Introduction
Acute myeloid leukaemia (AML) is a heterogeneous
haematopoietic malignancy, which is well-known for the rapid
accumulation and malignant proliferation of immature myeloid
progenitors in the bone marrow (BM) and peripheral blood (PB)
(1). For decades, without efficient
therapeutic strategy, AML quickly led to poor prognosis and became
fatal. Over the last few decades, AML treatment has improved in
risk assessment, post-remission chemotherapy and haematopoietic
stem-cell transplantation (2,3). The
phosphoinositide 3-kinase, AKT, and mammalian target of rapamycin
(PI3K/AKT/mTOR) signalling network controls proliferation,
differentiation, and survival of haematopoietic cells (4,5). The
fact that in 50–80% of AML cases, the PI3K/AKT/mTOR pathway is
abnormally activated indicates that the PI3K/AKT/mTOR pathway is a
promising therapeutic target (6,7), and
many agents have been developed. LY294002, which directly targets
PI3K, has revealed strong cytotoxic effects in preclinical AML
models (8–10). Ribavirin, an eIF4E inhibitor, was
studied in preclinical models and in a pilot study and presents
efficiency in 30% of AML patients (11,12).
Aberrant activation of AKT is the most common frequent cause in
AML, and dual PI3K/mTOR inhibitors (P-103 and BEZ235), mTOR1/2
inhibitors (OSI-027 and PP242), and AKT inhibitors (perifosine)
were also investigated in preclinical models (13–17).
Long non-coding RNAs (lncRNAs) are a class of RNA
molecules >200 nucleotides. They are transcribed and not protein
coded. A large number of this class of RNA molecules has been
identified, numbering >58,000 (18,19).
Accumulating evidence has revealed that lncRNAs play critical roles
not only in physiological processes in normal cells but also in a
multitude of biological processes that are central in tumorigenesis
and the progression of cancer (18,20),
including in AML. Wu et al reported that HOX transcript
antisense intergenic RNA (HOTAIR) was overexpressed in AML and was
a promising biomarker of poor prognosis (21). By performing large-scale sequencing,
it was determined via evaluation of the lncRNA profile that it is
possible to provide valuable information for improved risk
stratification of AML patients (22).
lncRNAs are reported to regulate the PI3K/AKT/mTOR
pathway in several types of cancer cells. Highly upregulated in
liver cancer (HULC) was revealed to act as an oncogene in gliomas
(23). The upregulated HULC
promoted malignant cell behaviours, including proliferation,
migration and invasion via activation of the PI3K/AKT/mTOR pathway
in glioma cells (23). A newly
identified lncRNA ENST00113, which is most upregulated in
atherosclerosis, regulated proliferation, survival and migration by
activating the PI3K/AKT/mTOR pathway, potentially by inhibiting the
process of phosphorylation (24).
lncRNA LUNAR1 was reported to play a central role in the interplay
of NOTCH1-IGF-1 interactions via activation of the PI3K/AKT/mTOR
pathway in AML cells (25).
Accordingly, these data revealed that lncRNAs are tightly
associated with activation or inactivation of the PI3K/AKT/mTOR
pathway and thus play a regulatory role in the malignant behaviours
of several types of cancers, including AML.
Long intergenic non-protein coding RNA 239
(LINC00239) is a newly identified lncRNA that has been revealed to
be aberrantly expressed in AML (26), without knowledge of its exact role
in AML. In the present study, the fundamental role of LINC00239 in
human AML cells was examined and the potential role in regulating
malignant behaviours via the PI3K/AKT/mTOR pathway was
investigated.
Materials and methods
Cell culture and treatment
The AML cell lines HL-60 (FAB M2) and KG-1
(erythroleukemia-FAB M6) were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA). Cells were maintained
in RPMI-1640 medium supplemented with 10% heat-inactivated fetal
bovine serum (FBS) (both from Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 1% antibiotic-antimycotic mixture (Thermo
Fisher Scientific, Inc.) in a humidified, 37°C and 5%
CO2 atmosphere. Medium was half-refreshed every 3
days.
Doxorubicin and dual pan-PI3K/mTOR kinase inhibitor
dactolisib (NVP-BEZ235) were purchased from (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). For the evaluation of chemosensitivity
to doxorubicin, a range concentration from 0.5 to 100 µM was
incubated with KG-1 or HL-60 cells for 24 h, respectively. For
inhibition of the PI3K/AKT/mTOR pathway, KG-1 or HL-60 cells were
pretreated with 0.5 Μm NVP-BEZ235 for 6 h.
RT-qPCR
Total RNA was isolated from culture cell using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Reverse transcription
of RNA was performed following the instructions of the MBI
Fermentas kit (Fermentas; Thermo Fisher Scientific, Inc.). Reaction
conditions were as follows: 70°C for 10 min, ice bath for 2 min,
42°C for 60 min and 70°C for 10 min. The cDNA was stored in a −80°C
freezer. The reaction system for qPCR was prepared following the
instructions of the kit (Fermentas; Thermo Fisher Scientific,
Inc.). The primer sequences were as follows: linc00239, forward
5′-AGGTACCAGCTGGGATGTTG-3′ and reverse 5′-CATGACCGTCTTCCTGTGTG-3′;
U6, forward 5′-GTGCTCGCTTCGGCAGCACA-3′ and reverse
5′-AAAATATGGAACGCTTCA-3′; β-actin, forward
5′-CATGTACGTTGCTATCCAGGC-3′ and reverse
5′-CTCCTTAATGTCACGCACGAT-3′; GAPDH, forward
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′. The reaction conditions were as
follows: pre-denaturation at 98°C for 2 min, denaturation at 98°C
for 5 sec, annealing and extension at 60°C for 1 min, 35 cycles in
total. snoU6, β-actin and GAPDH were used as the internal
references. The expression of the genes were calculated using
relative quantitative measurement, where 2−ΔΔCq
represented the relative expression of each target gene (27). Each experiment was repeated for 3
times.
Cell viability assays
Cells were seeded (5×103/well) into
96-well plates overnight. After treatment with doxorubicin (0.5–100
µM) for 24 h, 10 µl tetrazolium salt Cell Counting Kit-8 (CCK-8)
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) was added to
each well (final volume ratio, 10%). Optical density (OD) was
assessed at a wavelength of 450 nm (OD450).
Cell cycle distribution
Cells were pelleted and washed with pre-cold
phosphate-buffered saline (PBS) 3 times. Cells were fixed with 1 ml
of 75% ethyl alcohol, stored overnight at 4°C, washed with PBS 2
times and 100 µl RNase A and 400 µl propidium iodide (PI;
Sigma-Aldrich; Merck KGaA) were added in the dark and incubated for
30 min at room temperature. After 30-min incubation at 4°C, flow
cytometric analysis was performed using the FACS LSR II flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Colony formation on soft agar
KG-1 or HL-60 cells (5×103) were
suspended in serum-free RPMI-1640 medium and dissolved in 0.3%
low-melting agarose (Sigma-Aldrich; Merck KGaA), supplemented with
10% FBS and a 1% antibiotic-antimycotic mixture. Cells were allowed
to grow for 14 days and colonies were identified as having >50
cells. Cells were visualized using an Olympus CX41 microscope
(Olympus Corp., Tokyo, Japan).
Migration
Transwell permeable plates with 8.0 µM pores
(Corning Incorporated Life Sciences, Corning, NY, USA) were
employed to analyze cell migration. In the bottom chamber, 500 µl
of serum-free RPMI-1640 medium was added supplemented with 5% FBS.
In the upper chamber, 5×104 cells were added suspended
in serum-free medium. After 24 h of incubation, the amount of
viable migrated cells was determined by counting using trypan blue
staining. Cells were visualized using an Olympus CX41 microscope
(Olympus Corp.).
CFSE/PI staining
Cells were washed in serum-free RPMI-1640 medium 3
times and suspended in 500 µl of fresh medium. Carboxyfluorescein
diacetate succinimidyl ester (CFSE) fluorescent dye (100 µl) (5
µmol/l) was added and incubated at 37°C for 30 min. Then, the cells
were washed with fresh medium 3 times. Subsequently, 24 h later,
cells were collected and suspended in 500 µl fresh medium and
incubated with 5 µg/ml PI at room temperature for 10 min. Flow
cytometry was performed using the FACS LSR II flow cytometer (BD
Biosciences). Analysis of results was performed using FlowJo 7.6
software (Tree Star, Inc., Ashland, OR, USA). The experiments were
repeated at least 3 times.
Annexin V-FITC/PI staining
KG-1 or HL-60 cells were washed with 1 ml of
ice-cold PBS 3 times. Cells were suspended with 100 µl of binding
buffer, 5 µl of Annexin V-FITC (BD Biosciences) and 10 µl of PI
(Sigma-Aldrich; Merck KGaA) for 10 min avoiding light at room
temperature. Then, FITC and PI signals were assessed using FACS LSR
II flow cytometer (BD Biosciences). Analysis of results was
performed using FlowJo 7.6 software (Tree Star, Inc.). The
experiments were repeated at least 3 times.
TUNEL staining
Cells were fixed with 4% paraformaldehyde at room
temperature for 10 min. After 3 washes with PBS, cells were
permeabilized with 0.1% Triton X-100 in PBS for 10 min at room
temperature. Cells were stained with In Situ Cell Death
Detection Kit (Roche Diagnostics, Basel, Switzerland) following the
manufacturer's instructions. Cells were visualized under an X71
(U-RFL-T) fluorescence microscope (Olympus, Melville, NY, USA) and
analyzed with ImageJ software (version 1.46; National Institutes of
Health, Bethesda, MD, USA).
Western blotting
For the extraction of total protein from KG-1 or
HL-60 cells, 100 µl of lysis buffer containing 1% NP-40, 100 mM
Tris-HCl pH 7.5, 150 mM NaCl and 5 mM EDTA was employed. The
protein concentration was measured using Pierce BCA Protein Assay
Kit (Thermo Fisher Scientific, Inc.). Total protein (20 µg) was
fractionated in a 10% SDS gel and transferred to nitrocellulose
membranes. The membranes were blocked in PBS with 0.25% Tween-20
(PBS-T) containing 5% skim milk for 1 h at room temperature. The
blocked membranes were incubated with primary antibodies for 2 h at
room temperature. The primary antibodies used were as follows:
anti-γ-H2A.X antibody diluted in 1:1,000 (cat. no. ab2893),
anti-histone H2A.X antibody diluted in 1:2,500 (cat. no. ab11175),
anti-β-actin antibody diluted in 1:5,000 (cat. no. ab8226),
anti-AKT (phospho S473) antibody diluted in 1:1,000 (cat. no.
ab18206), anti-pan-AKT antibody diluted in 1:1,000 (cat. no.
ab8805), anti-mTOR (phospho S2448) diluted in 1:500 (cat. no.
ab109268), anti-mTOR diluted in 1:1,000 (cat. no. ab2732) and
anti-GAPDH antibody diluted in 1:5,000 (cat. no. ab8245). All
antibodies were purchased from Abcam (Cambridge, UK). After 3
washes with PBS-T, the membranes were incubated with the
corresponding following secondary antibodies: HRP-labeled goat
anti-mouse IgG antibody diluted in 1:5,000 (cat. no. ab97040) or
goat anti-rabbit IgG antibody diluted in 1:5,000 (cat. no. ab7090)
for 2 h at room temperature. Protein levels were detected after
incubation with SuperSignal Wester Femto Maximum Sensitivity
Substrate (Thermo Fisher Scientific, Inc.). Blots were imaged and
analyzed with Quantity One (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Statistical analysis
All data were presented as the means ± SD (SD).
Student's t-test was performed to assess statistical significance.
The Wilcoxon signed-rank test was used to compare paired samples,
whereas Kruskal-Wallis and Mann-Whitney U tests were used to
compare different groups followed by the Tukey's post hoc test.
P-values <0.05 were considered to indicate a statistically
significant difference.
Results
Confirmation of the existence of
lncRNA linc00239 in the AML cell lines KG-1 and HL-60
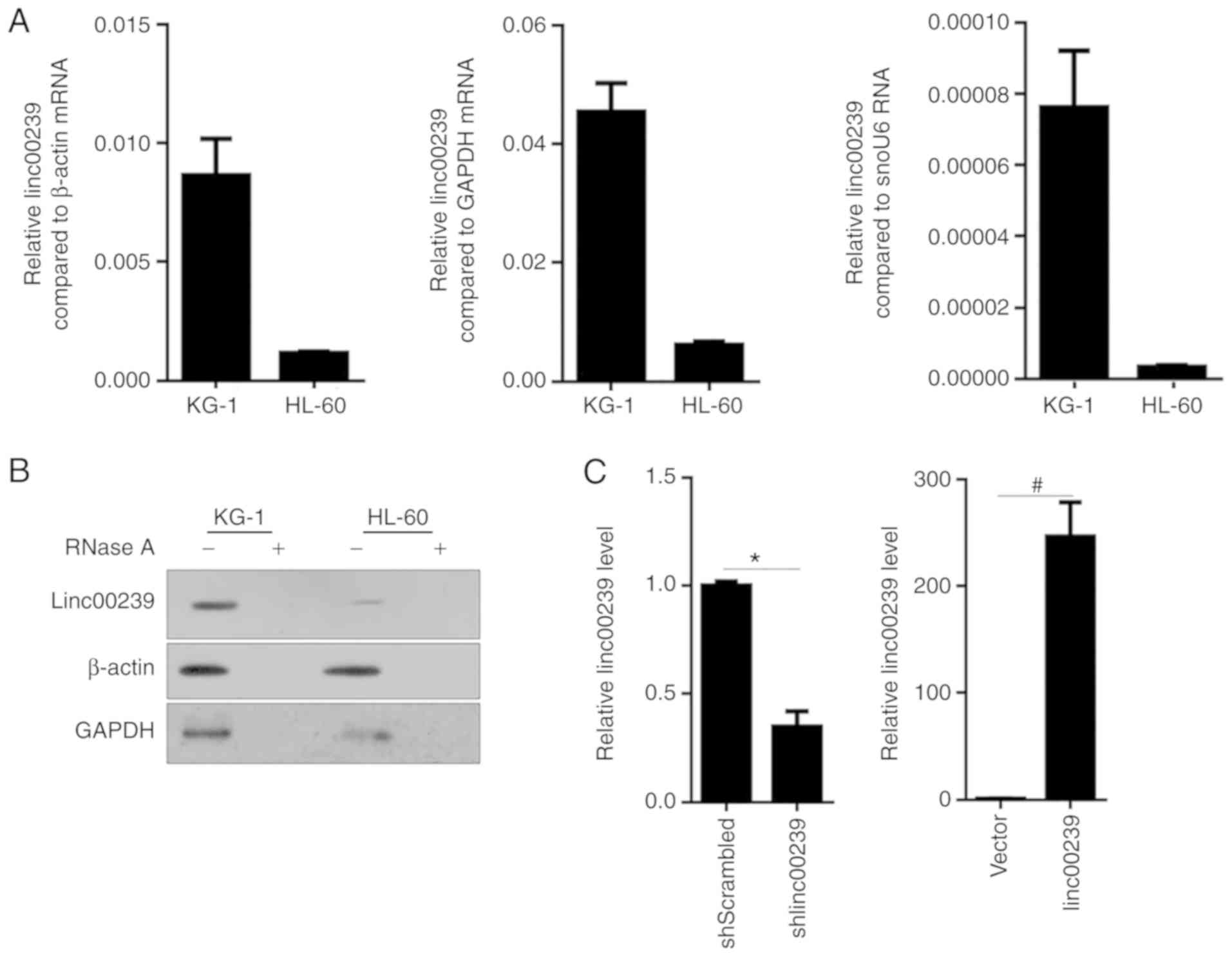
In order to identify the expression levels of
linc00239 in the AML cell lines KG-1 and HL-60, RT-qPCR was
performed, and the data were normalized to β-actin, GAPDH or snoU6
RNA. As revealed in Fig. 1A, the
expression of linc00239 was detected in both KG-1 and HL-60 cells,
and KG-1 presented an evidently higher expression compared to
HL-60. Northern blotting also confirmed the existence of linc00239
in both of these cell lines (Fig.
1B). shRNA targeted to linc00239 (shlinc00239) or scrambled
shRNA (shScrambled) was transiently transfected into KG-1 cells for
linc00239 knockdown. linc00239 expression vector (linc00239) or
empty vector (vector) was transiently transfected into HL-60 cells
for linc00239 overexpression analysis. Efficient overexpression by
expressing vector or knockdown by shlinc00239 transient
transfection was confirmed by RT-qPCR (Fig. 1C).
Effects of linc00239 on cell
viability, cell cycle distribution, colony formation and
migration
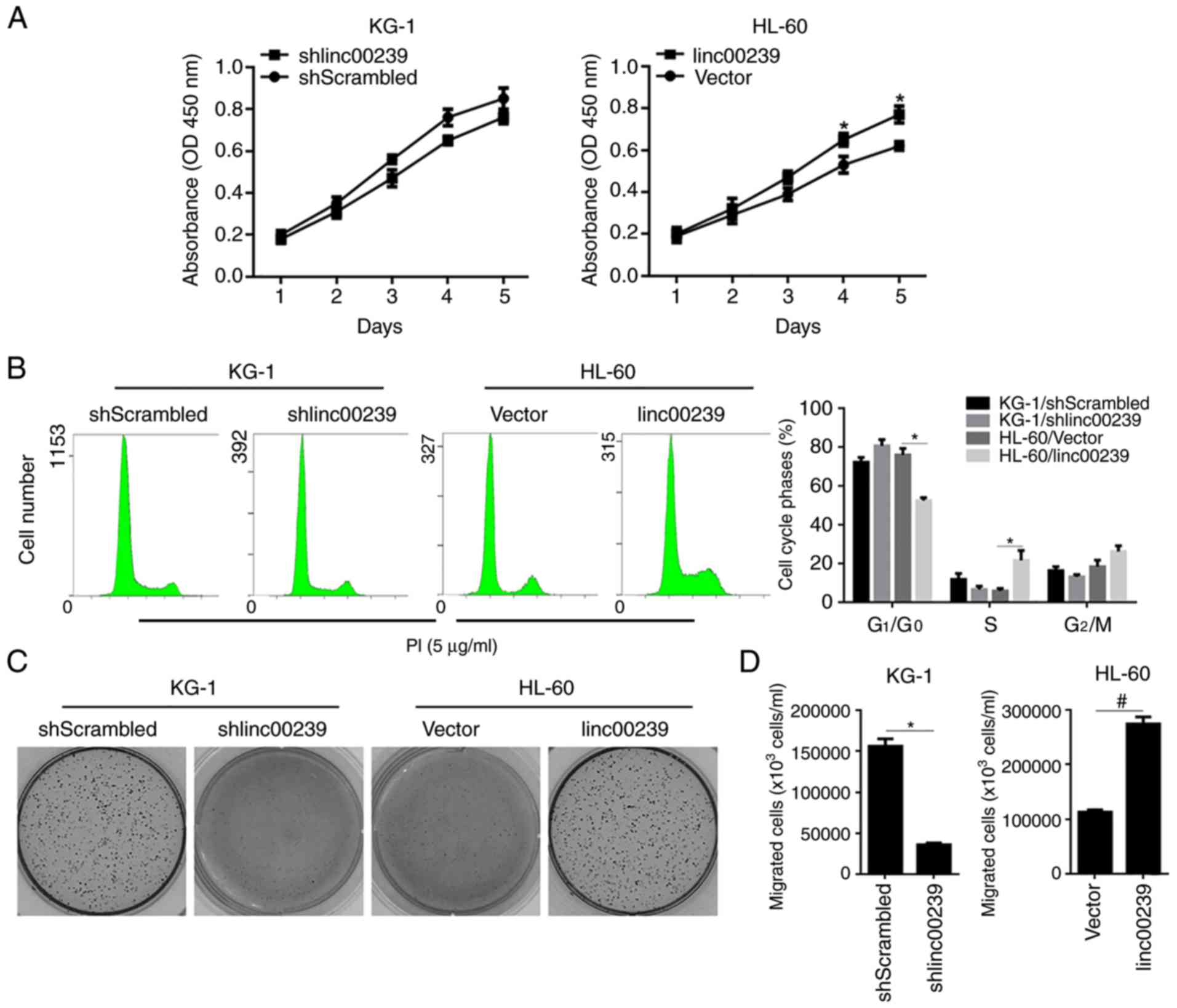
The accumulating evidence indicating that lncRNAs
may tightly regulate tumour development encouraged us to
investigate the exact role of linc00239 in AML cells (28,29).
By performing a CCK-8 assay and PI staining, followed by a flow
cytometric assay, the results revealed that although linc00239
knockdown slightly affected cell viability and cell cycle
distribution in KG-1 cells, linc00239 overexpression significantly
promoted cell viability potentially by promoting the cell cycle by
decreasing the G1/G0 phase in HL-60 cells
(Fig. 2A and B). The effects of
linc00239 on colony formation and migration were further detected
to illustrate the role of linc00239 in the regulation of malignant
behaviours in AML cells. As revealed in Fig. 2C and D, in both KG-1 and HL-60
cells, the presence of linc00239 positively regulated colony
formation and migration.
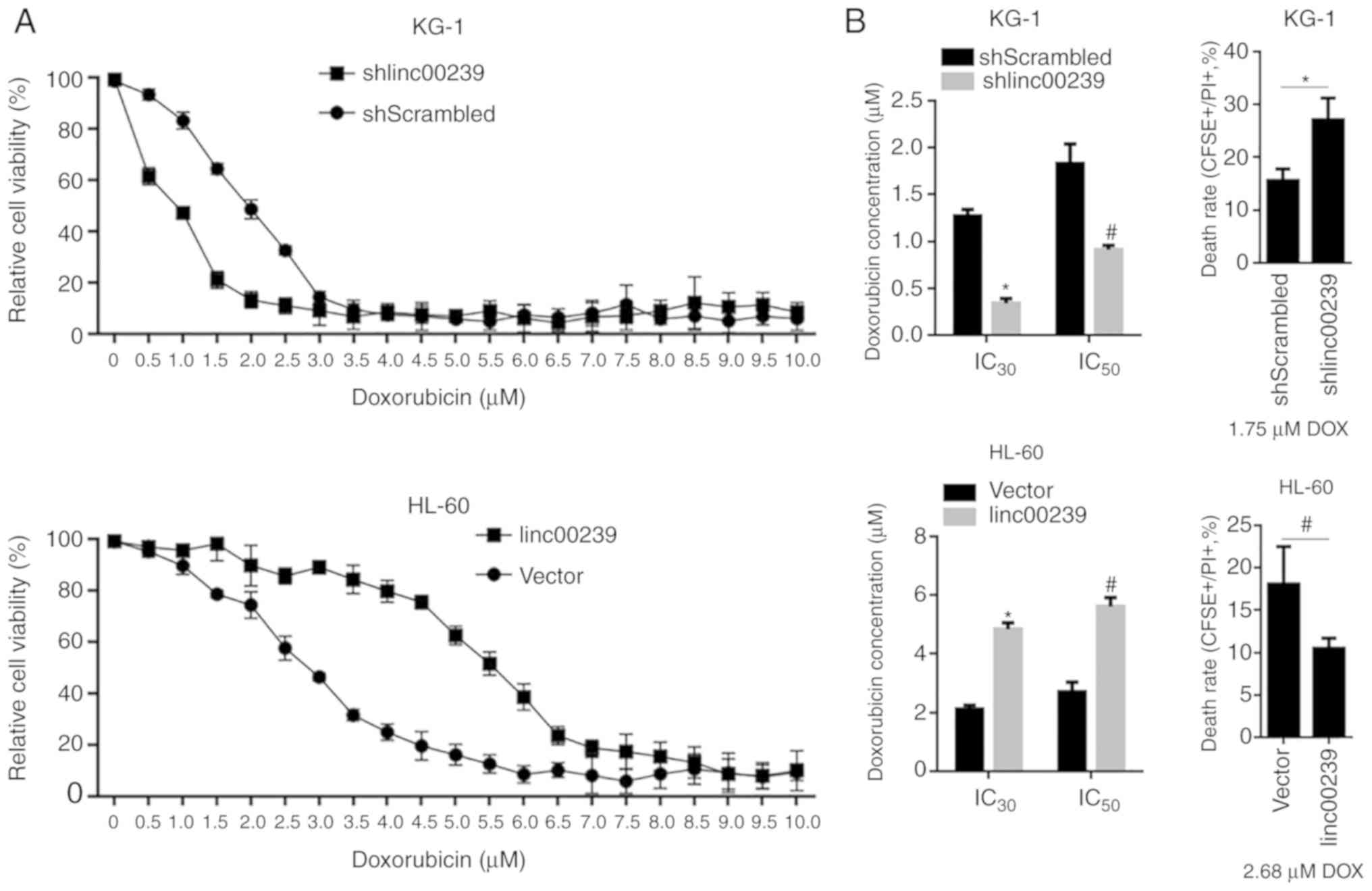
linc00239 regulates chemoresistance to
doxorubicin and exerts a protective effect against apoptotic cell
death
Various lncRNAs have been reported to regulate
chemosensitivity in cancer cells to therapeutic agents, including
doxorubicin, by modulating DNA damage response pathways (30,31).
This encouraged us to investigate the role of linc00239 in the
chemosensitivity to doxorubicin, which is a commonly used
therapeutic agent. By exposure to a concentration range of
doxorubicin, relative cell viability was assessed by performing a
CCK-8 assay. In KG-1 cells, knockdown of linc00239 significantly
decreased the IC30 and IC50 concentrations of
doxorubicin (Fig. 3A and B, upper
panel). After 1.75 µM treatment of doxorubicin, the rate of cell
death detected by CFSE/PI double staining illustrated consistently
that knockdown of linc00239 significantly increased the cell death
rate. In HL-60 cells, overexpression of linc00239 significantly
increased the IC30 and IC50 concentrations of
doxorubicin (Fig. 3A and B, lower
panel). As anticipated, overexpressed linc00239 decreased the rate
of cell death after 2.68 µM treatment of doxorubicin (Fig. 2B).
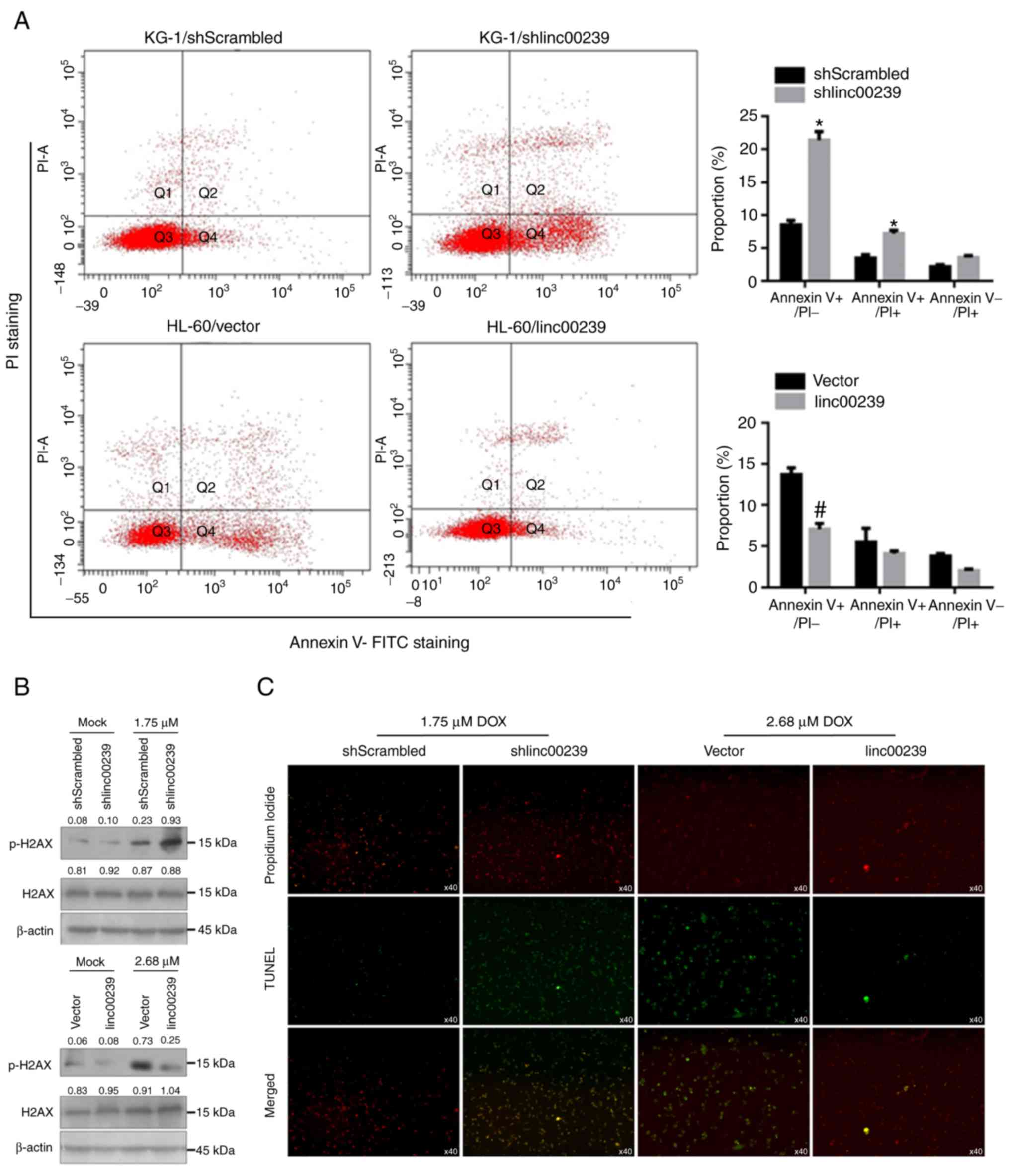
Doxorubicin exerts an anticancer effect mainly via
induction of apoptotic cell death (32). To further confirm the effects of
linc00239 on doxorubicin-induced apoptotic cell death and DNA
damage, apoptotic cell death and the level of phosphorylation
(Ser139) and total histone H2AX protein by Annexin V-FITC/PI double
staining were assessed, followed by flow cytometric assay and
western blotting. These data revealed that in KG-1, knockdown of
linc00239 significantly increased the proportion of Annexin
V-FITC+/PI− and Annexin
V-FITC+/PI+ (Fig.
4A). Since KG-1 cells presented a much higher expression level
of linc00239 compared to HL-60 cells, linc00239 was knocked down in
KG-1 and overexpressed linc00239 in HL-60 cells. In HL-60 cells,
overexpression of linc00239 significantly decreased the proportion
of Annexin V-FITC+/PI−. Detection of
phosphorylation (Ser139) and total histone H2AX protein illustrated
that the overexpression of linc00239 inhibited phosphorylation
(Ser139) of histone H2AX without disturbing total histone H2AX
(Fig. 4B). To elucidate whether
linc00239 had a protective effect against DNA damage in KG-1 and
HL-60 cells, a terminal dUTP nick-end labelling (TUNEL) assay was
performed. The results clearly revealed a decrease in the
percentage of TUNEL-positive cells, confirming the protective
effect against doxorubicin-induced DNA damage (Fig. 4C).
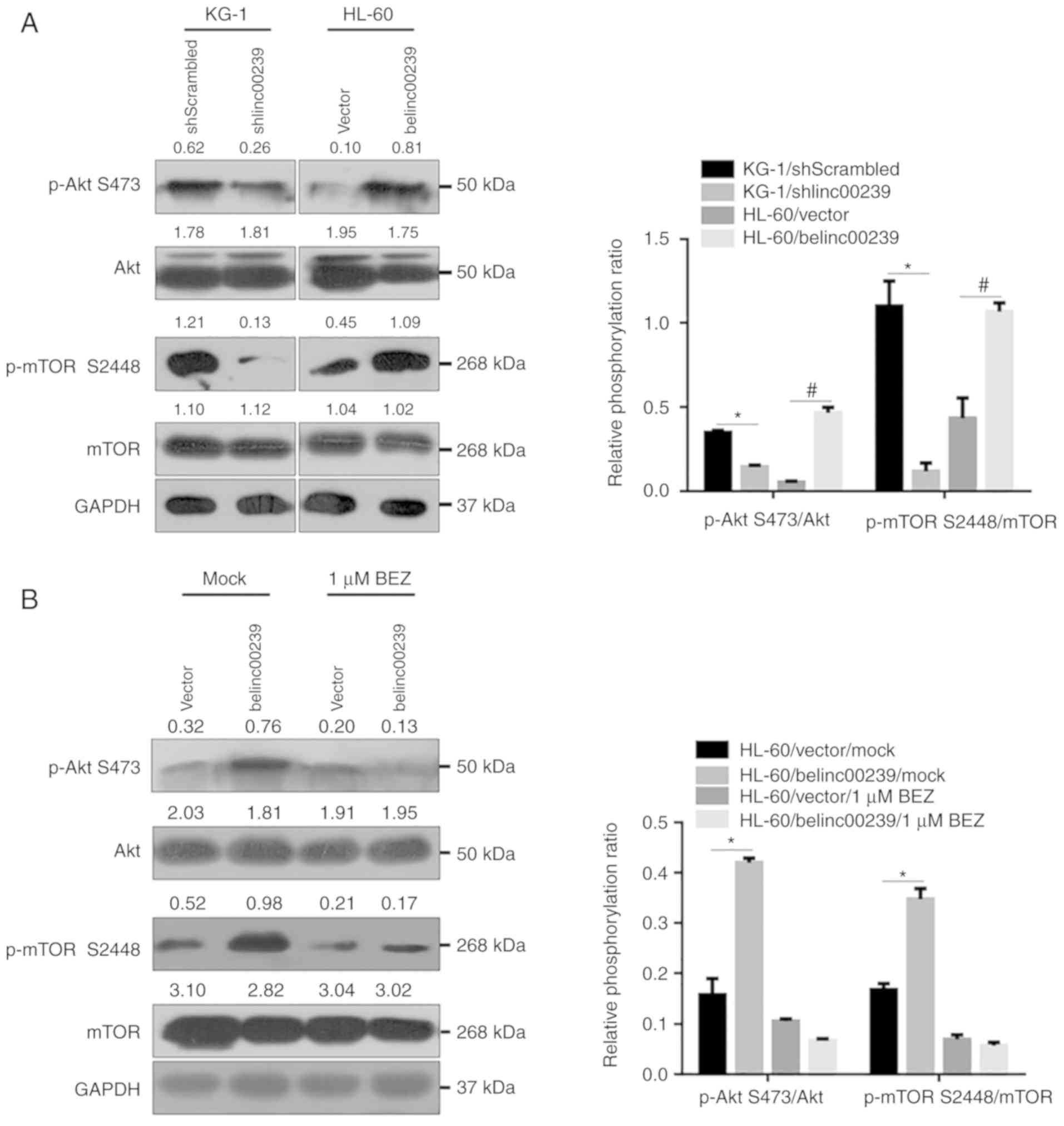
The presence of linc00239 is tightly
associated with activation of the PI3K/Akt/mTOR pathway
To assess whether the expression of linc00239 was
associated with activation of the PI3K/Akt/mTOR pathway, the
phosphorylation of Akt and mTOR was detected after linc00239
knockdown in KG-1 cells or overexpression in HL-60 cells.
Forty-eight hours after transfection, protein lysates were obtained
and analysed by western blotting. The results revealed that
knockdown of linc00239 reduced the phospho/total Akt ratio in KG-1
cells, and in contrast, overexpression of linc00239 significantly
increased the phospho/total Akt ratio in L-60 (Fig. 5A). Moreover, the expression level of
linc00239 also positively regulated the phosphorylation levels of
mTOR on S2448, thus affecting mTORC1 activity, without disturbing
mTOR total protein. To further confirm whether linc00239 regulated
the PI3K/Akt/mTOR pathway by promoting phosphorylation of Akt and
mTOR, the dual PI3K/Akt and mTOR inhibitor NVP-BEZ235 was employed.
After pretreatment with 0.5 µM NVP-BEZ235, phosphorylation of Akt
and mTOR were inhibited, and the phosphorylation-promoting effect
of linc00239 was abolished (Fig.
5B).
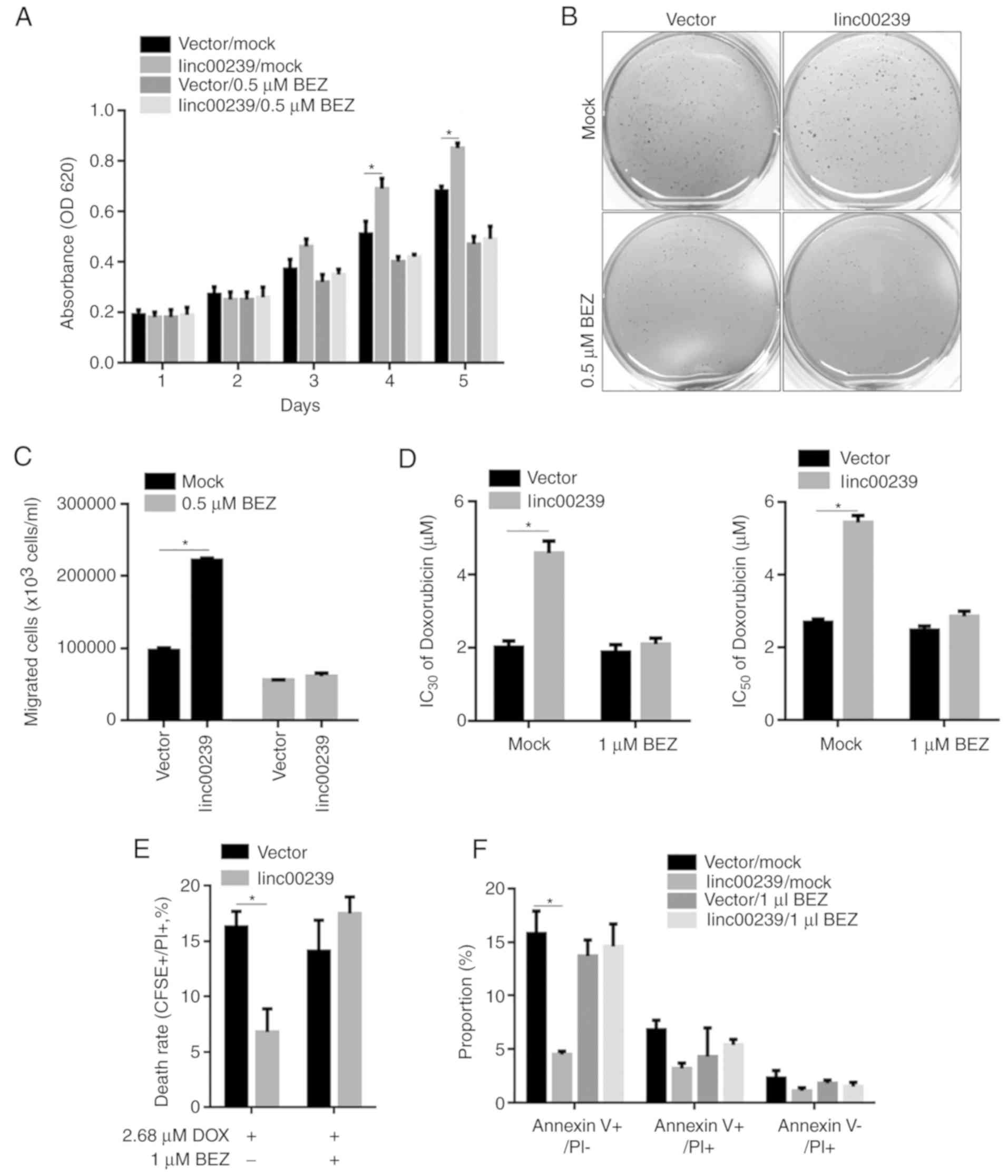
Effects of linc00239 on malignant
behaviours, including proliferation, colony formation, migration
and chemosensitivity, are abolished by NVP-BEZ235 treatment
As aforementioned (Fig.
5), activation of the PI3K/Akt/mTOR pathway by linc00239 was
eliminated by NVP-BEZ235 treatment. To demonstrate whether
linc00239 functioned via regulation of the PI3K/Akt/mTOR pathway
and thus regulated malignant behaviours in AML cells, the effects
of linc00239 on proliferation, colony formation, migration, and
chemosensitivity were detected after the PI3K/Akt/mTOR pathway was
inhibited by NVP-BEZ235 treatment in HL-60 cells. As anticipated,
after NVP-BEZ235 pretreatment, the regulatory effects of linc00239
on the proliferation, colony formation, migration and
chemosensitivity to doxorubicin were eliminated (Fig. 6). All of these data indicated that
linc00239 regulated malignant behaviours mainly via activation of
the PI3K/Akt/mTOR pathway.
Discussion
In the present study, by performing RT-qPCR and
northern blotting in the AML cell lines KG-1 and HL-60, we revealed
the existence of linc00239 RNA in these two cell lines was
revealed, which was an expected result. In order to compare the
relative amount of linc00239 in these two cell lines, RT-qPCR was
performed, and the data were normalized to β-actin mRNA, GAPDH
mRNA, or snoU6 RNA. These results indicated that the endogenous
linc00239 level in KG-1 cells was much higher than that of HL-60
cells. The expression difference between these cell lines allowed
us to investigate the roles of linc00239 in the malignant
behaviours of AML. To achieve this purpose, KG-1 cells were used to
downregulate linc00239; HL-60 cells were used to overexpress
linc00239. In this way, it was revealed that the presence of
linc00239 in AML cells was positively related to malignant
behaviours, including proliferation, colony formation and
migration.
Doxorubicin, a cytotoxic antiproliferative and
apoptosis-inducing drug, was clinically and widely used as a
cytotoxic agent for chemotherapy in several types of cancers,
including AML (33,34). It disturbs the DNA replication
processes, resulting in cell cycle arrest and cellular apoptosis
(35). Thus, the effects of
linc00239 on doxorubicin-induced cell cycle arrest and apoptotic
cell death were investigated. Notably, without an evident effect on
cell cycle arrest (data not shown), the presence of linc00239
significantly desensitized KG-1 and HL-60 cells to doxorubicin. To
confirm the effects of linc00239 on chemosensitivity to
doxorubicin, the apoptotic rate and the rate of cell death were
evaluated, and the expected results were obtained. No obvious
effects of linc00239 on cell cycle distribution were due to the
induced cell death. Moreover, the existence of linc00239 also
affected doxorubicin-induced cell death in an unknown manner.
It was also revealed that the presence of linc00239
was tightly related to the phosphorylation of AKT at S473 and mTOR
at S2448, without affecting total protein levels. After
pretreatment with an inhibitor of the PI3K/AKT/mTOR pathway,
BEZ235, the presence of linc00239 failed to affect the
PI3K/AKT/mTOR pathway and to exert protective effects against
doxorubicin treatment. Moreover, when pre-treated with BEZ235, the
regulatory roles of linc00239 on malignant behaviours in KG-1 or
HL-60 cells were eliminated. PI3K/AKT/mTOR is considered as the
most important factor in initiation and maintenance of AML, and the
abnormality of this pathway was found in most AML cases (36). Our data revealed that the presence
of linc00239 tightly affected the activation of the PI3K/ATK/mTOR
pathway, indicating that it is a potential therapeutic target in
AML. Moreover, in a future investigation, it is worth studying the
epigenetic regulating role of microRNA and long non-coding RNA on
this pathway.
In summary, it was revealed that linc00239 tightly
regulated the activation of the PI3K/AKT/mTOR pathway without
knowing the target in the PI3K/AKT/mTOR pathway, and by depending
on this, it desensitized KG-1 or HL-60 cells to the chemoagent. The
presence of linc00239 positively regulated the malignant
behaviours, including proliferation, colony formation and migration
in KG-1 and HL-60 cells. Collectively, linc00239 functioned as a
PI3K/AKT/mTOR pathway activator and may be considered as a
therapeutic target for AML treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chengdu
Scientific Benefiting Project (no. 2015-HM01-00461-SF).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
YY performed the cellular and molecular experiments.
WD contributed to the molecular experiments and was responsible for
the data collection. YS performed the cellular experiments. ZZ
performed the molecular experiments and was a major contributor in
writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Troy JD, Atallah E, Geyer JT and Saber W:
Myelodysplastic syndromes in the United States: An update for
clinicians. Ann Med. 46:283–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daver N and Cortes J: Molecular targeted
therapy in acute myeloid leukemia. Hematology. 17 (Suppl
1):S59–S62. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stone R, Sekeres M and Garcia-Manero G:
Evolving strategies in the treatment of MDS and AML. Clin Adv
Hematol Oncol. 7:1–14. 2009.PubMed/NCBI
|
|
4
|
Buitenhuis M and Coffer PJ: The role of
the PI3K-PKB signaling module in regulation of hematopoiesis. Cell
Cycle. 8:560–566. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martelli AM, Evangelisti C, Chiarini F and
McCubrey JA: The phosphatidylinositol 3-kinase/Akt/mTOR signaling
network as a therapeutic target in acute myelogenous leukemia
patients. Oncotarget. 1:89–103. 2010.PubMed/NCBI
|
|
7
|
Altman JK, Sassano A and Platanias LC:
Targeting mTOR for the treatment of AML. New agents and new
directions. Oncotarget. 2:510–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Birkenkamp KU, Geugien M, Schepers H,
Westra J, Lemmink HH and Vellenga E: Constitutive NF-kappaB
DNA-binding activity in AML is frequently mediated by a
Ras/PI3-K/PKB-dependent pathway. Leukemia. 18:103–112. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu Q, Simpson SE, Scialla TJ, Bagga A and
Carroll M: Survival of acute myeloid leukemia cells requires PI3
kinase activation. Blood. 102:972–980. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Neri LM, Borgatti P, Tazzari PL, Bortul R,
Cappellini A, Tabellini G, Bellacosa A, Capitani S and Martelli AM:
The phosphoinositide 3-kinase/AKT1 pathway involvement in drug and
all-trans-retinoic acid resistance of leukemia cells. Mol
Cancer Res. 1:234–246. 2003.PubMed/NCBI
|
|
11
|
Kentsis A, Topisirovic I, Culjkovic B,
Shao L and Borden KL: Ribavirin suppresses eIF4E-mediated oncogenic
transformation by physical mimicry of the 7-methyl guanosine mRNA
cap. Proc Natl Acad Sci USA. 101:18105–18110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Assouline S, Culjkovic B, Cocolakis E,
Rousseau C, Beslu N, Amri A, Caplan S, Leber B, Roy DC, Miller WH
Jr and Borden KL: Molecular targeting of the oncogene eIF4E in
acute myeloid leukemia (AML): A proof-of-principle clinical trial
with ribavirin. Blood. 114:257–260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chapuis N, Tamburini J, Green AS, Vignon
C, Bardet V, Neyret A, Pannetier M, Willems L, Park S, Macone A, et
al: Dual inhibition of PI3K and mTORC1/2 signaling by NVP-BEZ235 as
a new therapeutic strategy for acute myeloid leukemia. Clin Cancer
Res. 16:5424–5435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Altman JK, Sassano A, Kaur S, Glaser H,
Kroczynska B, Redig AJ, Russo S, Barr S and Platanias LC: Dual
mTORC2/mTORC1 targeting results in potent suppressive effects on
acute myeloid leukemia (AML) progenitors. Clin Cancer Res.
17:4378–4388. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Park S, Chapuis N, Bardet V, Tamburini J,
Gallay N, Willems L, Knight ZA, Shokat KM, Azar N, Viguié F, et al:
PI-103, a dual inhibitor of Class IA phosphatidylinositide 3-kinase
and mTOR, has antileukemic activity in AML. Leukemia. 22:1698–1706.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng Z, Shi YX, Tsao T, Qiu Y, Kornblau
SM, Baggerly KA, Liu W, Jessen K, Liu Y, Kantarjian H, et al:
Targeting of mTORC1/2 by the mTOR kinase inhibitor PP242 induces
apoptosis in AML cells under conditions mimicking the bone marrow
microenvironment. Blood. 120:2679–2689. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Papa V, Tazzari PL, Chiarini F, Cappellini
A, Ricci F, Billi AM, Evangelisti C, Ottaviani E, Martinelli G,
Testoni N, et al: Proapoptotic activity and chemosensitizing effect
of the novel Akt inhibitor perifosine in acute myelogenous leukemia
cells. Leukemia. 22:147–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garzon R, Volinia S, Papaioannou D,
Nicolet D, Kohlschmidt J, Yan PS, Mrózek K, Bucci D, Carroll AJ,
Baer MR, et al: Expression and prognostic impact of lncRNAs in
acute myeloid leukemia. Proc Natl Acad Sci USA. 111:18679–18684.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B
and Chen Y: Overexpression of long non-coding RNA HOTAIR predicts a
poor prognosis in patients with acute myeloid leukemia. Oncol Lett.
10:2410–2414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mer AS, Lindberg J, Nilsson C, Klevebring
D, Wang M, Grönberg H, Lehmann S and Rantalainen M: Expression
levels of long non-coding RNAs are prognostic for AML outcome. J
Hematol Oncol. 11:522018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu Y, Zhang X, Qi L, Cai Y, Yang P, Xuan
G and Jiang Y: HULC long noncoding RNA silencing suppresses
angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling
pathway in human gliomas. Oncotarget. 7:14429–14440.
2016.PubMed/NCBI
|
|
24
|
Yao X, Yan C, Zhang L, Li Y and Wan Q:
LncRNA ENST00113 promotes proliferation, survival, and migration by
activating PI3K/Akt/mTOR signaling pathway in atherosclerosis.
Medicine. 97:e04732018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Venter JC, Adams MD, Myers EW, Li PW,
Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al:
The sequence of the human genome. Science. 291:1304–1351. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
ChemMedChem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W,
De W, Wang Z and Wang R: The long noncoding RNA HOTAIR contributes
to cisplatin resistance of human lung adenocarcinoma cells via
downregualtion of p21WAF1/CIP1 expression. PLoS One.
8:e772932013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hou Z, Xu C, Xie H, Xu H, Zhan P, Yu L and
Fang X: Long noncoding RNAs expression patterns associated with
chemo response to cisplatin based chemotherapy in lung squamous
cell carcinoma patients. PLoS One. 9:e1081332014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Naci D, El Azreq MA, Chetoui N, Lauden L,
Sigaux F, Charron D, Al-Daccak R and Aoudjit F: α2β1 integrin
promotes chemoresistance against doxorubicin in cancer cells
through extracellular signal-regulated kinase (ERK). J Biol Chem.
287:17065–17076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lagadinou ED, Ziros PG, Tsopra OA, Dimas
K, Kokkinou D, Thanopoulou E, Karakantza M, Pantazis P,
Spyridonidis A and Zoumbos NC: c-Jun N-terminal kinase activation
failure is a new mechanism of anthracycline resistance in acute
myeloid leukemia. Leukemia. 22:1899–1908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kweon SH, Song JH and Kim TS:
Resveratrol-mediated reversal of doxorubicin resistance in acute
myeloid leukemia cells via downregulation of MRP1 expression.
Biochem Biophys Res Commun. 395:104–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao J, Lu Q, Wu D, Li P, Xu B, Qing W,
Wang M, Zhang Z and Zhang W: microRNA-21 modulates cell
proliferation and sensitivity to doxorubicin in bladder cancer
cells. Oncol Rep. 25:1721–1729. 2011.PubMed/NCBI
|
|
36
|
Fransecky L, Mochmann LH and Baldus CD:
Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell
Ther. 3:22015. View Article : Google Scholar : PubMed/NCBI
|