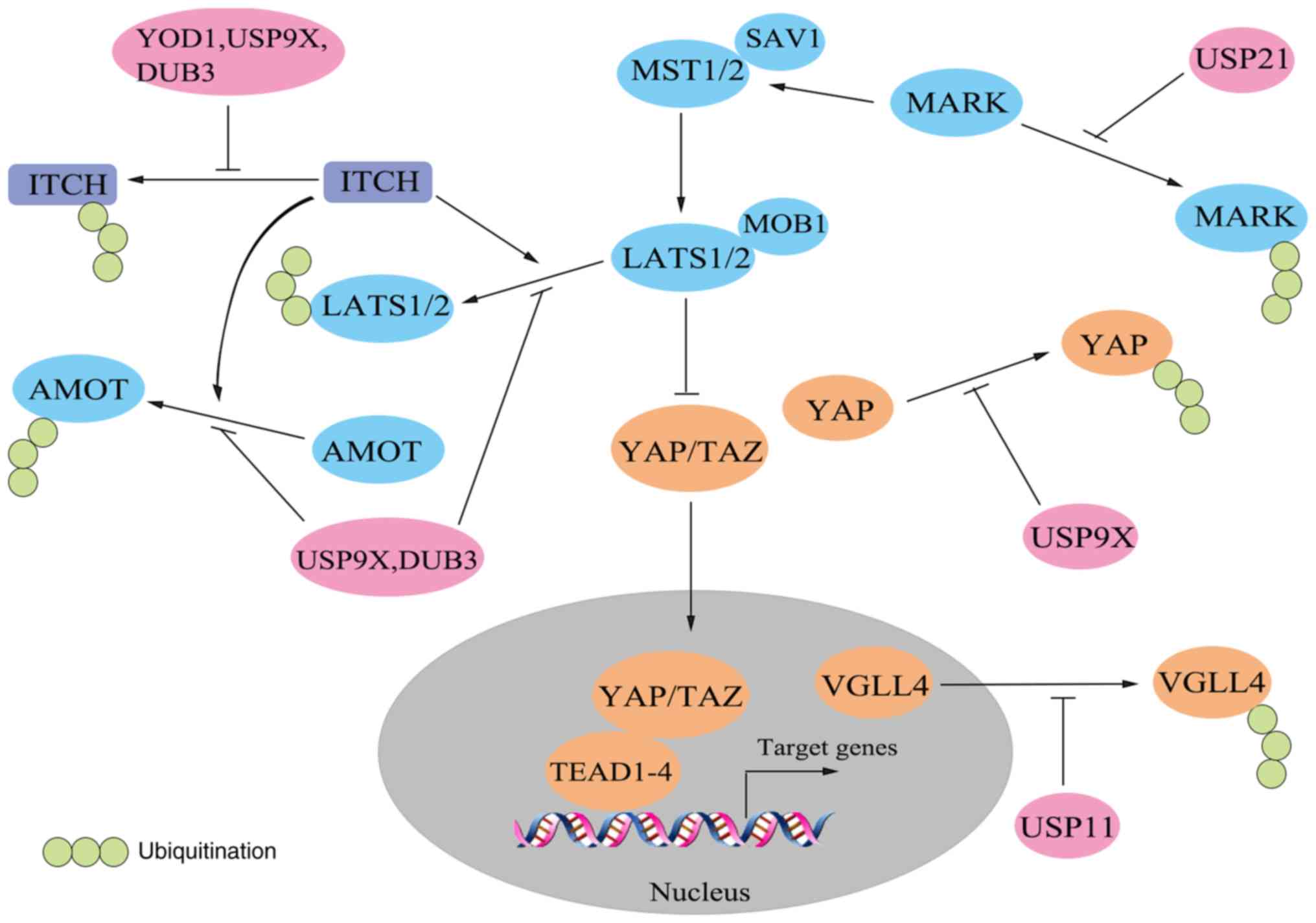 | Figure 3.Deubiquitination in the Hippo
pathway. Deubiquitinases (pink) counteract the ubiquitination of
ITCH and Hippo pathway components (blue). AMOT, angiomotin; DUB,
deubiquitinase; LATS, large tumor suppressor; MARK, microtubule
affinity-regulating kinase; MOB1, MOB kinase activator 1; MST,
mammalian STE20-like protein kinase SAV1, Salvador homologue 1;
TAZ, transcriptional co-activator with PDZ-binding motif; TEAD, TEA
domain-containing sequence-specific transcription factor; USP,
ubiquitin-specific protease; VGLL4, vestigial-like family member 4;
YAP, Yes-associated protein. |
 | Table I.Summary of the ubiquitination in the
Hippo pathway. |
Table I.
Summary of the ubiquitination in the
Hippo pathway.
| Author, year | Components of the
hippo pathway | E3 ubiquitin
ligases | Interaction | Effects | (Refs.) |
|---|
| Salah et al,
2011; Yeung et al, 2013 | LATS1 | ITCH, WWP1 | WW-PPxY | LATS1
ubiquitination and degradation, YAP/TAZ activation | (144,146) |
| Bae et al,
2015 |
| NEDD4 | Uncertain | LATS1
ubiquitination and degradation, YAP/TAZ activation | (147) |
| Ma et al,
2015 |
| SIAH2 | Uncertain | LATS1
ubiquitination and degradation, YAP activation | (150) |
| Li et al,
2014 |
|
CRL4DCAF1 | Ubiquitination
sites-DCAF1 | LATS1
poly-ubiquitination and degradation in the nucleus, YAP/TAZ
activation | (152) |
| Bae et al,
2015 | LATS2 | NEDD4 | Uncertain | LATS2
ubiquitination and degradation, YAP/TAZ activation | (147) |
| Ma et al,
2015 |
| SIAH2 | Lysine residues of
LATS2 are involved | LATS2
ubiquitination and degradation, YAP activation | (150) |
| Li et al,
2014 |
|
CRL4DCAF1 | Ubiquitination
sites-DCAF1 | LATS2
oligo-ubiquitination and inactivation in the nucleus, YAP/TAZ
activation | (152) |
| Bae et al,
2015 | WW45(SAV1) | NEDD4 | N-terminal
regions | WW45 ubiquitination
and degradation, the Hippo pathway inactivation | (147) |
| Zhao et al,
2010 | YAP |
SCFβ-Trcp | Phosphorylated
phosphodegron of YAP (S381 phosphorylation by LATS,
S384/387phosphory-lation by CK1)-β-Trcp | YAP ubiquitination
and degradation | (174) |
| Tu et al,
2014 |
| FBXW7 | Uncertain | YAP ubiquitination
and degradation | (185) |
| Hong et al,
2015 |
| Elongin
B/C-Cullin5- SOCS5/6 | YAP-SOCS5/6 | YAP ubiquitination
and degradation | (186) |
| Liu et al,
2010 | TAZ |
SCFβ-Trcp | Phosphorylated
phosphodegron of TAZ (S311 phosphorylation by LATS,S314
phosphorylation by CK1)-β-Trcp | TAZ ubiquitination
and degradation | (175) |
| Tian et al,
2007 |
|
SCFβ-Trcp | Phosphorylated S314
by NEK1-β-Trcp | TAZ serves as an
adaptor for β-TrCP to promote ubiquitination of PC2 | (180) |
| Huang et al,
2012 |
|
SCFβ-Trcp | Phosphorylated
S58/62 by GSK3-β-Trcp | TAZ ubiquitination
and degradation | (182) |
| Wang et al,
2012 | AMOT-p130 | NEDD4, NEDD4-2
(NEDD4L),ITCH | L/P-PxY-WW | AMOT-p130
ubiquitination and degradation | (201) |
| Adler et al,
2013 |
| AIP4(ITCH) | L/P-PxY-WW | AMOT-p130
ubiquitination and stabilization, YAP degradation in the YAP-AMOT
p130-AIP4 complex | (202) |
| Wang et al,
2012 | AMOTL1 | NEDD4, NEDD4-2
(NEDD4L), ITCH | L/P-PxY-WW | AMOTL1
ubiquitination and degradation | (201) |
| Choi et al,
2016 |
| NEDL2(HECW2) | K63 ubiquitination
site of AMOTL1 is | AMOTL1 increased
stability involved | (204) |
| Kim et al,
2016 | AMOTL2 | Uncertain | K347, K408
ubiquitination sites of AMOTL2 are involved. | AMOTL2
mono-ubiquitination, LATS activation and YAP inhibition in response
to cell confluence | (205) |
| Wang et al,
2012 | PTPN14 |
CRL2LRR1 | PTPN14-LRR1 | PTPN14
ubiquitination and degradation, YAP/TAZ activation | (218) |
| Lim et al,
2016 | WBP2 | ITCH | PPxY-WW | WBP2 ubiquitination
and degradation | (222) |
| Pefani et
al, 2016 | RASSF1A | ITCH | Uncertain | RASSF1A
ubiquitination and degradation, allowing YAP1 to interact with
SMAD2 and drive target genes transcription | (235) |
| Song et al,
2008 |
|
SCFSkp2 | Ser203 of RASSF1A
is involved | RASSF1A
ubiquitination and degradation, promoting the transition from
G1 to S phase | (238) |
| Jiang et al,
2011 |
| CUL4A-DDB1 | A region containing
amino acids 165–200-DDB1 | RASSF1A
ubiquitination and degradation during M phase, promoting cell cycle
progression | (239) |
| Zhou et al,
2012 | RASSF1C | Mule,
SCFβ-Trcp | Uncertain | RASSF1C
ubiquitination and degradation in response to stress signals | (240) |
| Lignitto et
al, 2013 | MOB1 | PRAJA2 | Uncertain | MOB1 ubiquitination
and degradation, attenuation of the Hippo pathway | (243) |
| Ren et al,
2008 | MST | CHIP | Uncertain | MST ubiquitination
and degradation, attenuation of the Hippo pathway | (245) |
 | Table II.Summary of the deubiquitination in
the Hippo pathway. |
Table II.
Summary of the deubiquitination in
the Hippo pathway.
| Author, year |
Deubiquitinases | Targets | Effects | (Refs.) |
|---|
| Kim et al,
2017 | YOD1 | ITCH | Stabilizes ITCH,
thereby promoting LATS degradation and YAP activation | (154) |
| Toloczko et
al, 2017 | USP9X | LATS | Deubiquitinates
LATS, thereby inhibiting YAP/TAZ activity | (156) |
| Li et al,
2018 |
| YAP1 | Deubiquitinates and
stabilizes YAP1, thereby promoting YAP1 activity | (189) |
| Thanh Nguyen et
al, 2016 |
| AMOT | Deubiquitinates
AMOT, thereby inhibiting YAP/TAZ activity | (157) |
| Mouchantaf et
al, 2006 |
| ITCH | Antagonizes ITCH
auto-ubiquitination | (206) |
| Nguyen et
al, 2017 | DUB3 | ITCH | Antagonizes ITCH
auto-ubiquitination | (159) |
| Nguyen et
al, 2017 |
| LATS | Stabilizes LATS and
increases LATS protein level, thereby inhibiting YAP/TAZ
activity | (159) |
| Nguyen et
al, 2017 |
| AMOT | Stabilizes AMOT,
thereby inhibiting YAP/TAZ activity | (159) |
| Zhang et al,
2016 | USP11 | VGLL4 | Stabilizes VGLL4,
thereby inhibiting YAP-TEAD interaction | (214) |
| Nguyen et
al, 2017 | USP21 | MARK | Stabilizes MARK,
thereby activating LATS and inhibiting YAP/TAZ activities | (242) |
LATS1/2
LATS kinases have gained much research interest
owing to their wide range of activities in numerous biological
processes. During mammalian evolution, a genomic duplication event
led to the emergence of two paralogs (LATS1 and LATS2) (129). These are serine/threonine kinases
of the AGC subfamily sharing extensive sequence similarity within
their kinase domain (85% similarity) situated at the C terminus of
the proteins, where they each contain a hydrophobic motif (130). In addition, the N terminus
contains two stretches of conserved amino-acid sequences (LCD1 and
LCD2) and ubiquitin-associated (UBA) domains that bind to
ubiquitinated proteins. LATS1 harbors a proline-rich domain,
whereas a PAPA repeat exists in LATS2. In addition, LATS2 contains
one PPxY (P, proline; X, any amino acid; Y, tyrosine) motif and
LATS1 contains two (131,132). There also exist a number of
phosphorylation, auto-phosphorylation and ubiquitination sites on
the two proteins at different locations. It is hypothesized that
the structural distinctions between LATS1 and LATS2 are conducive
to their divergent regulation and functions (133).
LATS1 and LATS2 are notable regulators of cell fate
and are associated to cancer initiation and progression by
mediating the functions of oncogenic or anti-tumor effectors during
the processes of the cell cycle, apoptosis, migration and EMT
(134). The most well-known
mechanism is that of LATS1/2 as tumor suppressors, which restrict
YAP/TAZ activity in the classical Hippo pathway (2). However, beyond the Hippo pathway,
studies have determined that LATS1 is able to interact with HSPA2
and FKBP5 to modulate the estrogen signaling pathway (135). In breast tissues, LATS2 controls
estrogen receptor (ER) activity (136); whereas, in the prostate, androgen
receptor chromatin binding and transcriptional activities are
restrained by LATS2 (137).
Furthermore, LATS kinases have been demonstrated to suppress the
activity of ER by promoting its degradation (138). All these findings support the
inhibitory roles of LATS in breast and prostate cancers. In
addition, LATS2 associates with p53, proliferating cell nuclear
antigen, Aurora A and other proteins that are involved in cell
cycle metabolism (135).
Strikingly, loss of LATS1/2 kinases has been reported to result in
the induction of anti-cancer immune responses that decrease tumor
growth and improve vaccine efficacy (139), revealing a critical role of the
Hippo pathway in modulating tumor immunogenicity. The wide range of
regulatory functions of LATS1/2 are of great importance in
biological homeostasis. More detailed studies of LATS1/2 may
provide effective methods for the detection and treatment of
cancer.
In recent years, ubiquitination has been identified
as a mode of post-translational modification shared by LATS1 and
LATS2 proteins. According to their structural characteristics, E3
ubiquitin ligases are divided into two main subfamilies, RING
finger and HECT (140). The
neural-precursor-cell-expressed developmentally downregulated 4
(NEDD4)-like ubiquitin ligase family belongs to the HECT subfamily,
consisting of E3 ubiquitin-protein ligase Itchy homolog (ITCH),
NEDD4, NEDD4-2, WW domain containing E3 ubiquitin protein ligase 1
(WWP1), WWP2, E3 ubiquitin-protein ligase SMURF1 (Smurf1), Smurf2,
NEDD4-like ubiquitin protein ligase (NEDL)1 and NEDL2 in humans
(141). Each member of this
subfamily contains a Ca2+/lipid-binding (C2) domain
associated with membrane localization, 2–4 WW domains conferring
substrate specificity, and a HECT-type ligase domain contributing
to the catalytic E3 activity (142,143). An increasing number of studies
have indicated that members of the NEDD4-like ubiquitin ligase
family interact with LATS1/2, thereby affecting the activity and
functional outcomes of the Hippo pathway.
The ITCH ligase contains four WW domains and
physically interacts with LATS1, primarily through binding of its
first WW domain (WW1) to the PPxY motifs of LATS1, leading to LATS1
ubiquitination and degradation by the 26S proteasome (144). ITCH-mediated turnover of LATS1
results in increased cell proliferation, decreased apoptosis,
greater tumorigenesis and increased EMT, which may be associated in
part with the accumulation of nuclear YAP and its enhanced
transcriptional coactivation function (144). Furthermore, activation of the
Hippo pathway, for example by MST activation, upregulates ITCH and
thereby promotes the ITCH-LATS1 interaction, which indicates the
presence of a negative feedback loop (144). Other studies have demonstrated
that overexpression of ITCH in breast cancer cells is associated
with increased tumor formation and progression, and that in cases
of invasive and metastatic breast cancer, ITCH expression is
strikingly elevated (145). Thus,
it may be inferred that ITCH exerts a tumor-boosting function by
triggering LATS1 degradation and YAP activation, and may be
considered a biological marker and therapeutic target in breast
cancer. Likewise, WWP1, another member of the NEDD4-like ubiquitin
ligase family, negatively regulates LATS1 turnover in a similar
manner, which is also crucial for enhanced cell proliferation in
breast cancer cells (146). In
contrast to the ITCH-LATS1 interaction, LATS1 primarily binds to WW
domains 1–3 of WWP1 through its two PPxY motifs (146). However, in another study, NEDD4
(also termed NEDD4-1) was reported to control intestinal stem cell
homeostasis by regulating WW45 (SAV1) and LATS1/2, which are
polyubiquitinated by NEDD4 and degraded by the 26S proteasome,
resulting in the inactivation of the Hippo pathway (147). Additionally, these findings reveal
that NEDD4 modulates LATS directly and indirectly. In general, WW45
associates with its binding partners through either the WW domains
or the C-terminal Sav-Ras association domain family (RASSF)-Hpo
(SARAH) domain (148). In one
study, NEDD4 and WW45 were demonstrated to interact with each other
directly through their N-terminal regions (147). In contrast to WW45, the C-terminal
region of NEDD4 likely serves important roles in its interaction
with LATS2 (147). Furthermore,
NEDD4 is capable of destabilizing LATS1 (147), although the underlying interaction
and mechanism remain unclear and merit additional
investigation.
In addition to the distinct E3 ligase NEDD4-like
ubiquitin ligase, seven in absentia homolog 2 (SIAH2), which is an
important component of the hypoxia response pathway (149), promotes LATS1 and LATS2
degradation in a UPS-dependent manner to stimulate YAP activity
(150). Hypoxia is known to be a
common feature of solid tumors (151). Low SIAH2 levels with stabilized
LATS2 and upregulated pYAP suppress tumorigenesis in a xenograft
mouse model (150), indicating
that the SIAH2-LATS2 axis likely serves a role in tumor formation.
Amino acids 403–480 and amino acids 667–720 are two critical
regions of LATS2 that are responsible for the SIAH2-LATS2
interaction and contain a relatively larger number of lysine
residues in LATS2, among which Lys 670 and Lys 672 are two key
sites that contribute to SIAH2-mediated ubiquitination and
proteolysis (150). According to
the structural similarity of LATS1 and LATS2, it may be
hypothesized that an analogous binding mechanism is involved in the
SIAH2-LATS1 interaction, although this requires further study.
In addition, the activity of the nuclear E3
ubiquitin ligase CRL4-DDB1 and CUL4 associated factor 1
(CRL4DCAF1), which belongs to the RING-finger subfamily,
is repressed by NF2, thereby inhibiting the proteasomal degradation
of LATS1 and the ubiquitination-induced conformational alteration
in LATS2 (152).
CRL4DCAF1 comprises the scaffold protein Cullin 4A
(CUL4A), the catalytic subunit Roc1/Rbx1, the adaptor DNA
damage-binding protein 1 (DDB1), and the substrate recognition
module DCAF1 (DDB1 and CUL4 associated factor 1), which directly
binds the ubiquitination sites of LATS via its WD40 domain
(153). The number and location of
ubiquitination sites differ between LATS1 and LATS2, which may be
one of the reasons for the distinct regulatory mechanisms employed
by CRL4DCAF1 and the consequences of
CRL4DCAF1 mediation.
Deubiquitination, the inverse process of
ubiquitination, is also involved in regulation of the Hippo
pathway. Kim et al (154)
demonstrated using unbiased small interfering RNA screening that
the DUB ubiquitin thioesterase OTU1 (YOD1) controls the biological
responses mediated by YAP/TAZ. YOD1 deubiquitinates and stabilizes
ITCH, contributing to the degradation of LATS1 and the activation
of YAP. Overexpression of YOD1 leads to enhanced hepatocyte
proliferation and hepatomegaly in a YAP-dependent manner in a
transgenic mouse model, and a strong connection exists between YOD1
and YAP expression in patients with liver cancer, which implies
that YOD1 may promote tumor initiation (155). In comparison, USP9X (FAM)
deubiquitinates LATS kinases to suppress tumor growth (156). The DUB3 family of deubiquitinating
enzymes, which includes 25 USP17-like proteins, was identified,
using a short hairpin RNA-based screening technique, to be a
mediator of YAP/TAZ activity (157,158). Notably, DUB3 antagonizes ITCH
auto-ubiquitination, protecting ITCH from degradation. Higher ITCH
expression is believed to decrease the levels of LATS kinase;
conversely LATS kinases are able to bind to DUB3, resulting in its
increased stability and elevated protein expression (159).
In conclusion, ITCH and WWP1 specifically
destabilize LATS1. NEDD4 and SIAH2 may target LATS1 and LATS2 for
destruction, leading in certain contexts to YAP/TAZ-driven
overgrowth and neoplasia. Given that YAP and TAZ are capable of
binding LATS1/2 through their WW domains (160), the WW-PPxY interaction between
these NEDD4-like E3 ligases and LATS1/2 may reduce LATS1/2
expression levels and displace YAP/TAZ from its PPxY-binding site,
possibly causing YAP/TAZ dephosphorylation and increased oncogenic
activity. The E3 ubiquitin ligases and DUBs involved in modifying
LATS act as oncogenes and/or tumor suppressors, suggesting a new
and attractive strategy for the prognosis and treatment of
tumors.
YAP/TAZ
YAP and TAZ are regarded as the prime mediators of
the major functional outputs of the Hippo pathway, and act as a
signaling nexus and integrators of numerous other signaling
pathways, including the Wnt, G protein-coupled receptor (GPCR),
epidermal growth factor and Notch pathways (17). TAZ shares ~50% sequence identity
with YAP (161) and they
structurally have much in common. The most prominent feature of YAP
or TAZ is the WW domain, which contains two conserved and
consistently-positioned tryptophan residues. These domains are
responsible for conferring signaling specificity and thereby
controlling YAP/TAZ localization and activity (160). In the C-terminal region, YAP and
TAZ each contain a PDZ-binding motif that interacts with the PDZ
domain of other proteins, including tight junction protein ZO-2 and
Na(+)/H(+) exchange regulatory cofactor NHE-RF2 to direct the
localization of YAP and TAZ (162,163). An unstructured transcriptional
activation domain exists in the extended C terminus of YAP and TAZ,
which is likely to control their transcriptional roles (164). They also possess a domain that
modulates TEAD transcription factor binding. However, in YAP, this
region possesses a PxxΦP motif (x, any amino acid; Φ, a hydrophobic
residue) that is absent in TAZ, resulting in differences between
YAP and TAZ interactions with TEAD transcription factors (165). Though YAP and TAZ share numerous
structural features, certain distinctions are apparent. For
example, YAP possesses two WW domains, whereas TAZ has only one.
YAP contains a proline-rich region in the N terminal region, whose
interplay with heterogeneous nuclear ribonuclear protein U is
involved in mRNA processing, although TAZ lacks this proline-rich
region (166). In addition, an
SH3-binding motif is present on YAP that does not exist in TAZ
(167).
Growing evidence supports the idea that YAP and TAZ
are oncogenes in mammalian cells. Overexpression of YAP/TAZ in
normal epithelial cells promotes cell transformation and confers a
cancer stem cell phenotype (168,169). In mice, upregulated YAP levels
enhance tissue hyperproliferation and lead to cancer development in
various epithelial tissues (43).
Furthermore, as a signaling nexus, YAP and TAZ may be influenced by
the aberrant activity of other pathways, including the GTPase KRAS
signaling pathway (170), which is
critical for tumor development. It is generally considered that YAP
and TAZ exert their oncogenic functions by regulating the
expression of target genes. Further investigation has suggested
that YAP in certain context acts as a tumor suppressor, in part by
dampening Wnt signaling (171) and
inducing DNA damage-induced apoptosis (172). Considering the fact that YAP and
TAZ are able to interact with various transcription factors that
have dissimilar functions, it may be inferred that their function
as oncogenes may depend on the cellular and signaling context and
varies in different types of cancer.
Like LATS, YAP and TAZ are also directly modulated
by ubiquitination and deubiquitination. YAP has five consensus
HXRXXS motifs in which Ser 127 is phosphorylated following the
activation of LATS, leading to 14-3-3 binding and the cytoplasmic
retention of YAP (173). This
results in YAP being sequestered from the nucleus and being unable
to drive target gene expression. In TAZ, the well-studied Ser 89 is
required for 14-3-3 binding and cytoplasmic retention (161). Apart from this spatial separation,
YAP and TAZ are similarly degraded in a β-Trcp-dependent manner
(174,175).
A member of the RING-finger E3 family,
Skp1-Cullin1-F-box (SCF), consists of two scaffold proteins (Skp1
and Cullin1), the RING-finger domain protein Rbx1 and an F-box
(176). There are a variety of
F-box proteins, which may be further subclassified into three
families, FBXL, FBXO and FBXW. The FBXW family comprises ten
proteins that contain the F-box motif in their N-terminal for
interacting with Skp1 (177), and
seven WD40 repeats in the C-terminal for substrate specificity and
promoting ubiquitination (178).
β-Trcp belongs to the FBXW family and is highly conserved with two
paralogs, β-Trcp1 (also termed FBXW-7) and β-Trcp2 (also termed
FBXW-11) in mammals (179).
SCFβ−Trcp functions by recognizing a DSGXXS motif in the
substrate. Only when the serine residues are phosphorylated in this
motif, which is termed phosphodegron, may SCFβ−Trcp bind
to the substrate and thus lead to ubiquitination and degradation of
the target protein (179). A
recent study has reported that YAP has a DSGXS motif, analogous to
the classical DSGXXS motif (174).
When the Hippo pathway is switched on, activated LATS kinases
phosphorylate YAP on Ser 381 in one of the HXRXXS motifs, priming
YAP for subsequent phosphorylation at Ser384 and Ser387 in the
C-terminal phosphodegron by casein kinase I (CK1). Phosphorylated
phosphodegron is recognized by SCFβ−Trcp, resulting in
the ubiquitination and proteasome-mediated degradation of YAP,
thereby inhibiting the oncogenic activity of YAP (174). In addition, Ser387 phosphorylation
is known to be crucial for YAP ubiquitination and is indispensable
(174). Likewise, TAZ is
recognized by SCFβ−Trcp and degraded via a similar
mechanism. LATS and CK1 phosphorylate TAZ at Ser 311 and Ser 314,
respectively (175). Furthermore
phosphorylation of TAZ at Ser314 by another kinase,
serine/threonine-protein kinase NEK1, also recruits β-TrCP;
however, under these circumstances, TAZ serves as an adaptor for
β-TrCP to promote the ubiquitination of the calcium-permeable
cation channel protein polycystin 2 (180), thereby mediating cilia-directed
signaling (181). Notably, the
N-terminal phosphodegron of TAZ, which is not shared by YAP, also
affects the regulation of TAZ protein abundance. It is
phosphorylated at Ser58/62 by GSK3, recruiting SCFβ−Trcp
and thus triggering TAZ ubiquitination and degradation, which
notably does not appear to require prior phosphorylation by LATS
(182). Elevated levels of TAZ
with increased activation of PI3K signaling has been observed in
cancer (183), and activated PI3K
may inhibit GSK3. It is therefore possible that TAZ works as a
downstream effector of the PI3K pathway to regulate tissue growth
and tumor development. Therefore, ubiquitination induced by the
either the C-terminal or N-terminal phosphodegron may serve roles
in modulating the activity and biological functions of TAZ.
Another FBXW family protein, FBXW7, is regarded as a
tumor suppressor in human cancer (184). A recent study demonstrated that
decreased expression of FBXW7 is associated with poor
clinicopathological features. It induces apoptosis and growth
arrest, at least in part, by targeting YAP for ubiquitination and
degradation in HCC (185).
However, further investigation is required to confirm whether the
interaction between FBXW7 and YAP is similar to that between
SCFβ−Trcp and YAP, and whether FBXW7 is able to target
TAZ for degradation. In addition, YAP protein turnover may be
mediated by RAS signaling through regulation of suppressor of
cytokine signaling (SOCS)5 and SOCS6 expression (186). SOCS5 and SOCS6 serve as substrate
recognition modules of the Elongin B/C-Cullin 5 ubiquitin ligase
complex (187) and are able to
recruit YAP for ubiquitination. Activated RAS signaling may
downregulate SOCS5 and SOCS6, promoting the stability of YAP. In
addition, the RAS/mitogen-activated protein kinase pathway acts via
phosphorylation of Ajuba to inactivate LATS kinases (188), which causes reduced YAP
phosphorylation, SCFβ−Trcp-dependent YAP proteolysis and
increased YAP activity. RAS exerts its oncogenic functions, at
least in part, by these mechanisms.
As for deubiquitination, a recent study determined
that USP9X targets YAP1 for deubiquitination and stabilization,
thereby promoting breast cancer cell survival and progression
(189). Elimination of USP9X
upregulates YAP1 degradation and renders cells more sensitive to
chemotherapy (189), suggesting an
oncogenic role for USP9X and identifying it as a potential
therapeutic target in breast cancer treatment.
Collectively, the turnover of YAP and TAZ is
primarily controlled by SCFβ−Trcp, which is of central
importance regarding the abundance of YAP/TAZ and the functional
outcomes of the Hippo pathway. In addition to SCFβ−Trcp,
FBXW7 and Elongin B/C-Cullin5-SOCS5/6, along with certain other E3
ubiquitin ligase enzymes, have been demonstrated to directly
modulate YAP/TAZ degradation and their activity. However, there is
still a need to identify whether there are other DUBs involved in
counteracting these E3 ligases and rescuing YAP and TAZ from
degradation.
Angiomotin (AMOT)
AMOT was originally identified as a protein that
interacted with angiostatin, an inhibitor of angiogenesis (190). The AMOT family is composed of
AMOT, which exists as AMOT-p130 or AMOT-p80, angiomotin-like 1
(AMOTL1), and angiomotin-like 2 (AMOTL2), which are characterized
by coiled-coil domains in the N terminus and a consensus
PDZ-binding domain in the C terminus (191). AMOT-p130 differs from AMOT-p80 in
its N-terminal cytoplasmic extension of 409 amino acids, which is
rich in glutamine and mediates the binding of AMOT-p130 to
filamentous (F)-actin and cell-cell tight junctions (192). The AMOT family members usually
serve as tight junction proteins that control endothelial cell (EC)
junction stability and permeability (190) and are expressed predominantly in
the endothelial cells of capillaries, in addition to angiogenic
tissues such as solid tumors (193). Furthermore, an increasing number
of studies have demonstrated that AMOT is able to interact with
LATS and YAP/TAZ to exert their regulatory roles in the Hippo
pathway.
On one hand, AMOT primarily utilizes its first and
second L/P-PxY motifs in the N terminus to directly associate with
the WW domains of YAP/TAZ, recruiting them to tight junctions and
causing the cytoplasmic retention and decreased activity of YAP/TAZ
(194,195). Since AMOT is an F-actin-binding
protein, YAP/TAZ and F-actin compete for binding to AMOT (196). On the other hand, the Crumbs
homolog (CRB) complex, localized to apical junctions, recruits
AMOT, which may directly bind and activate LATS, thereby leading to
the downregulation of YAP/TAZ activity (197,198). Additionally, activated LATS
kinases phosphorylate AMOT through a conserved HXRXXS consensus
site situated in the N-terminal regions of AMOT members, which
results in the separation of AMOT from F-actin at the junctions of
epithelial cells. This may enhance the interaction between AMOT and
YAP/TAZ in the cytoplasm, resulting in the suppression of cell
proliferation and tissue growth by AMOT (199). Taken together, AMOT downregulates
YAP/TAZ activity in LATS-independent and LATS-dependent manners,
and thereby functions as a tumor suppressor. However, a
contradictory report has demonstrated that AMOT-p130 enhances
YAP-mediated hepatic epithelial cell proliferation and
tumorigenesis by promoting the nuclear localization of YAP, and by
forming a functional complex with YAP and TEADs on target genes,
indicating an oncogenic role of AMOT (200). Different cellular and molecular
conditions may provide a possible explanation for this discrepancy,
although further analysis is necessary.
Due to the significant roles of AMOT in the Hippo
pathway, it is important to determine the modulatory mechanism of
AMOT by ubiquitination and deubiquitination. A study demonstrated
that three NEDD4-like ubiquitin ligases, NEDD4, NEDD4-2 (also known
as NEDD4L) and ITCH, mediate the polyubiquitination of AMOT-p130
in vivo (201).
Overexpression of NEDD4, NEDD4-2 or ITCH results in the efficient
ubiquitination and proteasomal degradation of AMOT-p130, which
depend on the interaction between the WW domains of NEDD4-like
ubiquitin ligase and the L/P-PxY motifs of AMOT-p130. The short
isoform AMOT-p80 cannot be ubiquitinated and degraded by NEDD4-like
ubiquitin ligase due to a lack of L/P-PxY motifs (201). According to the pattern similarity
between YAP/TAZ-AMOT and AMOT-NEDD4-like ubiquitin ligase, YAP/TAZ
may compete with NEDD4 for binding to AMOT-p130. Another study
demonstrated that atrophin-1 interacting protein 4 (AIP4; also
termed ITCH) ubiquitinates AMOT-p130 in an analogous manner,
although it reciprocally stabilizes AMOT-p130, which acts as a
scaffold to form a complex in combination with ITCH and YAP.
Consequently, ITCH degrades YAP, which is subsequently prevented
from binding to LATS1, leading to the inhibition of cell
proliferation and tissue growth (202). Collectively, ITCH may promote the
degradation and stabilization of AMOT-p130 through ubiquitination,
for which the precise mechanism requires investigation.
In addition, NEDD4-like ubiquitin ligases (NEDD4,
NEDD4-2, and ITCH) also promote the degradation of AMOTL1 through
the interaction with WW-L/P-PxY (201). Notably, cytoplasmic YAP1 is able
to recruit the tyrosine kinase c-Abl to phosphorylate NEDD4-2 to
maintain the cell tight junctions, thus hampering the
NEDD4-2-mediated degradation of AMOTL1, which suggests that a
feedback loop may exist between NEDD4-2 and YAP (203). On the other hand, NEDL2 (also
termed HECW2) increases the protein stability of AMOTL1 via
K63-linked polyubiquitination and enhances endothelial cell
junctions through the AMOTL1-YAP pathway (204). With respect to AMOTL2, the ligases
NEDD4, NEDD4-2 and ITCH are unable to influence AMOTL2 activity or
promote its degradation since a phenylalanine replaces the tyrosine
in the third L/P-P-X-Y motif of AMOTL2 compared with that of
AMOT-p130 and AMOTL1. This results in the loss of WW domain-binding
capacity (201); however, a recent
study indicated that AMOTL2 is mono-ubiquitinated at K347 and K408
by a certain, currently unidentified, E3 ubiquitin ligase (205). This ubiquitinated AMOTL2, which
may be counteracted by USP9X (157), binds to the LATS UBA domain,
activating LATS and leading to YAP inhibition in response to cell
confluency.
Regarding deubiquitination, using a cell-based RNA
interference screen for YAP/TAZ activity, Thanh Nguyen et al
(157) identified the DUB USP9X as
a negative regulator of YAP/TAZ activity. USP9X deubiquitinates
AMOT (AMOT-p130) at lysine 496, and thus protects AMOT from
degradation and decreases the activity of YAP and TAZ. With reduced
levels of USP9X, AMOT is unable to limit the activity of YAP and
TAZ, which may be one of the reasons why low USP9X expression is
associated with a number of cancer types; for instance, it is
associated with poor outcomes in renal clear cell carcinoma
(157). Another deubiquitinating
enzyme, DUB3, is a potent tumor suppressor that acts by
antagonizing ITCH auto-ubiquitination to prevent its degradation,
simultaneously stabilizing LAST and AMOT to inhibit the activity of
YAP and TAZ (159). In fact, USP9X
has also been reported to cleave ubiquitin from ITCH, acting as a
protective factor (206).
Therefore, from two reports (157,159), it appears that the protection of
AMOT mediated by DUB3 and USP9X offsets the ubiquitination of AMOT
on account of the stabilization and elevated levels of ITCH,
although it is unclear whether the consequences are similar in
other contexts.
Thus, AMOT regulates the functional outputs of the
Hippo pathway primarily by controlling YAP/TAZ activity. The
biological effects resulting from the ubiquitination and
deubiquitination of different members of the AMOT family vary in
diverse contexts, which possibly depends on the specific types of
cells and tissues, upstream stimuli, signal transduction, and other
factors. It is apparent that this is a complex network.
VGLL4 and other components of the
Hippo pathway
VGLL proteins are transcriptional cofactors in the
nucleus that are named after the Drosophila transcriptional
co-activator Vestigial (207).
VGLL1-VGLL4 proteins in mammals are able to interact with TEADs via
their similar sequences in the TEAD-interacting domain (TDU domain)
(207,208). Studies have demonstrated that VGLL
proteins are associated with tumorigenesis. For example, the
VGLL1-TEAD complex, like the YAP/TAZ-TEAD complex, promotes
anchorage-independent cell proliferation by increasing the
expression of proliferation-promoting genes, including such as
IGFBP-5 (209). Downregulation of
VGLL3 leads to a decrease in the proliferation and migration of
soft tissue sarcoma (210).
Furthermore, VGLL4 is considered to be a growth inhibitor and a
common tumor suppressor in various human cancer types, including
lung cancer, breast cancer and esophageal squamous cell carcinoma
(27,211,212). Mechanistically, VGLL4 contains an
extra TDU domain compared with that of VGLL1, VGLL2 and VGLL3, and
is able to compete with YAP and TAZ for binding to TEADs through
its two TDU domains (27). The
extra TDU domain particularly hinders the formation of YAP-TEAD
complexes and downregulates the expression of its target genes
(27). Furthermore, VGLL4 may
promote apoptosis by negatively regulating inhibitor of apoptosis
proteins (213).
With respect to the PTMs of VGLL4, deubiquitinating
enzyme USP11 is known to stabilize VGLL4 through binding of its USP
domain to the N-terminal region of VGLL4 and, in the absence of
USP11, cell proliferation and invasion is enhanced in a
YAP-dependent manner (214). This
suggests that USP11 may also function as a tumor suppressor.
However, no studies have currently identified the E3 ubiquitin
ligase of VGLL4. This E3 ubiquitin ligase may have an oncogenic
function by targeting VGLL4 for proteolysis.
Another Hippo pathway component, tyrosine-protein
phosphatase non-receptor type 14 (PTPN14; also known as Pez, PTPD2
and PTP36) is a classical non-transmembrane protein tyrosine
phosphatase, and was initially identified as a
cytoskeleton-associated protein that serves important roles in cell
adhesion and proliferation (215,216). PTPN14 has an N-terminal FERM
domain that mediates interactions with proteins at the plasma
membrane, a C-terminal phosphatase domain, and central PPxY motifs
(217). It has been reported that
PTPN14 utilizes its PPxY motifs to bind to the WW domains of YAP,
thereby negatively regulating the carcinogenic activity of YAP
(218). In addition, PTPN14 may be
ubiquitinated. The E3 ubiquitin ligase associated with PTPN14 is
termed cullin 2-RING ubiquitin ligase-leucine-rich repeat protein 1
(CRL2LRR1) and consists of the scaffold protein Cullin
2, the RING protein Roc1, the adaptor protein complex of Elongin B
and Elongin C, and the substrate-recognizing adaptor protein
peptidylprolyl isomerase-like 5 (also termed LRR1) (219,220). In response to low cell density,
CRL2LRR1 targets PTPN14 for degradation, thus promoting
YAP nuclear localization and its transactivation activity (218). Additionally, WBP2 acts as an
important co-factor of YAP and TAZ and enhances YAP/TEAD-mediated
and TAZ/TEAD-mediated gene transcription (40,221).
The E3 ligase ITCH mediates the proteasomal degradation of WBP2 to
serve as a tumor suppressor, which relies on the interaction
between its WW domains and the PPxY motifs of WBP2 (222). Noteworthy, it has been reported
that this mode of degradation may be inhibited by WNT3A,
contributing to the development of breast cancer (222). However, currently there are no
reports regarding the role of DUBs in reversing the ubiquitination
of PTPN14 and WBP2.
RASSF consists of two subclasses, C-RASSF and
N-RASSF (223,224). Accumulating evidence indicates
that C-RASSF proteins RASSF1-RASSF6 regulate the Hippo pathway
through interaction with MST via the SARAH domain, which N-RASSF
proteins RASSF7-RASSF10 lack (225). RASSF1A and RASSF1C, which are
ubiquitously expressed, are the principal transcripts of the seven
alternatively spliced variants of the RASSF1 gene, including
isoforms A-G (226). Structurally,
RASSF1A and RASSF1C contain Ras association and SARAH domains,
while RASSF1A also contains a cysteine-rich diacylglycerol-binding
C1 domain that is absent in RASSF1C (227). These differences between RASSF1A
and RASSF1C may result in their distinctive functions. Ectopic
expression of RASSF1A, by either deletions or promoter
hypermethylation of the RASSF1 gene, is associated with various
cancer types (226). RASSF1A is
considered to be a tumor suppressor due to its critical roles in
modulating apoptosis, microtubule stability and cell cycle arrest
(225). One of the notable and
well-known functions of RASSF1A is that it serves as an upstream
regulator of the Hippo pathway. By stabilizing MST (228), preventing the dephosphorylation of
MST (229), or releasing MST from
inhibition by RAF1 and promoting the interaction between MST and
LATS (230), RASSF1A activates
MST, with SARAH-SARAH interactions between RASSF1A and MST serving
a pivotal role (231). In
FAS-induced apoptosis, RASSF1A-activated MST2 phosphorylates LATS1,
leading to YAP1 phosphorylation and its release from LATS1 in the
cytoplasm (230). Consequently,
free YAP1 is able to translocate to the nucleus and form a complex
with p73, which drives the transcription of pro-apoptotic target
genes including BCL2 binding component 3 and BCL2-assocated X,
apoptosis regulator (230). It may
be assumed that this process is a possible explanation for the
tumor-suppressive function of YAP. However, it is notable that
RASSF1A is able to utilize its SARAH domain to associate directly
with the SARAH domain of SAV, stimulating p73 independently of the
canonical Hippo pathway (232).
This adds another layer of complexity to the interplay between
RASSF1A and p73. Additionally, studies have demonstrated that in
cells with methylated RASSF1A, RASSF1C expression is upregulated
and enhances the SRC/YES-mediated phosphorylation of E-cadherin,
and the tyrosine phosphorylation of β-catenin and YAP1, causing
instability in cell junctions and the transcriptional activation of
β-catenin/TBX-YAP/TEAD target genes in the nucleus (233). Also, increased RASSF1C expression
with lower expression of RASSF1A may be observed in breast tumors
(234). These findings, taken
together, indicate that RASSF1C functions as an oncogene.
As for the ubiquitination of RASSF1, it has been
demonstrated that in response to TGF-β, RASSF1A is recruited to
TGF-β receptor I at the cell membrane where it is ubiquitinated and
degraded by E3 ligase ITCH. As a result, YAP1 is able to interact
with mothers against decapentaplegic homolog 2, translocating to
the nucleus and driving the transcription of TGF-β target genes
(235). Furthermore, RASSF1A
represses TGF-β-induced cell invasion (235), serving as a tumor suppressor,
which is attenuated by ITCH-mediated proteolysis. ITCH targets
RASSF5 for degradation through WW-PPxY interactions (236), although the PPxY motif is not
present in RASSF1 (237). Thus, it
may be hypothesized that there is another mechanism of interaction
between RASSF1A and ITCH that requires investigation. Additionally,
RASSF1A may be either a positive or a negative modulator of cell
cycle progression by mediating the expression of cyclin,
cyclin-dependent kinase, cyclin-dependent kinase inhibitors and
other relevant cell-cycle components (225). A previous study determined that
Skp2, the F-box protein and substrate-recognition component of
SCFSkp2 ligase, targets RASSF1A for ubiquitination and
proteolysis at the G1/S transition, which requires prior
phosphorylation of RASSF1A at Ser203 by cyclin D-Cdk4 (238). With the reduction of RASSF1A, cell
cycle progression is accelerated from the G1 to the S
phase, which may contribute to tumorigenesis. Of note, during the
M-phase of the cell cycle, RASSF1A associates with the substrate
adaptor DDB1 via a region containing amino acids 165–200 and is
targeted for degradation by CUL4A-DDB1 E3 ligase, leading to the
promotion of cell cycle progression (239). Therefore, it appears that
SCFSkp2 and CUL4A-DDB1 are able to modulate the
expression of RASSF1A during the cell cycle; however, the factors
that determine the interaction of RASSF1A with either of the two E3
ligases remain unknown. Furthermore, how decreases in the levels of
RASSF1A may affect the Hippo pathway, including the impact on MST,
merits further study.
Another isoform, RASSF1C, is a highly unstable
protein and is primarily degraded in the nucleus. Ubiquitination
has been identified as an important post-translational modification
of RASSF1C. Exposure to stress signals, including those induced by
ultraviolet irradiation, may activate Mule, a HECT family E3
ligase, and SCFβ−Trcp to target RASSF1C for proteasomal
destruction (240). Since the
roles of RASSF1C in the Hippo pathway have been rarely reported, it
is difficult to assess whether RASSF1C ubiquitination affects the
pathway.
While the ubiquitination of RASSF1 has begun to be
defined, currently there are no reports regarding the
deubiquitination of RASSF1 or the DUBs that may be involved. With
deubiquitination being such an important counterbalance to
ubiquitination, more research is required to define this
process.
MARK family proteins are serine/threonine kinases
that have been reported to positively regulate MST and LATS
(241). USP21 is able to
deubiquitinate MARK proteins to control their stability. The
stabilized MARK proteins in turn activate LATS and thereby promote
the phosphorylation of YAP and TAZ (242). Furthermore, evidence has
demonstrated that USP21 restricts the anchorage-independent growth
of transformed primary cells and cancer cell lines, and that its
expression is lower in renal clear cell carcinoma samples compared
with normal renal cells (242),
suggesting that USP21 may be useful as an anti-cancer molecule and
a biomarker.
MOB1 is a regulator of LATS in the Hippo pathway
that contributes to the complete activation of LATS, and is
targeted and degraded by the RING ligase PRAJA2, which attenuates
Hippo signaling, enhances YAP-dependent gene transcription and
bolsters glioblastoma growth (243). In the canonical Hippo pathway,
MST, another core component, is an important upstream activator of
LATS. It is hypothesized that MST may also be regulated by
ubiquitination. Consistent with this, it has been reported that the
C terminus of an Hsp70 interacting protein [E3 ubiquitin-protein
ligase CHIP (CHIP)], which is an E3 ubiquitin ligase of the U-box
protein family (244), is able to
target MST for proteasomal degradation (245). CHIP and its targeting of MST may
be repressed during oxidative stress responses by the protein
kinase c-Abl (246). The turnover
and stability of the upstream regulators of ubiquitination and
deubiquitination of MST, including KIBRA and
serine/threonine-protein kinase TAO, likely also affects MST and
leads to various biological outputs. In general, the ubiquitination
and deubiquitination of MST require further study.
Conclusions and future perspectives
Ubiquitination and deubiquitination are widespread
and important post-translational modifications associated with
multiple biological processes. Numerous components of the Hippo
pathway are modulated by these two PTMs, whose imbalance is
conducive to tumor formation and metastasis. Therefore, it is
necessary for cells to strike a balance between ubiquitination and
deubiquitination for maintaining homeostasis. While much has been
determined about ubiquitination and deubiquitination, a number of
issues remain to be resolved.
For instance, LATS1 may be degraded by all
NEDD4-like family member ligases, depending on the dosage; however,
only the loss of endogenous ITCH and WWP1 increases the protein
stability of LATS1, indicating that only ITCH and WWP1 are
essential to the maintenance of LATS1 stability (146). However, the underlying mechanism
and the effect on LATS2 are currently unknown. Furthermore, it is
unclear whether there are additional types of E3 ligases and DUBs
that modify Hippo pathway components (including TEAD1-4, NF2, FAT4
and CRB). Certain questions remain, including whether the E3
ligases and DUBs regulate the temporal and spatial organization of
the Hippo pathway, and how the Hippo pathway may be used
therapeutically in cancer.
Deregulation of the Hippo pathway is associated
with cancer, allowing its targeting to be a promising therapeutic
strategy. The pivotal roles of YAP and TAZ make them the most
attractive targets, and studies have demonstrated that inhibiting
YAP and TAZ may be effective in treating a variety of cancer types
that are predisposed to YAP/TAZ activation. However, the long-term
consequences of YAP/TAZ inhibition on normal and cancerous tissues
require further investigation. Therefore, a better method may be to
selectively target the YAP/TAZ-TEAD complex in order to decrease
side effects. For example, dobutamine, a β-adrenergic receptor
antagonist, is able to recruit YAP from the nucleus to the cytosol,
thereby repressing YAP-induced gene transcription. However, this
drug has not yet been placed into clinical trials (247).
Verteporfin, a clinical photosensitizer used in
photocoagulation therapy for macular degeneration (248), has been reported to interfere in
the interaction between YAP and TEAD, and thus to inhibit
YAP-induced transcription. Notably, verteporfin has been approved
by the US Food and Drug Administration and is capable of blocking
mouse hepatic tumorigenesis driven by either YAP1 overexpression or
loss of NF2 (249), making it a
promising drug in cancer therapy. To date, there has been a phase
I/II study of verteporfin photodynamic therapy in locally advanced
pancreatic cancer and it has exhibited a partial response (250).
miR-375 is an anti-oncogenic molecule in gastric
cancer (GC) that acts, at least in part, by directly targeting
YAP1, TEAD4 and CTGF, which may be exploited for treating GC
(251). In addition, disruption of
the YAP/TAZ-TEAD interaction by stimulating and enhancing VGLL4
expression may be a useful strategy against YAP/TAZ-driven tumors.
In line with this concept, a peptide mimicking the function of
VGLL4, which acts as a YAP antagonist, has recently been reported
to inhibit tumor development in a Helicobacter pylori mouse
model of GC (252). Such peptides
may also be applicable to the treatment of other cancer types,
including lung, breast and esophageal cancer. Additionally, based
on the immune suppressing effect of LATS, targeting LATS1/2 in
cancer immunotherapy may be considered. Furthermore, GPCR signaling
regulates the Hippo pathway in multiple ways. Sphingosine
1-phosphate (S1P), serum-borne lysophosphatidic acid and thrombin
each work through G12/G13-coupled receptors to inhibit LATS1/2 and
activate YAP and TAZ. This has led to attempts to antagonize this
signaling in an effort to repress the carcinogenic activities of
YAP/TAZ (253). For instance, the
S1P-blocking antibody sphinaomab has been reported to decrease lung
tumor metastasis (254).
Sphingosine kinase 1 (SPHK1) generates S1P. Phenoxodiol is an
isoflavone-derived SPHK1 inhibitor that is currently in clinical
trial in patients with platinum/taxane-refractory/resistant ovarian
cancer, fallopian tube cancer or primary peritoneal cancers
(255). MST and other components
of the Hippo pathway may also be targeted; however, the therapeutic
approach must be designed in light of their specific roles in
different contexts.
Research into ubiquitination-deubiquitination in
the Hippo pathway has provided additional effective methods of
early detection, prognosis and treatment of cancer. Proteasome
inhibitors, including bortezomib, have been used in the treatment
of relapsed multiple myeloma and mantle cell lymphoma (256); however, they are associated with
substantial toxicity due to the inhibition of overall protein
degradation. Therefore, in comparison, the targeting of a specific
E3 ligase may be a more ideal approach, with a higher level of
specificity and potentially less associated toxicity. For example,
NEDD4-like ubiquitin ligases are regarded as oncogenes due to their
triggering of LATS degradation and promotion of YAP/TAZ-driven gene
expression. Thus, drugs may be developed to interfere with the
formation of NEDD4-like ubiquitin ligases or disrupt their
interaction with LATS. Numerous types of miRNAs have been
discovered to target NEDD4-like ubiquitin ligases. For instance,
miR-497 exerts an anti-metastatic effect by targeting Smurf1
(257) and miR-1 directly
regulates NEDD4/NEDD4L (258).
These data imply that miRNAs may be applicable to mediating the
function of NEDD4-like ubiquitin ligases and be used in drug
development. However, currently there are very few studies on drug
development targeting E3 ligases or DUBs. Thus, additional
investigation is warranted.
Each type of E3 ligase or DUB usually has multiple
substrates, and thus their specificity must be taken into
consideration during drug development and targeted therapy for
tumors. Beyond therapy, E3 ligases and DUBs may also be useful
biomarkers allowing for improved diagnosis and detection of certain
cancers. The Hippo pathway has a complex network of crosstalk with
other signaling pathways that may also be regulated by
ubiquitination-deubiquitination, and that may indirectly affect the
Hippo pathway. Furthermore, cancer initiation and progression
involve numerous interacting biological processes, of which a
number remain unclear. All these facts add to the complexity of
tumorigenesis. The field of ubiquitination-deubiquitination as it
relates to the Hippo pathway remains in its infancy and further
research in this area will undoubtedly contribute to the
improvement of cancer diagnosis and therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81760432 and
81660405), the JiangXi Province Talent 555 Project (grant no.
20152ACB20024), the National Natural Science Foundation of JiangXi
Province (grant nos. 20152ACB20024 and 20151BBG70228), the JiangXi
Province General Project (grant no. 20171BBG70121), the Youth
Scientific Funds-Youth Fund Project (grant no. 20171BAB215041), the
JiangXi Province Education Fund Project (grant no. 700653002), and
the Department of Health of JiangXi Province Project (grant no.
20181041).
Availability of data and materials
Not applicable.
Authors' contributions
YL and JD were both involved in the conception of
the study. YL was a major contributor in writing the manuscript and
JD revised the manuscript. Both authors read and approved the final
manuscript, and agreed to be accountable for all aspects of the
work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AIP4
|
atrophin-1 interacting protein 4
|
|
AMOT-p130
|
angiomotin-p130
|
|
AMOTL
|
angiomotin-like
|
|
β-Trcp
|
β-transducin repeat-containing E3
ubiquitin protein ligase
|
|
CHIP
|
C terminus of Hsp70 interacting
protein
|
|
CRB
|
Crumbs homolog
|
|
CRL2
|
cullin 2-RING ubiquitin ligase
|
|
CRL4
|
cullin 4-RING ubiquitin ligase
|
|
DCAF1
|
DDB1 and CUL4 associated factor 1
|
|
DDB1
|
DNA damage-binding protein 1
|
|
DUB
|
deubiquitinase
|
|
KIBRA
|
kidney and brain protein
|
|
LATS
|
large tumor suppressor
|
|
LRR1
|
leucine-rich repeat protein 1
|
|
MARK
|
microtubule affinity-regulating
kinase
|
|
MOB1
|
MOB kinase activator 1
|
|
MST
|
mammalian STE20-like protein
kinase
|
|
NEDD4
|
neural-precursor-cell-expressed
developmentally downregulated 4
|
|
NEDD4L
|
NEDD4-like
|
|
NEDL2
|
NEDD4-like ubiquitin protein ligase
2
|
|
NF2
|
neurofibromin 2
|
|
PTPN14
|
protein tyrosine phosphatase
non-receptor type 14
|
|
RASSF
|
Ras association domain family
|
|
RUNX
|
RUNT-related transcription factor
|
|
SAV1
|
Salvador homolog 1
|
|
SCF
|
Skp1-Cullin1-F-box
|
|
SIAH2
|
seven in absentia homolog 2
|
|
TAO
|
thousand and one amino acid
protein
|
|
TAZ
|
transcriptional co-activator with
PDZ-binding motif
|
|
TEAD
|
TEA domain-containing
sequence-specific transcription factor
|
|
USP
|
ubiquitin-specific protease
|
|
VGLL4
|
vestigial-like family member 4
|
|
WBP2
|
WW domain-binding protein 2
|
|
WWP1
|
WW domain containing E3 ubiquitin
protein ligase 1
|
|
YAP
|
Yes-associated protein
|
References
|
1
|
Udan RS, Kangosingh M, Nolo R, Tao C and
Halder G: Hippo promotes proliferation arrest and apoptosis in the
Salvador/Warts pathway. Nat Cell Biol. 5:914–920. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Varelas X: The Hippo pathway effectors TAZ
and YAP in development, homeostasis and disease. Development.
141:1614–1626. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Halder G and Johnson RL: Hippo signaling:
Growth control and beyond. Development. 138:9–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu FX and Guan KL: The Hippo pathway:
Regulators and regulations. Genes Dev. 27:355–371. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Harvey KF, Zhang X and Thomas DM: The
Hippo pathway and human cancer. Nat Rev Cancer. 13:246–257. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu FX, Zhao B and Guan KL: Hippo pathway
in organ size control, tissue homeostasis, and cancer. Cell.
163:811–828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wierzbicki PM and Rybarczyk A: The Hippo
pathway in colorectal cancer. Folia Histochem Cytobiol. 53:105–119.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi P, Feng J and Chen C: Hippo pathway in
mammary gland development and breast cancer. Acta Biochim Biophys
Sin (Shanghai). 47:53–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Ding W, Chen C, Niu Z, Pan M and
Zhang H: Roles of Hippo signaling in lung cancer. Indian J Cancer.
52 Suppl 1:e1–e5. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen M, Wang M, Xu S, Guo X and Jiang J:
Upregulation of miR-181c contributes to chemoresistance in
pancreatic cancer by inactivating the Hippo signaling pathway.
Oncotarget. 6:44466–44479. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mo JS, Park HW and Guan KL: The Hippo
signaling pathway in stem cell biology and cancer. EMBO Rep.
15:642–656. 2014.PubMed/NCBI
|
|
13
|
Herrmann J, Lerman LO and Lerman A:
Ubiquitin and ubiquitin-like proteins in protein regulation. Circ
Res. 100:1276–1291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hanpude P, Bhattacharya S, Dey AK and
Maiti TK: Deubiquitinating enzymes in cellular signaling and
disease regulation. IUBMB Life. 67:544–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wilkinson KD: Ubiquitination and
deubiquitination: Targeting of proteins for degradation by the
proteasome. Semin Cell Dev Biol. 11:141–148. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hong W and Guan KL: The YAP and TAZ
transcription co-activators: Key downstream effectors of the
mammalian Hippo pathway. Semin Cell Dev Biol. 23:785–793. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen CG, Moroishi T and Guan KL: YAP and
TAZ: A nexus for Hippo signaling and beyond. Trends Cell Biol.
25:499–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu
J, Lin JD, Wang CY, Chinnaiyan AM, et al: TEAD mediates
YAP-dependent gene induction and growth control. Genes Dev.
22:1962–1971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Strano S, Monti O, Baccarini A, Sudol M,
Sacchi A, Oren M, Sudol M, Cesareni G and Blandino G: Physical
interaction with yes-associated protein enhances p73
transcriptional activity. J Biol Chem. 37:15164–15173. 2001.
View Article : Google Scholar
|
|
20
|
Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu
J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ,
et al: Nuclear CDKs drive Smad transcriptional activation and
turnover in BMP and TGF-beta pathways. Cell. 139:757–769. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Varelas X, Sakuma R, Samavarchitehrani P,
Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW and Wrana JL:
TAZ controls Smad nucleocytoplasmic shuttling and regulates human
embryonic stem-cell self-renewal. Nat Cell Biol. 10:837–848. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ferrigno O, Lallemand F, Verrecchia F,
L'Hoste S, Camonis J, Atfi A and Mauviel A: Yes-associated protein
(YAP65) interacts with Smad7 and potentiates its inhibitory
activity against TGF-beta/Smad signaling. Oncogene. 21:4879–4884.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kelleher FC and O'Sullivan H: FOXM1 in
sarcoma: Role in cell cycle, pluripotency genes and stem cell
pathways. Oncotarget. 7:42792–42804. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Murakami M, Nakagawa M, Olson EN and
Nakagawa O: A WW domain protein TAZ is a critical coactivator for
TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc
Natl Acad Sci USA. 102:18034–18039. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rosenbluh J, Nijhawan D, Cox AG, Li X,
Neal JT, Schafer EJ, Zack TI, Wang X, Tsherniak A, Schinzel AC, et
al: β-Catenin-driven cancers require a YAP1 transcriptional complex
for survival and tumorigenesis. Cell. 151:1457–1473. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yagi R, Chen LF, Shigesada K, Murakami Y
and Ito Y: A WW domain-containing Yes-associated protein (YAP) is a
novel transcriptional co-activator. EMBO J. 18:2551–2562. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F,
Han X, Feng Y, Zheng C, Wang Z, et al: VGLL4 functions as a new
tumor suppressor in lung cancer by negatively regulating the
YAP-TEAD transcriptional complex. Cell Res. 24:331–343. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao B, Lei QY and Guan KL: The Hippo-YAP
pathway: New connections between regulation of organ size and
cancer. Curr Opin Cell Biol. 20:638–646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ahn EY, Kim JS, Kim GJ and Park YN:
RASSF1A-mediated regulation of AREG via the Hippo pathway in
hepatocellular carcinoma. Mol Cancer Res. 11:748–758. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma K, Xu Q, Wang S, Zhang W, Liu M, Liang
S, Zhu H and Xu N: Nuclear accumulation of Yes-Associated Protein
(YAP) maintains the survival of doxorubicin-induced senescent cells
by promoting survivin expression. Cancer Lett. 375:84–91. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Staley BK and Irvine KD: Hippo signaling
in Drosophila: Recent advances and insights. Dev Dyn.
241:3–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Boggiano JC, Vanderzalm PJ and Fehon RG:
Tao-1 phosphorylates Hippo/MST kinases to regulate the
Hippo-Salvador-Warts tumor suppressor pathway. Dev Cell.
21:888–895. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang HL, Wang S, Yin MX, Dong L, Wang C,
Wu W, Lu Y, Feng M, Dai C, Guo X, et al: Par-1 regulates tissue
growth by influencing Hippo phosphorylation status and
Hippo-salvador association. PLoS Biol. 11:e10016202013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Genevet A, Wehr MC, Brain R, Thompson BJ
and Tapon N: Kibra is a regulator of the Salvador/Warts/Hippo
signaling network. Dev Cell. 18:300–308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Meng Z, Moroishi T, Mottierpavie V,
Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, et al:
MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2
in the Hippo pathway. Nat Commun. 6:83572015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yin F, Yu J, Zheng Y, Chen Q, Zhang N and
Pan D: Spatial organization of Hippo signaling at the plasma
membrane mediated by the tumor suppressor Merlin/NF2. Cell.
154:1342–1355. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hirate Y, Hirahara S, Inoue K, Suzuki A,
Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al:
Polarity-dependent distribution of angiomotin localizes Hippo
signaling in preimplantation embryos. Curr Biol. 23:1181–1194.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huntoon CJ, Nye MD, Geng L, Peterson KL,
Flatten KS, Haluska P, Kaufmann SH and Karnitz LM: Heat shock
protein 90 inhibition depletes LATS1 and LATS2, two regulators of
the mammalian hippo tumor suppressor pathway. Cancer Res.
70:8642–8650. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Das Thakur M, Feng Y, Jagannathan R, Seppa
MJ, Skeath JB and Longmore GD: Ajuba LIM proteins are negative
regulators of the Hippo signaling pathway. Curr Biol. 20:657–662.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chan SW, Lim CJ, Huang C, Chong YF,
Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF and Hong W: WW
domain-mediated interaction with Wbp2 is important for the
oncogenic property of TAZ. Oncogene. 30:600–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu X, Yang N, Figel SA, Wilson KE,
Morrison CD, Gelman IH and Zhang J: PTPN14 interacts with and
negatively regulates the oncogenic function of YAP. Oncogene.
32:1266–1273. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lian I, Kim J, Okazawa H, Zhao J, Zhao B,
Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, et al:
The role of YAP transcription coactivator in regulating stem cell
self-renewal and differentiation. Genes Dev. 24:1106–1118. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Camargo FD, Gokhale S, Johnnidis JB, Fu D,
Bell GW, Jaenisch R and Brummelkamp TR: YAP1 increases organ size
and expands undifferentiated progenitor cells. Curr Biol.
17:2054–2060. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xin M, Kim Y, Sutherland LB, Murakami M,
Qi X, Mcanally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et
al: Hippo pathway effector Yap promotes cardiac regeneration. Proc
Natl Acad Sci USA. 110:13839–13844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schlegelmilch K, Mohseni M, Kirak O,
Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J,
Brummelkamp TR and Camargo FD: Yap1 acts downstream of α-catenin to
control epidermal proliferation. Cell. 144:782–795. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Heallen T, Zhang M, Wang J,
Bonilla-Claudio M, Klysik E, Johnson RL and Martin JF: Hippo
pathway inhibits Wnt signaling to restrain cardiomyocyte
proliferation and heart size. Science. 332:458–461. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang H, Pasolli HA and Fuchs E:
Yes-associated protein (YAP) transcriptional coactivator functions
in balancing growth and differentiation in skin. Proc Natl Acad Sci
USA. 108:2270–2275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
von Gise A, Lin Z, Schlegelmilch K, Honor
LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD and Pu
WT: YAP1, the nuclear target of Hippo signaling, stimulates heart
growth through cardiomyocyte proliferation but not hypertrophy.
Proc Natl Acad Sci USA. 109:2394–2399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Harvey KF, Pfleger CM and Hariharan IK:
The Drosophila Mst ortholog, hippo, restricts growth and
cell proliferation and promotes apoptosis. Cell. 114:457–467. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jia J, Zhang W, Wang B, Trinko R and Jiang
J: The Drosophila Ste20 family kinase dMST functions as a
tumor suppressor by restricting cell proliferation and promoting
apoptosis. Genes Dev. 17:2514–2519. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu
Q, Wang Y, Halder G, Finegold MJ, Lee JS and Johnson RL: Hippo
signaling is a potent in vivo growth and tumor suppressor pathway
in the mammalian liver. Proc Natl Acad Sci USA. 107:1437–1442.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xin M, Kim Y, Sutherland LB, Qi X,
Mcanally J, Schwartz RJ, Richardson JA, Basselduby R and Olson EN:
Regulation of insulin-like growth factor signaling by Yap governs
cardiomyocyte proliferation and embryonic heart size. Sci Signal.
4:ra702011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Johnson R and Halder G: The two faces of
Hippo: Targeting the Hippo pathway for regenerative medicine and
cancer treatment. Nat Rev Drug Discov. 13:63–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhou D, Conrad C, Xia F, Park JS, Payer B,
Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J and Bardeesy N: Mst1
and Mst2 maintain hepatocyte quiescence and suppress hepatocellular
carcinoma development through inactivation of the Yap1 oncogene.
Cancer Cell. 16:425–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu Q, Le ES, Jahchan N, Ji X, Xu A and
Luo K: SnoN antagonizes the Hippo kinase complex to promote TAZ
signaling during breast carcinogenesis. Dev Cell. 37:399–412. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jing Z, Wang G, Chu SJ, Zhu JS, Rui Z, Lu
WW, Xia LQ, Lu YM, Wei D and Sun Q: Loss of large tumor suppressor
1 promotes growth and metastasis of gastric cancer cells through
upregulation of the YAP signaling. Oncotarget. 7:16180–16193.
2016.PubMed/NCBI
|
|
57
|
Strazisar M, Mlakar V and Glavac D: LATS2
tumour specific mutations and down-regulation of the gene in
non-small cell carcinoma. Lung Cancer. 64:257–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ishizaki K, Fujimoto J, Kumimoto H,
Nishimoto Y, Shimada Y, Shinoda M and Yamamoto T: Frequent
polymorphic changes but rare tumor specific mutations of the LATS2
gene on 13q11-12 in esophageal squamous cell carcinoma. Int J
Oncol. 21:1053–1057. 2002.PubMed/NCBI
|
|
59
|
Bonilla X, Parmentier L, King B, Bezrukov
F, Kaya G, Zoete V, Seplyarskiy VB, Sharpe HJ, Mckee T, Letourneau
A, et al: Genomic analysis identifies new drivers and progression
pathways in skin basal cell carcinoma. Nat Genet. 48:398–406. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lee JH, Kavanagh JJ, Wildrick DM, Wharton
JT and Blick M: Frequent loss of heterozygosity on chromosomes 6q,
11, and 17 in human ovarian carcinomas. Cancer Res. 50:2724–2728.
1990.PubMed/NCBI
|
|
61
|
Theile M, Seitz S, Arnold W, Jandrig B,
Frege R, Schlag PM, Haensch W, Guski H, Winzer KJ, Barrett JC and
Scherneck S: A defined chromosome 6q fragment (at D6S310) harbors a
putative tumor suppressor gene for breast cancer. Oncogene.
13:677–685. 1996.PubMed/NCBI
|
|
62
|
Noviello C, Courjal F and Theillet C: Loss
of heterozygosity on the long arm of chromosome 6 in breast cancer:
Possibly four regions of deletion. Clin Cancer Res. 2:1601–1606.
1996.PubMed/NCBI
|
|
63
|
Chen KH, He J, Wang DL, Cao JJ, Li MC,
Zhao XM, Sheng X, Li WB and Liu WJ: Methylation-associated
inactivation of LATS1 and its effect on demethylation or
overexpression on YAP and cell biological function in human renal
cell carcinoma. Int J Oncol. 45:2511–2521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Van Hateren NJ, Das RM, Hautbergue GM,
Borycki AG, Placzek M and Wilson SA: FatJ acts via the Hippo
mediator Yap1 to restrict the size of neural progenitor cell pools.
Development. 138:1893–1902. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Qi C, Zhu YT, Hu L and Zhu YJ:
Identification of Fat4 as a candidate tumor suppressor gene in
breast cancers. Int J Cancer. 124:793–798. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zang ZJ, Cutcutache I, Poon SL, Zhang SL,
Mcpherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mitamura T, Watari H, Wang L, Kanno H,
Miyazaki M, Kitagawa M, Hassan MK, Dong P, Kimura T, Tanino M and
Sakuragi N: MicroRNA 31 functions as an endometrial cancer oncogene
by suppressing Hippo tumor suppressor pathway. Mol Cancer.
13:972014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Tan G, Cao X, Dai Q, Zhang B, Huang J,
Xiong S, Zhang Y, Chen W, Yang J and Li H: A novel role for
microRNA-129-5p in inhibiting ovarian cancer cell proliferation and
survival via direct suppression of transcriptional co-activators
YAP and TAZ. Oncotarget. 6:8676–8686. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shen S, Guo X, Yan H, Lu Y, Ji X, Li L,
Liang T, Zhou D, Feng XH, Zhao JC, et al: A miR-130a-YAP positive
feedback loop promotes organ size and tumorigenesis. Cell Res.
25:997–1012. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhu G, Wang Y, Mijiti M, Wang Z, Wu PF and
Jiafu D: Upregulation of miR-130b enhances stem cell-like phenotype
in glioblastoma by inactivating the Hippo signaling pathway.
Biochem Biophys Res Commun. 465:194–199. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Riquelme I, Ili C, Roa JC and Brebi P:
Long non-coding RNAs in gastric cancer: Mechanisms and potential
applications. Oncotarget. 2016.doi: 10.18632/oncotarget.9396.
|
|
74
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen Y, Wei G, Xia H, Yu H, Tang Q and Bi
F: Down regulation of lincRNA-p21 contributes to gastric cancer
development through Hippo-independent activation of YAP.
Oncotarget. 8:63813–63824. 2017.PubMed/NCBI
|
|
76
|
Qu S, Yue Z, Shang R, Xuan Z, Song W,
Kjems J and Li H: The emerging landscape of circular RNA in life
processes. RNA Biol. 14:992–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
He M, Zhou Z, Shah AA, Yang H, Chen Q and
Wan Y: New insights into posttranslational modifications of Hippo
pathway in carcinogenesis and therapeutics. Cell Div. 11:42016.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tomlinson V, Gudmundsdottir K, Luong P,
Leung KY, Knebel A and Basu S: JNK phosphorylates Yes-associated
protein (YAP) to regulate apoptosis. Cell Death Dis. 1:e292010.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jang SW, Yang SJ, Srinivasan S and Ye K:
Akt phosphorylates MstI and prevents its proteolytic activation,
blocking FOXO3 phosphorylation and nuclear translocation. J Biol
Chem. 282:30836–30844. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Collak FK, Yagiz K, Luthringer DJ, Erkaya
B and Cinar B: Threonine-120 phosphorylation regulated by
phosphoinositide-3-kinase/Akt and mammalian target of rapamycin
pathway signaling limits the antitumor activity of mammalian
sterile 20-like kinase 1. J Biol Chem. 287:23698–23709. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Bi W, Xiao L, Jia Y, Wu J, Xie Q, Ren J,
Ji G and Yuan Z: c-Jun N-terminal kinase enhances MST1-mediated
pro-apoptotic signaling through phosphorylation at serine 82. J
Biol Chem. 285:6259–6264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Hata S, Hirayama J, Kajiho H, Nakagawa K,
Hata Y, Katada T, Furutaniseiki M and Nishina H: A novel
acetylation cycle of transcription co-activator Yes-associated
protein that is downstream of Hippo pathway is triggered in
response to SN2 alkylating agents. J Biol Chem. 287:22089–22098.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Mao B, Hu F, Cheng J, Wang P, Xu M, Yuan
F, Meng S, Wang Y, Yuan Z and Bi W: SIRT1 regulates YAP2-mediated
cell proliferation and chemoresistance in hepatocellular carcinoma.
Oncogene. 33:1468–1474. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Oudhoff MJ, Freeman SA, Couzens AL,
Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B,
Barsyte-Lovejoy D, Vedadi M, et al: Control of the hippo pathway by
Set7-dependent methylation of Yap. Dev Cell. 26:188–194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Kreppel LK, Blomberg MA and Hart GW:
Dynamic glycosylation of nuclear and cytosolic proteins. Cloning
and characterization of a unique O-GlcNAc transferase with multiple
tetratricopeptide repeats. J Biol Chem. 272:9308–9315. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Singh JP, Zhang K, Wu J and Yang X:
O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer
Lett. 356:244–250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Starska K, Forma E, Brzezińska-Błaszczyk
E, Lewy-Trenda I, Bryś M, Jóźwiak P and Krześlak A: Gene and
protein expression of O-GlcNAc-cycling enzymes in human laryngeal
cancer. Clin Exp Med. 15:455–468. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Itkonen HM, Minner S, Guldvik IJ, Sandmann
MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T and Mills IG:
O-GlcNAc transferase integrates metabolic pathways to regulate the
stability of c-MYC in human prostate cancer cells. Cancer Res.
73:5277–5287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang X, Qiao Y, Wu Q, Chen Y, Zou S, Liu
X, Zhu G, Zhao Y, Chen Y, Yu Y, et al: The essential role of YAP
O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat
Commun. 8:152802017. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Jennissen HP: Ubiquitin and the enigma of
intracellular protein degradation. Eur J Biochem. 231:1–30. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Hicke L: Protein regulation by
monoubiquitin. Nat Rev Mol Cell Biol. 2:195–201. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Berndsen CE and Wolberger C: New insights
into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol.
21:301–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Pickart CM: Mechanisms underlying
ubiquitination. Annu Rev Biochem. 70:503–533. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Thrower JS, Hoffman L, Rechsteiner M and
Pickart CM: Recognition of the polyubiquitin proteolytic signal.
EMBO J. 19:94–102. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang X, Yang J, Han L, Zhao K, Wu Q, Bao
L, Li Z, Lv L and Li B: TRAF5-mediated Lys-63-linked
polyubiquitination plays an essential role in positive regulation
of RORγt in promoting IL-17A expression. J Biol Chem.
290:29086–29094. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Yang WL, Wu CY, Wu J and Lin HK:
Regulation of Akt signaling activation by ubiquitination. Cell
Cycle. 9:487–497. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Mukhopadhyay D and Riezman H:
Proteasome-independent functions of ubiquitin in endocytosis and
signaling. Science. 315:201–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Terrell J, Shih S, Dunn R and Hicke L: A
function for monoubiquitination in the internalization of a G
protein-coupled receptor. Mol Cell. 1:193–202. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Rome S, Meugnier E and Vidal H: The
ubiquitin-proteasome pathway is a new partner for the control of
insulin signaling. Curr Opin Clin Nutr Metab Care. 7:249–254. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Izzi L and Attisano L: Regulation of the
TGFbeta signalling pathway by ubiquitin-mediated degradation.
Oncogene. 23:2071–2078. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lan Q, Gao Y, Li Y, Hong X and Xu P:
Progress in ubiquitin, ubiquitin chain and protein ubiquitination.
Sheng Wu Gong Cheng Xue Bao. 32:14–30. 2016.(In Chinese).
PubMed/NCBI
|
|
104
|
He M, Zhou Z, Shah AA, Zou H, Tao J, Chen
Q and Wan Y: The emerging role of deubiquitinating enzymes in
genomic integrity, diseases, and therapeutics. Cell Biosci.
6:622016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Nijman SM, Luna-Vargas MP, Velds A,
Brummelkamp TR, Dirac AM, Sixma TK and Bernards R: A genomic and
functional inventory of deubiquitinating enzymes. Cell.
123:773–786. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Gao C, Huang W, Kanasaki K and Xu Y: The
role of ubiquitination and sumoylation in diabetic nephropathy.
Biomed Res Int 2014. 1606922014.
|
|
107
|
Chen Z and Lu W: Roles of ubiquitination
and SUMOylation on prostate cancer: Mechanisms and clinical
implications. Int J Mol Sci. 16:4560–4580. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Popovic D, Vucic D and Dikic I:
Ubiquitination in disease pathogenesis and treatment. Nat Med.
20:1242–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Hong L, Huang HC and Jiang ZF:
Relationship between amyloid-beta and the ubiquitin-proteasome
system in Alzheimer's disease. Neurol Res. 36:276–282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Debald M, Schildberg FA, Linke A,
Walgenbach K, Kuhn W, Hartmann G and Walgenbach-Brünagel G:
Specific expression of k63-linked ubiquitination of calmodulin-like
protein 5 in breast cancer of premenopausal patients. J Cancer Res
Clin Oncol. 139:2125–2132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Zarrizi R, Menard JA, Belting M and
Massoumi R: Deubiquitination of gamma-tubulin by BAP1 prevents
chromosome instability in breast cancer cells. Cancer Res.
74:6499–6508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Li X, Stevens PD, Yang H, Gulhati P, Wang
W, Evers BM and Gao T: The deubiquitination enzyme USP46 functions
as a tumor suppressor by controlling PHLPP-dependent attenuation of
Akt signaling in colon cancer. Oncogene. 32:471–478. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bhattacharya S and Ghosh MK: Cell death
and deubiquitinases: Perspectives in cancer. Biomed Res Int 2014.
4351972014.
|
|
114
|
Burger AM and Seth AK: The
ubiquitin-mediated protein degradation pathway in cancer:
Therapeutic implications. Eur J Cancer. 40:2217–2229. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hussain S, Zhang Y and Galardy PJ: DUBs
and cancer: The role of deubiquitinating enzymes as oncogenes,
non-oncogenes and tumor suppressors. Cell Cycle. 8:1688–1697. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Surget S, Khoury MP and Bourdon JC:
Uncovering the role of p53 splice variants in human malignancy: A
clinical perspective. Oncotargets Ther. 7:57–68. 2013.
|
|
117
|
Canner JA, Sobo M, Ball S, Hutzen B,
Deangelis S, Willis W, Studebaker AW, Ding K, Wang S, Yang D and
Lin J: MI-63: A novel small-molecule inhibitor targets MDM2 and
induces apoptosis in embryonal and alveolar rhabdomyosarcoma cells
with wild-type p53. Br J Cancer. 101:774–781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Meng X, Franklin DA, Dong J and Zhang Y:
MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res.
74:7161–7167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Brooks CL, Li M, Hu M, Shi Y and Gu W: The
p53-Mdm2-HAUSP complex is involved in p53 stabilization by HAUSP.
Oncogene. 26:7262–7266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Hock AK, Vigneron AM, Carter S, Ludwig RL
and Vousden KH: Regulation of p53 stability and function by the
deubiquitinating enzyme USP42. EMBO J. 30:4921–4930. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Dolcet X, Llobet D, Pallares J and
Matias-Guiu X: NF-κB in development and progression of human
cancer. Virchows Arch. 446:475–482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Häcker H and Karin M: Regulation and
function of IKK and IKK-related Kinases. Sci STKE 2006.
re132006.
|
|
123
|
Fuchs S: Activation of β-Trcp ubiquitin
ligases in cancers: Mechanisms and outcomes. Cancer Res.
67:2007.
|
|
124
|
Schweitzer K, Bozko PM, Dubiel W and
Naumann M: CSN controls NF-κB by deubiquitinylation of IκBα. EMBO
J. 26:1532–1541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Harhaj EW and Dixit VM: Regulation of
NF-κB by deubiquitinases. Immunol Rev. 246:107–124. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Luise C, Capra M, Donzelli M, Mazzarol G,
Jodice MG, Nuciforo P, Viale G, Di Fiore PP and Confalonieri S: An
atlas of altered expression of deubiquitinating enzymes in human
cancer. PLoS One. 6:e158912011. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Priolo C, Tang D, Brahamandan M, Benassi
B, Sicinska E, Ogino S, Farsetti A, Porrello A, Finn S, Zimmermann
J, et al: The isopeptidase USP2a protects human prostate cancer
from apoptosis. Cancer Res. 66:8625–8632. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Metzig M, Nickles D, Falschlehner C,
Lehmann-Koch J, Straub BK, Roth W and Boutros M: An RNAi screen
identifies USP2 as a factor required for TNF-α-induced NF-κB
signaling. Int J Cancer. 129:607–618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Hilman D and Gat U: The evolutionary
history of YAP and the hippo/YAP pathway. Mol Biol Evol.
28:2403–2417. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Avruch J, Zhou D, Fitamant J, Bardeesy N,
Mou F and Barrufet LR: Protein kinases of the Hippo pathway:
Regulation and substrates. Semin Cell Dev Biol. 23:770–784. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Yabuta N, Fujii T, Copeland NG, Gilbert
DJ, Jenkins NA, Nishiguchi H, Endo Y, Toji S, Tanaka H, Nishimune Y
and Nojima H: Structure, expression, and chromosome mapping of
LATS2, a mammalian homologue of the Drosophila tumor
suppressor gene lats/warts. Genomics. 63:263–270. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Nishiyama Y and Al E: A human homolog of
Drosophila warts tumor suppressor, h-warts, localized to
mitotic apparatus and specifically phosphorylated during mitosis.
FEBS Lett. 459:159–165. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Visser S and Yang X: LATS tumor
suppressor: A new governor of cellular homeostasis. Cell Cycle.
9:3892–3903. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Furth N and Aylon Y: The LATS1 and LATS2
tumor suppressors: Beyond the Hippo pathway. Cell Death Differ.
24:1488–1501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Wang W, Li X, Huang J, Feng L, Dolinta KG
and Chen J: Defining the protein-protein interaction network of the
human hippo pathway. Mol Cell Proteomics. 13:119–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Lit LC, Scott S, Zhang H, Stebbing J,
Photiou A and Giamas G: LATS2 is a modulator of estrogen receptor
alpha. Anticancer Res. 33:53–63. 2013.PubMed/NCBI
|
|
137
|
Powzaniuk M, Mcelwee-Witmer S, Vogel RL,
Hayami T, Rutledge SJ, Chen F, Harada S, Schmidt A, Rodan GA,
Freedman LP and Bai C: The LATS2/KPM tumor suppressor is a negative
regulator of the androgen receptor. Mol Endocrinol. 18:2011–2023.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Britschgi A, Duss S, Kim S, Couto JP,
Brinkhaus H, Koren S, De Silva D, Mertz KD, Kaup D, Varga Z, et al:
The Hippo kinases LATS1 and 2 control human breast cell fate via
crosstalk with ERα. Nature. 541:541–545. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Moroishi T, Hayashi T, Pan WW, Fujita Y,
Holt MV, Qin J, Carson DA and Guan KL: The Hippo pathway kinases
LATS1/2 suppress cancer immunity. Cell. 167:1525–1539.e17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hershko A and Ciechanover A: The ubiquitin
system. Annu Rev Biochem. 67:425–79. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Boase NA and Kumar S: NEDD4: The founding
member of a family of ubiquitin-protein ligases. Gene. 557:113–122.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bernassola F, Karin M, Ciechanover A,
Melino and Gerry: The HECT family of E3 ubiquitin ligases: Multiple
players in cancer development. Cancer Cell. 14:10–21. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Yang B and Kumar S: Nedd4 and Nedd4-2:
Closely related ubiquitin-protein ligases with distinct
physiological functions. Cell Death Differ. 17:68–77. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Salah Z, Melino G and Aqeilan RI: Negative
regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is
sufficient to promote tumorigenicity. Cancer Res. 71:2010–2020.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Salah Z, Itzhaki E and Aqeilan RI: The
ubiquitin E3 ligase ITCH enhances breast tumor progression by
inhibiting the Hippo tumor suppressor pathway. Oncotarget.
5:10886–10900. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Yeung B, Ho KC and Yang X: WWP1 E3 ligase
targets LATS1 for ubiquitin-mediated degradation in breast cancer
cells. PLoS One. 8:e610272013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Bae SJ, Kim M, Kim SH, Kwon YE, Lee JH,
Kim J, Chung CH, Lee WJ and Seol JH: NEDD4 controls intestinal stem
cell homeostasis by regulating the Hippo signalling pathway. Nat
Commun. 6:63142015. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wu S, Huang J, Dong J and Pan D: Hippo
encodes a Ste-20 family protein kinase that restricts cell
proliferation and promotes apoptosis in conjunction with salvador
and warts. Cell. 114:445–456. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Nakayama K, Qi J and Ronai Z: The
ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res.
7:443–451. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Ma B, Chen Y, Chen L, Cheng H, Mu C, Li J,
Gao R, Zhou C, Cao L, Liu J, et al: Hypoxia regulates Hippo
signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol.
17:95–103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Brown JM and Wilson WR: Exploiting tumour
hypoxia in cancer treatment. Nat Rev Cancer. 4:437–447. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Li W, Cooper J, Zhou L, Yang C,
Erdjument-Bromage H, Zagzag D, Snuderl M, Ladanyi M, Hanemann CO,
Zhou P, et al: Merlin/NF2 loss-driven tumorigenesis linked to
CRL4(DcAF1)-mediated inhibition of the hippo pathway kinases Latsl
and 2 in the nucleus. Cancer Cell. 26:48–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Lee J and Zhou P: DCAFs, the missing link
of the CUL4-DDB1 ubiquitin ligase. Mol Cell. 26:775–780. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Kim Y, Kim W, Song Y, Kim JR, Cho K, Moon
H, Ro SW, Seo E, Ryu YM, Myung SJ and Jho EH: Deubiquitinase YOD1
potentiates YAP/TAZ activities through enhancing ITCH stability.
Proc Natl Acad Sci USA. 114:4691–4696. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Kim Y and Jho EH: Deubiquitinase YOD1: The
potent activator of YAP in hepatomegaly and liver cancer. BMB Rep.
50:281–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Toloczko A, Guo F, Yuen HF, Wen Q, Wood
SA, Ong YS, Chan PY, Alli SA, Gunaratne J, Dunne MJ, et al:
Deubiquitinating enzyme USP9X suppresses tumor growth via LATS
kinase and core components of the hippo pathway. Cancer Res.
77:4921–4933. 2017.PubMed/NCBI
|
|
157
|
Thanh Nguyen H, Andrejeva D, Gupta R,
Choudhary C, Hong X, Eichhorn PJ, Loya AC and Cohen SM:
Deubiquitylating enzyme USP9× regulates hippo pathway activity by
controlling angiomotin protein turnover. Cell Discov. 2:160012016.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Okada T, Gondo Y, Goto J, Kanazawa I,
Hadano S and Ikeda JE: Unstable transmission of the RS447 human
megasatellite tandem repetitive sequence that contains the USP17
deubiquitinating enzyme gene. Hum Genet. 110:302–313. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Nguyen HT, Kugler JM and Cohen SM: DUB3
deubiquitylating enzymes regulate hippo pathway activity by
regulating the stability of ITCH, LATS and AMOT proteins. PLoS One.
12:e01695872017. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Salah Z, Alian A and Aqeilan RI: WW
domain-containing proteins: Retrospectives and the future. Front
Biosci (Landmark Ed). 17:331–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Kanai F, Marignani PA, Sarbassova D, Yagi
R, Hall RA, Donowitz M, Hisaminato A, Fujiwara T, Ito Y, Cantley LC
and Yaffe MB: TAZ: A novel transcriptional co-activator regulated
by interactions with 14-3-3 and PDZ domain proteins. EMBO J.
19:6778–6791. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Ye F and Zhang M: Structures and target
recognition modes of PDZ domains: Recurring themes and emerging
pictures. Biochem J. 455:1–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Remue E, Meerschaert K, Oka T, Boucherie
C, Vandekerckhove J, Sudol M and Gettemans J: TAZ interacts with
zonula occludens-1 and −2 proteins in a PDZ-1 dependent manner.
FEBS Lett. 584:4175–4180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Jang EJ, Jeong H, Han KH, Kwon HM, Hong JH
and Hwang ES: TAZ suppresses NFAT5 activity through tyrosine
phosphorylation. Mol Cell Biol. 32:4925–4932. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang
H, Guan KL and Xu Y: Structural insights into the YAP and TEAD
complex. Genes Dev. 24:235–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Howell M, Borchers C and Milgram SL:
Heterogeneous nuclear ribonuclear protein U associates with YAP and
regulates its co-activation of Bax transcription. J Biol Chem.
279:26300–26306. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Sudol M: Yes-associated protein (YAP65) is
a proline-rich phosphoprotein that binds to the SH3 domain of the
Yes proto-oncogene product. Oncogene. 9:2145–2152. 1994.PubMed/NCBI
|
|
168
|
Cordenonsi M, Zanconato F, Azzolin L,
Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR,
Poletti A, et al: The Hippo transducer TAZ confers cancer stem
cell-related traits on breast cancer cells. Cell. 147:759–772.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Overholtzer M, Zhang J, Smolen GA, Muir B,
Li W, Sgroi DC, Deng CX, Brugge JS and Haber DA: Transforming
properties of YAP, a candidate oncogene on the chromosome 11q22
amplicon. Proc Natl Acad Sci USA. 103:12405–12410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Kapoor A, Yao W, Ying H, Hua S, Liewen A,
Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al: Yap1 activation
enables bypass of oncogenic Kras addiction in pancreatic cancer.
Cell. 158:185–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Attisano L and Wrana JL: Signal
integration in TGF-β, WNT, and Hippo pathways. F1000prime Rep.
5:172013. View
Article : Google Scholar : PubMed/NCBI
|
|
172
|
Cottini F, Hideshima T, Xu C, Sattler M,
Dori M, Agnelli L, ten Hacken E, Bertilaccio MT, Antonini E, Neri
A, et al: Rescue of Hippo co-activator yap1 triggers DNA
damage-induced apoptosis in hematological cancers. Nat Med.
20:599–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim
J, Xie J, Ikenoue T, Yu J, Li L, et al: Inactivation of YAP
oncoprotein by the Hippo pathway is involved in cell contact
inhibition and tissue growth control. Genes Dev. 21:2747–2761.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Zhao B, Li L, Tumaneng K, Wang CY and Guan
KL: A coordinated phosphorylation by Lats and CK1 regulates YAP
stability through SCFβ-TRCP. Genes Dev. 24:72–85. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Liu CY, Zha ZY, Zhou X, Zhang H, Huang W,
Zhao D, Li T, Chan SW, Lim CJ, Hong W, et al: The hippo tumor
pathway promotes TAZ degradation by phosphorylating a phosphodegron
and recruiting the SCF{beta}-TrCP E3 ligase. J Biol Chem.
285:37159–37169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Deshaies RJ: SCF and Cullin/Ring H2-based
ubiquitin ligases. Annu Rev Cell Dev Biol. 15:435–467. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Bai C, Sen P, Hofmann K, Ma L, Goebl M,
Harper J and Elledge SJ: SKP1 connects cell cycle regulators to the
ubiquitin proteolysis machinery through a Novel motif, the F-Box.
Cell. 86:263–274. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Hart M, Concordet J, Lassot I, Albert I,
del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F,
Benarous R and Polakis P: The F-box protein β-TrCP associates with
phosphorylated beta-catenin and regulates its activity in the cell.
Curr Biol. 9:207–210. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Fuchs SY, Spiegelman VS and Kumar KG: The
many faces of beta-TrCP E3 ubiquitin ligases: Reflections in the
magic mirror of cancer. Oncogene. 23:2028–2036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Tian Y, Kolb R, Hong JH, Carroll J, Li D,
You J, Bronson R, Yaffe MB, Zhou J and Benjamin T: TAZ promotes PC2
degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell
Biol. 27:6383–6395. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Yim H, Sung CK, You J, Tian Y and Benjamin
T: Nek1 and TAZ interact to maintain normal levels of polycystin 2.
J Am Soc Nephrol. 22:832–837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Huang W, Lv X, Liu C, Zha Z, Zhang H,
Jiang Y, Xiong Y, Lei QY and Guan KL: The N-terminal phosphodegron
targets TAZ/WWTR1 protein for SCFβ-TrCP-dependent degradation in
response to phosphatidylinositol 3-Kinase inhibition. J Biol Chem.
287:26245–26253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Zhao JJ and Roberts TM: PI3 kinases in
cancer: From oncogene artifact to leading cancer target. Sci STKE
2006. pe522006.
|
|
184
|
Wang Z, Inuzuka H, Zhong J, Wan L,
Fukushima H, Sarkar FH and Wei W: Tumor suppressor functions of
FBW7 in cancer development and progression. FEBS Lett.
586:1409–1418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Tu K, Yang W, Li C, Zheng X, Lu Z, Guo C,
Yao Y and Liu Q: Fbxw7 is an independent prognostic marker and
induces apoptosis and growth arrest by regulating YAP abundance in
hepatocellular carcinoma. Mol Cancer. 13:1102014. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Hong X, Nguyen HT, Chen Q, Zhang R, Hagman
Z, Voorhoeve PM and Cohen SM: Opposing activities of the Ras and
Hippo pathways converge on regulation of YAP protein turnover. EMBO
J. 33:2447–2457. 2015. View Article : Google Scholar
|
|
187
|
Linossi EM and Nicholson SE: The SOCS
box-adapting proteins for ubiquitination and proteasomal
degradation. IUBMB Life. 64:316–323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Reddy BV and Irvine K: Regulation of Hippo
signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev
Cell. 24:459–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Li L, Liu T, Li Y, Wu C, Luo K, Yin Y,
Chen Y, Nowsheen S, Wu J, Lou Z and Yuan J: The deubiquitinase
USP9X promotes tumor cell survival and confers chemoresistance
through YAP1 stabilization. Oncogene. 37:2422–2431. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Troyanovsky B, Levchenko T, Månsson G,
Matvijenko O and Holmgren L: Angiomotin: An angiostatin binding
protein that regulates endothelial cell migration and tube
formation. J Cell Biol. 152:1247–1254. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Bratt A, Wilson WJ, Troyanovsky B, Aase K,
Kessler R, Van Meir EG and Holmgren L: Angiomotin belongs to a
novel protein family with conserved coiled-coil and PDZ binding
domains. Gene. 298:69–77. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Ernkvist M, Aase K, Ukomadu C,
Wohlschlegel J, Blackman R, Veitonmäki N, Bratt A, Dutta A and
Holmgren L: p130-angiomotin associates to actin and controls
endothelial cell shape. FEBS J. 273:2000–2011. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Zetter BR: Hold that line. Angiomotin
regulates endothelial cell motility. J Cell Biol. 152:F35–F36.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q
and Guan KL: Angiomotin is a novel Hippo pathway component that
inhibits YAP oncoprotein. Genes Dev. 25:51–63. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Chan SW, Lim CJ, Chong YF, Pobbati AV,
Huang C and Hong W: Hippo pathway-independent restriction of TAZ
and YAP by angiomotin. J Biol Chem. 286:7018–7026. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Mana-Capelli S, Paramasivam M, Dutta S and
Mccollum D: Angiomotins link F-actin architecture to Hippo pathway
signaling. Mol Biol Cell. 25:1676–1685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Varelas X, Samavarchi-Tehrani P, Narimatsu
M, Weiss A, Cockburn K, Larsen BG, Rossant J and Wrana JL: The
Crumbs complex couples cell density sensing to Hippo-dependent
control of the TGF-β-SMAD pathway. Dev Cell. 19:831–844. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Paramasivam M, Sarkeshik A, Yates JR III,
Fernandes MJ and Mccollum D: Angiomotin family proteins are novel
activators of the LATS2 kinase tumor suppressor. Mol Biol Cell.
22:3725–3733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Chan SW, Lim CJ, Guo F, Tan I, Leung T and
Hong W: Actin-binding and cell proliferation activities of
angiomotin family members are regulated by Hippo pathway-mediated
phosphorylation. J Biol Chem. 288:37296–37307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Yi C, Shen Z, Stemmer-Rachamimov A, Dawany
N, Troutman S, Showe LC, Liu Q, Shimono A, Sudol M, Holmgren L, et
al: The p130 isoform of angiomotin is required for Yap-mediated
hepatic epithelial cell proliferation and tumorigenesis. Sci
Signal. 6:ra772013. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Wang C, An J, Zhang P, Xu C, Gao K, Wu D,
Wang D, Yu H, Liu JO and Yu L: The Nedd4-like ubiquitin E3 ligases
target angiomotin/p130 to ubiquitin-dependent degradation. Biochem
J. 444:279–289. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Adler JJ, Heller BL, Bringman LR, Ranahan
WP, Cocklin RR, Goebl MG, Oh M, Lim HS, Ingham RJ and Wells CD:
Amot130 adapts atrophin-1 interacting protein 4 to inhibit
yes-associated protein signaling and cell growth. J Biol Chem.
288:15181–15193. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Skouloudaki K and Walz G: YAP1 recruits
c-Abl to protect angiomotin-like 1 from Nedd4-mediated degradation.
PLoS One. 7:e357352012. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Choi KS, Choi HJ, Lee JK, Im S, Zhang H,
Jeong Y, Park JA, Lee IK, Kim YM and Kwon YG: The endothelial E3
ligase HECW2 promotes endothelial cell junctions by increasing
AMOTL1 protein stability via K63-linked ubiquitination. Cell
Signal. 28:1642–1651. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Kim M, Kim M, Park SJ, Lee C and Lim DS:
Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition.
EMBO Rep. 17:64–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Mouchantaf R, Azakir BA, McPherson PS,
Millard SM, Wood SA and Angers A: The ubiquitin ligase itch is
auto-ubiquitylated in vivo and in vitro but is protected from
degradation by interacting with the deubiquitylating enzyme
FAM/USP9X. J Biol Chem. 281:38738–38747. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Chen HH, Mullett SJ and Stewart AF: Vgl-4,
a novel member of the vestigial-like family of transcription
cofactors, regulates alpha1-adrenergic activation of gene
expression in cardiac myocytes. J Biol Chem. 279:30800–30806. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Pobbati AV and Hong W: Emerging roles of
TEAD transcription factors and its coactivators in cancers. Cancer
Biol Ther. 14:390–398. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Pobbati A, Chan SW, Lee I, Song H and Hong
W: Structural and functional similarity between the Vgll1-TEAD and
the YAP-TEAD complexes. Structure. 20:1135–1140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Hélias-Rodzewicz Z, Pérot G, Chibon F,
Ferreira C, Lagarde P, Terrier P, Coindre JM and Aurias A: YAP1 and
VGLL3, encoding two cofactors of TEAD transcription factors, are
amplified and overexpressed in a subset of soft tissue sarcomas.
Genes Chromosom Cancer. 49:1161–1171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Zhang Y, Shen H, Withers HG, Yang N,
Denson KE, Mussell AL, Truskinovsky A, Fan Q, Gelman IH, Frangou C
and Zhang J: VGLL4 selectively represses YAP-dependent gene
induction and tumorigenic phenotypes in breast cancer. Sci Rep.
7:61902017. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Jiang W, Yao F and He J, Lv B, Fang W, Zhu
W, He G, Chen J and He J: Downregulation of VGLL4 in the
progression of esophageal squamous cell carcinoma. Tumour Biol.
36:1289–1297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Jin HS, Park HS, Shin JH, Kim DH, Jun SH,
Lee CJ and Lee TH: A novel inhibitor of apoptosis protein
(IAP)-interacting protein, Vestigial-like (Vgl)-4, counteracts
apoptosis-inhibitory function of IAPs by nuclear sequestration.
Biochem Biophys Res Commun. 412:454–459. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Zhang E, Shen B, Mu X, Qin Y, Zhang F, Liu
Y, Xiao J, Zhang P, Wang C, Tan M and Fan Y: Ubiquitin-specific
protease 11 (USP11) functions as a tumor suppressor through
deubiquitinating and stabilizing VGLL4 protein. Am J Cancer Res.
6:2901–2909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Smith AL, Mitchell PJ, Shipley J,
Gusterson BA, Rogers MV and Crompton MR: Pez: A novel human cDNA
encoding protein tyrosine phosphatase- and ezrin-like domains.
Biochem Biophys Res Commun. 209:959–965. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Ogata M, Takada T, Mori Y, Oh-hora M,
Uchida Y, Kosugi A, Miyake K and Hamaoka T: Effects of
overexpression of PTP36, a putative protein tyrosine phosphatase,
on cell adhesion, cell growth, and cytoskeletons in HeLa cells. J
Biol Chem. 274:12905–12909. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Tonks NK: Protein tyrosine phosphatases:
From genes, to function, to disease. Nat Rev Mol Cell Biol.
7:833–846. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Wang W, Huang J, Wang X, Yuan J, Li X,
Feng L, Park JI and Chen J: PTPN14 is required for the
density-dependent control of YAP1. Genes Dev. 26:1959–1971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Bosu DR and Kipreos ET: Cullin-RING
ubiquitin ligases: Global regulation and activation cycles. Cell
Div. 3:72008. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Jang LK, Lee ZH, Kim HH, Hill JM, Kim JD
and Kwon BS: A novel leucine-rich repeat protein (LRR-1): Potential
involvement in 4-1BB-mediated signal transduction. Mol Cells.
12:304–312. 2001.PubMed/NCBI
|
|
221
|
Walko G, Woodhouse S, Pisco AO, Rognoni E,
Liakath-Ali K, Lichtenberger BM, Mishra A, Telerman SB, Viswanathan
P, Logtenberg M, et al: A genome-wide screen identifies YAP/WBP2
interplay conferring growth advantage on human epidermal stem
cells. Nat Commun. 8:147442017. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Lim SK, Lu SY, Kang SA, Tan HJ, Li Z,
Adrian Wee ZN, Guan JS, Reddy Chichili VP, Sivaraman J, Putti T, et
al: Wnt signaling promotes breast cancer by blocking ITCH-mediated
degradation of YAP/TAZ transcriptional coactivator WBP2. Cancer
Res. 76:6278–6289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Avruch J, Xavier R, Bardeesy N, Zhang X,
Praskova M, Zhou D and Xia F: Rassf family of tumor suppressor
polypeptides. J Biol Chem. 284:11001–11005. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Volodko N, Gordon M, Salla M, Ghazaleh HA
and Baksh S: RASSF tumor suppressor gene family: Biological
functions and regulation. FEBS Lett. 588:2671–2684. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Iwasa H, Hossain S and Hata Y: Tumor
suppressor C-RASSF proteins. Cell Mol Life Sci. 75:1773–1787. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Agathanggelou A, Cooper WN and Latif F:
Role of the Ras-association domain family 1 tumor suppressor gene
in human cancers. Cancer Res. 65:3497–3508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Richter AM, Pfeifer GP and Dammann RH: The
RASSF proteins in cancer; from epigenetic silencing to functional
characterization. Biochim Biophys Acta 1796. 114–128. 2009.
|
|
228
|
Oh HJ, Lee KK, Song SJ, Jin MS, Song MS,
Lee JH, Im CR, Lee JO, Yonehara S and Lim DS: Role of the tumor
suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res.
66:2562–2569. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Guo C, Zhang X and Pfeifer GP: The tumor
suppressor RASSF1A prevents dephosphorylation of the mammalian
STE20-like kinases MST1 and MST2. J Biol Chem. 286:6253–6261. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Matallanas D, Romano D, Yee K, Meissl K,
Kucerova L, Piazzolla D, Baccarini M, Vass JK, Kolch W and O'Neill
E: RASSF1A elicits apoptosis through an MST2 pathway directing
proapoptotic transcription by the p73 tumor suppressor protein. Mol
Cell. 27:962–975. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Bitra A, Sistla S, Mariam J, Malvi H and
Anand R: Rassf proteins as modulators of Mst1 kinase activity. Sci
Rep. 7:450202017. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Donninger H, Allen N, Henson A, Pogue J,
Williams A, Gordon L, Kassler S, Dunwell T, Latif F and Clark GJ:
Salvador protein is a tumor suppressor effector of RASSF1A with
hippo pathway-independent functions. J Biol Chem. 286:18483–18491.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
233
|
Vlahov N, Scrace S, Soto MS, Grawenda AM,
Bradley L, Pankova D, Papaspyropoulos A, Yee KS, Buffa F, Goding
CR, et al: Alternate RASSF1 transcripts control SRC activity,
E-cadherin contacts, and YAP-mediated invasion. Curr Biol.
25:3019–3034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Burbee D, Forgacs E, ZöchbauerMüller S,
Shivakumar L, Fong K, Gao B, Randle D, Kondo M, Virmani A, Bader S,
et al: Epigenetic inactivation of RASSF1A in lung and breast
cancers and malignant phenotype suppression. J Natl Cancer Inst.
93:691–699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
235
|
Pefani DE, Pankova D, Abraham A, Grawenda
A, Vlahov N, Scrace S and O'Neill E: TGF-β targets the Hippo
pathway scaffold RASSF1A to facilitate YAP/SMAD2 nuclear
translocation. Mol Cell. 63:156–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Suryaraja R, Anitha M, Anbarasu K, Kumari
G and Mahalingam S: The E3 ubiquitin ligase Itch regulates tumor
suppressor protein RASSF5/NORE1 stability in an
acetylation-dependent manner. Cell Death Dis. 4:e5652013.
View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Kumari G, Singhal PK, Rao MR and
Mahalingam S: Nuclear transport of Ras-associated tumor suppressor
proteins: Different transport receptor binding specificities for
arginine-rich nuclear targeting signals. J Mol Biol. 367:1294–1311.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Song MS, Song SJ, Kim SJ, Nakayama K,
Nakayama KI and Lim D: Skp2 regulates the antiproliferative
function of the tumor suppressor RASSF1A via ubiquitin-mediated
degradation at the G1-S transition. Oncogene. 27:3176–3185. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
239
|
Jiang L, Rong R, Sheikh MS and Huang Y:
Cullin-4A·DNA damage-binding protein 1 E3 ligase complex targets
tumor suppressor RASSF1A for degradation during mitosis. J Biol
Chem. 286:6971–6978. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Zhou X, Li TT, Feng X, Hsiang E, Xiong Y,
Guan KL and Lei QY: Targeted polyubiquitylation of RASSF1C by the
mule and SCFβ-TrCP ligases in response to DNA damage. Biochem J.
441:227–236. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
241
|
Mohseni M, Sun J, Lau A, Curtis S,
Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, et al: A
genetic screen identifies an LKB1-MARK signalling axis controlling
the Hippo-YAP pathway. Nat Cell Biol. 16:108–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Nguyen HT, Kugler JM, Loya AC and Cohen
SM: USP21 regulates Hippo pathway activity by mediating MARK
protein turnover. Oncotarget. 8:64095–64105. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
243
|
Lignitto L, Arcella A, Sepe M, Rinaldi L,
Delle Donne R, Gallo A, Stefan E, Bachmann VA, Oliva MA, Tiziana
Storlazzi C, et al: Proteolysis of MOB1 by the ubiquitin ligase
praja2 attenuates Hippo signalling and supports glioblastoma
growth. Nat Commun. 4:18222013. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Ballinger CA, Connell P, Wu Y, Hu Z,
Thompson LJ, Yin LY and Patterson C: Identification of CHIP, a
novel tetratricopeptide repeat-containing protein that interacts
with heat shock proteins and negatively regulates chaperone
functions. Mol Cell Biol. 19:4535–4545. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Ren A, Yan G, You B and Sun J:
Down-regulation of mammalian sterile 20-like kinase 1 by heat shock
protein 70 mediates cisplatin resistance in prostate cancer cells.
Cancer Res. 68:2266–2274. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
246
|
Xiao L, Chen D, Hu P, Wu J, Liu W, Zhao Y,
Cao M, Fang Y, Bi W, Zheng Z, et al: The c-Abl-MST1 signaling
pathway mediates oxidative stress-induced neuronal cell death. J
Neurosci. 31:9611–9619. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
247
|
Bao Y, Nakagawa K, Yang Z, Ikeda M,
Withanage K, Ishigami-Yuasa M, Okuno Y, Hata S, Nishina H and Hata
Y: A cell-based assay to screen stimulators of the Hippo pathway
reveals the inhibitory effect of dobutamine on the YAP-dependent
gene transcription. J Biochem. 150:199–208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Michels S and Schmidt-Erfurth U:
Photodynamic therapy with verteporfin: A new treatment in
ophthalmology. Semin Ophthalmol. 16:201–206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Liu-Chittenden Y, Huang B, Shim JS, Chen
Q, Lee SJ, Anders RA, Liu JO and Pan D: Genetic and pharmacological
disruption of the TEAD-YAP complex suppresses the oncogenic
activity of YAP. Genes Dev. 26:1300–1305. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
250
|
Huggett MT, Jermyn M, Gillams A, Illing R,
Mosse S, Novelli M, Kent E, Bown SG, Hasan T, Pogue BW and Pereira
SP: Phase I/II study of verteporfin photodynamic therapy in locally
advanced pancreatic cancer. Br J Cancer. 110:1698–1704. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
251
|
Kang W, Huang T, Zhou Y, Zhang J, Lung
RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F, et al: miR-375 is
involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in
gastric carcinogenesis. Cell Death Dis. 9:922018. View Article : Google Scholar : PubMed/NCBI
|
|
252
|
Jiao S, Wang H, Shi Z, Dong A, Zhang W,
Song X, He F, Wang Y, Zhang Z, Wang W, et al: A peptide mimicking
VGLL4 function acts as a YAP antagonist therapy against gastric
cancer. Cancer Cell. 25:166–180. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
253
|
Yu FX, Zhao B, Panupinthu N, Jewell J,
Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al:
Regulation of the Hippo-YAP pathway by G-protein-coupled receptor
signaling. Cell. 150:780–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
254
|
Ponnusamy S, Selvam SP, Mehrotra S,
Kawamori T, Snider AJ, Obeid LM, Shao Y, Sabbadini R and Ogretmen
B: Communication between host organism and cancer cells is
transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate
signalling to regulate tumour metastasis. EMBO Mol Med. 4:761–775.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
255
|
Kelly MG, Mor G, Husband A, O'Malley DM,
Baker L, Azodi M, Schwartz PE and Rutherford TJ: Phase II
evaluation of phenoxodiol in combination with cisplatin or
paclitaxel in women with platinum/taxane-refractory/resistant
epithelial ovarian, fallopian tube, or primary peritoneal cancers.
Int J Gynecol Cancer. 21:633–639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
256
|
Laubach JP, Mitsiades CS, Roccaro AM,
Ghobrial IM, Anderson KC and Richardson PG: Clinical challenges
associated with bortezomib therapy in multiple myeloma and
Waldenstroms Macroglobulinemia. Leuk Lymphoma. 50:694–702. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
257
|
Wang W, Ren F, Wu Q, Jiang D, Li H, Peng
Z, Wang J and Shi H: MicroRNA-497 inhibition of ovarian cancer cell
migration and invasion through targeting of SMAD specific E3
ubiquitin protein ligase 1. Biochem Biophys Res Commun.
449:432–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
258
|
Zhu JY, Heidersbach A, Kathiriya IS, Garay
BI, Ivey KN, Srivastava D, Han Z and King IN: The E3 ubiquitin
ligase Nedd4/Nedd4L is directly regulated by microRNA 1.
Development. 144:866–875. 2017. View Article : Google Scholar : PubMed/NCBI
|

















