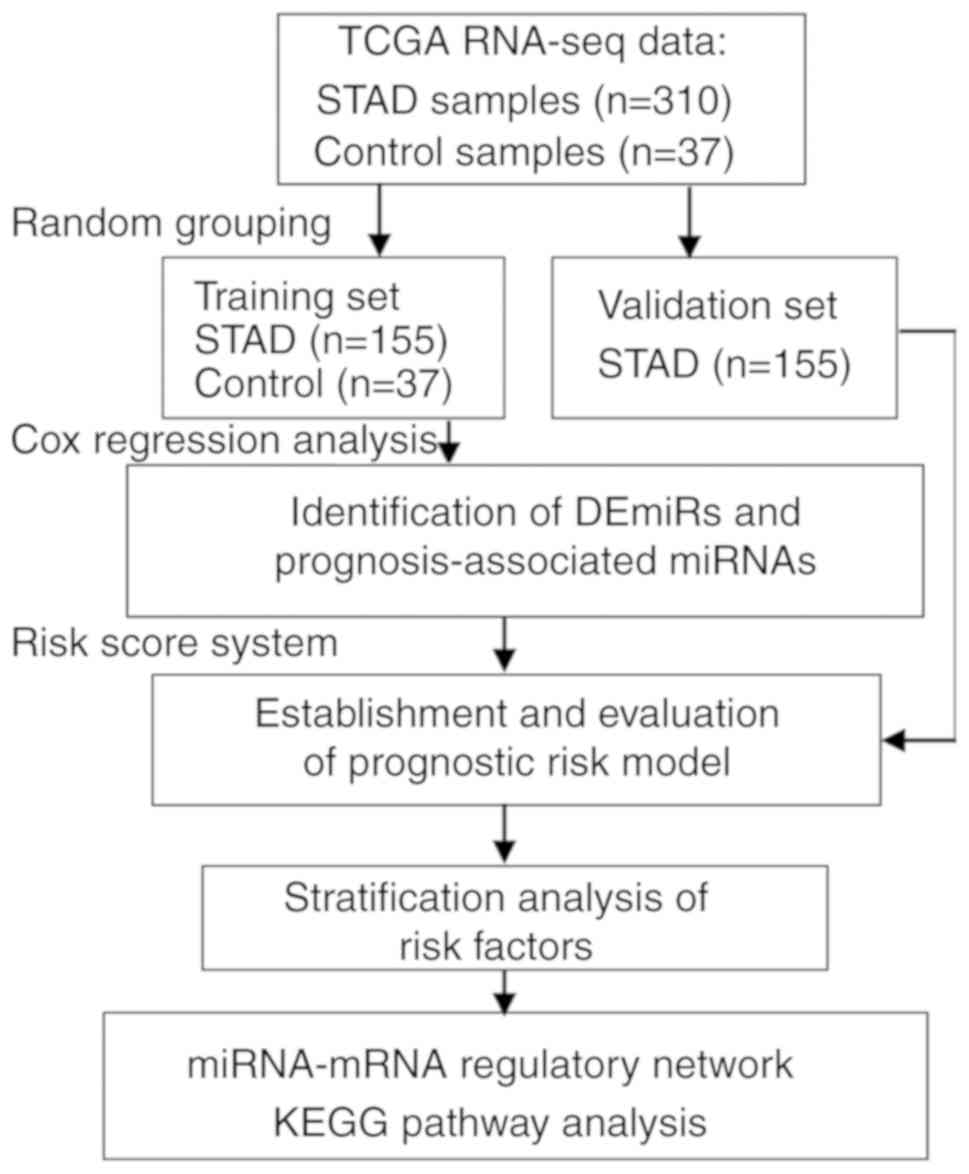
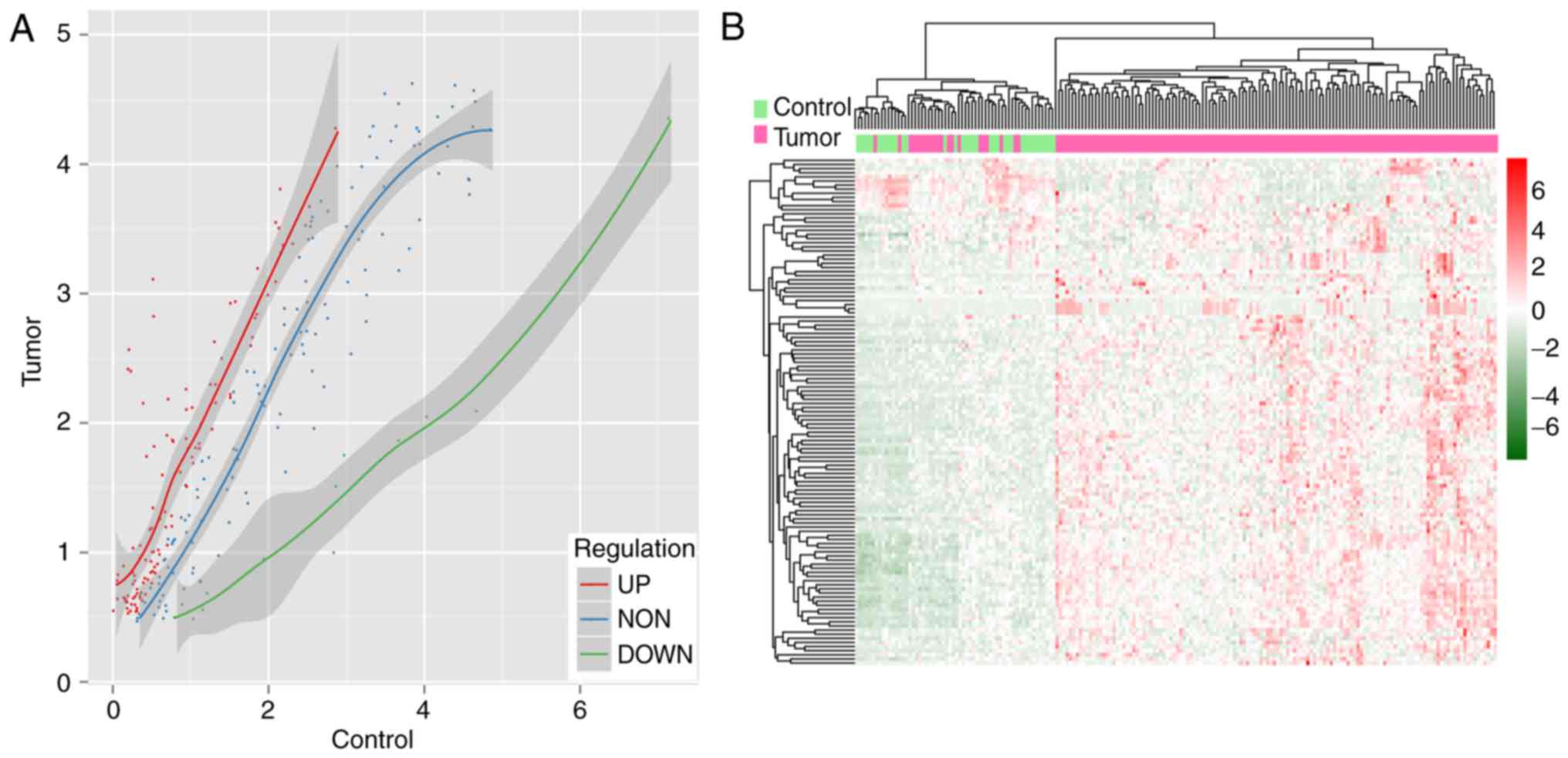
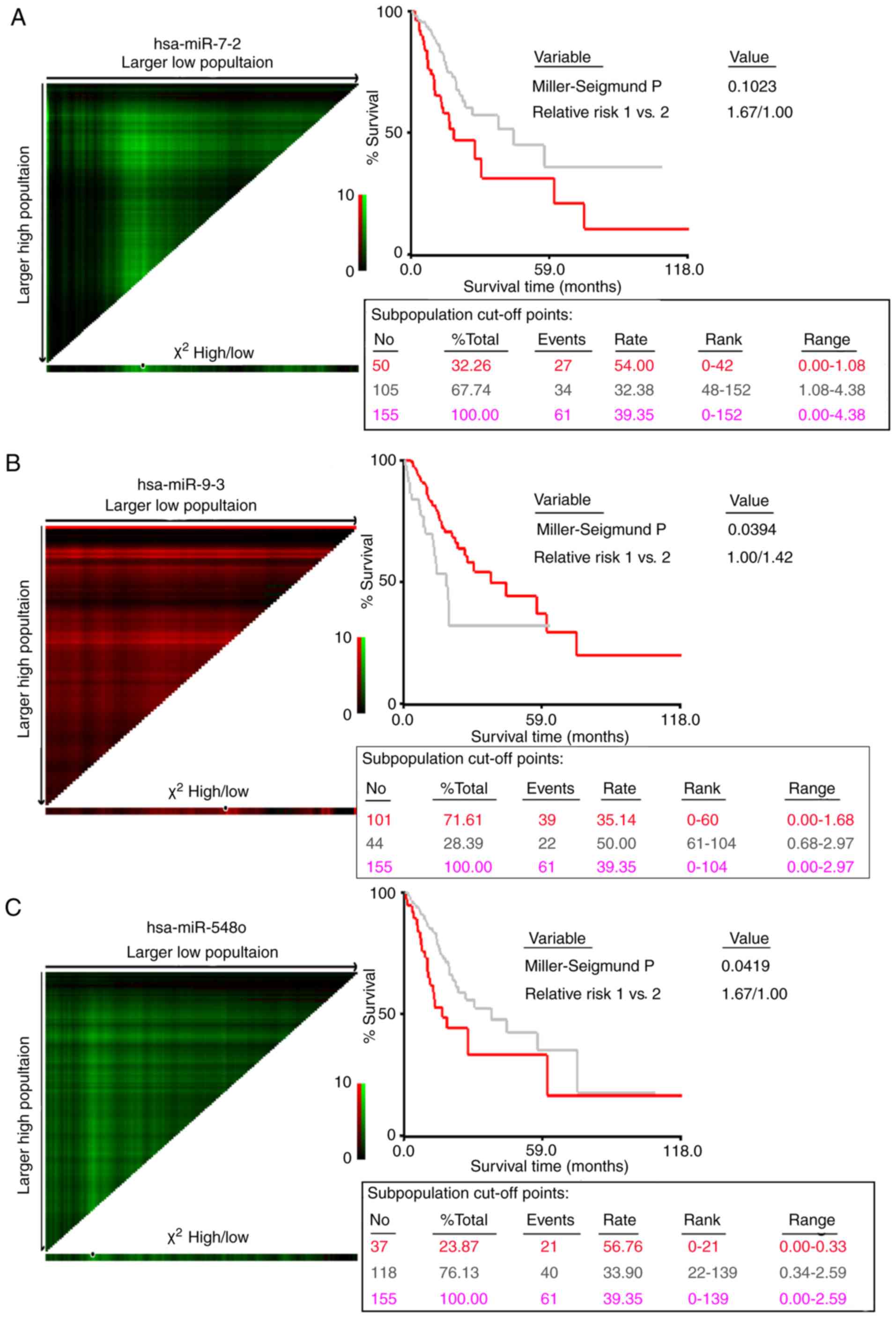
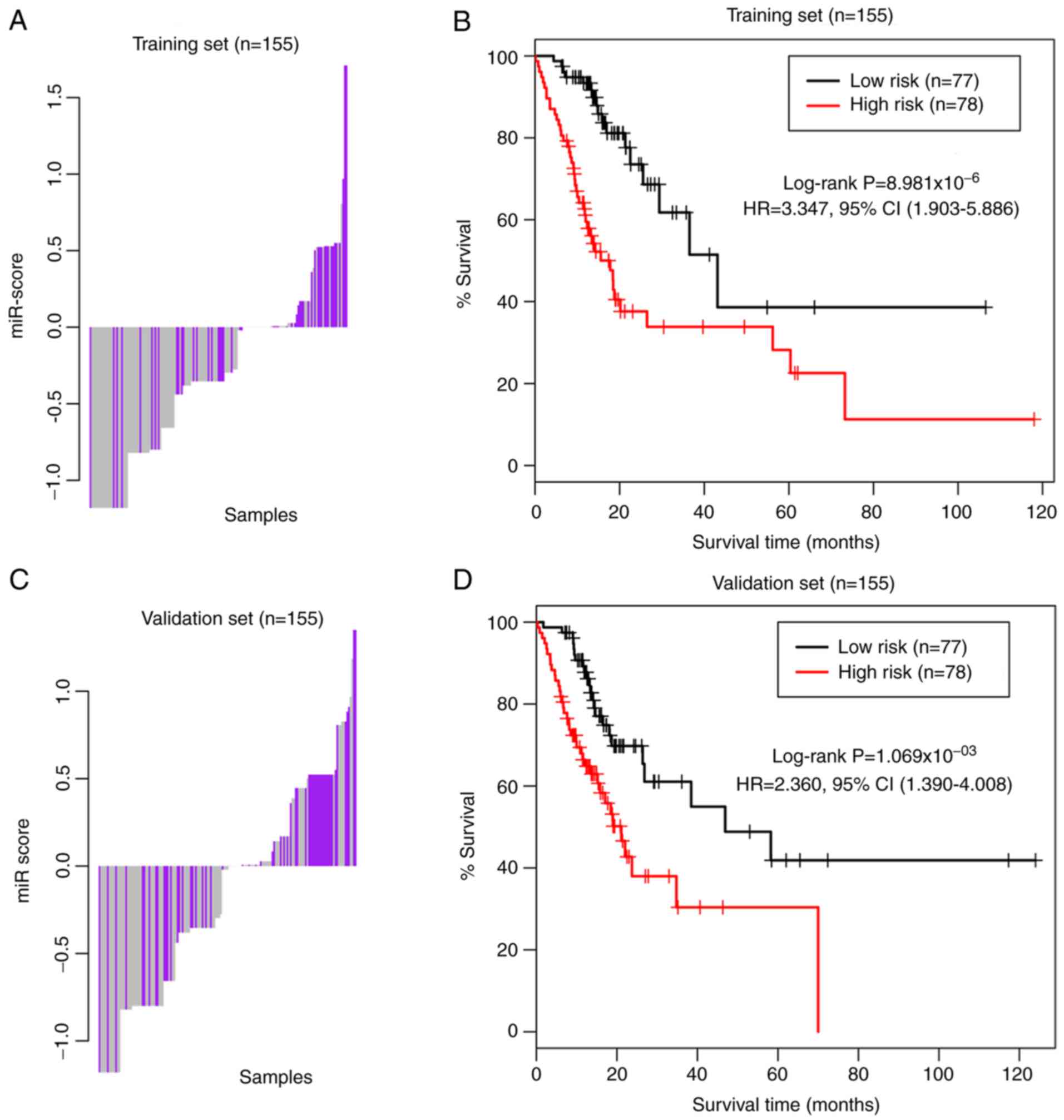
|
1
|
Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang
L, Zhang H, Wang W, Zhu J, Cheng W, et al: Six serum-based miRNAs
as potential diagnostic biomarkers for gastric cancer. Cancer
Epidemiol Biomarkers Prev. 26:188–196. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rugge M, Meggio A, Pravadelli C,
Barbareschi M, Fassan M, Gentilini M, Zorzi M, Pretis G, Graham DY
and Genta RM: Gastritis staging in the endoscopic follow-up for the
secondary prevention of gastric cancer: A 5-year prospective study
of 1755 patients. Gut. 68:11–17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohtsu A, Shah MA, Van Cutsem E, Rha SY,
Sawaki A, Park SR, Lim HY, Yamada Y, Wu J, Langer B, et al:
Bevacizumab in combination with chemotherapy as first-line therapy
in advanced gastric cancer: A randomized, double-blind,
placebo-controlled phase III study. J Clin Oncol. 29:3968–3976.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen S, Bai J, Wei Y, Wang G, Li Q, Zhang
R, Duan W, Yang S, Du M, Zhao Y, et al: A seven-gene prognostic
signature for rapid determination of head and neck squamous cell
carcinoma survival. Oncol Rep. 38:3403–3411. 2017.PubMed/NCBI
|
|
8
|
Meng Z, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
9
|
Parsonnet J, Friedman GD, Vandersteen DP,
Chang Y, Vogelman JH, Orentreich N and Sibley RK: Helicobacter
pylori Infection and the risk of gastric carcinoma. N Engl J
Med. 325:1127–1131. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Neugut AI, Hayek M and Howe G:
Epidemiology of gastric cancer. World J Gastroenterol.
23:2812006.
|
|
11
|
Kwon YH, Heo J, Lee HS, Cho CM and Jeon
SW: Failure of Helicobacter pylori eradication and age are
independent risk factors for recurrent neoplasia after endoscopic
resection of early gastric cancer in 283 patients. Aliment
Pharmacol Ther. 39:609–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alexandrov LB, Nik-Zainal S, Siu HC, Leung
SY and Stratton MR: A mutational signature in gastric cancer
suggests therapeutic strategies. Nat Commun. 6:86832015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu C, Ren C, Han J, Ding Y, Du J, Dai N,
Dai J, Ma H, Hu Z, Shen H, et al: A five-microRNA panel in plasma
was identified as potential biomarker for early detection of
gastric cancer. Br J Cancer. 110:2291–2299. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ellmark P, Ingvarsson J, Carlsson A,
Lundin BS, Wingren C and Borrebaeck CA: Identification of protein
expression signatures associated with Helicobacter pylori
infection and gastric adenocarcinoma using recombinant antibody
microarrays. Mol Cell Proteomics. 5:1638–1646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu X, Tian X, Yu C, Shen C, Yan T, Hong
J, Wang Z, Fang JY and Chen H: A long non-coding RNA signature to
improve prognosis prediction of gastric cancer. Mol Cancer.
15:602016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shin VY, Ng EK, Chan VW, Kwong A and Chu
KM: A three-miRNA signature as promising non-invasive diagnostic
marker for gastric cancer. Mol Cancer. 14:2022015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y
and Sun H: MicroRNA-25 promotes gastric cancer proliferation,
invasion, and migration by directly targeting F-box and WD-40
Domain Protein 7, FBXW7. Tumor Biol. 36:7831–7840. 2015. View Article : Google Scholar
|
|
18
|
Ren C, Wang W, Han C, Chen H, Fu D, Luo Y,
Yao H, Wang D, Ma L, Zhou L, et al: Expression and prognostic value
of miR-92a in patients with gastric cancer. Tumor Biol.
37:9483–9491. 2016. View Article : Google Scholar
|
|
19
|
Zhang Y, Wang B, Jin W and Hu C: Reduction
of miR-132-3p contributes to gastric cancer proliferation mainly by
targeting MUC13. Int J Clin Experimental Med. 9:8023–8030.
2016.
|
|
20
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Oncogene. 33:783–793. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gorur A, Balci Fidanci S, Dogruer Unal N,
Ayaz L, Akbayir S, Yildirim Yaroglu H, Dirlik M, Serin MS and Tamer
L: Determination of plasma microRNA for early detection of gastric
cancer. Mol Biol Rep. 40:2091–2096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bandres E, Bitarte N, Arias F, Agorreta J,
Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ,
et al: microRNA-451 regulates macrophage migration inhibitory
factor production and proliferation of gastrointestinal cancer
cells. Clin Cancer Res. 15:2281–2290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen H, Ren C, Han C, Wang D, Chen Y and
Fu D: Expression and prognostic value of miR-486-5p in patients
with gastric adenocarcinoma. PLoS One. 10:e01193842015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Robinson MD, Mccarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H, et al: RNA-seq analyses of multiple
meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang P, Wang Y, Hang B, Zou X and Mao JH:
A novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
28
|
Camp RL, Dolledfilhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Wen Y, Zhang J, Sun W, Lui VW, Wei
Y, Chen F and Wen W: Prediction of radiotherapy response with a
5-microRNA signature-based nomogram in head and neck squamous cell
carcinoma. Cancer Med. 7:726–735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang K, Sun H, Li X, Hu T, Yang R, Wang
S, Jia Y, Chen Z, Tang F, Shen J, et al: Prognostic risk model
development and prospective validation among patients with cervical
cancer stage IB2 to IIB submitted to neoadjuvant chemotherapy. Sci
Rep. 6:275682016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 42015.doi: 10.7554/eLife.05005.
|
|
32
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J, et al: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Spentzos D, Levine DA, Ramoni MF, Joseph
M, Gu X, Boyd J, Libermann TA and Cannistra SA: Gene expression
signature with independent prognostic significance in epithelial
ovarian cancer. J Clin Oncol. 22:4700–4710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ong CW, Maxwell P, Alvi MA, McQuaid S,
Waugh D, Mills I and Salto-Tellez M: A gene signature associated
with PTEN activation defines good prognosis intermediate risk
prostate cancer cases. J Pathol Clin Res. 4:103–113. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weiler S, Wolf T, Pinna F, Roessler S,
Lutz T, Wan S, Marquardt J, Lang H, Schirmacher P and Breuhahn K:
Abstract 4269: A gene signature defines chromosomal instability
(CIN) and poor survival in liver cancer patients. Cancer Res.
75:4269. 2015. View Article : Google Scholar
|
|
38
|
Daniyal M, Ahmad S, Ahmad M, Asif HM,
Akram M, Ur Rehman S and Sultana S: Risk factors and epidemiology
of gastric cancer in Pakistan. Asian Pac J Cancer Prev.
16:4821–4824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ong HS and Smithers BM: Epidemiology of
gastric cancer. Cancer Rev Asia Pacific. 2:1–7. 2012. View Article : Google Scholar
|
|
40
|
Uemura N, Mukai T, Okamoto S, Yamaguchi S,
Mashiba H, Taniyama K, Sasaki N, Haruma K, Sumii K and Kajiyama G:
Effect of Helicobacter pylori eradication on subsequent development
of cancer after endoscopic resection of early gastric cancer.
Cancer Epidemiol Biomarkers Prev. 6:639–642. 1997.PubMed/NCBI
|
|
41
|
Lee YC, Chiang TH, Chou CK, Tu YK, Liao
WC, Wu MS and Graham DY: Association between Helicobacter
pylori eradication and gastric cancer incidence: A systematic
review and Meta-analysis. Gastroenterology. 150:1113–1124.e5. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bae SE, Jung HY, Kang J, Park YS, Baek S,
Jung JH, Choi JY, Kim MY, Ahn JY, Choi KS, et al: Effect of
Helicobacter pylori eradication on metachronous recurrence
after endoscopic resection of gastric neoplasm. Am J Gastroenterol.
109:60–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Viganò L, Capussotti L, Lapointe R,
Barroso E, Hubert C, Giuliante F, Ijzermans JN, Mirza DF, Elias D
and Adam R: Early recurrence after liver resection for colorectal
metastases: Risk factors, prognosis, and treatment. A
LiverMetSurvey-based study of 6,025 patients. Ann Surg Oncol.
21:1276–1286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Van GM, Verslegers I, Biltjes I and
Parizel PM: MR is/is not a useful diagnostic tool for breast cancer
management. Acta Chir Belg. 107:267–270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shan J, Zhang S, Wang Z, Fu Y, Li L and
Wang X: Breast malignant phyllodes tumor with rare pelvic
metastases and long-term overall survival: A case report and
literature review. Medicine. 95:e49422016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hohaus K, Kostler EJ, Klemm E and Wollina
U: Merkel cell carcinoma-a retrospective analysis of 17 cases. J
Eur Acad Dermatol Venereol. 17:20–24. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang
C, Huang D, Chen X, Zhang H, Zhuang R, et al: A five-microRNA
signature identified from genome-wide serum microRNA expression
profiling serves as a fingerprint for gastric cancer diagnosis. Eur
J Cancer. 47:784–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bandres E, Agirre X, Bitarte N, Ramirez N,
Zarate R, Roman-Gomez J, Prosper F and Garcia-Foncillas J:
Epigenetic regulation of microRNA expression in colorectal cancer.
Int J Cancer. 125:2737–2743. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Roth P, Wischhusen J, Happold C, Chandran
PA, Hofer S, Eisele G, Weller M and Keller A: A specific miRNA
signature in the peripheral blood of glioblastoma patients. J
Neurochem. 118:449–457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang X, Jhiang S, Fernandez S and Coombes
K: miR-551b and SEMA3D as potential therapeutic targets in
papillary thyroid cancer. Translational Data Analytics @ Ohio
State. November 14–2016http://hdl.handle.net/1811/79454
|
|
52
|
Fittipaldi S, Vasuri F, Bonora S,
Degiovanni A, Santandrea G, Cucchetti A, Gramantieri L, Bolondi L
and D'Errico A: miRNA signature of hepatocellular carcinoma
vascularization: How the controls can influence the signature. Dig
Dis Sci. 62:2397–2407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fussek S, Rönnau C, Span PN, Burchardt M,
Verhaegh GW and Schalken JA: 246 The castration-resistant prostate
cancer-associated miRNAs, miR-3687 and miR-4417, are involved in
tumour cell hypoxia response and tumour cell migration. Eur Urol
Suppl. 15:e246. 2016. View Article : Google Scholar
|
|
54
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gong J, Tian J, Lou J, Wang X, Ke J, Li J,
Yang Y, Gong Y, Zhu Y, Zou D, et al: A polymorphic MYC response
element in KBTBD11 influences colorectal cancer risk, especially in
interaction with a MYC regulated SNP rs6983267. Ann Oncol.
29:632–639. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dang CV, Resar LM, Emison E, Kim S, Li Q,
Prescott JE, Wonsey D and Zeller K: Function of the c-Myc oncogenic
transcription factor. Exp Cell Res. 253:63–77. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bergamaschi A, Kim YH, Kwei KA, La Choi Y,
Bocanegra M, Langerød A, Han W, Noh DY, Huntsman DG, Jeffrey SS, et
al: CAMK1D amplification implicated in epithelial-mesenchymal
transition in basal-like breast cancer. Mol Oncol. 2:327–339. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Marimuthu A: Identification and validation
of differentially expressed genes in gastric Adenocarcinoma.
Manipal University. 2012, http://shodhganga.inflibnet.ac.in/handle/10603/4994
|
|
59
|
Nakamura Y, Migita T, Hosoda F, Okada N,
Gotoh M, Arai Y, Fukushima M, Ohki M, Miyata S, Takeuchi K, et al:
Krüppel-like factor 12 plays a significant role in poorly
differentiated gastric cancer progression. Int J Cancer.
125:1859–1867. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen Q, Chen X, Zhang M, Fan Q, Luo S and
Cao X: miR-137 is frequently down-regulated in gastric cancer and
is a negative regulator of Cdc42. Dig Dis Sci. 56:2009–2016. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Du Y, Chen Y, Wang F and Gu L: miR-137
plays tumor suppressor roles in gastric cancer cell lines by
targeting KLF12 and MYO1C. Tumour Biol. 37:13557–13569. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mak CS, Yung MM, Hui LM, Leung LL, Liang
R, Chen K, Liu SS, Qin Y, Leung TH, Lee KF, et al: MicroRNA-141
enhances anoikis resistance in metastatic progression of ovarian
cancer through targeting KLF12/Sp1/survivin axis. Mol Cancer.
16:112017. View Article : Google Scholar : PubMed/NCBI
|