Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common cancer in the world. The annual global incidence
of this disease exceeds 630,000 with an annual mortality of
~350,000 (1–4). Asia accounts for 57.5% of global HNSCC
occurrence. In India, HNSCC is the most common cancer in males,
which may be attributed to profound exposure of the population to
lifestyle risk factors, including tobacco, cigarettes, areca nuts,
and alcohol (3,5,6).
Recently, human papillomavirus (HPV) infection has also been
suggested to be a risk factor for HNSCC (3,5,6). In
India, patients that are diagnosed at early and advanced stages of
HNSCC have 5-year survival rates of 82 and 27%, respectively
(7).
A better understanding of molecular and biological
characteristics of cancer cells may help in designing strategies
for the prevention of or finding alternative drug targets for HNSCC
treatment. Notably, the cell lines established from primary tumors
retain the heterogeneity of the cell population as displayed by the
parent tumors (8). Such
characterization requires a constant and reliable source of cells
belonging to a particular HNSCC. There have been a plethora of
reports on the establishment of HNSCC cell lines. In the USA,
initial attempts were made to establish HNSCC cell lines (9,10);
however, these cell lines were reported to be cross-contaminated
with the HeLa cell line (11).
Subsequently, in the UK, 10 HNSCC cell lines were established from
tongue and larynx of patients who had undergone chemotherapy and
radiotherapy. Among the 10 cell lines, 9 exhibited secretion of
immune-reactive β human chorionic gonadotropin (12). Another study demonstrated
differences in the differentiation status, number of desmosomes,
and expression of tonofilaments between primary and recurrent
tumors (13). Heo et al
established 21 HNSCC cell lines that were aneuploid and resistant
to natural killer cells; however, these cell lines were efficiently
lysed by lymphokine-activated killer cells (14). In the United States, establishment
of 85 cell lines from Head and Neck tumor site was reported. The
origins of these cell lines included head and neck squamous cell
carcinoma, thyroid cancer, cutaneous squamous cell carcinoma,
adenoid cystic carcinoma, oral leukoplakia, immortalized primary
keratinocytes and normal epithelium (15). White et al (16) reported establishment of 52 HNSCC
cell lines; the tendency of tumors with poor prognosis to
successfully form cell lines was reported. In addition, HNSCC cell
lines were able to form spheroids that exhibited a higher
expression of the cancer stem cell (CSC) markers cluster of
differentiation (CD) 44, CD133, sex determining region Y-box 2 and
BMI1, as compared with normal epithelial oral cells (17). Recently, 16 cell lines were
established from the tongue (10 cell lines), alveolus (4 cell
lines), buccal mucosa (1 cell line) and hard palate (1 cell line)
(18). Whole exome sequencing
revealed upregulation of FAT1 and CASP8 in these cell
lines as compared with blood samples of the matched patients.
Furthermore, the individual or simultaneous knockdown of
FAT1 and CASP8 genes exhibited reduced intercellular
adhesion (18). Additionally, an
oral tongue squamous cell carcinoma cell line was established from
a non-smoker human papillomavirus (HPV)-negative patient (19). Whole exome sequencing of this cell
line revealed mutations in the genes CDKN2A, TP53, SPTBN5,
NOTCH2 and FAM136A (19).
In addition, two buccal mucosa carcinoma cell lines
have been established from Chinese patients with one positive for
HPV (20). Furthermore, a Japanese
group have reported the establishment of 2 tongue-derived squamous
carcinoma cell lines (one Node-positive and one Node-negative cell
line). The node-positive cell line exhibited higher expression of
keratins 8/18 as compared with the node-negative cell line.
Furthermore, the downregulation of keratins 13, 14, and 16 in these
cell lines was associated with the invasive and metastatic
abilities of cancer cells (21).
Subsequently, 6 HNSCC cell lines were established in Korea. The
study revealed that the cell lines exhibited a P53
transversion mutation in exon 7 and a transition mutation in the
exon 8 (22). Furthermore, in
Malaysia, cell lines were established from oral cavity cancers,
which exhibited mutations in the P53 gene; one of the cell
lines exhibited mouse double minute 2 homolog overexpression and
the other exhibited epidermal growth factor receptor overexpression
(23).
The establishment of HNSCC cell lines from Indian
patients has previously been achieved from carcinomas of tongue,
alveolus and retromolar trigone (24,25);
however, these cell lines did not possess the ability to give rise
to tumors when injected into immune-compromised mice (24,25).
Furthermore, the establishment of a cell line from carcinoma of the
upper aerodigestive tract, which can give rise to tumors when
subcutaneously xenografted in nude mice was reported (26). In the present study, the
establishment of three cell lines is reported, namely ACOSC3,
ACOSC4 and ACOSC16, from advanced-stage treatment-naive squamous
cell carcinomas of the buccal mucosa. The epithelial nature of the
cell lines was confirmed by performing immunofluorescence assay
(IFA) for keratins 8 and 14. The ploidy of the cell lines was
assessed by flow cytometry and karyotyping. The novelty of the cell
lines was also validated by determining their short tandem repeat
(STR) profiles. These cell lines also formed orospheres (spheroids)
when they were exposed to low-adherence conditions. The primary
orospheres obtained from the cell lines also produced secondary
orospheres. Notably, the cell lines were able to give rise to
tumors when administered subcutaneously into non-obese
diabetic/severe combined immune deficiency (NOD/SCID) mice.
Materials and methods
Patient sample collection and
processing
A total of 16 advanced stage treatment naive oral
squamous cell carcinoma (OSCC) samples were collected from patients
at the department of Head and Neck Oncology, Tata Memorial Centre
(Mumbai, India; TMH and ACTREC biorepositories) between May and
October 2016. The ages of the patients ranged between 33 and 70
years. Of the 16 patients, 15 were males and 1 was female. Patients
with human immunodeficiency virus or hepatitis B virus infections
were excluded from the study. Fresh tumor samples were removed and
collected by surgical resection from advanced-stage treatment-naive
oral cancer and were stored in sterile vials. These samples were
transported on ice and were treated with a 10% povidone-iodine
solution (Wokadine™) for disinfection, followed by washing with
sterile PBS to remove any traces of the disinfectant.
Explant culture
The samples were minced into fine pieces (2–3 mm) by
using surgical blades. These pieces were then placed on 35-mm
(diameter) tissue culture plates (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing medium for explant culture. The medium
contained minimum essential medium (MEM; Thermo Fisher Scientific,
Inc.) supplemented with 10% horse serum (Thermo Fisher Scientific,
Inc.) and 10% fetal bovine serum (HiMedia Laboratories, Mumbai,
India). The explants were maintained at 37°C with 5% CO2
atmosphere. The explant cultures were regularly monitored for cell
growth. Excessive fibroblast growth was avoided by passaging with
differential trypsinization, and keratinocytes were passaged when
they reached 75–80% confluency.
Cell culture and storage
The cell lines were trypsinized using 0.25% trypsin
(EDTA) with glucose. The cell monolayers were washed twice with PBS
and trypsinized for 2–3 min. Trypsin was neutralized using complete
medium. The cells were frozen and stored in liquid nitrogen in
medium containing 10% dimethyl sulfoxide (DMSO). The vials
containing the cell suspension and DMSO were cooled to −80°C at a
rate of 1°C/min prior to being transferred to liquid nitrogen.
Determination of doubling time
Cell lines were inoculated in 60-mm wide tissue
culture plates (cat. no. 353002; Corning Incorporated, Corning, NY,
USA). The cells were incubated at 37°C at 5% CO2
atmosphere for up to 96 h. The cells were counted at 24-h
intervals. Doubling time was calculated using the formula:
T.ln2/ln(Xe/Xb) where T=time of incubation, Xe=number of cells at
the end of incubation time and Xb=number of cells at the beginning
of the incubation time.
Immunocytochemical staining
Immunocytochemistry was performed on all three cell
lines using primary antibodies against keratin 8 (1:100; cat. no.
C5301; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and keratin
14 (1:250; cat. no. MCA890; Serotech; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Briefly, the cells were fixed at −20°C for 20
min using absolute methanol and were permeabilized using a 0.3%
solution of Triton in methanol. This was followed by blocking with
5% fetal bovine serum for 1 h at room temperature. Furthermore, the
cells were incubated with the primary antibodies overnight at 4°C,
followed by incubation with a fluorescein isothiocyanate-conjugated
(1:400; cat. no. 115095003) and cyanine-3-conjugated (1:400; cat.
no. 715165151; both Jackson ImmunoReseach Laboratories, Inc., West
Grove, PA, USA) secondary antibody for 1 h at room temperature.
Immunocytochemistry for CD44 was conducted using a mouse monoclonal
antibody (1:600; cat. no. 156-3C11; Cell Signaling Technology,
Inc., Danvers, MA, USA). The cells were fixed at room temperature
with 4% formaldehyde for 20 min, permeabilized via 0.3% Triton
X-100 in PBS, and blocked using 5% normal goat serum (cat. no.
005000121; Jackson ImmunoReseach Laboratories, Inc.) for 1 h at
room temperature. Following blocking, the cells were incubated with
the primary antibody overnight at 4°C. The cells were then treated
with the cyanine-3-conjugated secondary antibody for 1 h at room
temperature. The images were recorded using a confocal microscope
at ×400 magnification.
Determination of ploidy
Cells from all lines were harvested following
trypsinization and resuspended in PBS containing RNase (cat. no.
R5000; Sigma-Aldrich; Merck KGaA) (0.2 mg/ml) and propidium iodide
(0.08 mg/ml) and incubated at 37°C for 30 min for DNA staining, and
the DNA content was compared with that of human mononuclear cells
from peripheral blood, (cat. no. 690PB-100A; Sigma-Aldrich; Merck
KGaA), which served as a control for diploid human genomic DNA
content. The cells were analyzed using a BD FACSCalibur system to
determine their fluorescence. The data was analyzed by using ModFit
2.0 software (Verity Software House, Inc., Topsham, ME, USA). The
mean channel of cells that were in the G0 phase was
divided by that of the lymphocytes to determine DNA index of each
cell line. The DNA index was then used to predict ploidy number of
the cells.
HPV typing
DNA was extracted from each cell line using a lysis
buffer containing 0.4 M Tris (pH 8.0), 0.1 M NaCl, 0.005 M EDTA,
0.2% SDS and proteinase K. The isolated DNA was screened for HPV
infection by performing polymerase chain reaction (PCR), using a
KAPA Taq PCR kit (cat. no. KK1015; Sigma-Aldrich; Merck KGaA) with
MY09 (5′-CGTCCMARRGGAWACTGATC-3′) and MY11
(5′-GCMCAGGGWCATAAYAATGG-3′) primers (27). The following thermocycling
conditions were used: Initial denaturation at 95°C for 5 min was
followed by 40 cycles of denaturation at 95°C for 1 min, annealing
at 55°C for 1 min and elongation at 72°C for 1 min. At the end of
40 cycles, the final elongation was carried out at 72°C for 10 min.
The MY09 and MY11 primers were used to amplify a 450-bp region in
the HPV genome. DNA isolated from the HeLa cell line was obtained
from Dr Manoj Mahimkar (ACTREC, Tata Memorial Center, Mumbai,
India) to be used as the positive control for the HPV detection
PCR. DNA isolated from MCF7 cell line (obtained from Dr Santosh
Kumar, Bhaba Atomic Reseach Centre, Mumbai, India) was used as the
negative control for the assay.
Karyotping of the cell lines
Chromosome analysis was performed by using a
modification of the standard procedures used for karyotyping from
fibroblast cultures (28). The
metaphases tended to overspread. Therefore, the trypsinization time
was reduced for G-banding. For cytogenetic analysis, 50 metaphase
spreads were screened for ploidy in all three cell lines.
Orosphere formation assay
To evaluate the capability of the three oral cancer
cell lines to produce spheroids in vitro, each cell line was
subjected to the spheroid formation assay. In total, 5,000 cells
were grown in complete MammoCult medium (MammoCult™ kit; basal
human medium; cat. no. 05621; Stemcell Technologies, Inc.,
Vancouver, BC, Canada) in 6-well flat bottom ultra-low attachment
plates (cat. no. 3471; Costar; Corning Incorporated) and were
incubated at 37°C in a CO2 incubator. Primary orospheres
were obtained following 3–4 days of culture; these primary
orospheres were further trypsinized and subjected to secondary
spheroid culture to assess the ability of cells to form secondary
orospheres.
Tumorigenesis assay
To assess the tumorigenic potential of the three
OSCC cell lines, 2 million cells of each cell line were resuspended
in 200 µl culture medium supplemented with 25% Matrigel (cat. no.
356230; Corning Incorporated) and were xenografted subcutaneously
into the NOD/SCID mice. Animal protocols were approved by the
Institutional Animal Ethics Committee Advanced Centre for
Treatment, Education and Research in Cancer (ACTREC), Tata Memorial
Centre (Navi Mumbai, India). NOD/SCID mice for tumorigenesis assay
were procured from the Institutional Animal Facility at ACTREC. For
the tumorigenesis assay, 15 female NOD/SCID mice with 5 replicates
(n=5) per group for ACOSC3, ACOSC4 and ACOSC16 were used. The mice
were aged 6–8 weeks and weighed 20–22 g. The measurements of tumor
diameters and volumes were recorded once per week using a Vernier
caliper. Mice were sacrificed when the tumors volume reached a
maximum diameter of ~20 mm. The dynamics of subcutaneous tumor
growth were evaluated in ACOSC4 (n=5) and ACOSC16 (n=3) cell lines.
The histograms of tumor volume were plotted by using GraphPad Prism
version 5 software (GraphPad Software, Inc., La Jolla, CA, USA).
Mice were maintained at a temperature of 22±2°C, humidity of 50±5%,
and under positive air pressure with 12–15 air changes/h. A
light/dark cycle of 12-h was maintained. Mice were provided food
and water ad libitum.
Cell line authentication by STR
profiling
STR profiles of the cell lines were determined using
16 STR markers. The cells of the three OSCC cell lines were
centrifuged at 162 × g for 5 min, and their DNA was extracted using
GenElute™ Mammalian Genomic DNA Miniprep Kit (cat. no. G1N350;
Sigma-Aldrich; Merck KGaA). To protect the identity of the
patients, only eight of these markers have been provided in the
data. Data analysis was performed using GeneMapper® ID-X
software version 1.4 (Thermo Fisher Scientific, Inc.). The STR
profiling was performed using PowerPlex® 16 HS System
(Promega Corporation, Madison, WI, USA). The profiles were compared
to the DSMZ STR profiles database (29).
Results
Establishment of the cell lines from
the advanced stage treatment naive oral cancer tissues
Tissue samples from advanced-stage treatment-naive
patients with oral cancer were used for explant culture and
establishing the cell lines (Table
I). The processed tissue samples were inoculated in complete
MEM, which was changed every 2 days. Over 2–3 weeks, the explants
exhibited the growth of keratinocytes with a compact, clustered
cell morphology and fibroblasts with an elongated morphology
surrounding the explant tissues. The selective removal of the
fibroblasts was performed by differential trypsinization to enrich
the keratinocytes. During the initial passages, the growth of the
cells was observed in discrete patches. Following several passages,
the pure keratinocyte cultures were obtained from 3 different
tissue samples, which were sub-cultured for >40 passages.
Importantly, all three cell lines exhibited and maintained the
polygonal cell shape, which is typical of epithelial cell
morphology (Fig. 1). Furthermore,
the rate of cell division of all three cell lines was measured by
calculating their doubling time. The data indicated that the
doubling times of the cell lines ACOSC3, ACOSC4, and ACOSC16 were
45.42, 33.78, and 34.00 h respectively. These results suggest that
the established OSCC cell lines had long-term expansion potential
and stable cell morphology.
 | Table I.Clinical and pathological findings of
established oral cancer cell lines. |
Table I.
Clinical and pathological findings of
established oral cancer cell lines.
| Cell line | Patient age
(years) | Sex | Origin of
tumor | Pathological
staging (TNM) | Tumor
diagnosis | Oral habit |
|---|
| ACOSC3 | 40 | Male | Buccal mucosa | pT2 N2b M0 | Squamous cell
carcinoma; moderately differentiated | Tobacco chewer |
| ACOSC4 | 70 | Female | Buccal mucosa | pT4 N2b M0 | Squamous cell
carcinoma; moderately differentiated | Tobacco chewer |
| ACOSC16 | 34 | Male | Buccal mucosa | pT4a N2b M0 | Squamous cell
carcinoma; moderately differentiated | Tobacco chewer |
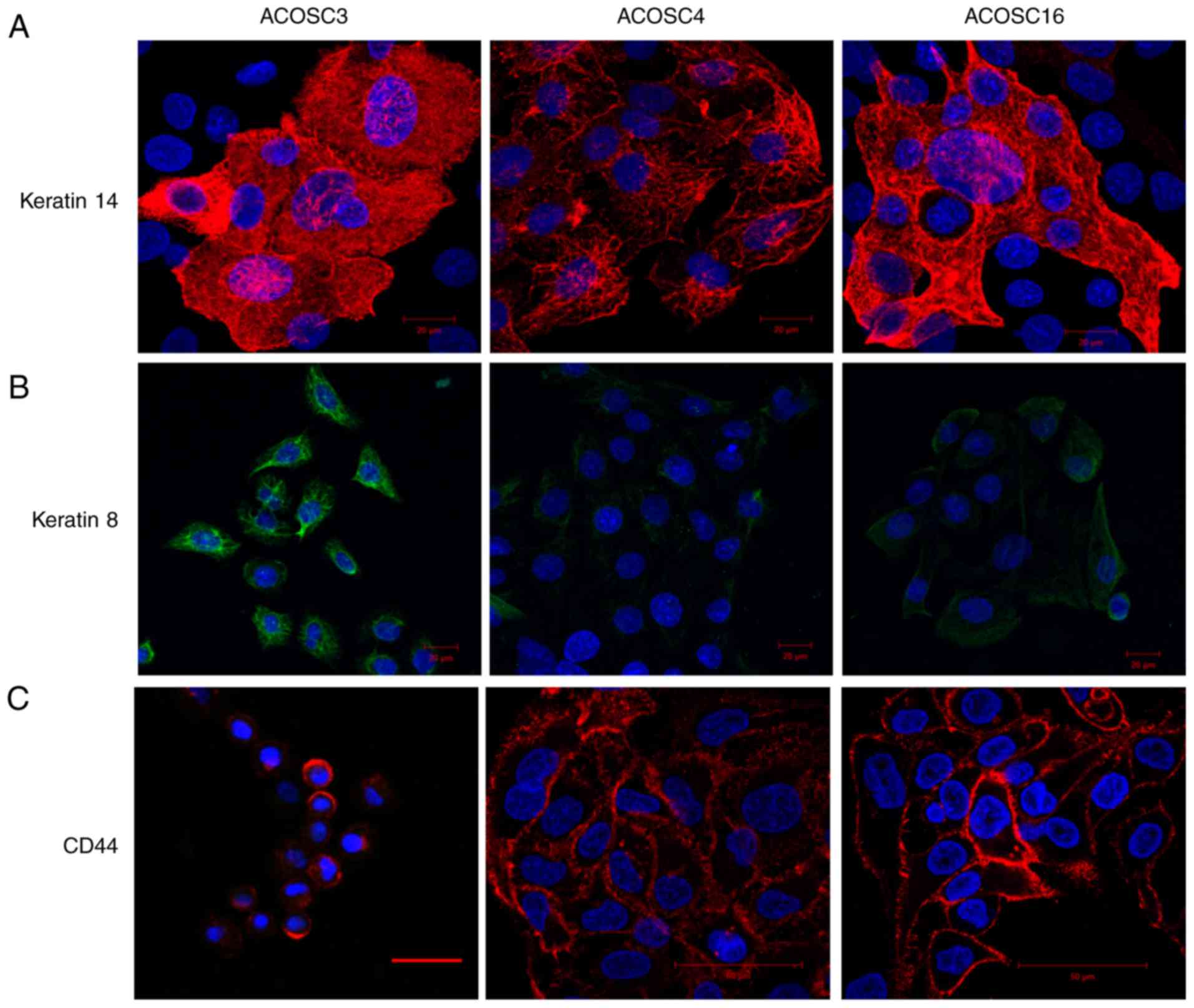
Evaluation of the keratin and CD44
expression in the established cell lines
Epithelial cells derived from the different
epithelial tissues displayed the distinct expression pattern of the
cytokeratin isoforms. In addition, cancer cells originating from
epithelial tissues exhibited abnormal expression of different
cytokeratins. Immunocytochemistry was performed on the OSCC cell
lines using the keratins 14 and 8, which are expressed in the basal
cells of the epithelium and transformed keratinocytes, respectively
(30,31). The data indicated that the ACOSC4
cell line exhibited uniform and bright staining for keratin 14,
whereas the cell lines ACOSC3 and ACOSC16 exhibited heterogeneity
in keratin 14 expression; certain cells exhibited bright staining
for keratin 14, whereas other cells were completely negative for
keratin 14 (Fig. 2A). In addition,
keratin 8 expression analysis indicated that the three OSCC cell
lines expressed different levels of keratin 8. ACOSC3 cells had the
brightest staining for keratin 8, and ACOSC16 cells exhibited only
slightly brighter staining for keratin 8 than ACOSC4 cells
(Fig. 2B). IFA was performed using
CD44 (CSC marker), which was expressed in all three cell lines
(Fig. 2C). Collectively, these data
suggest that all three OSCC cell lines are derived from basal
keratinocytes (epithelial origin), have abnormal expression of the
tumor-associated protein keratin 8, and contain a CSC
subpopulation.
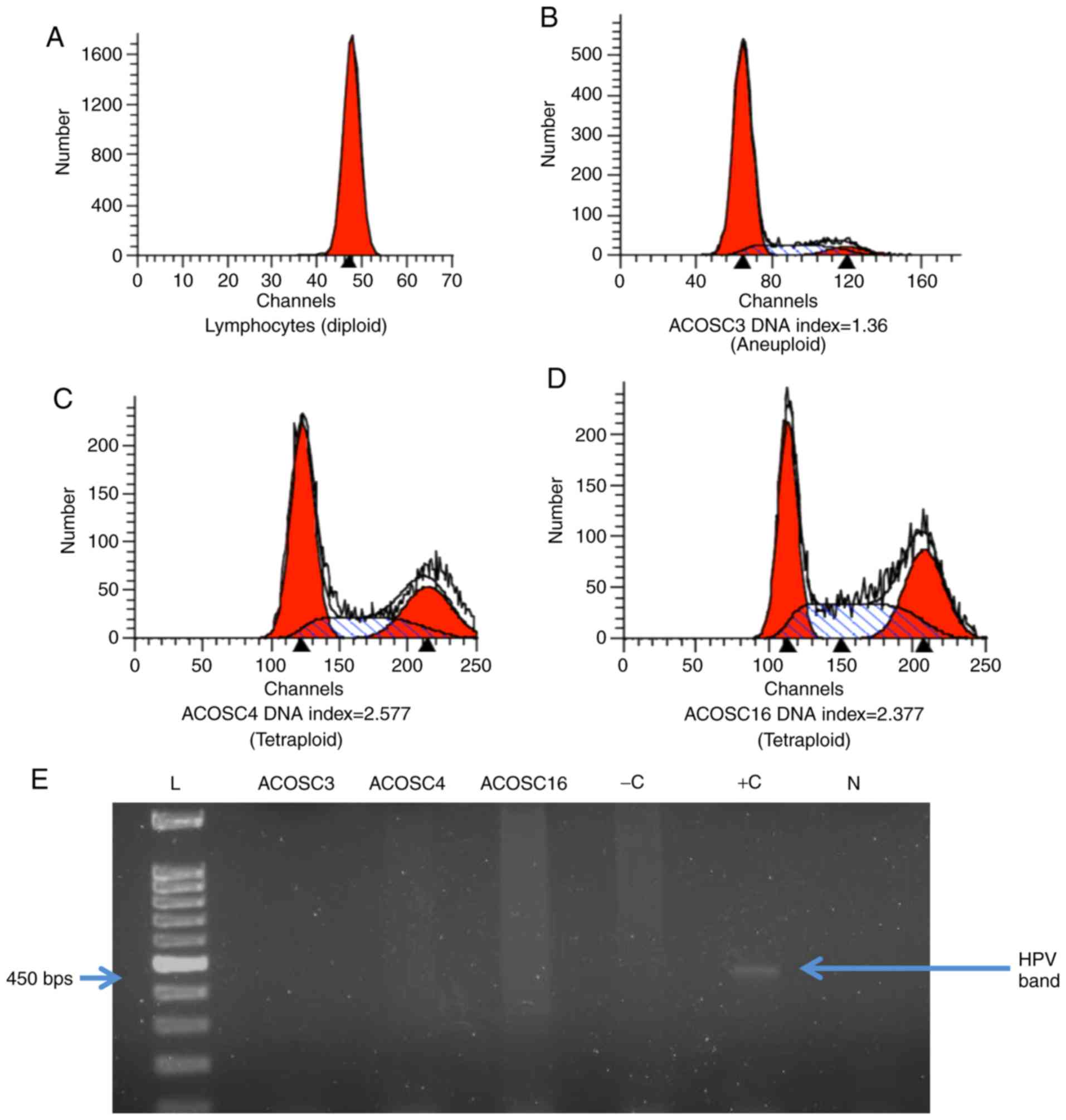
Determination of ploidy
Abnormal DNA content in tumor cells due to defective
mitosis or amplification of different DNA fragments is a leading
hallmark of cancer. DNA content in all three OSCC cell lines was
evaluated by performing ploidy analysis. Normal human lymphocytes
were used as a control for diploid DNA content. The DNA index
number was calculated by dividing mean channel for the cells in the
G0 phase by that of the diploid lymphocytes. The DNA
indices for the ACOSC3, ACOSC4, and ACOSC16 cell lines were
calculated to be 1.360, 2.577 and 2.377, respectively (Fig. 3A-D). ACOSC3 cells had slightly
higher than diploid DNA content, whereas both ACOSC4 and ACOSC16
cell lines exhibited slightly higher than tetraploid DNA content.
These results indicated that all three cell lines have abnormal DNA
content; which may be further investigated to identify the
potential role of abnormal DNA content in the transformation of the
cells.
HPV typing
Infection by HPV 16 and 18 has raised concern as a
risk factor for oral cancers. Approximately 25% of oropharyngeal
cancers are attributed to HPV 16 infection, whereas 1–3% of all
oropharyngeal cancers are attributed to HPV 18 infection (32). HPV-positive cancers are associated
with a more favorable prognosis as compared with HPV-negative
cancers (33). To examine the HPV
status in all three oral cancer cell lines, PCR was performed. DNA
from the HeLa cell line was used as the positive control for HPV
detection, whereas DNA isolated from the MCF7 cell line was used as
the negative control. All three OSCC cell lines were found to be
HPV negative by PCR using the MY09/MY11 primer sets (Fig. 3E).
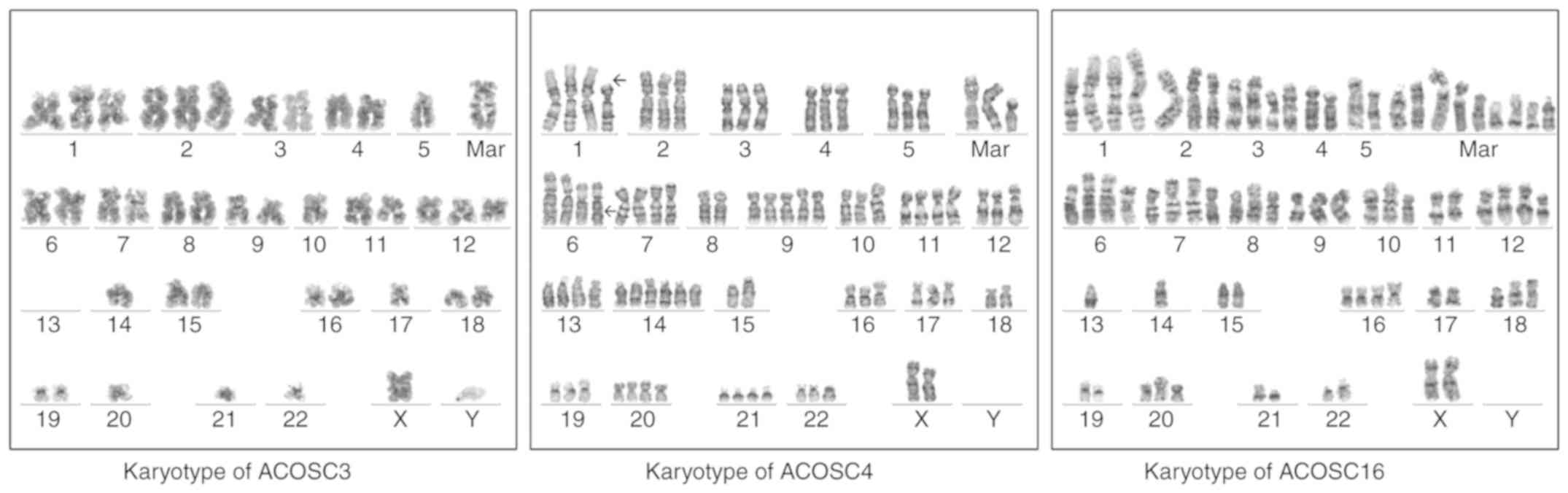
Karyotyping of the cell lines
To precisely determine ploidy and gain insight into
the chromosomal abnormalities in the cell lines, karyotyping of all
three cell lines was performed. None of the cell lines exhibited
metaphases with a normal karyotype (Fig. 4).
The ACOSC3 cell line was aneuploid, with
hyperdiploidy (64–80 chromosomes) and near-diploidy (40–41
chromosomes) in 70 and 30% of the metaphases, respectively. The
loss of the Y chromosome and two or three distinctive clonal
markers was observed. One of these was a large submetacentric
marker with sharp, equally spaced bands, which suggested
amplification. Gains of chromosomes 1, 2, 3, 12, 19 and X were
observed, whereas consistent losses of chromosomes 4, 7, 8, 13, 14,
21, 22 and Y were observed. A derivative chromosome 16 with very
lightly stained short arms, suggesting t(16;19), was also observed.
Seven metaphases were karyotyped.
The ACOSC4 cell line was aneuploid, with
hyperdiploidy (63–83 chromosomes) in 85% of the metaphases and
near-diploidy or pseudodiploidy in ~15% of the metaphases. Clonal
structural anomalies, including a derivative chromosome 1 with the
loss of the ‘p’ arm and a probable unbalanced t(1;12)(p11;p11), a
suspicion of isochromosome 9q, deletion 6q and a Robertsonian
t(15;21) were detected. Five or six copies of chromosomes 9 and 14
were observed in some metaphases. Frequent losses of chromosomes 18
and 19 was also observed. Six metaphases were karyotyped.
ACOSC16 was aneuploid, with hyperdiploidy (55–71
chromosomes) and loss of the Y chromosome in all metaphases. Clonal
structural anomalies included additional material on 1p, deletion
3p, isochromosome 9q, additional material on 14q32 (immunoglobulin
heavy chain locus rearrangement) and 11q23 (MLL rearrangement)
together with medium-sized markers. Loss of chromosomes 4, 5, 7, 8,
13, 17 and 21 was frequently observed. Five metaphases were
karyotyped.
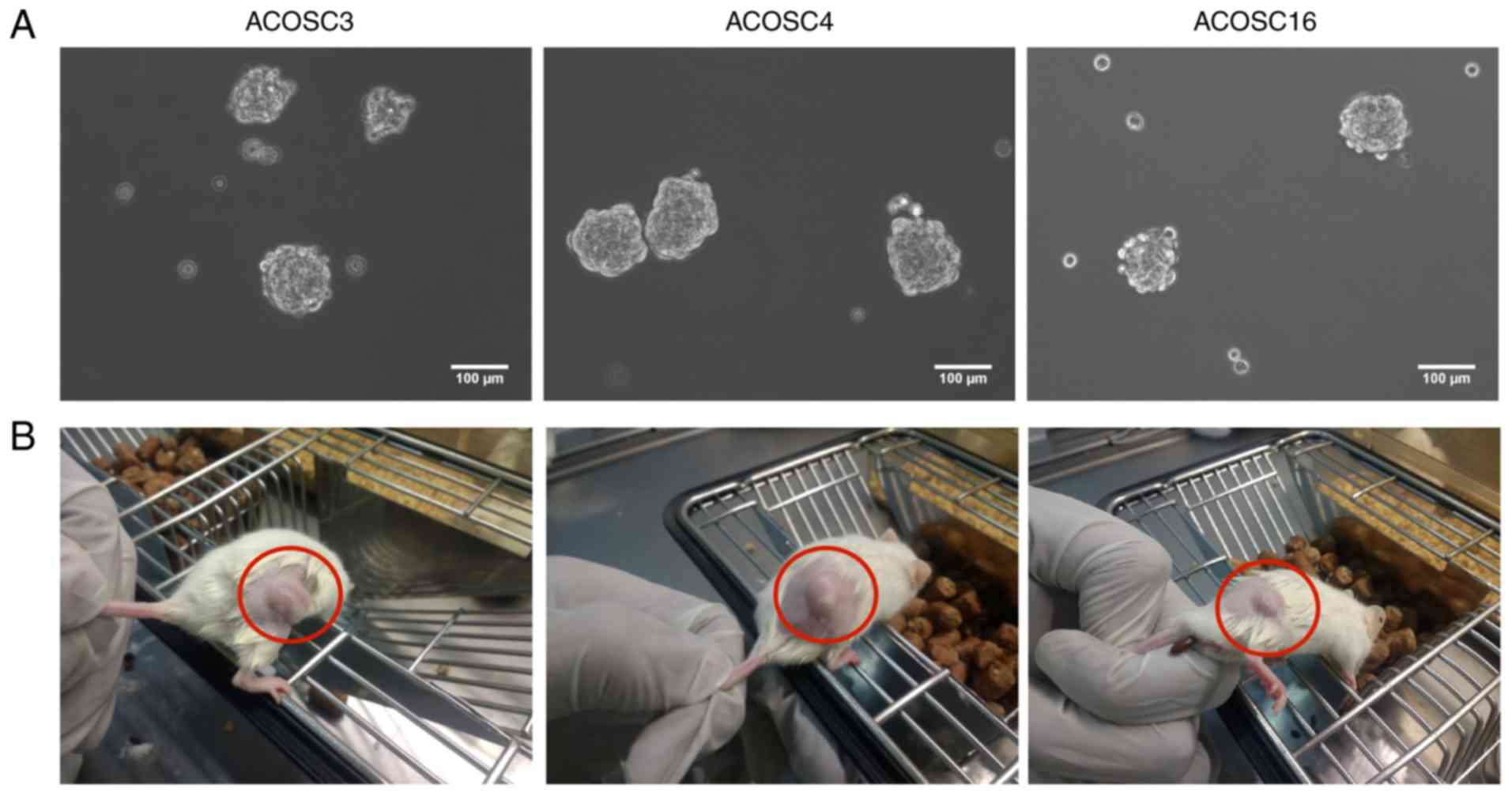
Orosphere formation assay
Cancer cells exhibit anchorage-independent cell
growth. To evaluate the ability of these cells to form spheres
in vitro, equal numbers (10,000) of cells were cultured on a
low-adherence plate for 5 days. The data suggested that the ACOSC3,
ACOSC4, and ACOSC16 cell lines formed compactly rounded spheres,
with high sphere-formation efficiency (Fig. 5A). Furthermore, these primary
spheres were analyzed for their ability to give rise to secondary
spheres. The primary spheres were trypsinized to prepare a
single-cell suspension and were cultured in the low-adherence
plate. Notably, when grown for 3–4 days, all three cell lines
produced secondary orospheres. Collectively, these data suggest
that the cells of all three OSCC cell lines are
anchorage-independent, which is enhanced in stem cell
populations.
Evaluation of the in vivo tumorigenic
potential
Furthermore, to investigate the in vivo tumor
formation efficiency of these cells, the NOD/SCID mice were
subcutaneously injected with 2×106 cells of all three
cell lines (ACOSC3, ACOSC4, and ACOSC16) with Matrigel. The results
indicated that all three cell lines exhibited capability to give
rise to tumors in immunocompromised mice. This experiment was
performed in 5 replicates for each cell line, and all the mice
injected with these cells exhibited tumor formation within 10–15
days. Each injection of the cells resulted in the formation of
single, irregularly shaped tumors (Fig.
5B). Specifically, the ACOSC4 cell line was more aggressive as
compared with ACOSC3 and ACOSC16 cell lines as it produced tumors
in a relatively decreased duration of time (Fig. 6). These data confirm the tumorigenic
potential of all three OSCC cell lines.
Cell line authentication through STR
profiling
STR profiling was performed on all three OSCC cell
lines to confirm that their novelty and genetic distinctness from
previously established cell lines (Table II). The STR profiles of the three
cell lines were compared with the DSMZ database. The profiles of
the three cell lines did not exhibit any significant match with
those of any previously established cell lines. The STR profiles of
the ACOSC3, ACOSC4 and ACOSC16 cell lines were distinct, thus
confirming their uniqueness and the absence of cross-contamination.
Collectively, these data confirm the novelty of the cell lines and
that they are derived from the previously unreported tissue
samples. It was confirmed that the cell lines were derived from the
tumors donated by patients by determining the STR profiles of the
tumors and matching them with the profiles of their respective cell
lines.
 | Table II.Analysis of STR markers in all three
cell lines. |
Table II.
Analysis of STR markers in all three
cell lines.
| Marker | ACOSC3 | ACOSC4 | ACOSC16 |
|---|
| TH01 | 6 | 6,8 | 7,9.3 |
| D5S818 | 12,13 | 10,12 | 11 |
| D13S317 | 8,11 | 8,11 | 11 |
| D7S820 | 9,10 | 11 | 7,12 |
| D16S539 | 11 | 11,12 | 9,10 |
| CSF1PO | 10,12 | 10,11 | 11,12 |
| vWA | 14,15 | 16,29 | 15,29 |
| TPOX | 8,9 | 10,11 | 8 |
| Amel | X,Y | X,X | X,Y |
Discussion
OSCC is the second most common cancer in India
(3,5,6).
However, only a few cell lines of OSCC have been derived from
Indian patients. Buccal mucosa carcinoma-derived cell lines were
previously established using tissue samples from Indian patients
could not form tumors when injected into immunocompromised mice
(24). The establishment of cell
lines from patients with oral cancers is crucial because studies on
these cell lines have contributed considerably to the current
understanding of tumor heterogeneity, and highlight any differences
between the Indian (Asian) and Western populations at a molecular
level. OSCC presents a wide variation in epidemiology in
populations from Western countries and Southeast Asian regions,
which may be due to genetic differences between these populations
(4). The cell lines established
from patients with OSCC can provide a source of cancer cells that
retain the heterogeneity of the cell populations present in the
parental tumor. Therefore, they can be used as model systems to
study pharmacokinetics and pharmacogenomics of newly discovered
anticancer drugs. Notably, they can be used to understand the
molecular mechanism involved in the resistance of cancer cells to
chemotherapy and radiotherapy.
Recently, 16 HNSCC cell lines were established by
using oral mucosa cancer tissue samples, and whole exome sequencing
was performed along with patient-matched blood (18). The present study intended to
identify the mutations present in the tumor and distinguish them
from the mutations accumulated during sub-culturing the cell lines.
A similar study on cell lines derived from Indian patients may
reveal considerable information on cancer development and mode of
action of the carcinogens frequently encountered by Indian
patients, including tobacco, betel quid and areca nut.
A previous study demonstrated that cancer cells
possess stem-like characteristics. These CSCs can be enriched the
spheroid formation assay. Furthermore,
CD44+/CD66− is a reliable combination of
markers for the isolation of CSCs (17). Similarly, the presently established
OSCC cell lines can also be used to identify a combination of
markers for the identification of true CSCs in Indian patients,
which can be useful in developing effective therapeutic strategies
for these patients. CSCs can be isolated from the OSCC cell lines,
and the molecular validation of the genes specifically deregulated
in the CSCs can be performed.
The cell lines were established from treatment-naive
advanced-stage cancer patients directly by using primary tumors,
thus eliminating any genotypic or phenotypic changes arising
because of patient-derived xenograft generation. These cell lines
grow in an anchorage-independent manner and do not require any
feeder cells to grow. The polygonal shape of the cells and their
tendency to grow in discrete patches confirm the epithelial nature
of the cell lines. The evidence of keratins 8 and 14 expression
additionally supports this inference as keratins are exclusively
expressed by epithelial cells (34).
DNA ploidy analysis of cancer cells can provide an
understanding of the aggressiveness, metastatic potential, and
prognosis of the disease (35–37).
The DNA content of all three cell lines was higher than the diploid
DNA content of the lymphocytes. The DNA index of ACOSC3 (1.36)
implied hyperdiploid DNA content, whereas the DNA indices of ACOSC4
(2.577) and ACOSC16 (2.377) implied that the DNA content exceeded
the tetraploid DNA content. This may be explained by the rapid and
frequent divisions of cancer cells. Uncontrolled cell divisions
result in improper karyokinesis, which results in an increase in
the ploidy level of the cells (38).
Chromosomal analysis suggested that all three cell
lines had abnormal karyotypes. In the ACOSC3 cell line, two
subpopulations were present; the larger subpopulation (70%) was
hyperdiploid (64–80 chromosomes), whereas the other 30% metaphases
were near-diploid (40–41 chromosomes). In the ACOSC4 cell line, a
hyperdiploid (63–83 chromosomes) subpopulation was observed, which
constituted 85% of the karyotyped metaphases, whereas ~15% of the
metaphases were near-diploid or pseudodiploid. All 50 analyzed
metaphases of the ACOSC16 cell line were hyperdiploid (55–71
chromosomes). All three cell lines exhibited gains, losses, and
translocations of various chromosomal regions or entire
chromosomes. These results underline the genomic instability of
these cells.
HPV (high-risk HPV genotypes 16, 18, 31, 33, and 35)
is one of the dominant risk factors for the development of OSCC.
Evidence from the literature suggests that HPV infection is a risk
factor for oral cancer (32). The
present cell lines were evaluated for the presence of HPV markers
by performing PCR using MY09/MY11 primers for the capsid protein of
the virus. The present results indicated that all three of the cell
lines were found to be free of HPV infection.
One of the notable properties of cancer stem cells
(CD44+/ALDH+ cells) is the formation of
spheres when cultured on low-adherence plates (39). An orosphere assay was performed on
all three cell lines to detect the presence of
anchorage-independent CSCs, which were plated onto low-adherence
plates; this confirmed the presence of CSCs within these cell
lines. Additionally, immunofluorescence exhibited the expression of
CD44, which is one of the CSC markers.
In vivo tumorigenesis assay is the gold
standard assay to detect the presence of CSCs from a heterogeneous
cell population. Tumorigenicity is a very important property of the
cell lines as this property enables in-depth studies on the
development of tumors and the development of therapy resistance in
them to be conducted. Furthermore, all three cell lines produced
tumors when injected into NOD/SCID mice. These cell lines can be
used to generate xenografts, which can be used as model systems for
testing effectiveness of therapeutic agents on tumors of human
origin.
The novelty of the present OSCC cell lines was
confirmed through STR profiling. The STR profiles of the three cell
lines were compared with the DSMZ database of STR profiles. The STR
profiles of the present cell lines did not significantly match with
any of the cell lines present in the STR profile database or
amongst each other. Also, the cell lines had differences in their
growth patterns, keratin expression patterns and DNA content and
STR profiles, thus confirming their unique nature (40).
Recently, a number of studies have been carried out
in Western populations by using patient-derived tumor cell lines,
which include exome sequencing that reveals a large amount of
information regarding the mutations in several genes across the
genome (16,18,19).
These studies enable deduction of molecular dysfunctions that may
be associated with carcinogenesis and therapy resistance. These
cell lines allow for extensive studies using cells from Indian
patients. The cell lines may be used to further delineate the
signaling networks, genetic, epigenetic, transcriptomic and
proteomic characterization of the oral cancer cells derived from
Indian patients, contributing towards a better understanding of the
biology.
In conclusion, the present study reports the
establishment of three novel OSCC cell lines from the
advanced-stage treatment-naive oral cancer derived from buccal
mucosa carcinoma patients of Indian ethnicity; all of them with a
habit of tobacco consumption. These cell lines have a wide range of
potential application for basic and translational research in oral
cancer.
Acknowledgements
The authors would like to thank Ms. Sayoni Roy for
helping in formatting the manuscript and Mr. Raghava R. Sunkara for
his help in the discussion. We also thank Dr Prochi Madon
(Department of Assisted Reproduction and Genetics, Jaslok Hospital
and Research Centre, Mumbai, India) for her help in the karyotype
determination and interpretation for the present study.
Funding
SSN is supported by CSIR fellowship. GLC is
supported by ACTREC fellowship. The present study was supported
from the grant of ACTREC-TMC intramural fund (grant no. 3542).
Availability of data and materials
All data generated and/or analyzed during this study
are included in this published article.
Authors' contributions
SKW conceived and designed the study, and analyzed
and interpreted the data. NPG and SSN performed the experiments. PC
provided the tumor samples, and analyzed and interpreted the data.
GLC analyzed and interpreted the data. SSN, GLC and SKW wrote the
manuscript. NPG and SSN prepared the images. SKW and GLC reviewed
the data. SKW reviewed the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of Advanced Centre for Treatment, Education and
Research in Cancer, Tata Memorial Centre (Navi Mumbai, India).
Informed consent for the study was obtained from all the patients
involved in the present study.
Patient consent for publication
Informed consent for the study was obtained from all
the patients involved in the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vigneswaran N and Williams MD:
Epidemiologic trends in head and neck cancer and aids in diagnosis.
Oral Maxillofac Surg Clin North Am. 26:123–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kulkarni M: Head and neck cancer burden in
India. Int J Head Neck Surg. 4:29–35. 2013. View Article : Google Scholar
|
|
6
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iype EM, Pandey M, Mathew A, Thomas G,
Sebastian P and Nair MK: Oral cancer among patients under the age
of 35 years. J Postgrad Med. 47:171–176. 2001.PubMed/NCBI
|
|
8
|
Beck B and Blanpain C: Unravelling cancer
stem cell potential. Nat Rev Cancer. 13:727–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eagle H: Propagation in a fluid medium of
a human epidermoid carcinoma, strain KB. Proc Soc Exp Biol Med.
89:362–364. 1955. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin CJ, Grandis JR, Carey TE, Gollin SM,
Whiteside TL, Koch WM, Ferris RL and Lai SY: Head and neck squamous
cell carcinoma cell lines: Established models and rationale for
selection. Head Neck. 29:163–188. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lacroix M: Persistent use of ‘false’ cell
lines. Int J Cancer. 122:1–4. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Easty DM, Easty GC, Carter RL, Monaghan P
and Butler LJ: Ten human carcinoma cell lines derived from squamous
carcinomas of the head and neck. Br J Cancer. 43:772–785. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Easty DM, Easty GC, Carter RL, Monaghan P,
Pittam MR and James T: Five human tumour cell lines derived from a
primary squamous carcinoma of the tongue, two subsequent local
recurrences and two nodal metastases. Br J Cancer. 44:363–370.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heo DS, Snyderman C, Gollin SM, Pan S,
Walker E, Deka R, Barnes EL, Johnson JT, Herberman RB and Whiteside
TL: Biology, cytogenetics, and sensitivity to immunological
effector cells of new head and neck squamous cell carcinoma lines.
Cancer Res. 49:5167–5175. 1989.PubMed/NCBI
|
|
15
|
Zhao M, Sano D, Pickering CR, Jasser SA,
Henderson YC, Clayman GL, Sturgis EM, Ow TJ, Lotan R, Carey TE, et
al: Assembly and initial characterization of a panel of 85
genomically validated cell lines from diverse head and neck tumor
sites. Clin Cancer Res. 17:7248–7264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
White JS, Weissfeld JL, Ragin CC, Rossie
KM, Martin CL, Shuster M, Ishwad CS, Law JC, Myers EN, Johnson JT
and Gollin SM: The influence of clinical and demographic risk
factors on the establishment of head and neck squamous cell
carcinoma cell lines. Oral Oncol. 43:701–712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kaseb HO, Fohrer-Ting H, Lewis DW, Lagasse
E and Gollin SM: Identification, expansion and characterization of
cancer cells with stem cell properties from head and neck squamous
cell carcinomas. Exp Cell Res. 348:75–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayes TF, Benaich N, Goldie SJ, Sipilä K,
Ames-Draycott A, Cai W, Yin G and Watt FM: Integrative genomic and
functional analysis of human oral squamous cell carcinoma cell
lines reveals synergistic effects of FAT1 and CASP8 inactivation.
Cancer Lett. 383:106–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SJ, Asthana S, van Zante A, Heaton
CM, Phuchareon J, Stein L, Higuchi S, Kishimoto T, Chiu CY, Olshen
AB, et al: Establishment and characterization of an oral tongue
squamous cell carcinoma cell line from a never-smoking patient.
Oral Oncol. 69:1–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wong DY, Chang KW, Chen CF and Chang RC:
Characterization of two new cell lines derived from oral cavity
human squamous cell carcinomas-OC1 and OC2. J Oral Maxillofac Surg.
48:385–390. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morifuji M, Taniguchi S, Sakai H,
Nakabeppu Y and Ohishi M: Differential expression of cytokeratin
after orthotopic implantation of newly established human tongue
cancer cell lines of defined metastatic ability. Am J Pathol.
156:1317–1326. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee EJ, Kim J, Lee SA, Kim EJ, Chun YC,
Ryu MH and Yook JI: Characterization of newly established oral
cancer cell lines derived from six squamous cell carcinoma and two
mucoepidermoid carcinoma cells. Exp Mol Med. 37:379–390. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamid S, Lim KP, Zain RB, Ismail SM, Lau
SH, Mustafa WM, Abraham MT, Nam NA, Teo SH and Cheong SC:
Establishment and characterization of Asian oral cancer cell lines
as in vitro models to study a disease prevalent in Asia. Int J Mol
Med. 19:453–460. 2007.PubMed/NCBI
|
|
24
|
Tatake RJ, Rajaram N, Damle RN, Balsara B,
Bhisey AN and Gangal SG: Establishment and characterization of four
new squamous cell carcinoma cell lines derived from oral tumors. J
Cancer Res Clin Oncol. 116:179–186. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Patil TT, Kowtal PK, Nikam A, Barkume MS,
Patil A, Kane SV, Juvekar AS, Mahimkar MB and Kayal JJ:
Establishment of a tongue squamous cell carcinoma cell line from
indian gutka chewer. J Oral Oncol. 2014:92014.
|
|
26
|
Mulherkar R, Goud AP, Wagle AS, Naresh KN,
Mahimkar MB, Thomas SM, Pradhan SA and Deo MG: Establishment of a
human squamous cell carcinoma cell line of the upper aero-digestive
tract. Cancer Lett. 118:115–121. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gravitt PE, Peyton CL, Alessi TQ, Wheeler
CM, Coutlée F, Hildesheim A, Schiffman MH, Scott DR and Apple RJ:
Improved amplification of genital human papillomaviruses. J Clin
Microbiol. 38:357–361. 2000.PubMed/NCBI
|
|
28
|
Rooney DE: Human Cytogenetics:
Constitutional Analysis. A Practical Approach. Oxford University
Press. (Oxford). 2001.
|
|
29
|
Dirks WG, MacLeod RA, Nakamura Y, Kohara
A, Reid Y, Milch H, Drexler HG and Mizusawa H: Cell line
cross-contamination initiative: An interactive reference database
of STR profiles covering common cancer cell lines. Int J Cancer.
126:303–304. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Coulombe PA, Kopan R and Fuchs E:
Expression of keratin K14 in the epidermis and hair follicle:
Insights into complex programs of differentiation. J Cell Biol.
109:2295–2312. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matthias C, Mack B, Berghaus A and Gires
O: Keratin 8 expression in head and neck epithelia. BMC Cancer.
8:2672008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghittoni R, Accardi R, Chiocca S and
Tommasino M: Role of human papillomaviruses in carcinogenesis.
Ecancermedicalscience. 9:5262015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nagel R, Martens-de Kemp SR, Buijze M,
Jacobs G, Braakhuis BJ and Brakenhoff RH: Treatment response of
HPV-positive and HPV-negative head and neck squamous cell carcinoma
cell lines. Oral Oncol. 49:560–566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Herrmann H, Bar H, Kreplak L, Strelkov SV
and Aebi U: Intermediate filaments: From cell architecture to
nanomechanics. Nat Rev Mol Cell Biol. 8:562–573. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Stell PM: Ploidy in head and neck cancer:
A review and meta-analysis. Clin Otolaryngol Allied Sci.
16:510–516. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cooke LD, Cooke TG, Bootz F, Forster G,
Helliwell TR, Spiller D and Stell PM: Ploidy as a prognostic
indicator in end stage squamous cell carcinoma of the head and neck
region treated with cisplatinum. Br J Cancer. 61:759–762. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Slootweg PJ, Rutgers DH and Wils IS: DNA
ploidy analysis of squamous cell head and neck cancer to identify
distant metastasis from second primary. Head Neck. 14:464–466.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vitale I, Galluzzi L, Senovilla L, Criollo
A, Jemaà M, Castedo M and Kroemer G: Illicit survival of cancer
cells during polyploidization and depolyploidization. Cell Death
Differ. 18:1403–1413. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Krishnamurthy S and Nor JE: Orosphere
assay: A method for propagation of head and neck cancer stem cells.
Head Neck. 35:1015–1021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Parson W, Kirchebner R, Muhlmann R, Renner
K, Kofler A, Schmidt S and Kofler R: Cancer cell line
identification by short tandem repeat profiling: Power and
limitations. FASEB J. 19:434–436. 2005. View Article : Google Scholar : PubMed/NCBI
|