Most breast cancers (BC), over 2/3 of cases, express
estrogen (ER) and progesterone (PR) receptors (1). This is extremely important since these
are used as biomarkers for subtype classification, with
implications in choice of treatment and prognosis in BC patients
(2). Notably, endocrine therapies
(ET) have been successfully used for treating ER positive BC
patients with significant impact in patient outcome. Several
endocrine drugs are approved for BC treatment, most notably
tamoxifen, toremifene, anastrozole, letrozole, exemestane and
fulvestrant, which may be used in different clinical contexts, such
as chemoprophylaxis, neoadjuvant, adjuvant and palliative
treatments. However, the effectiveness of ET is limited as up to
40% of patients may experience disease recurrence while on ET
adjuvant treatment (1,3). Moreover, in the metastatic setting,
acquired resistance to ET is virtually an universal feature, and is
clinically defined in accordance to the 3rd ESO-ESMO International
Consensus Guidelines (4) and many
efforts have been made to understand the mechanisms involved in
acquisition of acquired resistance to ET. These, however, remain
mostly elusive and no biomarkers have been validated in this
setting despite intense drug development and approval.
Epigenetics may be defined as mechanisms that
regulate cell fate specifications, while the DNA remains unchanged
(5). Some of these mechanisms
include DNA methylation, non-coding RNAs, chromatin remodeling and
histone post-translational modifications or variants. Collectively,
these components constitute the epigenome machinery whose role is
to define which information is available for transcription and for
translation (5). DNA methylation is
performed by specific enzymes, the DNA methyltransferases (DNMTs)
that introduce a methyl group at the 5′ position of a cytosine ring
inside CpG dinucleotides (6).
Globally, promoter methylation of genes is associated with
transcription inhibition (6).
Furthermore, the N-terminal tails of histones may undergo
post-translation modifications that subsequently impact the
chromatin structure (7). The most
well-studied histone post-translation modifications are histone
acetylation and histone methylation. Histone acetylation is
associated with gene expression and is carried out by histone
acetyltransferases (HATs), while histone deacetylation is
accomplished by histone deacetylases (HDACs) (7). Histone methylation, which depending on
the residue and the number of methyl groups may lead either to
transcription repression or activation (8), is catalyzed by histone
methyltransferases (HMTs), while histone demethylation is performed
by histone demethylases (HDMs) (7).
In addition to post-translational histone modifications, histone
variants that can replace canonical histones are an additional
level of epigenetic complexity, and contribute to the shaping of
the chromatin structure.
Non-coding RNAs (ncRNAs) comprise a hidden layer of
internal signals that control various levels of gene expression
(9). Among these, microRNAs
(miRNAs) and long non-coding RNAs (lncRNAs) are the most frequently
reported in BC. lncRNAs are ncRNA molecules usually longer than 200
nucleotides that do not fit into known classes of small or
structural RNAs (9) and may act as
protein-DNA or protein-protein scaffolds, miRNA sponges, protein
decoys, or regulators of translation (10). miRNAs are endogenous, small
non-coding single-stranded RNAs with ~22 nucleotides in length,
that exert a finely tuned regulation of gene expression at the
post-transcriptional level (11) by
binding to mRNA targets, inducing its cleavage or repressing its
translation (11).
Over the last few years, convincing data has
suggested that altered epigenetic regulation may be involved in
tumor initiation, progression and cancer resistance to therapy,
including endocrine resistance, particularly in BC. For instance,
ER expression is currently one of the foremost predictive
biomarkers of response to ET, and altered expression of ER may be
due to hypermethylation of CpG islands within its promoter,
increased histone deacetylase activity in the ESR1 promoter or
translational repression by miRNAs (12). Since ER was found to be deleted in
only 15–20% of endocrine-resistant BC, several epigenetic
mechanisms may be involved in the development of endocrine
treatment-resistance (3), and some
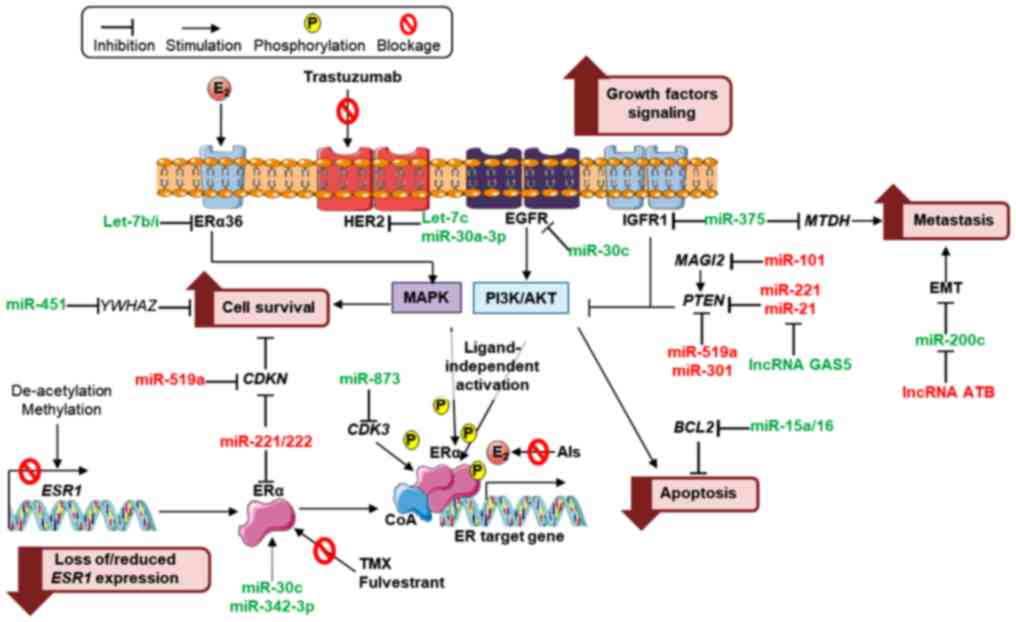
of these are depicted in Fig.
1.
Our objective was to review the published evidence
regarding epigenetic mechanisms associated to ET resistance in BC,
as it may be considered an emerging subject and worth special
focus.
DNA methylation is one of the most common epigenetic
changes and has been reported in multiple tumors, including BC
(9,13). This epigenetic alteration is
inherently stable and has been proposed as a promising cancer
biomarker in multiple cancers since it can be sampled from less
invasive sources such as liquid biopsies (plasma or urine)
(13–15). Thus, the role of DNA methylation as
a predictor of ET resistance is a field of growing interest and has
become the focus of several research teams (16–18)
since it may improve BC patients' risk stratification.
As previously mentioned, decreased ER expression may
be due to post-transcription regulation of miRNAs, including that
of miR-221/222, whose overexpression has been associated with
resistance to tamoxifen (23,24)
and fulvestrant (25). Conversely,
miR-342-3p levels were revealed to be positively correlated with ER
mRNA expression in human BC and associated with tamoxifen
sensitivity (26,27). miRNAs that regulate growth,
survival, apoptosis, epithelial-mesenchymal transition (EMT) and
metastasis of BC cells may be implicated in loss of responsiveness
to ET. In particular, PTEN downregulation due to specific miRNAs,
permitting abnormal Pi3K/Akt pathway activation, promote
estrogen-independent growth and survival of BC cells leading to
endocrine treatment resistance (28,29).
Several clinical trials are currently ongoing to
evaluate the role of miRNAs as predictive biomarkers in BC.
Specifically, trials such as NCT01231386 and NCT01722851, aim to
identify circulating miRNAs aiding at the identification of
biomarkers of early response to neoadjuvant therapy, including ET,
which may be used as potential targets for personalized therapies.
Conversely, the NCT01612871 trial was set to explore a panel of
circulating miRNAs that could aid to monitor the disease status of
the patient while on adjuvant ET (30–32).
lncRNAs have also been associated with endocrine
treatment resistance. Particularly, lncRNAs, breast cancer
anti-estrogen resistance 4 (BRCAAR4) overexpression (33,34)
and DSCAM antisense RNA 1 (DSCAM-AS1) (35), which contains an ER promoter binding
motif, have been revealed to predict tamoxifen resistance in
primary BC (Table II and Fig. 1).
Histone post-translation modifications induce
chromatin landscape changes that subsequently favor ER repression,
thus promoting other signaling pathways that could lead to
endocrine resistance, as exemplified by Magnani et al that
revealed how the genome's accessibility is altered in
drug-resistant vs. drug-responsive BC cells (36). Recently, expression of the H3K36
methyltransferase NSD2 was found to be higher in
tamoxifen-resistant BC cell lines, associated with disease
recurrence and worse survival (37). Furthermore, H3K37me3 profiles
enabled the identification of patients with poor outcome after
aromatase inhibitor (AI) treatment (38).
Furthermore, it was recently demonstrated that
transcription repression performed by ER co-repressors confer
tamoxifen sensitivity through recruitment of HDACs to DNA (39). This evidence suggests that loss of
ER co-repressors may sensitize BC cells to the cytotoxic effects of
HDACs inhibitors (HDACi). Notably, some clinical trials have
demonstrated that HDACi appears to re-establish sensitivity to
anti-estrogens in a subset of endocrine treated-resistant tumors
(40,41). In addition, the ENCORE-301, a
randomized phase II trial (41)
tested entinostat, an oral HDACi, in the endocrine-resistance, more
specifically AI in post-menopausal women. The results revealed
modest improvement in PFS but much greater improvement in overall
survival (OS)-median OS improved to 28.1 months in the experimental
arm vs. 19.8 months (HR, 0.59; 95% CI, 0.36 to 0.97; P=0.036).
Ongoing clinical trials are further testing entinostat in
monotherapy or in combination. Moreover, in custom-generated
tamoxifen resistant cell lines, treatment with HDACi re-established
sensitivity to tamoxifen through significant Bcl-2 downregulation,
growth arrest and apoptosis (42).
Histone variants, such as H2A.Z, an H2A variant,
have been shown to be intimately linked to estrogen signaling
(43). Notably, a study has already
provided a link (yet uncharacterized) between H2A.Z and endocrine
resistance by revealing that H2A.Z overexpression led to increased
estrogen-independent proliferation (44). Furthermore, another study
demonstrated that the histone HIST1H2BE, an H2B variant, was
overexpressed not only in endocrine-resistant cell lines, but also
in AI-treated tumors from patients which relapsed compared to those
that did not (45).
Furthermore, an emerging class of transcription
factors named ‘pioneer factors’, appear to be key players in
shaping chromatin structure through binding to chromatin prior to
transcription factors, making it accessible for transcription
factors, together with histone post-translation modifications and
histone variants [68–70]. PBX1 is an example of this class-its
expression levels have been associated with reduced metastasis-free
survival in ER-positive BC patients (46). Furthermore, a gene expression
signature based on NOTCH-PBX1 activity was found to discriminate BC
patients that are responsive to ET from those which are not.
Notably, PBX1 knockdown was sufficient to arrest ER-resistant BC
cell growth (36).
These and other chromatin remodeling complexes
associated with endocrine resistance are summarized in Table III along with their putative role
and the biological samples in which they were characterized.
Notwithstanding the prevalence of endocrine
treatment resistance in BC, predictive and diagnostic biomarkers in
this setting are markedly lacking in clinical practice. In this
review, we summarized emerging evidence that epigenetic mechanisms
may prove useful for this purpose. These would perform as
non-invasive predictive biomarkers of treatment-resistance,
providing affordable and sequential monitoring during the course of
treatment. The concept of early detection (preclinical) of therapy
resistance is compelling, as it could assist clinicians in choosing
the most appropriate individualized therapeutic strategy.
Furthermore, some epigenetic modifications in
addition to conveying information concerning prediction of
response, are also appealingly targetable, in particular due to
their reversible nature. The clinical usefulness of these findings,
however, is still elusive, mostly due to lack of standardization in
methodology, limiting reproducibility.
Promising results have been arising in clinically
meaningful trials, such as ENCORE-301. A useful approach would be
the integration of the candidate biomarkers into a panel, enabling
its validation in a clinical trial setting. Hopefully, this will be
accomplished in the near future.
Not applicable.
The present study was supported by a grant from the
Research Center of Portuguese Oncology Institute of Porto (PI
74-CI-IPOP-19-2016) and the Portuguese Society of Oncology-YOuR
Project. SS was supported by a PhD fellowship IPO/ESTIMA-1
NORTE-01-0145-FEDER-000027.
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
MFS, SPDS, RH and CJ conceived and designed the
review. MFS, MA, SS performed the literature search and wrote the
manuscript. SPDS, RH and CJ reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Magnani L, Brunelle M, Gévry N and Lupien
M: Chromatin landscape and endocrine response in breast cancer.
Epigenomics. 4:675–683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheang MC, van de Rijn M and Nielsen TO:
Gene expression profiling of breast cancer. Annu Rev Pathol.
3:67–97. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Normanno N, Di Maio M, De Maio E, De Luca
A, de Matteis A, Giordano A and Perrone F; NCI-Naple Breast Cancer
Group, : Mechanisms of endocrine resistance and novel therapeutic
strategies in breast cancer. Endocr Relat Cancer. 12:721–747. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cardoso F, Costa A, Senkus E, Aapro M,
André F, Barrios CH, Bergh J, Bhattacharyya G, Biganzoli L, Cardoso
MJ, et al: 3rd ESO-ESMO international consensus guidelines for
Advanced Breast Cancer (ABC 3). Breast. 31:244–259. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodríguez-Paredes M and Esteller M: Cancer
epigenetics reaches mainstream oncology. Nat Med. 17:330–339. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bannister AJ and Kouzarides T: Regulation
of chromatin by histone modifications. Cell Res. 21:381–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zentner GE and Henikoff S: Regulation of
nucleosome dynamics by histone modifications. Nat Struct Mol Biol.
20:259–266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amorim M, Salta S, Henrique R and Jerónimo
C: Decoding the usefulness of non-coding RNAs as breast cancer
markers. J Transl Med. 14:2652016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharma D, Blum J, Yang X, Beaulieu N,
Macleod AR and Davidson NE: Release of methyl CpG binding proteins
and histone deacetylase 1 from the estrogen receptor α (ER)
promoter upon reactivation in ER-negative human breast cancer
cells. Mol Endocrinol. 19:1740–1751. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: Applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jerónimo C and Henrique R: Epigenetic
biomarkers in urological tumors: A systematic review. Cancer Lett.
342:264–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Costa-Pinheiro P, Montezuma D, Henrique R
and Jerónimo C: Diagnostic and prognostic epigenetic biomarkers in
cancer. Epigenomics. 7:1003–1015. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Widschwendter M, Siegmund KD, Müller HM,
Fiegl H, Marth C, Müller-Holzner E, Jones PA and Laird PW:
Association of breast cancer DNA methylation profiles with hormone
receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan M, Yan PS, Hartman-Frey C, Chen L,
Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al:
Diverse gene expression and DNA methylation profiles correlate with
differential adaptation of breast cancer cells to the antiestrogens
tamoxifen and fulvestrant. Cancer Res. 66:11954–11966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stone A, Zotenko E, Locke WJ, Korbie D,
Millar EK, Pidsley R, Stirzaker C, Graham P, Trau M, Musgrove EA,
et al: DNA methylation of oestrogen-regulated enhancers defines
endocrine sensitivity in breast cancer. Nat Commun. 6:77582015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Williams KE, Anderton DL, Lee MP,
Pentecost BT and Arcaro KF: High-density array analysis of DNA
methylation in Tamoxifen-resistant breast cancer cell lines.
Epigenetics. 9:297–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maier S, Nimmrich I, Koenig T,
Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen
C, Mueller V, Nährig J, et al: European Organisation for Research
and Treatment of Cancer (EORTC) PathoBiology group: DNA-methylation
of the homeodomain transcription factor PITX2 reliably predicts
risk of distant disease recurrence in tamoxifen-treated,
node-negative breast cancer patients: Technical and clinical
validation in a multi-centre setting in collaboration with the
European Organisation for Research and Treatment of Cancer (EORTC)
PathoBiology group. Eur J Cancer. 43:1679–1686. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Harbeck N, Nimmrich I, Hartmann A, Ross
JS, Cufer T, Grützmann R, Kristiansen G, Paradiso A, Hartmann O,
Margossian A, et al: Multicenter study using paraffin-embedded
tumor tissue testing PITX2 DNA methylation as a marker for
outcome prediction in tamoxifen-treated, node-negative breast
cancer patients. J Clin Oncol. 26:5036–5042. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting
p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma
X, Coppola D and Cheng JQ: MicroRNA-221/222 negatively regulates
estrogen receptor α and is associated with tamoxifen resistance in
breast cancer. J Biol Chem. 283:31079–31086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cittelly DM, Das PM, Spoelstra NS,
Edgerton SM, Richer JK, Thor AD and Jones FE: Downregulation of
miR-342 is associated with tamoxifen resistant breast tumors. Mol
Cancer. 9:3172010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R
and Tang JH: miR-342 is associated with estrogen receptor-α
expression and response to tamoxifen in breast cancer. Exp Ther
Med. 5:813–818. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ward A, Shukla K, Balwierz A, Soons Z,
König R, Sahin O and Wiemann S: MicroRNA-519a is a novel oncomir
conferring tamoxifen resistance by targeting a network of
tumour-suppressor genes in ER+ breast cancer. J Pathol.
233:368–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
ClinicalTrials.gov: MIRNA Profiling of
Breast Cancer in Patients Undergoing Neoadjuvant or Adjuvant
Treatment for Locally Advanced & Inflammatory Breast Cancer.
ClinicalTrials.gov2016. https://clinicaltrials.gov/ct2/show/NCT01231386
|
|
31
|
ClinicalTrials.gov: Circulating miRNAs.
ICORG 10–11, V2. ClinicalTrials.gov2017. https://clinicaltrials.gov/ct2/show/NCT01722851
|
|
32
|
ClinicalTrials.gov: Circulating miRNAs as
Biomarkers of Hormone Sensitivity in Breast Cancer (MIRHO).
ClinicalTrials.gov. 2014, https://clinicaltrials.gov/ct2/show/NCT01612871
|
|
33
|
Meijer D, van Agthoven T, Bosma PT, Nooter
K and Dorssers LC: Functional screen for genes responsible for
tamoxifen resistance in human breast cancer cells. Mol Cancer Res.
4:379–386. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Godinho MF, Sieuwerts AM, Look MP, Meijer
D, Foekens JA, Dorssers LC and Van Agthoven T: Relevance of BCAR4
in tamoxifen resistance and tumour aggressiveness of human breast
cancer. Br J Cancer. 103:1284–1291. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Niknafs YS, Han S, Ma T, Speers C, Zhang
C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al:
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1
in breast cancer progression. Nat Commun. 7:127912016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Magnani L, Stoeck A, Zhang X, Lánczky A,
Mirabella AC, Wang TL, Gyorffy B and Lupien M: Genome-wide
reprogramming of the chromatin landscape underlies endocrine
therapy resistance in breast cancer. Proc Natl Acad Sci USA.
110:E1490–E1499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Duan Z, Nugent Z, Zou JX, Borowsky
AD, Zhang Y, Tepper CG, Li JJ, Fiehn O, Xu J, et al: Reprogramming
metabolism by histone methyltransferase NSD2 drives endocrine
resistance via coordinated activation of pentose phosphate pathway
enzymes. Cancer Lett. 378:69–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jansen MP, Knijnenburg T, Reijm EA, Simon
I, Kerkhoven R, Droog M, Velds A, van Laere S, Dirix L, Alexi X, et
al: Hallmarks of aromatase inhibitor drug resistance revealed by
epigenetic profiling in breast cancer. Cancer Res. 73:6632–6641.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Légaré S and Basik M: Minireview: The link
between ERα corepressors and histone deacetylases in tamoxifen
resistance in breast cancer. Mol Endocrinol. 30:965–976. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Munster PN, Thurn KT, Thomas S, Raha P,
Lacevic M, Miller A, Melisko M, Ismail-Khan R, Rugo H, Moasser M,
et al: A phase II study of the histone deacetylase inhibitor
vorinostat combined with tamoxifen for the treatment of patients
with hormone therapy-resistant breast cancer. Br J Cancer.
104:1828–1835. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yardley DA, Ismail-Khan RR, Melichar B,
Lichinitser M, Munster PN, Klein PM, Cruickshank S, Miller KD, Lee
MJ and Trepel JB: Randomized phase II, double-blind,
placebo-controlled study of exemestane with or without entinostat
in postmenopausal women with locally recurrent or metastatic
estrogen receptor-positive breast cancer progressing on treatment
with a nonsteroidal aromatase inhibitor. J Clin Oncol.
31:2128–2135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Raha P, Thomas S, Thurn KT, Park J and
Munster PN: Combined histone deacetylase inhibition and tamoxifen
induces apoptosis in tamoxifen-resistant breast cancer models, by
reversing Bcl-2 overexpression. Breast Cancer Res. 17:262015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gévry N, Hardy S, Jacques P-É, Laflamme L,
Svotelis A, Robert F and Gaudreau L: Histone H2A.Z is essential for
estrogen receptor signaling. Genes Dev. 23:1522–1533. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Svotelis A, Gévry N, Grondin G and
Gaudreau L: H2A.Z overexpression promotes cellular proliferation of
breast cancer cells. Cell Cycle. 9:364–370. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nayak SR, Harrington E, Boone D, Hartmaier
R, Chen J, Pathiraja TN, Cooper KL, Fine JL, Sanfilippo J, Davidson
NE, et al: A role for histone H2B variants in endocrine-resistant
breast cancer. Horm Cancer. 6:214–224. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Magnani L, Ballantyne EB, Zhang X and
Lupien M: PBX1 genomic pioneer function drives ERα signaling
underlying progression in breast cancer. PLoS Genet.
7:e10023682011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Phuong NT, Kim SK, Lim SC, Kim HS, Kim TH,
Lee KY, Ahn SG, Yoon JH and Kang KW: Role of PTEN promoter
methylation in tamoxifen-resistant breast cancer cells. Breast
Cancer Res Treat. 130:73–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hiken JF, McDonald JI, Decker KF, Sanchez
C, Hoog J, VanderKraats ND, Jung KL, Akinhanmi M, Rois LE, Ellis
MJ, et al: Epigenetic activation of the prostaglandin receptor EP4
promotes resistance to endocrine therapy for breast cancer.
Oncogene. 36:2319–2327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Iorns E, Turner NC, Elliott R, Syed N,
Garrone O, Gasco M, Tutt AN, Crook T, Lord CJ and Ashworth A:
Identification of CDK10 as an important determinant of resistance
to endocrine therapy for breast cancer. Cancer Cell. 13:91–104.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pathiraja TN, Nayak SR, Xi Y, Jiang S,
Garee JP, Edwards DP, Lee AV, Chen J, Shea MJ, Santen RJ, et al:
Epigenetic reprogramming of HOXC10 in endocrine-resistant
breast cancer. Sci Transl Med. 6:229ra412014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang Y, Zhang B, Fang J and Cao X:
Hypomethylation of DNA-binding inhibitor 4 serves as a potential
biomarker in distinguishing acquired tamoxifen-refractory breast
cancer. Int J Clin Exp Pathol. 8:9500–9505. 2015.PubMed/NCBI
|
|
52
|
Kim SJ, Kang HS, Jung SY, Min SY, Lee S,
Kim SW, Kwon Y, Lee KS, Shin KH and Ro J: Methylation patterns of
genes coding for drug-metabolizing enzymes in tamoxifen-resistant
breast cancer tissues. J Mol Med. 88:1123–1131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nimmrich I, Sieuwerts AM, Meijer-van
Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, Koenig T,
Hartmann O, Kluth A, Dietrich D, et al: DNA hypermethylation of
PITX2 is a marker of poor prognosis in untreated lymph
node-negative hormone receptor-positive breast cancer patients.
Breast Cancer Res Treat. 111:429–437. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Pathiraja TN, Shetty PB, Jelinek J, He R,
Hartmaier R, Margossian AL, Hilsenbeck SG, Issa JP and Oesterreich
S: Progesterone receptor isoform-specific promoter methylation:
Association of PRA promoter methylation with worse outcome
in breast cancer patients. Clin Cancer Res. 17:4177–4186. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Martens JW, Nimmrich I, Koenig T, Look MP,
Harbeck N, Model F, Kluth A, Bolt-de Vries J, Sieuwerts AM,
Portengen H, et al: Association of DNA methylation of phosphoserine
aminotransferase with response to endocrine therapy in patients
with recurrent breast cancer. Cancer Res. 65:4101–4117. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cui J, Yang Y, Li H, Leng Y, Qian K, Huang
Q, Zhang C, Lu Z, Chen J, Sun T, et al: MiR-873 regulates ERα
transcriptional activity and tamoxifen resistance via targeting
CDK3 in breast cancer cells. Oncogene. 34:3895–3907. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lü M, Ding K, Zhang G, Yin M, Yao G, Tian
H, Lian J, Liu L, Liang M, Zhu T, et al: MicroRNA-320a sensitizes
tamoxifen-resistant breast cancer cells to tamoxifen by targeting
ARPP-19 and ERRγ. Sci Rep. 4:87352015. View Article : Google Scholar
|
|
59
|
Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M,
Recker RR, Gatalica Z, Wang Z and Xiao GG: let-7 microRNAs
induce tamoxifen sensitivity by downregulation of estrogen receptor
α signaling in breast cancer. Mol Med. 17:1233–1241. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bergamaschi A and Katzenellenbogen BS:
Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes
breast cancer cell survival and endocrine resistance. Oncogene.
31:39–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yu Z, Xu Z, Disante G, Wright J, Wang M,
Li Y, Zhao Q, Ren T, Ju X, Gutman E, et al: miR-17/20 sensitization
of breast cancer cells to chemotherapy-induced apoptosis requires
Akt1. Oncotarget. 5:1083–1090. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen MJ, Cheng YM, Chen CC, Chen YC and
Shen CJ: MiR-148a and miR-152 reduce tamoxifen resistance in
ER+ breast cancer via downregulating ALCAM. Biochem
Biophys Res Commun. 483:840–846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manavalan TT, Teng Y, Litchfield LM,
Muluhngwi P, Al-Rayyan N and Klinge CM: Reduced expression of
miR-200 family members contributes to antiestrogen resistance in
LY2 human breast cancer cells. PLoS One. 8:e623342013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cittelly DM, Das PM, Salvo VA, Fonseca JP,
Burow ME and Jones FE: Oncogenic HER2{Delta}16 suppresses
miR-15a/16 and deregulates BCL-2 to promote endocrine resistance of
breast tumors. Carcinogenesis. 31:2049–2057. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jansen MP, Reijm EA, Sieuwerts AM,
Ruigrok-Ritstier K, Look MP, Rodríguez-González FG, Heine AA,
Martens JW, Sleijfer S, Foekens JA, et al: High miR-26a and low
CDC2 levels associate with decreased EZH2 expression and with
favorable outcome on tamoxifen in metastatic breast cancer. Breast
Cancer Res Treat. 133:937–947. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rodríguez-González FG, Sieuwerts AM, Smid
M, Look MP, Meijer-van Gelder ME, de Weerd V, Sleijfer S, Martens
JW and Foekens JA: MicroRNA-30c expression level is an independent
predictor of clinical benefit of endocrine therapy in advanced
estrogen receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hoppe R, Achinger-Kawecka J, Winter S,
Fritz P, Lo WY, Schroth W and Brauch H: Increased expression of
miR-126 and miR-10a predict prolonged relapse-free time of primary
oestrogen receptor-positive breast cancer following tamoxifen
treatment. Eur J Cancer. 49:3598–3608. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ahmad A, Ginnebaugh KR, Yin S,
Bollig-Fischer A, Reddy KB and Sarkar FH: Functional role of
miR-10b in tamoxifen resistance of ER-positive breast cancer cells
through downregulation of HDAC4. BMC Cancer. 15:5402015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y,
Li H, Zhu X, Yao L and Zhang J: Exosomal miR-221/222 enhances
tamoxifen resistance in recipient ER-positive breast cancer cells.
Breast Cancer Res Treat. 147:423–431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shen R, Wang Y, Wang CX, Yin M, Liu HL,
Chen JP, Han JQ and Wang WB: MiRNA-155 mediates TAM resistance by
modulating SOCS6-STAT3 signalling pathway in breast cancer. Am J
Transl Res. 7:2115–2126. 2015.PubMed/NCBI
|
|
71
|
Rothé F, Ignatiadis M, Chaboteaux C,
Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H,
Desmedt C, Harris AL, et al: Global microRNA expression profiling
identifies MiR-210 associated with tumor proliferation, invasion
and poor clinical outcome in breast cancer. PLoS One. 6:e209802011.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shibahara Y, Miki Y, Onodera Y, Hata S,
Chan MS, Yiu CC, Loo TY, Nakamura Y, Akahira J, Ishida T, et al:
Aromatase inhibitor treatment of breast cancer cells increases the
expression of let-7f, a microRNA targeting CYP19A1. J
Pathol. 227:357–366. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bailey ST, Westerling T and Brown M: Loss
of estrogen-regulated microRNA expression increases HER2 signaling
and is prognostic of poor outcome in luminal breast cancer. Cancer
Res. 75:436–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Masri S, Liu Z, Phung S, Wang E, Yuan YC
and Chen S: The role of microRNA-128a in regulating TGFbeta
signaling in letrozole-resistant breast cancer cells. Breast Cancer
Res Treat. 124:89–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hayes EL and Lewis-Wambi JS: Mechanisms of
endocrine resistance in breast cancer: An overview of the proposed
roles of noncoding RNA. Breast Cancer Res. 17:402015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang Z, Yamashita H, Toyama T, Sugiura H,
Omoto Y, Ando Y, Mita K, Hamaguchi M, Hayashi S and Iwase H: HDAC6
expression is correlated with better survival in breast cancer.
Clin Cancer Res. 10:6962–6968. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Saji S, Kawakami M, Hayashi S, Yoshida N,
Hirose M, Horiguchi S, Itoh A, Funata N, Schreiber SL, Yoshida M,
et al: Significance of HDAC6 regulation via estrogen signaling for
cell motility and prognosis in estrogen receptor-positive breast
cancer. Oncogene. 24:4531–4539. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Svotelis A, Bianco S, Madore J, Huppé G,
Nordell-Markovits A, Mes-Masson AM and Gévry N: H3K27 demethylation
by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2
determines ERα ligand dependency. EMBO J. 30:3947–3961. 2011.
View Article : Google Scholar : PubMed/NCBI
|