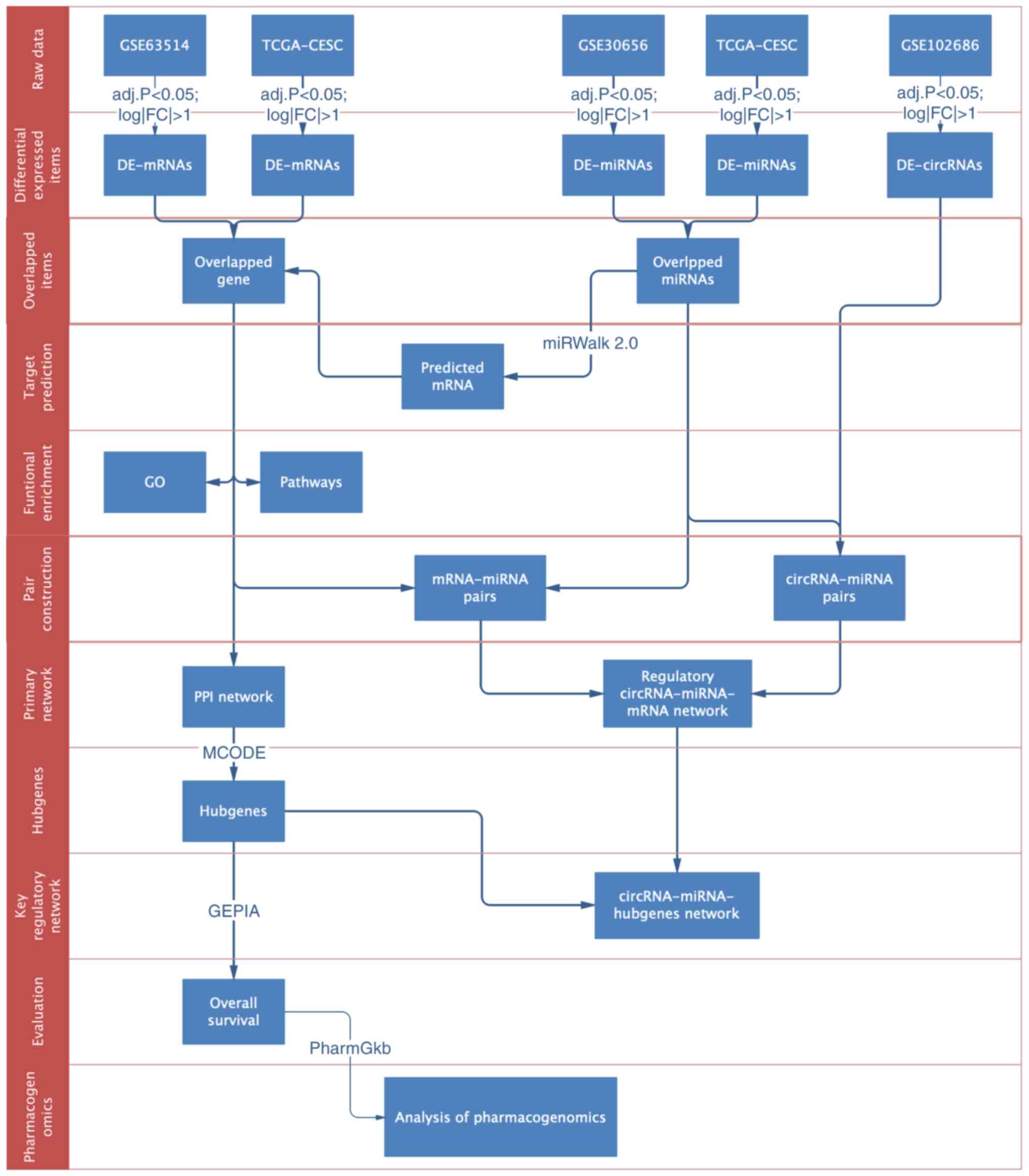
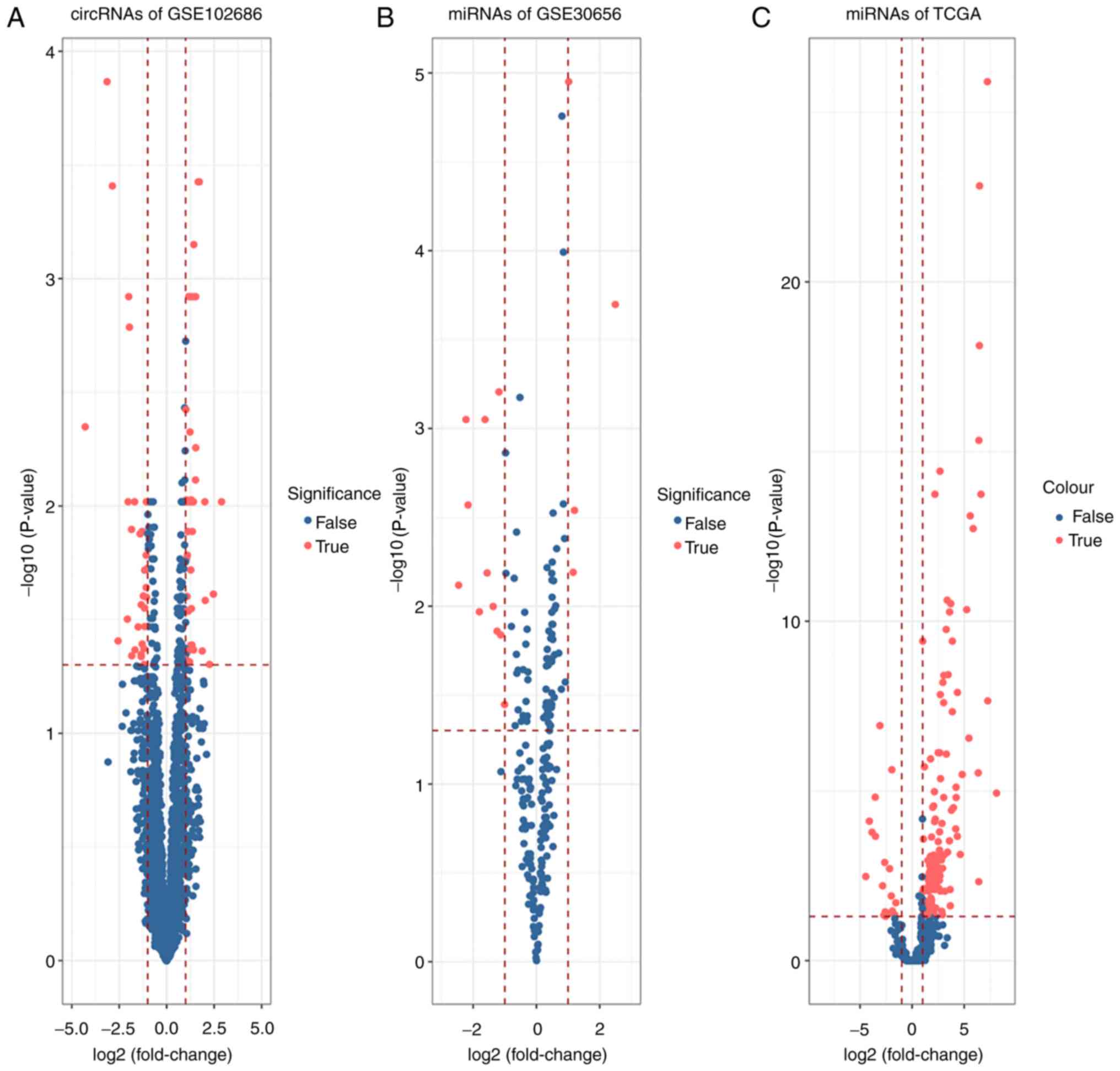
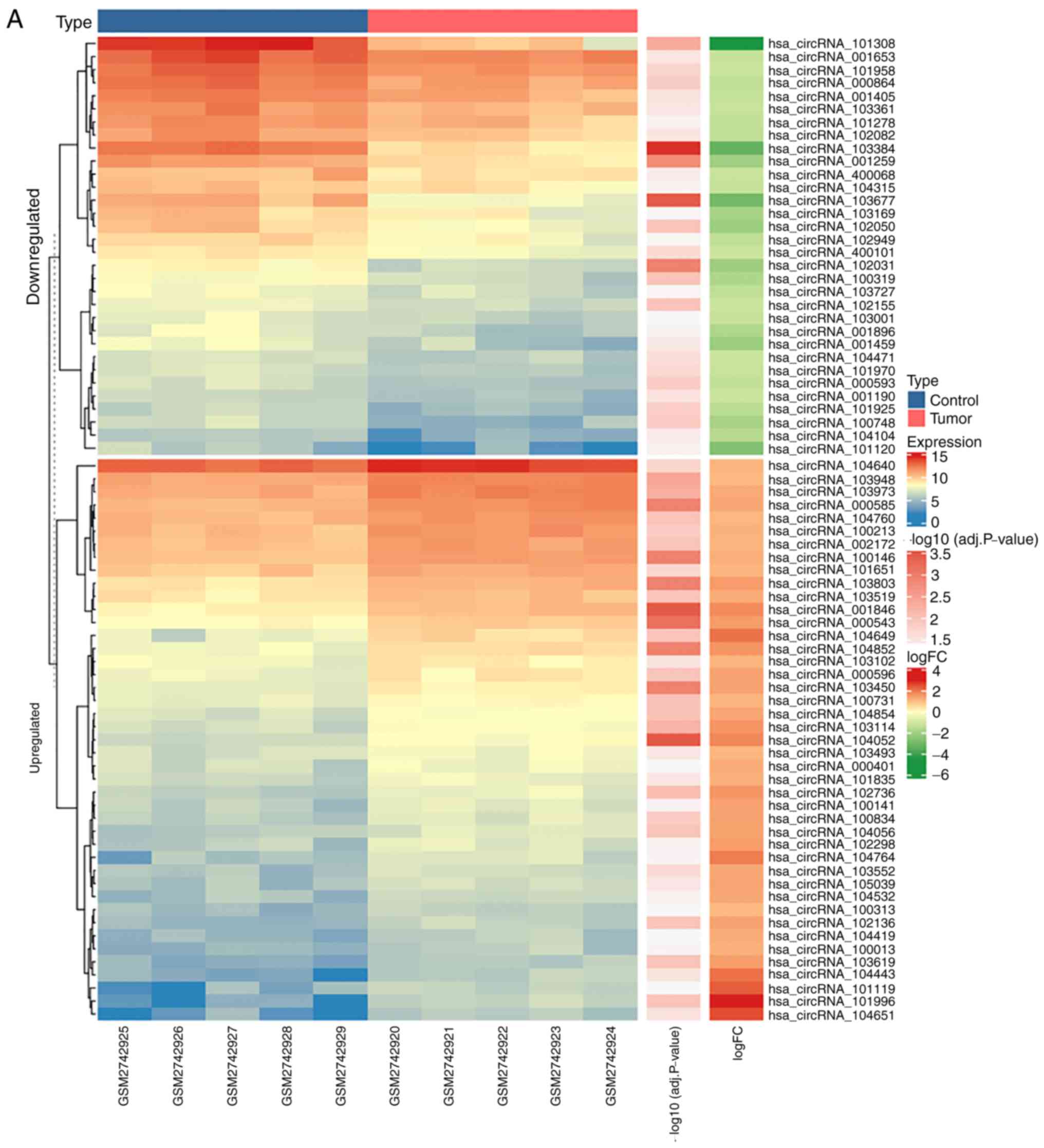
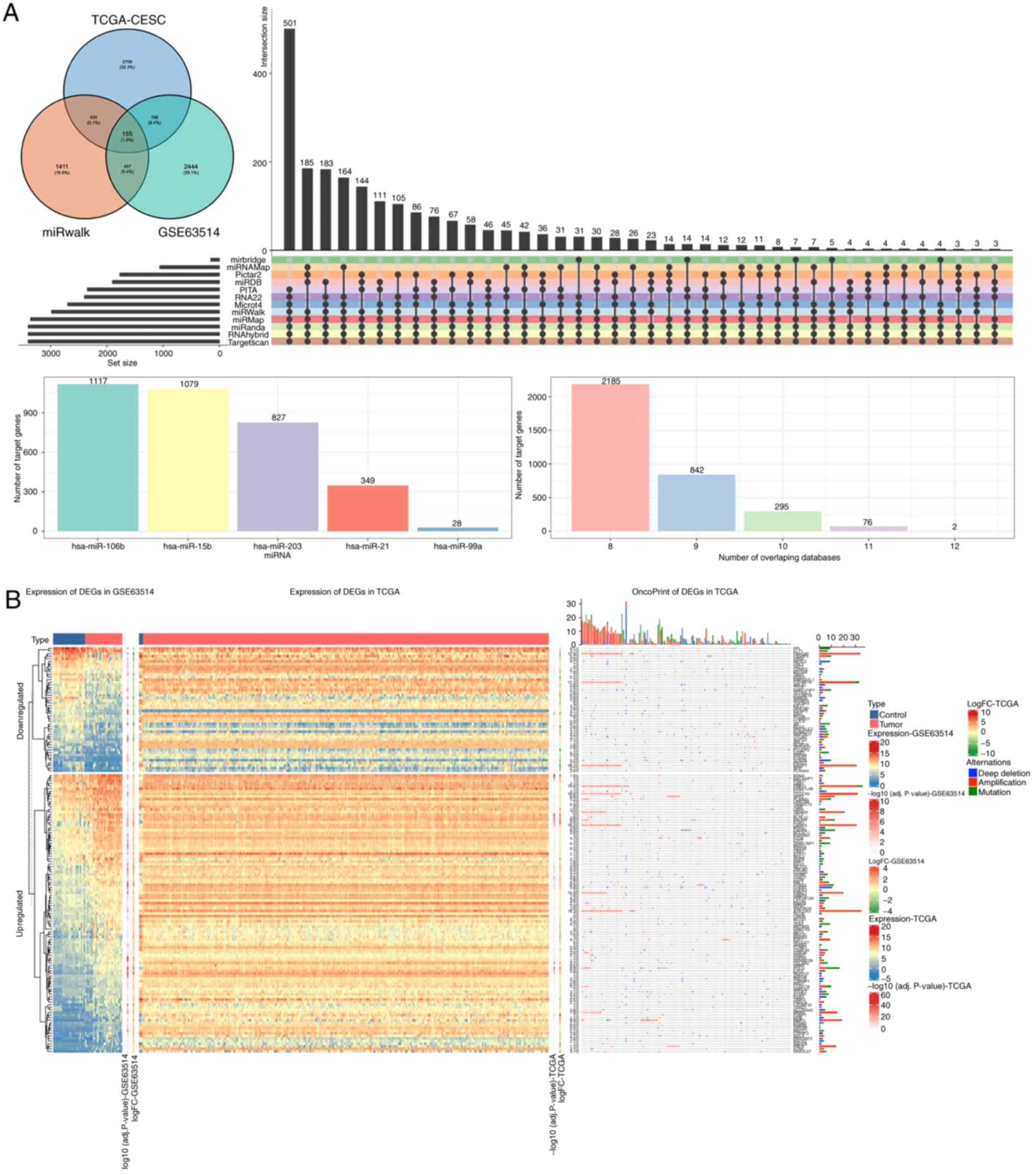
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marth C, Landoni F, Mahner S, McCormack M,
Gonzalez-Martin A and Colombo N; ESMO Guidelines Committee, :
Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 28 (Suppl 4):iv72–iv83. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
GuYu Z, YiMin Z, ChongDong L, GuangMing C,
Ran C and ZhenYu Z: Current status and future of targeted therapy
for patients with local advanced cervical cancer. Chinese J Pract
Gynecol Obstet. 34:1216–1220. 2018.
|
|
4
|
Fleming ND, Frumovitz M, Schmeler KM, dos
Reis R, Munsell MF, Eifel PJ, Soliman PT, Nick AM, Westin SN and
Ramirez PT: Significance of lymph node ratio in defining risk
category in node-positive early stage cervical cancer. Gynecol
Oncol. 136:48–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kapranov P, Cawley SE, Drenkow J,
Bekiranov S, Strausberg RL, Fodor SP and Gingeras TR: Large-scale
transcriptional activity in chromosomes 21 and 22. Science.
296:916–919. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kogo R, How C, Chaudary N, Bruce J, Shi W,
Hill RP, Zahedi P, Yip KW and Liu FF: The microRNA-218~Survivin
axis regulates migration, invasion, and lymph node metastasis in
cervical cancer. Oncotarget. 6:1090–1100. 2014.
|
|
8
|
Hou T, Ou J, Zhao X, Huang X, Huang Y and
Zhang Y: MicroRNA-196a promotes cervical cancer proliferation
through the regulation of FOXO1 and p27Kip1. Br J
Cancer. 110:1260–1268. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Q, Qin J, Chen A, Zhou J, Liu J,
Cheng J, Qiu J and Zhang J: Downregulation of microRNA-145 is
associated with aggressive progression and poor prognosis in human
cervical cancer. Tumor Biol. 36:3703–3708. 2015. View Article : Google Scholar
|
|
10
|
Fang H, Shuang D, Yi Z, Sheng H and Liu Y:
Up-regulated microRNA-155 expression is associated with poor
prognosis in cervical cancer patients. Biomed Pharmacother.
83:64–69. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bumrungthai S, Ekalaksananan T, Evans MF,
Chopjitt P, Tangsiriwatthana T, Patarapadungkit N, Kleebkaow P,
Luanratanakorn S, Kongyingyoes B, Worawichawong S, et al:
Up-regulation of miR-21 is associated with cervicitis and human
papillomavirus infection in cervical tissues. PLoS One.
10:e01271092015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:2018. View Article : Google Scholar
|
|
17
|
Liu Q, Zhang X, Hu X, Yuan L, Cheng J,
Jiang Y and Ao Y: Emerging roles of circRNA related to the
mechanical stress in human cartilage degradation of osteoarthritis.
Mol Ther Nucleic Acids. 7:223–230. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen
Z, Song W, Zhou J, Tang ZY and Huang XY: Comprehensive circular RNA
profiling reveals the regulatory role of the
circRNA-100338/MIR-141-3p pathway in hepatitis B-related
hepatocellular carcinoma. Sci Rep. 7:54282017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Huang L, Li D, Shao J, Xiong S,
Wang C and Lu S: Hsa_circ_0101996 combined with hsa_circ_0101119 in
peripheral whole blood can serve as the potential biomarkers for
human cervical squamous cell carcinoma. Int J Clin Exp Pathol.
10:11924–11931. 2017.
|
|
21
|
Gao YL, Zhang MY, Xu B, Han LJ, Lan SF,
Chen J, Dong YJ and Cao LL: Circular RNA expression profiles reveal
that hsa_circ_0018289 is up-regulated in cervical cancer and
promotes the tumorigenesis. Oncotarget. 8:86625–86633. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma HB, Yao YN, Yu JJ, Chen XX and Li HF:
Extensive profiling of circular RNAs and the potential regulatory
role of circRNA-000284 in cell proliferation and invasion of
cervical cancer via sponging miR-506. Am J Transl Res. 10:592–604.
2018.PubMed/NCBI
|
|
23
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression
analyses for RNA-sequencing and microarray studies. Nucleic Acids
Res. 43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dweep H and Gretz N: MiRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
28
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Conway JR, Lex A and Gehlenborg N: UpSetR:
An R package for the visualization of intersecting sets and their
properties. Bioinformatics. 33:2938–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kozomara A and Griffiths-Jones S: MiRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res 42 (Database Issue). D68–D73. 2014. View Article : Google Scholar
|
|
31
|
Krüger J and Rehmsmeier M: RNAhybrid:
MicroRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34:W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang H, Ma R, Zou S, Wang Y, Li Z and Li
W: Reconstruction and analysis of the lncRNA-miRNA-mRNA network
based on competitive endogenous RNA reveal functional lncRNAs in
rheumatoid arthritis. Mol Biosyst. 13:1182–1192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiao X, Sherman BT, Huang da W, Stephens
R, Baseler MW, Lane HC and Lempicki RA: DAVID-WS: A stateful web
service to facilitate gene/protein list analysis. Bioinformatics.
28:1805–1806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46:D925–D929.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kerpedjiev P, Hammer S and Hofacker IL:
Forna (force-directed RNA): Simple and effective online RNA
secondary structure diagrams. Bioinformatics. 31:3377–3379. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schoen MW, Woelich SK, Braun JT, Reddy DV,
Fesler MJ, Petruska PJ, Freter CE and Lionberger JM: Acute myeloid
leukemia induction with cladribine: Outcomes by age and leukemia
risk. Leuk Res. 68:72–78. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Johnston JB: Mechanism of action of
pentostatin and cladribine in hairy cell leukemia. Leuk Lymphoma.
52 (Suppl 2):S43–S45. 2011. View Article : Google Scholar
|
|
43
|
Mulligan SP, Karlsson K, Strömberg M,
Jønsson V, Gill D, Hammerström J, Hertzberg M, McLennan R, Uggla B,
Norman J, et al: Cladribine prolongs progression-free survival and
time to second treatment compared to fludarabine and high-dose
chlorambucil in chronic lymphocytic leukemia. Leuk Lymphoma.
55:2769–2777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mego M, Sycova-Mila Z, Obertova J, Rajec
J, Liskova S, Palacka P, Porsok S and Mardiak J: Intrathecal
administration of trastuzumab with cytarabine and methotrexate in
breast cancer patients with leptomeningeal carcinomatosis. Breast.
20:478–480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rusch VW, Figlin R, Godwin D and
Piantadosi S: Intrapleural cisplatin and cytarabine in the
management of malignant pleural effusions: A Lung Cancer Study
Group trial. J Clin Oncol. 9:313–319. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Baker JAR, Wickremsinhe ER, Li CH,
Oluyedun OA, Dantzig AH, Hall SD, Qian YW, Ring BJ, Wrighton SA and
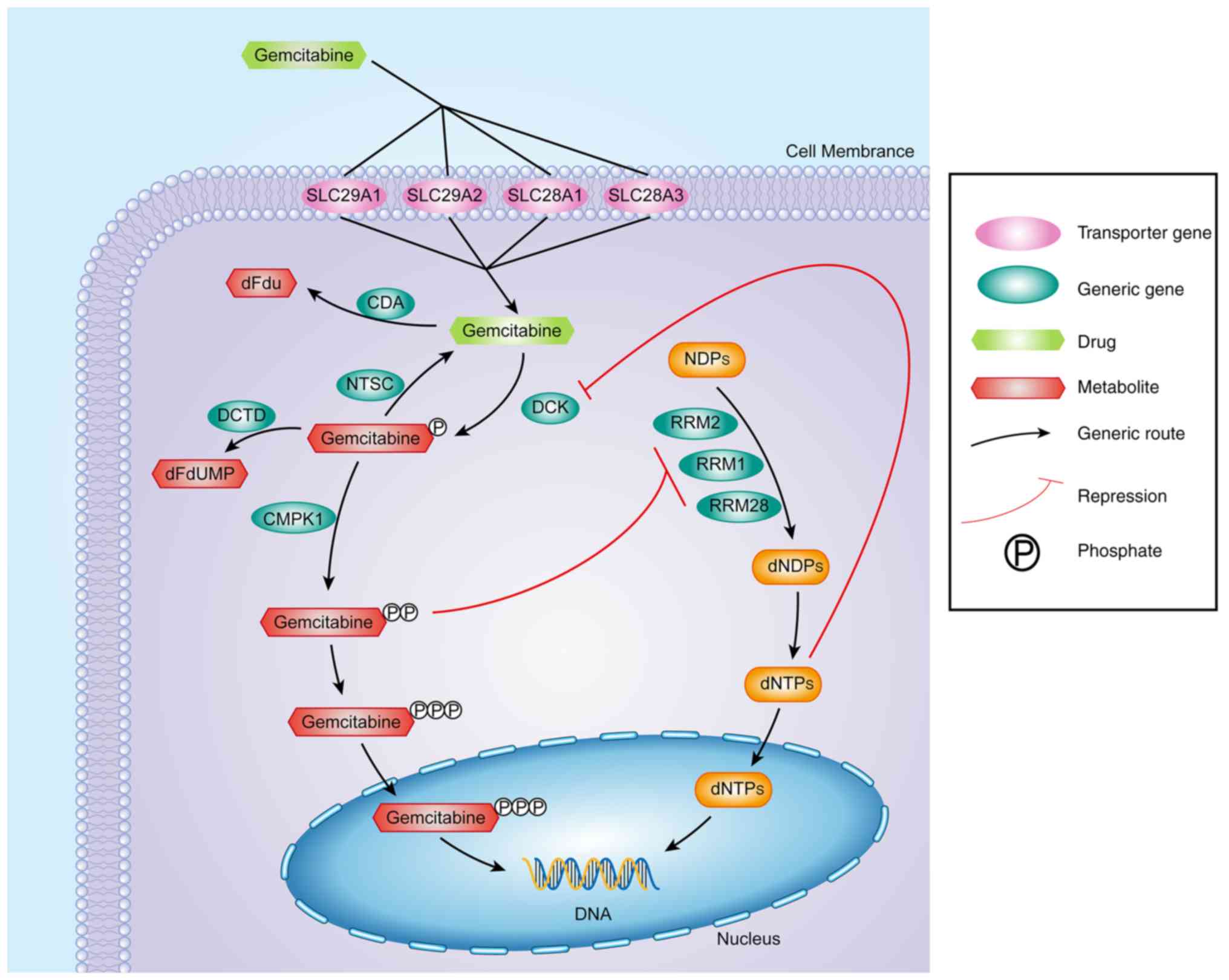
Guo Y: Pharmacogenomics of gemcitabine metabolism: Functional
analysis of genetic variants in cytidine deaminase and
deoxycytidine kinase. Drug Metab Dispos. 41:541–545. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lamba JK: Genetic factors influencing
cytarabine therapy. Pharmacogenomics. 10:1657–1674. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walboomers JM, Jacobs MV, Manos MM, Bosch
FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ and Muñoz N:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Du WW, Zhang C, Yang W, Yong T, Awan FM
and Yang BB: Identifying and characterizing circRNA-protein
interaction. Theranostics. 7:4183–4191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xie B, Zhao Z, Liu Q, Wang X, Ma Z and Li
H: CircRNA has_circ_0078710 acts as the sponge of microRNA-31
involved in hepatocellular carcinoma progression. Gene.
683:253–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xiong D, Dang Y, Lin P, Wen DY, He RQ, Luo
DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network identification
for exploring underlying pathogenesis and therapy strategy of
hepatocellular carcinoma. J Transl Med. 16:2202018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res. 27:626–641.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015.PubMed/NCBI
|
|
64
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
65
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mitra A, Pfeifer K and Park KS: Circular
RNAs and competing endogenous RNA (ceRNA) networks. Transl Cancer
Res. 7 (Suppl 5):S624–S628. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular functions and specific roles of circRNAs in the
cardiovascular system. Non-coding RNA Res. 3:75–98. 2018.
View Article : Google Scholar
|
|
68
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J
and Ao Y: Circular RNA related to the chondrocyte ECM regulates
MMP13 expression by functioning as a miR-136 ‘Sponge’ in human
cartilage degradation. Sci Rep. 6:225722016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J,
Lu W, Wan X, Ma D and Xie X: Progressive miRNA expression profiles
in cervical carcinogenesis and identification of HPV-related target
genes for miR-29. J Pathol. 224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yi Y, Liu Y, Wu W, Wu K and Zhang W: The
role of miR-106p-5p in cervical cancer: From expression to
molecular mechanism. Cell death Discov. 4:362018.PubMed/NCBI
|
|
72
|
Park S, Eom K, Kim J, Bang H, Wang HY, Ahn
S, Kim G, Jang H, Kim S, Lee D, et al: MiR-9, miR-21, and miR-155
as potential biomarkers for HPV positive and negative cervical
cancer. BMC Cancer. 17:6582017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xu L, Xu Q, Li X and Zhang X: MicroRNA-21
regulates the proliferation and apoptosis of cervical cancer cells
via tumor necrosis factor-α. Mol Med Rep. 16:4659–4663. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhu X, Er K, Mao C, Yan Q, Xu H, Zhang Y,
Zhu J, Cui F, Zhao W and Shi H: miR-203 suppresses tumor growth and
angiogenesis by targeting VEGFA in cervical cancer. Cell Physiol
Biochem. 32:64–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Melar-New M and Laimins LA: Human
papillomaviruses modulate expression of MicroRNA 203 upon
epithelial differentiation to control levels of p63 proteins. J
Virol. 84:5212–5221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Mao L, Zhang Y, Mo W, Yu Y and Lu H:
BANF1 is downregulated by IRF1-regulated microRNA-203 in
cervical cancer. PLoS One. 10:e01170352015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Reshmi G and Pillai MR: Beyond HPV:
Oncomirs as new players in cervical cancer. FEBS Lett.
582:4113–4116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhao S, Yao D, Chen J and Ding N:
Circulating miRNA-20a and miRNA-203 for screening
lymph node metastasis in early stage cervical cancer. Genet Test
Mol Biomarkers. 17:631–636. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang N, Li Y and Zhou J: Downregulation of
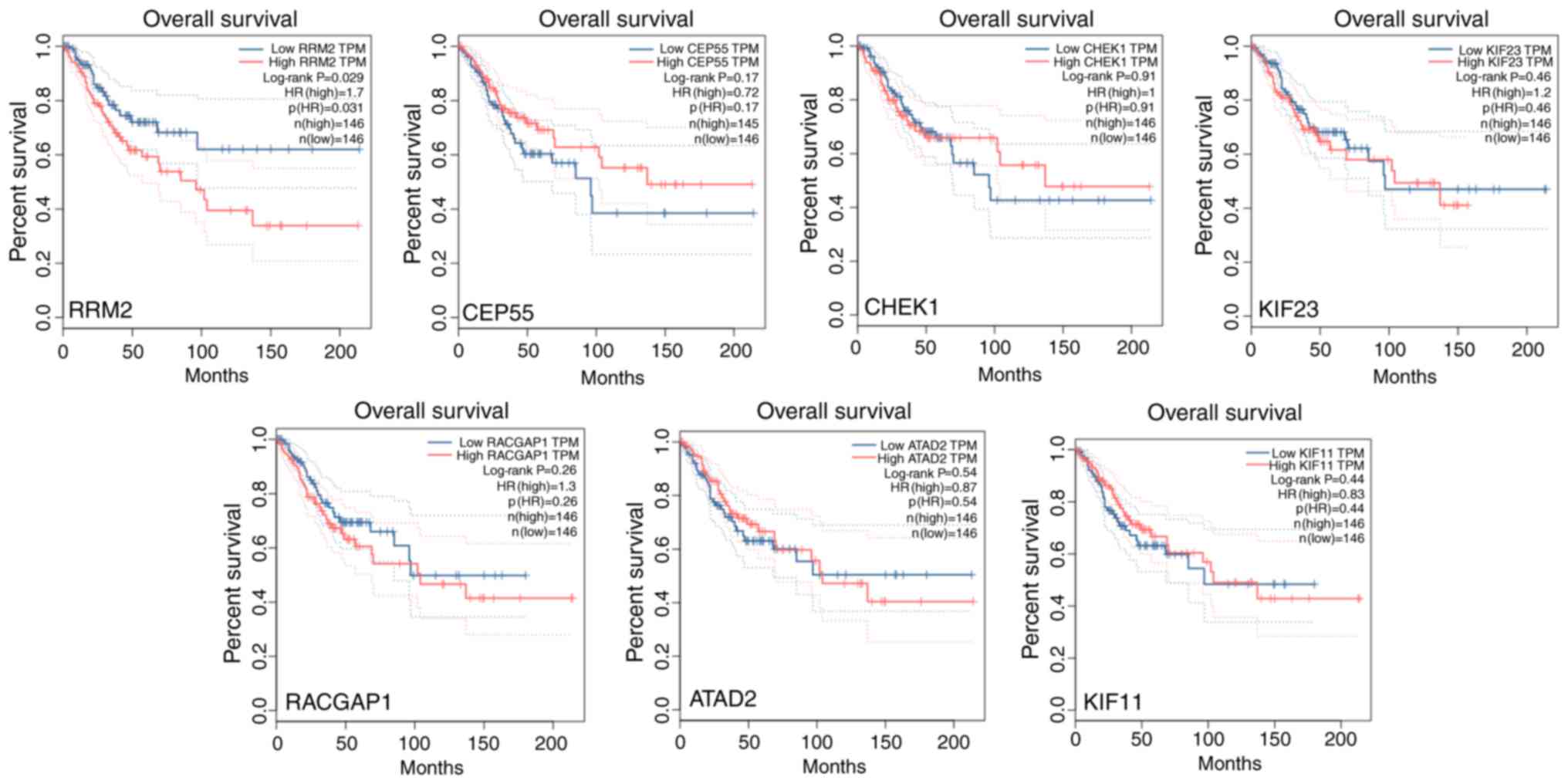
ribonucleotide reductase subunits M2 induces apoptosis and G1
arrest of cervical cancer cells. Oncol Lett. 15:3719–3725.
2018.PubMed/NCBI
|
|
80
|
Mazumder Indra D, Mitra S, Singh RK, Dutta
S, Roy A, Mondal RK, Basu PS, Roychoudhury S and Panda CK:
Inactivation of CHEK1 and EI24 is associated with the
development of invasive cervical carcinoma: Clinical and prognostic
implications. Int J cancer. 129:1859–1871. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zheng L, Li T, Zhang Y, Guo Y, Yao J, Dou
L and Guo K: Oncogene ATAD2 promotes cell proliferation, invasion
and migration in cervical cancer. Oncol Rep. 33:2337–2344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Cheng J, Lu X, Wang J, Zhang H, Duan P and
Li C: Interactome analysis of gene expression profiles of cervical
cancer reveals dysregulated mitotic gene clusters. Am J Transl Res.
9:3048–3059. 2017.PubMed/NCBI
|
|
83
|
Mutch DG and Bloss JD: Gemcitabine in
cervical cancer. Gynecol Oncol. 90:S8–S15. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Roy S, Devleena, Maji T, Chaudhuri P,
Lahiri D and Biswas J: Addition of gemcitabine to standard therapy
in locally advanced cervical cancer: A randomized comparative
study. Indian J Med Paediatr Oncol. 32:133–138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kalaghchi B, Abdi R, Amouzegar-Hashemi F,
Esmati E and Alikhasi A: Concurrent chemoradiation with weekly
paclitaxel and cisplatin for locally advanced cervical cancer.
Asian Pac J Cancer Prev. 17:287–291. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Seidman AD: Gemcitabine as single-agent
therapy in the management of advanced breast cancer. Oncology 15 (2
Suppl 3). S11–S14. 2001.
|
|
87
|
Wang E, Gulbis A, Hart JW and Nieto Y: The
emerging role of gemcitabine in conditioning regimens for
hematopoietic stem cell transplantation. Biol Blood Marrow
Transplant. 20:1382–1389. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hu XC, Zhang J, Xu BH, Cai L, Ragaz J,
Wang ZH, Wang BY, Teng YE, Tong ZS, Pan YY, et al: Cisplatin plus
gemcitabine versus paclitaxel plus gemcitabine as first-line
therapy for metastatic triple-negative breast cancer (CBCSG006): A
randomised, open-label, multicentre, phase 3 trial. Lancet Oncol.
16:436–446. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Telli ML, Jensen KC, Vinayak S, Kurian AW,
Lipson JA, Flaherty PJ, Timms K, Abkevich V, Schackmann EA, Wapnir
IL, et al: Phase II study of gemcitabine, carboplatin, and iniparib
as neoadjuvant therapy for triple-negative and BRCA1/2
mutation-associated breast cancer with assessment of a tumor-based
measure of genomic instability: PrECOG 0105. J Clin Oncol.
33:1895–1901. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Park YH, Im SA, Kim SB, Sohn JH, Lee KS,
Chae YS, Lee KH, Kim JH, Im YH, Kim JY, et al: Phase II,
multicentre, randomised trial of eribulin plus gemcitabine versus
paclitaxel plus gemcitabine as first-line chemotherapy in patients
with HER2-negative metastatic breast cancer. Eur J Cancer.
86:385–393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Niwińska A, Rudnicka H and Murawska M:
Breast cancer leptomeningeal metastasis: The results of combined
treatment and the comparison of methotrexate and liposomal
cytarabine as intra-cerebrospinal fluid chemotherapy. Clin Breast
Cancer. 15:66–72. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Laakmann E, Witzel I and Müller V:
Efficacy of Liposomal Cytarabine in the treatment of leptomeningeal
metastasis of breast cancer. Breast Care. 12:165–167. 2017.
View Article : Google Scholar : PubMed/NCBI
|