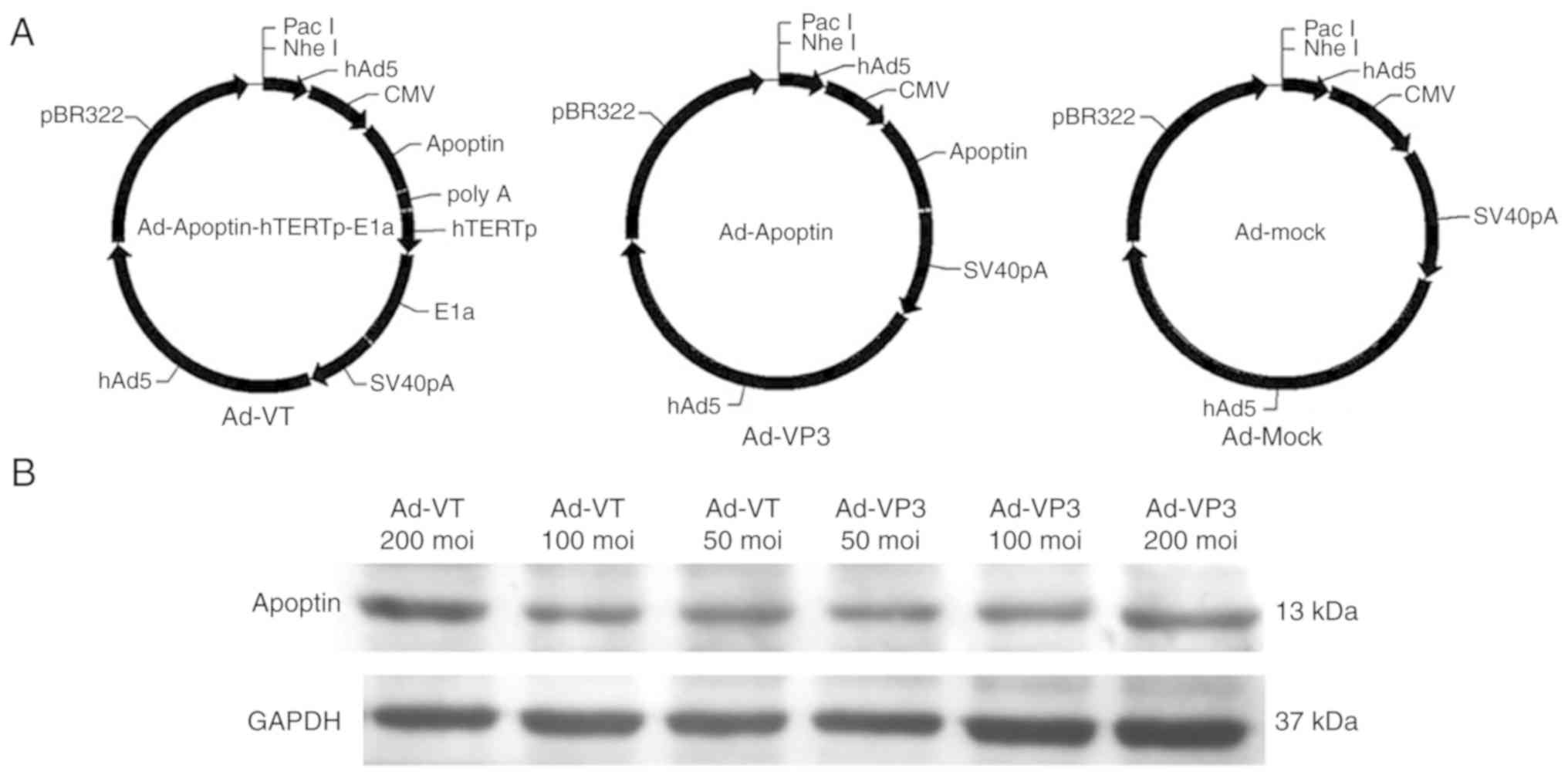
Introduction
Apoptin was originally identified as an
apoptosis-inducing protein derived from chicken anemia virus (CAV),
which is a single-stranded DNA virus of the Gyrovirus genus
(1). The CAV genome contains three
partially overlapping open reading frames encoding viral proteins
from a single polycistronic mRNA: VP1 (capsid protein), VP2
(protein phosphatase, scaffold protein) and the death-inducing
protein VP3 (2). The expression of
VP3 alone has been reported to be sufficient to trigger cell death
in chicken lymphoblastoid T cells and myeloid cells, but not in
chicken fibroblasts; therefore, this protein has been renamed
apoptin (3). The gene encoding
apoptin was among the first tumor-selective anticancer genes to be
isolated, and has become a focus of cancer research due to its
ability to induce apoptosis of various human tumor cells, including
melanoma, lymphoma, colon carcinoma and lung cancer, while leaving
normal cells relatively unharmed (4–7). It
may be hypothesized that apoptin senses an early event in oncogenic
transformation and induces cancer-specific apoptosis, regardless of
tumor type; therefore, it represents a potential future anticancer
therapeutic agent.
The length and viability of human telomerase reverse
transcriptase (hTERT) are associated with cell senescence and
immortalization. Telomerase is a ribonucleoprotein that can process
telomere repeats (TTAGGG) at the ends of chromosomes (8). Telomerase activity is regulated by the
signal transduction system and the apoptotic pathway, and its
activity is a marker of immature cell differentiation and
immortalization. The hTERT promoter is inactive in most normal
cells; however, it exhibits high activity in several types of human
cancer (9). Previous studies
revealed that targeting to tumor cells and efficient expression of
the protein of interest is also dependent on the high efficiency
and specificity of the hTERT promoter, thus providing novel
prospects for tumor therapy (10,11).
In our previous study, using the characteristics of apoptin and the
hTERT promoter, a tumor-specific replication recombinant adenovirus
expressing apoptin (Ad-Apoptin-hTERTp-E1a; Ad-VT) was constructed
(12), which allows the adenovirus
to specifically replicate in tumor cells, and enables the apoptin
protein to be expressed in a large amount in tumor cells, thereby
playing an effective role in tumor cell death. Our previous studies
have demonstrated the marked tumor-killing effect of the
recombinant adenovirus on various tumor cells (13–16).
Autophagy, which is described as ‘self-eating’,
constitutes a self-degradation process, and is a critical mechanism
underlying the cytoprotection of eukaryotic cells (17). It is a catabolically driven process
whereby stressed cells form cytoplasmic, double-layered,
crescent-shaped membranes, known as phagophores, which mature into
complete autophagosomes. The autophagosomes engulf long-lived
proteins and damaged cytoplasmic organelles, in order to provide
cellular energy and building blocks for biosynthesis (18). However, in the context of cancer,
autophagy appears to serve an ambiguous role. In association with
apoptosis, autophagy can act as a tumor suppressor. Conversely,
defects in autophagy, alongside abnormal apoptosis, may trigger
tumorigenesis and therapeutic resistance (19,20).
The role of autophagy as an alternative cell death
mechanism remains a controversial issue. It was previously reported
that dying cells exhibit autophagic vacuolization (21), which led to the suggestion that cell
death is mediated by autophagy. However, to the best of our
knowledge, there is no concrete evidence that autophagy is a direct
mechanism used to execute cell death. Numerous studies have
suggested that autophagy may lead to apoptosis or necroptosis as a
result of a failure to adapt to starvation (22–24).
Therefore, autophagy may constitute an adaptive response to
counteract cell death under lethal stress conditions, rather than a
cell death mechanism (21,25). The autophagic response is activated
in response to ATP depletion to restore the metabolic state and to
prevent necroptosis (26–28). In addition, autophagy ensures cell
metabolism, promotes tumor cell survival, and prevents cancer cells
from accumulating dysfunctions (29). Therefore, inhibition of autophagy
could induce a metabolic crisis that leads to the induction of
necroptosis (30,31).
The recombinant adenovirus Ad-VT can specifically
kill tumor cells; however, it remains to be determined as to
whether Ad-VT affects another form of programmed cell death from
apoptosis, such as autophagy. In addition, if it does affect
autophagy, it is unclear how. In the present study, MCF-7 breast
cancer cells and MCF-10A normal breast epithelial cells were used.
The levels of autophagy were detected using monodansylcadaverine
(MDC). Western blotting and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) were used to assess the
expressions levels of autophagy-associated proteins, including
Beclin-1, microtubule-associated protein 1A/1B-light chain 3 (LC3),
autophagy-related 4B cysteine peptidase (ATG4b), autophagy-related
5 (ATG5), P62 and BIF-1. Cell morphology was observed by
transmission electron microscopy (TEM) following infection with
recombinant adenoviruses expressing apoptin. A proteomics
experiment was also conducted to observe alterations in the levels
of autophagy pathway proteins. The results revealed that the
recombinant adenoviruses expressing apoptin altered autophagy in
the MCF-7 breast cancer cell line, but had no effect on MCF-10A
normal mammary epithelial cells. These findings suggested that
autophagy in MCF-7 cells may exert protection against the cell
death-inducing effects of apoptin.
Materials and methods
Cells and viruses
The MCF-7 human breast cancer cells and MCF-10A
human normal mammary epithelial cells were cryopreserved cells
purchased from the Shanghai Institute of Biology Cell Bank
(Shanghai, China). MCF-7 cells were maintained in RPMI 1640 medium
supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin
and 50 U/ml streptomycin at 37°C in an atmosphere containing 5%
CO2. MCF-10A cells were maintained in Dulbecco's
modified Eagle's medium supplemented with 10% FBS, 50 U/ml
penicillin and 50 U/ml streptomycin at 37°C in an atmosphere
containing 5% CO2. All reagents for cell culture were
purchased from HyClone; GE Healthcare Life Sciences (Logan, UT,
USA).
Recombinant adenoviruses Ad-VT, Ad-Apoptin (Ad-VP3)
and Ad-MOCK were constructed and preserved in our laboratory
(Laboratory of Molecular Virology and Immunology, Institute of
Military Veterinary Medicine, Academy of Military Medical Science,
Changchun, China) (Fig. 1)
(12).
Cell viability assay
MCF-7 and MCF-10A cells were seeded in 96-well
plates (5×103 cells/well). After 24 h of culture, the
cells were infected with various concentrations of the recombinant
adenoviruses [50, 100 and 200 multiplicity of infection (MOI)], and
blank control wells were set. After 6, 12, 24, and 48 h, 10 µl
WST-1 solution (cell proliferation assay reagent; cat. no.
11644807001; Roche Applied Science, Mannheim, Germany) was mixed
with 100 µl cell culture medium and was added to each well; cells
were then incubated at 37°C in an atmosphere containing 5%
CO2 for 1 h and 20 min. Subsequently, absorbance was
measured at 450 nm. Cell viability was calculated as follows: Cell
viability = 100 × (absorbance of virus-treated wells/absorbance of
control wells).
Measurement of apoptosis by flow
cytometry
MCF-7 and MCF-10A cells were infected with the
recombinant adenoviruses (100 MOI). After 24 h, the cells
(2×105) were harvested, resuspended in binding buffer
and stained with fluorescein isothiocyanate (FITC)-labeled Annexin
V (Annexin V-FITC Apoptosis Detection kit; BioVision, Inc.,
Milpitas, CA, USA), according to the manufacturer's protocol. To
exclude late apoptotic and necrotic cells, propidium iodide (PI; 50
µg/ml) was added to the FITC-Annexin V-stained samples and
incubated at room temperature for 20 min. The samples were then
examined by flow cytometry (FACSCalibur; BD Biosciences, Franklin
Lakes, NJ, USA) for apoptosis analysis (Cell Quest Pro 5.2.1; BD
Biosciences).
3-Methyladenine (3-MA; cat. no. M9281;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was dissolved in PBS
and stored at −20°C. Prior to use, 3-MA solution was heated to 56°C
to completely dissolve it, and was then added to MCF-7 and MCF-10A
cell culture media at a final concentration of 50 mmol/l and
incubated for 2 h. Following 3-MA treatment, MCF-7 cells underwent
flow cytometry and western blotting, whereas MCF-10A cells
underwent flow cytometry.
Cell cycle analysis
Cells were infected with recombinant adenoviruses
(100 MOI) for 24 h. After washing with PBS, the cells were
harvested and fixed overnight in 70–80% ethanol. The next day,
cells were washed with 1X PBS and were centrifuged at 175 × g for
10 min at room temperature, after which, the supernatant was
discarded. Cells were then mixed with 0.09% NaN3 Stain Buffer (2%
FBS), washed and centrifuged at 175 × g for 10 min at room
temperature, and the supernatant was discarded. Finally, 500 µl
PI/RNase (PI/RNase Staining Buffer; cat. no. 550825; BD Pharmingen;
BD Biosciences) staining solution was used to stain the cells for 1
h at room temperature. Flow cytometry was performed using a
FACSCalibur instrument and Cell Quest Pro 5.2.1 software.
MDC detection
Cells (2×105 MCF-7 cells) were placed in
each well of 6-well plates for 24 h. Following infection with
recombinant adenoviruses (100 MOI) for 6 h, autophagic vacuoles
were labeled with 0.05 mmol/l MDC (cat. no. 30432; Sigma-Aldrich;
Merck KGaA) in cell culture medium at 37°C for 15 min. The cells
were then washed three times with PBS. Autophagic vacuoles in MCF-7
cells were observed under a fluorescence microscope (BX-60; Olympus
Corporation, Tokyo, Japan) and analyzed using MetaMorph 6.2.6
software (Molecular Devices, LLC, Sunnyvale, CA, USA). Notably,
>5 fluorescent spots were considered to indicate positive
fluorescence, and 100 cells were counted to calculate the rate of
autophagy.
Enhanced green fluorescent protein-LC3
plasmid (p-EGFP- LC3) transfection
MCF-7 cells (2×105/well) in 12-well
plates with microscope cover glasses (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) were transfected with 5 µg pEGFP-LC3 (2.92
µg/µl; cat. no. 24920; Addgene, Inc., Cambridge, MA, USA) using 5
µl X-tremeGENE™ HP DNA Transfection Reagent (cat. no. 6366236001;
Roche Applied Science) for 24 h. Subsequently, cells were fixed
with 4% paraformaldehyde in PBS and cellular images were obtained
under a fluorescence microscope (BX-60; Olympus Corporation) using
MetaMorph 6.2.6 software.
TEM
TEM is considered the most reliable approach to
monitor autophagy (32). MCF-7
cells were cultured in 6-well plates for 24 h and infected with
recombinant adenoviruses (100 MOI) for 6 h. Cells were then
collected and cells were fixed in 2.5% glutaraldehyde at 4°C for 8
h. Ultrathin sections (50 nm) were cut using an ultramicrotome,
stained with 2% (w/v) uranyl acetate for 15 min and with lead
citrate for 10 min at room temperature, and examined under a
JEM-100cxII electron microscope (JEM-1010; JEOL, Ltd., Tokyo,
Japan).
RNA isolation and RT-qPCR
analysis
RNA was isolated from cells using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed
using RT reagents (M-MLV Reverse Transcriptase; cat. no. M1705;
Promega Corporation, Madison, WI, USA), as follows: 42°C for 50 min
followed by 75°C for 15 min. The mRNA expression levels of human
LC3, Beclin-1, GAPDH and P62 were measured by qPCR using the GoTaq
qPCR Master Mix (cat. no. A6002; Promega Corporation) on a StepOne
Real-Time PCR System (7500 Real-Time PCR system; Applied
Biosystems; Thermo Fisher Scientific, Inc.). All protocols were
performed according to the manufacturers' protocols. The PCR
reaction conditions were as follows: 95°C for 5 min followed by 40
cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec, and
a final extension step at 72°C for 10 min. The primers used in the
present study were as follows (5′-3′): Beclin-1, forward (F)
CCGTGGTAACCTTGTTCATCC, reverse (R) GCTCTGTCTTCAGCGACTTCC; LC3-II,
FAATAGAAGGCGCTTACAG, RGACAATTTCATTCCCGAAC; P62,
FGGACAAACGGCTCACTCT, and RTGCCAGTTCCCTCATTCT; and GAPDH,
FAACGGATTTGGTCGTATTG and RGGAAGATGGTGATGGGATT. The
2−ΔΔCq method was used to quantify the PCR results
(33).
Cell extracts and western
blotting
Certain autophagy-associated proteins were detected
by immunoblotting. Briefly, whole cell lysates were prepared from
treated MCF-7 cells at the indicated time points using 1X
radioimmunoprecipitation assay (RIPA) buffer (cat. no. P0013;
Beyotime Institute of Biotechnology, Haimen, China), according to
the manufacturer's protocol. In addition, 10 mM
phenylmethanesulfonyl fluoride (PMSF; cat. no. ST506-2; Beyotime
Institute of Biotechnology) was generated and stored at −20°C.
Prior to cell lysis, PMSF was added RIPA buffer to a final
concentration of 1 mM PMSF. Four-fold volume of RIPA cleaved cells.
Cleaved cells were sonicated at 4°C (15 cycles, 30 sec/cycle, 60
Hz) using a Bioruptor UCD-300 device (Diagenode SA, Seraing,
Belgium). The protein concentration was measured using the
bicinchoninic acid protein assay (cat. no. P0010s; Beyotime
Institute of Biotechnology). Proteins were loaded into each lane
and separated by 10% SDS-PAGE, and were then electrotransferred
onto nitrocellulose (NC) membranes (0.45 µm NC; cat. no. 10600002;
Amersham; GE Healthcare, Chicago, IL, USA). The membranes were
blocked with 5% non-fat dried milk in PBS supplemented with 0.1%
Tween 20 (PBST; cat. no. 170653; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) at room temperature for 2 h, and were then
incubated overnight with the corresponding primary antibodies
(1:1,000) at 4°C overnight. The blots were then incubated with a
secondary antibody (1:2,000) for 2 h at room temperature after
three washes with PBST. The membranes were further washed three
times with PBST before being developed using Clarity Western ECL
Blotting Substrate (cat. no. 32209; Pierce; Thermo Fisher
Scientific, Inc.). Images were collected following development of
photographic paper (X-OMAT BT Film; cat. no. 045602811; Carestream
Health, Inc., Rochester, NY, USA).
Antibodies for western blotting
The following primary antibodies were used: Rabbit
anti-GAPDH (cat. no. 5174), rabbit anti-B-cell lymphoma 2 (Bcl-2;
cat. no. 3498), rabbit anti-Bcl-2-associated X protein (Bax; cat.
no. 5023), rabbit anti-LC3 (cat. no. 12741), rabbit anti-Beclin-1
(cat. no. 3495), rabbit anti-ATG4b (cat. no. 13507) rabbit
anti-BIF-1 (cat. no. 4467), rabbit anti-ATG5 (cat. no. 12994), and
rabbit anti-tubulin (cat. no. 2148) (all from Cell Signaling
Technology, Inc., Danvers MA, USA), and rabbit anti-sqstm1/P62
(cat. no. p0067; Sigma-Aldrich; Merck KGaA). The following
secondary antibody was used: Peroxidase-conjugated goat anti-rabbit
immunoglobulin G (H+L) (cat. no. ZB2301; OriGene Technologies,
Inc., Beijing, China).
Total (phospho) proteome workflow
Approximately 2×104 adenovirus-infected
MCF-7 cells were rinsed with PBS and lysed with RIPA buffer. Cells
were collected in a 1.5 ml centrifuge tube and were centrifuged at
4°C and 1,950 × g for 5 min. The supernatant was discarded and
4-fold volume RIPA [8 M urea + phosphatase inhibitor (10X)] was
added to the sample, according to the volume of the precipitate.
Cells were then sonicated on ice (20 kHz; ultrasound for 1 sec,
stop for 2 sec; total, 60 sec), maintained for 20 min at 4°C and
centrifuged at 16,000 × g for 5 min. Proteins in the supernatant
were assessed using the Bradford reagent. After protein
quantification, 1 mg protein solution was treated with
dithiothreitol (10 mM; cat. no. 0281; Amresco, LLC, Solon, OH, USA)
in a water bath was 30 min. Subsequently, iodoacetamide (20 mM;
cat. no. I1149; Sigma-Aldrich; Merck KGaA) was added at room
temperature was 30 min and alkylation was carried out. After
alkylation, the protein solution was centrifuged for 15 min at
14,000 × g, and the waste liquid was discarded. The protein sample
was washed with 8 M urea, centrifuged at 14,000 × g for 15 min and
the waste liquid was discarded; this step was repeated twice. For
enzymatic hydrolysis, 50 mM sodium bicarbonate and 1.6 µg trypsin
(mass ratio 1/50) were added, the sample was agitated and digestion
was conducted at 37°C for 16 h. For peptide segment collection, the
sample was centrifuged for 10 min at 14,000 × g and the peptide
solution was collected; subsequently, 200 µl high-performance
liquid chromatography-H2O was added to the solution,
which was centrifuged for a further 10 min at 14,000 × g and this
second peptide solution was collected. Finally, the collected
peptide segment solutions were combined, freeze-dried and stored at
−80°C.
(Phospho) proteomic data mining
The choice of database was based on the required
species, and the completeness and reliability of the database
annotation. In the present study, the Uniprot database (www.uniprot.org) was used. MaxQuant 1.5.8.3 software
(Max-Planck-Institute of Biochemistry, Planegg, Germany) was used
to retrieve and quality control the tandem mass spectrum, remove
contaminating proteins and conduct reverse decoy database analysis,
normalize data and provide log2 values. The numerical values were
inputted directly into The R Project for Statistical Computing
(version 3.5.1; www.r-project.org/), and the pheatmap function was
selected for classification analysis, resulting in the generation
of a thermal graph and hierarchical clustering. The pathways
associated with the differential proteins were determined using
Kyoto Encyclopedia of Genes and Genomes (KEGG; www.kegg.jp/kegg) pathway analysis.
Statistical analysis
Statistical analysis was conducted using data from
at least three independent experiments. SPSS 20.0 (SPSS, Inc.,
Chicago, IL, USA) or SigmaStat 3.5 (Systat Software, Inc., San
Jose, CA, USA) were used for statistical analysis. The results were
statistically analyzed by Student's t-test or one-way analysis of
variance (ANOVA); when one-way ANOVA results were P<0.05,
further multiple comparisons were performed using the
Student-Newman-Keuls test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Antiproliferative effect of
recombinant adenoviruses on MCF-7 and MCF-10A cells
The antiproliferative activity of recombinant
adenoviruses on MCF-7 and MCF-10A cells was investigated using the
WST-1 assay, which is a sensitive colorimetric assay to determine
cell viability that measures dehydrogenase activity in living cells
(Fig. 2). Post-infection with
Ad-VT, Ad-VP3 and Ad-MOCK for 6, 12, 24 and 48 h, MCF-7 cell
proliferation was inhibited in a time- and dose-dependent manner
(Fig. 2A and C); however, the
recombinant adenoviruses exhibited no obvious inhibitory effect on
MCF-10A cells (Fig. 2A). In
addition, Annexin V-FITC/PI double staining was conducted to detect
the levels of apoptosis (Fig.
3A-D). The apoptotic rate of MCF-7 tumor cells was
significantly increased in response to the recombinant adenoviruses
Ad-VT and Ad-VP3 over 24 h (Fig. 3A and
C). When the recombinant adenoviruses Ad-VT or Ad-VP3 were
combined with the autophagy inhibitor 3-MA, the apoptotic rate of
tumor cells was increased compared with when cells were infected
with Ad-VT or Ad-VP3 alone; therefore, it was suggested that
inhibition of autophagy may increase the apoptosis of MCF-7 cells
infected with recombinant adenoviruses over 24 h (Fig. 3A and C). In addition, no significant
alterations in apoptosis were detected in MCF-10A normal human
breast cells (Fig. 3B and D). In
the present study, the early stage of apoptosis (24 h) was
selected, which was not very strong; in order to more clearly
discover the effect of autophagy. The results revealed that the
addition of an autophagy inhibitor increased the rate of apoptosis,
and it was hypothesized that apoptin-induced autophagy in the early
stage of apoptosis may inhibit the occurrence of apoptosis.
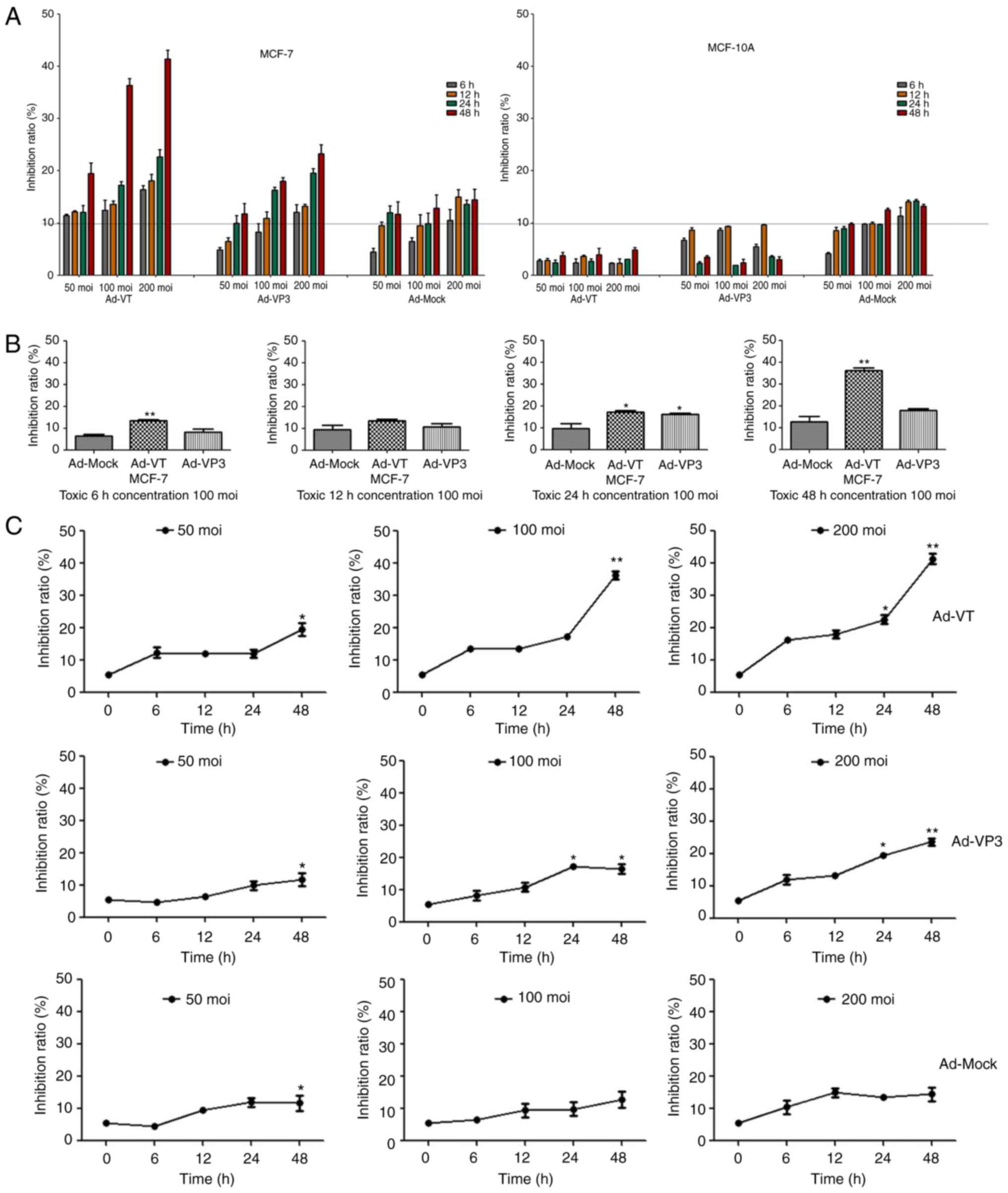 | Figure 2.Recombinant adenoviruses inhibited
the proliferation of MCF-7 cells. (A) MCF-10A and MCF-7 cell
viability was determined using the WST-1 assay post-infection with
various concentrations of Ad-VT, Ad-VP3 and Ad-MOCK (50, 100, and
200 MOI) for 6, 12, 24 and 48 h. (B) MCF-7 cell viability was
determined using WST-1 assay after post-infection with Ad-VT,
Ad-VP3 and Ad-MOCK (100 MOI) for various durations (6, 12, 24 and
48 h). Data are presented as the means ± standard deviation,
representative of three independent experiments (n=3). *P<0.05,
**P<0.01, compared with Ad-MOCK. (C) MCF-7 cell viability was
determined using the WST-1 assay post-infection with various MOIs
of Ad-VT, Ad-VP3 and Ad-MOCK for different durations (6, 12, 24 and
48 h). Data are presented as the means ± standard deviation,
representative of three independent experiments (n=3). *P<0.05,
**P<0.01, compared with the control (0 h). Ad-VP3, Ad-Apoptin;
Ad-VT, Ad-Apoptin-hTERTp-E1a; MOI, multiplicity of infection. |
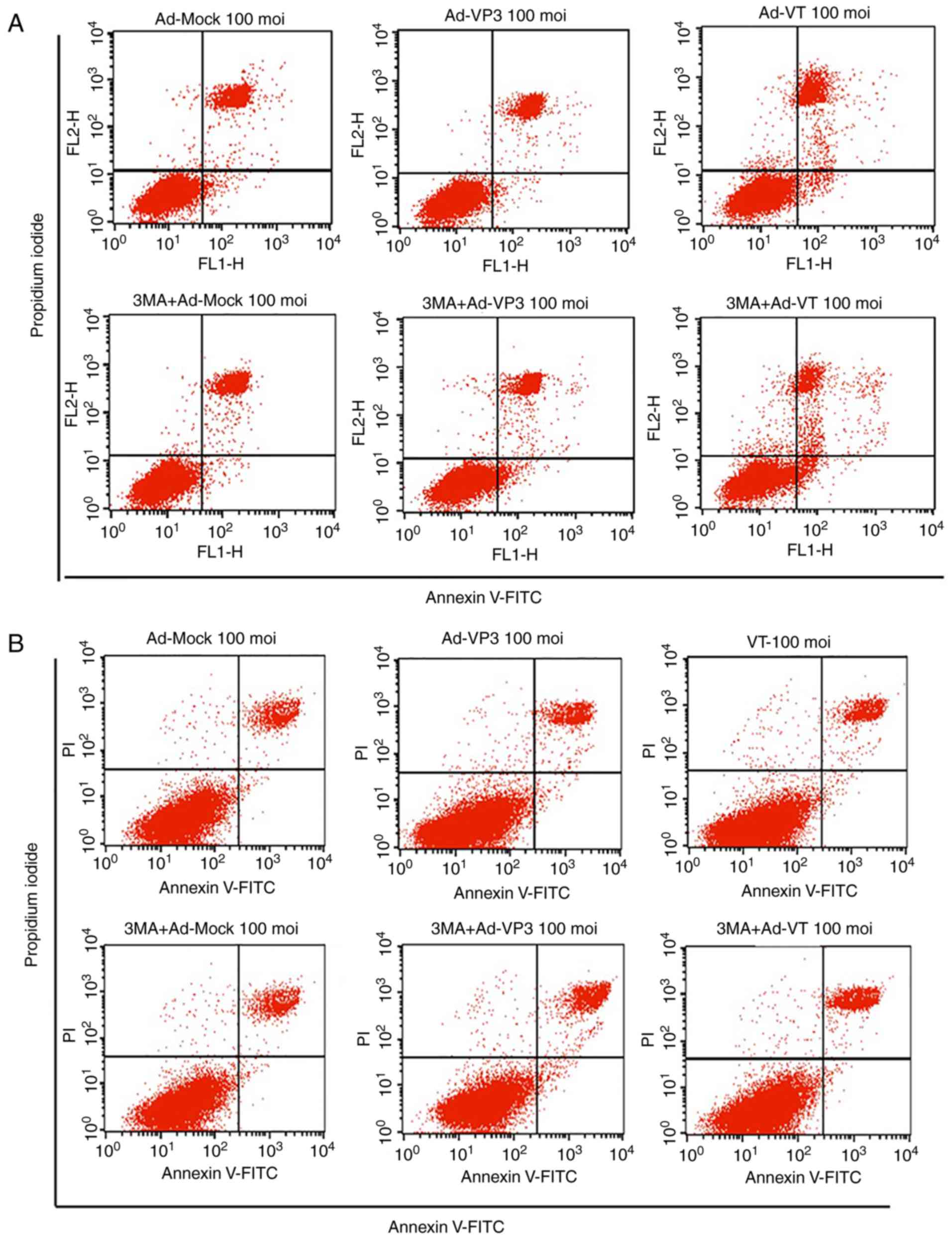 | Figure 3.Analysis of cell death and cell cycle
distribution of MCF-7 cells. (A) Scatter plot of Annexin V-FITC/PI
staining, which was used to evaluate apoptosis of MCF-7 cells
infected with Ad-VT and Ad-VP3 (100 MOI), in the presence or
absence 3-MA for 24 h. (B) Scatter plot of Annexin V-FITC/PI
staining, which was used to evaluate apoptosis of MCF-10A cells
infected with Ad-VT and Ad-VP3 (100 MOI), in the presence or
absence of 3-MA for 24 h. Analysis of cell death and cell cycle
distribution of MCF-7 cells. (C) Apoptotic ratio, using Annexin
V-FITC/PI staining, which was used to evaluate apoptosis of MCF-7
cells infected with Ad-VT and Ad-VP3 (100 MOI), in the presence or
absence 3-MA for 24 h. (D) Apoptotic ratio, using Annexin V-FITC/PI
staining, to evaluate apoptosis of MCF-10A cells infected with
Ad-VT and Ad-VP3 (100 MOI), in the presence or absence of 3-MA for
24 h. (E) Western blotting of MCF-7 cell extracts for Bcl-2 and
Bax. MCF-7 cells were infected with Ad-VT and Ad-VP3 (100 MOI)
alone, or, after 2 h treatment with 3-MA, Ad-VT and Ad-VP3 was
added to the cells for 12 h. (F) Flow cytometric analysis of cell
cycle distribution of MCF-7 cells, illustrating the effects of
infection with Ad-VT, Ad-VP3 and Ad-MOCK (100 MOI) for 24 h. Data
are presented as the means ± standard deviation, representative of
three independent experiments (n=3). *P<0.05, **P<0.01,
compared with Ad-MOCK. Ad-VP3, Ad-Apoptin; Ad-VT,
Ad-Apoptin-hTERTp-E1a; Bax, Bcl-2-associated X protein; Bcl-2,
B-cell lymphoma 2; FITC, fluorescein isothiocyanate; MOI,
multiplicity of infection; PI, propidium iodide. |
Inhibition of autophagy increases the
Bax/Bcl-2 ratio
Following inhibition of autophagy with 3-MA, MCF-7
cells were infected with Ad-VT and Ad-VP3 at 100 MOI for 12 h, and
the expression levels of apoptosis-associated proteins Bcl-2 and
Bax were detected by western blotting. As shown in Fig. 3E, the autophagy inhibitor 3-MA was
used to investigate the association between autophagy and apoptosis
following infection of MCF-7 cells with recombinant adenoviruses.
Compared with in the blank control group, the expression levels of
Bcl-2 were decreased in the Ad-VP3, Ad-VT, Ad-VP3 + 3-MA and Ad-VT
+ 3-MA groups, and it was suggested that Ad-VP3 and Ad-VT gradually
induced apoptosis at 12 h. The expression levels of Bcl-2 in the
Ad-VP3 + 3-MA and Ad-VT + 3-MA groups were significantly lower than
in the Ad-VP3 and Ad-VT groups (P<0.01), thus indicating that
inhibition of autophagy can further induce apoptosis of tumor cells
induced by the recombinant adenoviruses. In addition, compared with
in the Ad-VP3 and Ad-VT groups, the expression levels of the
proapoptotic protein Bax were significantly increased in the Ad-VP3
+ 3-MA and Ad-VT + 3-MA groups (P<0.01 and P<0.05), and the
ratio of Bax/Bcl-2 was increased. These findings indicated that
inhibition of autophagy, alongside infection with the recombinant
adenoviruses, promoted the occurrence of apoptosis at 12 h. This
early autophagy may protect MCF-7 cells from apoptosis.
Recombinant adenoviruses induce cell
cycle arrest at G1 phase in MCF-7 cells
The cell cycle is a repeating series of events that
take place in a cell leading to its division and DNA replication,
thus resulting in the production of two daughter cells. Disturbance
of the cancer cell cycle may inhibit cell growth and activate
autophagy. To explore the potential tumor suppressive mechanism of
recombinant adenoviruses, their effects on cell cycle progression
were determined. Representative results for MCF-7 cells infected
with the control and recombinant adenoviruses are shown in Fig. 3F. Flow cytometry revealed that,
compared with in the control group (uninfected cells), Ad-VT
infection significantly decreased the number of MCF-7 human breast
cancer cells in G2/M phase after 24 h (from 9.93 to
0.88%; P<0.01; Fig. 3F), and it
was hypothesized that Ad-VT may arrest MCF-7 cells at a certain
cell cycle phase. Notably, it was observed that cells infected with
Ad-VT and Ad-VP3 were delayed in S phase; however, this was not
significant. In addition, the amount of Ad-VT-infected cells in the
G1 phase was higher. Therefore, it was suggested that
Ad-VT infection for 24 h may block MCF-7 cells in G1
phase, whereas Ad-VP3 and Ad-MOCK had no effect on the proportion
of cells in G1 phase.
Recombinant adenoviruses induce
autophagy in MCF-7 cells
Autophagy is a lysosomal degradation process for
cytoplasmic constituents under stress conditions. To examine
whether the recombinant adenoviruses affected autophagy in MCF-7
cells, several autophagic assays were performed. Firstly, MDC
staining was used to detect autophagic vacuoles. MDC is a specific
dye that emits green fluorescence, which is absorbed by cells and
selectively binds to autophagic vacuoles; therefore, it is known as
a tracer for autophagic vesicles (34). When autophagy occurs in cells, the
fluorescence of MDC staining is enhanced, and the dot-like
structure (autophagic vacuoles) in a single cell is increased.
Using fluorescence microscopy, it was revealed that the
fluorescence intensity of MDC staining was significantly enhanced
post-infection with Ad-VT or Ad-VP3, and punctate MDC-positive
fluorescent particles were observed in the cytoplasm. Notably,
>5 fluorescent spots were considered to indicate positive
fluorescence, and 100 cells were counted to calculate the rate of
autophagy. As shown in Fig. 4A,
apoptin induced the accumulation of MDC-labeled vacuoles in the
cytoplasm. The accumulation of autophagic corpuscles in the
cytoplasm was increased in the Ad-VP3 and Ad-VT groups compared
with in the control group and the Ad-MOCK group. The percentage of
autophagic corpuscles in 100 MCF-7 cells after 6 h is
presented.
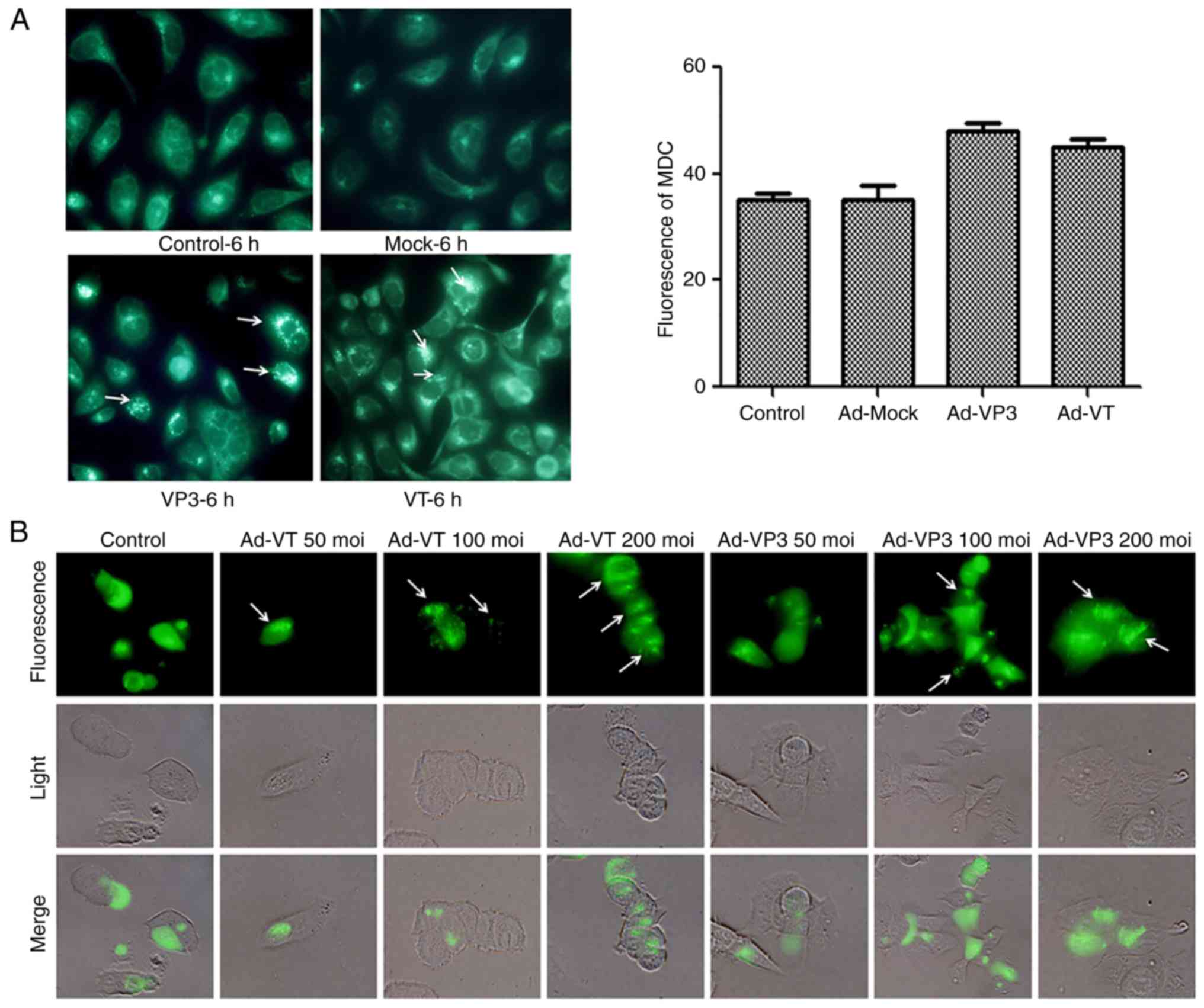 | Figure 4.Apoptin induces autophagy in MCF-7
cells. (A) To evaluate autophagic vacuoles, MCF7 cells were
infected with Ad-MOCK, Ad-VT, and Ad-VP3 (100 MOI) for 6 h and
stained with MDC. The graph shows the percentage of MDC-stained
autophagic corpuscles in MCF-7 cells in the various groups after 6
h (magnification, ×40). (B) MCF-7 cells were transfected with
pEGFP-LC3B for 24 h and then infected with carious concentrations
Ad-VT or Ad-VP3 (50, 100, and 200 MOI) for 6 h. pEGFP-LC3 signals
were observed by fluorescence microscopy (magnification, ×40).
Apoptin induces autophagy in MCF-7 cells. (C) Representative images
of immunocytochemistry. Visualization of LC3 by fluorescence
microscopy. MCF-7 cells were infected with Ad-MOCK, Ad-VT and
Ad-VP3 (100 MOI) for 6 h and stained with an antibody recognizing
LC3 for 18 h post-treatment. The control cells exhibited diffuse
cytosolic LC3 distribution. Ad-VP3-infected cells exhibited an
increase in the number of cells with LC3 dots, whereas the number
of cells with LC3 dots was further increased upon Ad-VT exposure.
Red fluorescence indicates the presence of the LC3 protein
(magnification, ×40). (D) Transmission electron microscopy revealed
autophagosome ultrastructures in the enlarged images following
infection of cells with Ad-VT or Ad-VP3 for 6 h. Control:
magnification, ×5,000; Ad-VT and Ad-VP3: magnification, ×10,000.
Ad-VP3, Ad-Apoptin; Ad-VT, Ad-Apoptin-hTERTp-E1a; LC3,
microtubule-associated protein 1A/1B-light chain 3; MDC,
monodansylcadaverine; MOI, multiplicity of infection; pEGFP-LC3,
enhanced green fluorescence protein-LC3 plasmid. |
The LC3 protein is widely distributed in cells. When
autophagy occurs, LC3 accumulates on the surface of autophagosomes,
and LC3-I proteins are cleaved into LC3-II proteins. In the present
study, LC3 was detected using pEGFP-LC3. When autophagy is induced,
the majority of autophagosomes exhibit a punctate distribution.
Conversely, when autophagy is inhibited, GFP-LC3 diffuses
throughout the cell. Therefore, transfected GFP plasmid was used to
observe the aggregation of GFP, in order to determine whether
alterations in autophagy had occurred. Ad-VT and Ad-VP3 were tested
at three concentrations (50, 100, and 200 MOI) for 6 h. No obvious
green fluorescent spots were detected in the cells infected with
Ad-VT and Ad-VP3 at 50 MOI. Green fluorescent dot aggregation in
the 100 and 200 MOI groups was obvious, and the number of
aggregated GFP points was increased in a concentration-dependent
manner. Ad-VT and Ad-VP3 at 200 MOI exhibited the most obvious
punctate green fluorescence. Conversely, GFP staining in the
negative control group was diffuse. These results indicated that
the recombinant adenoviruses could cause marked alterations in
autophagy in MCF-7 cells, in a concentration-dependent manner
(Fig. 4B). Similarly, an indirect
immunofluorescence assay using fluorescence microscopy was
conducted to detect the autophagy marker protein LC3-II; the
present study also verified that Ad-VT and Ad-VP3 increased the
expression levels of the key protein LC3-II (Fig. 4C).
TEM observation assays
TEM is the gold standard for detecting autophagosome
formation, since autophagosomes have a characteristic double
membrane or multi-membrane structures. These structures were
notably accumulated in MCF-7 cells infected with 100 MOI Ad-VT and
Ad-VP3, indicating the formation of autophagosomes (Fig. 4D). As shown in the electron
micrographs, markedly more autophagic vesicles containing
subcellular materials were observed following Ad-VT and Ad-VP3
infection for 6 h compared with in the control group. The size of
the autophagic vacuoles containing remnants of organelles was ~2 µm
in diameter.
Alterations in autophagy at the mRNA
level
The results of the RT-qPCR analysis demonstrated
that the recombinant adenoviruses induced significant alterations
in autophagy in MCF-7 cells. The results revealed that the mRNA
expression levels of the autophagy-associated proteins LC3 and
Beclin-1 were increased after 6 and 12 h of infection with Ad-VT
and Ad-VP3. In particular, LC3 expression was significantly
increased after 6 h (P<0.001; Fig.
5A). After 24 h, the expression levels of LC3 and Beclin-1 were
decreased compared with in the blank control group. After 48 h, the
expression levels of Beclin-1 were markedly increased again,
whereas LC3 exhibited the opposite trend (Fig. 5A). In addition, post-infection with
Ad-VP3, the mRNA expression levels of P62 exhibited the opposite
trend to LC3 at different time periods (Fig. 5A).
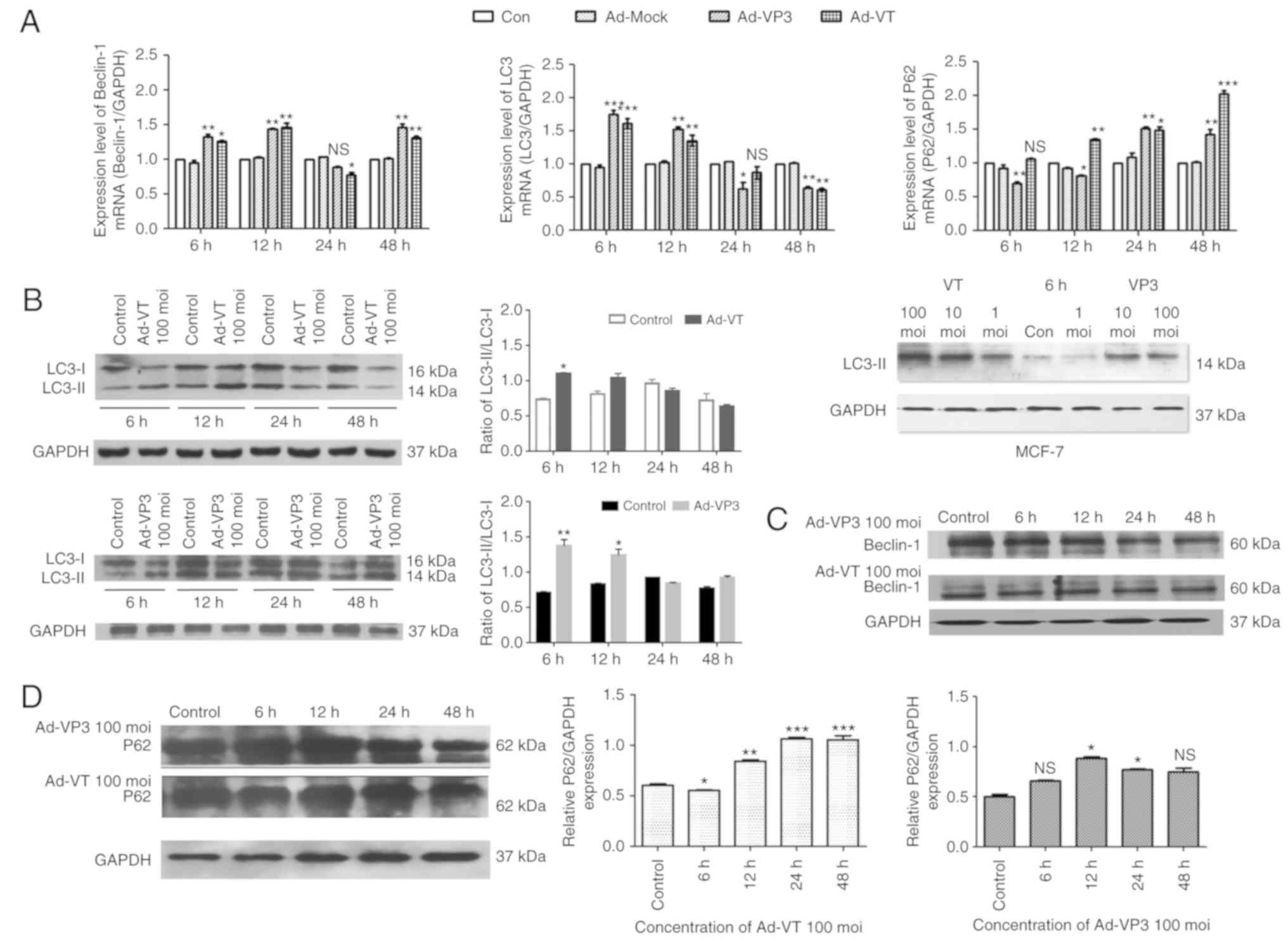 | Figure 5.RT-qPCR and western blotting of MCF-7
cell extracts. GAPDH was used as a loading control. (A) Ad-VT and
Ad-VP3 inhibited autophagy-mediated gene expression. The mRNA
expression level of the genes encoding Beclin-1, LC3 and P62 were
detected in MCF-7 cells using RT-qPCR. (B) Western blot analysis of
LC3 expression in MCF-7 cells following infection with adenoviruses
at various durations (6, 12, 24 and 48 h), or with different
concentrations of adenoviruses (1, 10, and 100 MOI) for 6 h. (C)
Western blot analysis of Beclin-1 expression in MCF-7 cells. (D)
Western blot analysis of P62 expression in MCF-7 cells. Data are
presented as the means ± standard deviation, n=3. *P<0.05,
**P<0.01, ***P<0.001 vs. the control group. Ad-VP3,
Ad-Apoptin; Ad-VT, Ad-Apoptin-hTERTp-E1a; LC3,
microtubule-associated protein 1A/1B-light chain 3; MOI,
multiplicity of infection; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Alterations in autophagy at the
protein level
The results of western blotting also indicated that
the expression levels of LC3-II were increased in response to 100
MOI Ad-VT and Ad-VP3 compared with 1 and 10 MOI adenoviruses
(Fig. 5B). The amount of LC3-II/I
has been reported to be proportional to the number of autophagic
vacuoles (35). In the present
study, the ratio of LC3-II/I gradually decreased from 6 h and
reached its lowest point at 48 h (Fig.
5B). Beclin-1 protein expression was not significantly altered
in response to Ad-VT and Ad-VP3 (Fig.
5C). P62 is a polyubiquitin-binding protein that contains an
LC3-interacting motif and ubiquitin-binding domain. By linking
ubiquitinated substrates with the autophagic machinery, P62 is
incorporated into complete autophagosomes and is then degraded in
autolysosomes, together with its bound proteins (36). Therefore, protein levels of P62 show
the opposite trend to the protein levels of LC3. At 6 h, this
phenomenon was noticeable in the present study (Fig. 5B and D). Therefore, it may be
hypothesized that at 6 h, infection of MCF-7 tumor cells with Ad-VT
and Ad-VP3 may promote autophagy. It was suggested that
apoptin-induced autophagy may be closely related to the autophagy
pathway involving the LC3 protein, and may also be associated with
Beclin-1 and P62 protein levels.
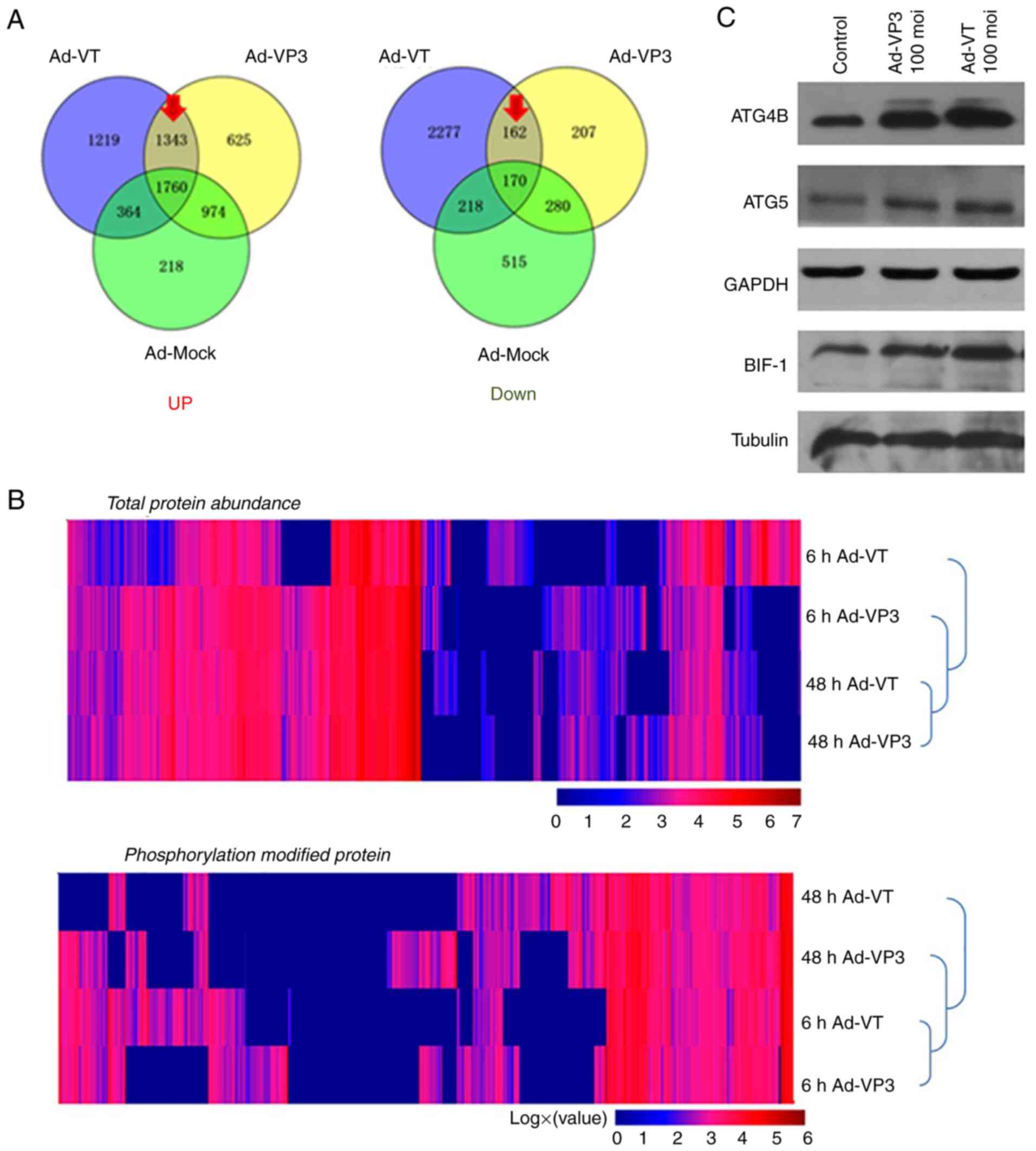
Proteomics analysis of the pathways
involved in apoptin-induced autophagy
Post-infection of MCF-7 cells with recombinant
adenoviruses, the quantity of numerous proteins was altered
compared with in the control group (Fig. 6). The present study focused on the
upregulated and downregulated proteins (marked by a red arrow in
Fig. 6A). The results of cluster
analysis revealed that for total protein abundance, 48 h Ad-VT and
48 h Ad-VP3 were initially clustered together, which were then
clustered together with 6 h Ad-VP3 and finally clustered with 6 h
Ad-VT. These findings indicated that the total protein abundance of
the 48 h Ad-VT and 48 h Ad-VP3 samples were relatively closer than
the total protein abundance of the 6 h Ad-VT and 6 h Ad-VP3
samples. With regards to levels of phosphorylation, 6 h Ad-VT and 6
h Ad-VP3 were initially clustered together, which were then
clustered with 48 h Ad-VP3 and finally clustered with 48 h Ad-VT.
These findings indicated that the phosphorylation modification
levels of the 6 h Ad-VT and 6 h Ad-VP3 samples were closer than the
phosphorylation modification levels of the of the 48 h Ad-VT and 48
h Ad-VP3 samples. Therefore, it may be suggested that the
recombinant adenovirus Ad-VP3, which only contains apoptin,
promoted autophagy at 6 h in MCF-7 tumor cells via the same
activation pathway as Ad-VT, which contains apoptin plus the
tumor-specific promoter hTERT. These findings are mainly due to the
effect of apoptin on tumor cells (Fig.
6B). In addition, the elevated proteins in the autophagy
pathway were identified by KEGG (Fig.
6D) and were further verified using western blotting. The
results revealed that ATG4B, ATG5 and BIF-1 (Fig. 6C), and LC3 (Fig. 5C) were markedly increased, and the
levels of phosphorylated ATG4 and synaptosome associated protein 29
(SNAP29) were obviously increased (Fig.
6C and D).
Discussion
Autophagy is a process of self-degradation that may
serve a dual role in cancer as a way of inducing cell death.
Autophagy can inhibit cancer in the early stages, and can lead to
drug resistance in cancer cells in the late stage (37–39).
Increasing evidence has suggested that autophagy and apoptosis can
be synergistic or antagonistic in determining cell fate. The
observed cancer-selective toxicity of apoptin (4,40) has
intrigued researchers, as this viral protein targets the molecular
basis of the transformed phenotype. Targeted cancer therapies
interfere with malignant cell growth and division at various points
during the development, growth and spread of cancer. The present
study examined the effects of apoptin on MCF-7 human breast cancer
cells and MCF-10A normal mammary epithelial cells. The present
study investigated whether recombinant adenoviruses expressing
apoptin could affect autophagy in MCF-7 human breast cancer cells.
Firstly, the results revealed that infection with adenoviruses
containing apoptin led to significant apoptosis of MCF-7 cells, as
verified by Annexin V-FITC/PI staining, which has also been
observed in another study (41).
However, these adenoviruses had no effect on MCF-10A normal mammary
gland cells. In addition, Ad-VT only induced G1 arrest
in MCF-7 human breast cancer cells, whereas Ad-VP3 and Ad-MOCK had
no significant effect on cell cycle progerssion.
In the present study two apoptin-containing
recombinant adenoviruses, Ad-VT and Ad-VP3, were constructed. In
Ad-VT, apoptin was stably and highly expressed and the tumor
specific promoter hTERT was also expressed in a human type 5
adenovirus; therefore, Ad-VT can replicate in tumor cells.
Conversely, Ad-VP3 did not have the ability to continuously
replicate in cells, since it only contained apoptin. In the process
of autophagy, a phagophore from the endoplasmic reticulum or plasma
membrane forms autophagosomes, which depend on Beclin-1 and LC3-II
(42,43). In the present study, the formation
of autophagosomes was determined by TEM, and autophagic vesicles
containing subcellular materials were observed post-infection with
Ad-VT and Ad-VP3 for 6 h. In addition, an increase in Beclin-1 and
LC3-II protein levels and a decrease in P62 protein levels was
observed by RT-qPCR and western blotting, particularly
post-infection with Ad-VP3 at 6 h. Autophagy decreased cell
survival by destroying the cytosol and organelles, leading to cell
death. In addition, proteomics analysis revealed a significant
increase in the phosphorylation of ATG4B in response to apoptin.
The biosynthesis of autophagospores involves two successive steps,
one involving ATG12-ATG5 and the other involving
ATG8-phosphatidyl-ethanolamine (PE) (44,45).
For the latter, the ATG4 cysteine protease cleaves the newly
synthesized ATG8 to reveal glycine residues and membrane-bound PE
(lipid binding) binding. ATG4 can also remove PE from ATG8
(delipidation). In humans, LC3B is the best characteristic ATG8
subtype (44,46–48).
In addition, ATG4B, but not three other ATG4 subtypes, exhibit a
highly selective preference for LC3B (49). A mouse model of genetic engineering
was used to study the importance of ATG4B in normal development and
disease. The results indicated that an ATG4B deficiency may lead to
a reduction in autophagy (50). In
cancer, ATG4B and LC3 have been implicated with the regulation of
autophagy, as a biomarker and potential therapeutic target.
Apoptosis and autophagy are two different forms of
cell death, which are independent of each other and restrict each
other. Numerous genes serve an important role in the mutual
transformation of the two different forms, including Beclin-1 and
Bcl-2. Members of the Bcl-2 family have an important role in the
regulation of mammalian cell apoptosis, targeting upstream of
irreversible cell damage, and working primarily at the
mitochondrial level. According to the structure and function of
family members, they are divided into anti-apoptotic proteins,
including Bcl-2 and Bcl-extra large, and proapoptotic proteins,
including Bax. A recent study revealed that dihydroartemisinin has
antitumor activity in cholangiocarcinoma and can reduce the
interaction of Beclin-N1 with Bcl-2 while promoting its interaction
with Phosphatidylinositol 3-kinase catalytic subunit type 3
(51). Similar results were
revealed in the present study. To investigate the role of
apoptin-induced autophagy in apoptosis, the autophagy-specific
inhibitor 3-MA was used to suppress autophagy and to investigate
the relationship between autophagy and apoptosis in MCF-7 cells. It
has previously been reported that the autophagy inhibitor 3-MA can
enhance endoplasmic reticulum stress-induced apoptosis of
nasopharyngeal carcinoma cells (52). In the present study, western
blotting results revealed that the expression levels of the
apoptosis-associated protein Bcl-2 were significantly decreased
when Ad-VP3 or Ad-VT infection was combined with 3-MA treatment,
whereas the expression levels of the proapoptotic protein Bax were
significantly increased. There was no significant difference
between cells treated with 3-MA alone and the control group.
Therefore, it was hypothesized that inhibition of autophagy could
increase the apoptosis of MCF-7 cells infected with recombinant
adenoviruses.
To the best of our knowledge, the present study is
the first to reveal that recombinant adenoviruses expressing
apoptin may significantly affect autophagy in MCF-7 breast cancer
cells. The results indicated that the recombinant adenoviruses may
enhance protective autophagy at the early stage (6 h) in MCF-7
breast cancer cells, such that cell damage by apoptosis is
inhibited. However, with the prolongation of apoptin treatment
duration, autophagy began to decrease. It was therefore
hypothesized that apoptosis may gradually dominate following
extended treatment with apoptin. It remains to be determined how
apoptin interacts with autophagy and apoptosis of MCF-7 cells. In
conclusion, the present study demonstrated that recombinant
adenoviruses expressing apoptin inhibited the early growth of
breast cancer cells via a mechanism associated with the induction
of autophagy. Our future study aims to analyze more breast cancer
cell lines alongside mammary epithelial cells as a control. These
results may lead to novel strategies to prevent and treat breast
cancer.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Key
Research and Development Program of China (grant no.
2016YFC1200900) and the Major Technological Program of Changchun
City (grant no. 16ss11).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
NYJ and XL designed the study and provided funding.
QGZ coordinated the study. SC designed and performed the
experiments, and analyzed the data. YQL, XZY, YLZ, YYF, WJL, SZL,
YLC and JZ performed the experiments. SC wrote the manuscript. QGZ
and YQL participated in editing the manuscript. QGZ was involved in
the conception and design of the study. All authors reviewed the
results and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Noteborn MH, de Boer GF, van Roozelaar DJ,
Karreman C, Kranenburg O, Vos JG, Jeurissen SH, Hoeben RC, Zantema
A and Koch G: Characterization of cloned chicken anemia virus DNA
that contains all elements for the infectious replication cycle. J
Virol. 65:3131–3139. 1991.PubMed/NCBI
|
|
2
|
Noteborn MH and Koch G: Chicken anaemia
virus infection: Molecular basis of pathogenicity. Avian Pathol.
24:11–31. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Noteborn MH, Todd D, Verschueren CA, de
Gauw HW, Curran WL, Veldkamp S, Douglas AJ, McNulty MS, van der EB
AJ and Koch G: A single chicken anemia virus protein induces
apoptosis. J Virol. 68:346–351. 1994.PubMed/NCBI
|
|
4
|
Danen-Van Oorschot AA, Fischer DF,
Grimbergen JM, Klein B, Zhuang S, Falkenburg JH, Backendorf C, Quax
PH, van der Eb AJ and Noteborn MH: Apoptin induces apoptosis in
human transformed and malignant cells but not in normal cells. Proc
Natl Acad Sci USA. 94:5843–5847. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guelen L, Paterson H, Gäken J, Meyers M,
Farzaneh F and Tavassoli M: TAT-apoptin is efficiently delivered
and induces apoptosis in cancer cells. Oncogene. 23:1153–1165.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pietersen AM, van der Eb MM, Rademaker HJ,
van den Wollenberg DJ, Rabelink MJ, Kuppen PJ, van Dierendonck JH,
van Ormondt H, Masman D, van de Velde CJ, et al: Specific
tumor-cell killing with adenovirus vectors containing the apoptin
gene. Gene Ther. 6:882–892. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhuang SM, Landegent JE, Verschueren CA,
Falkenburg JH, van Ormondt H, van der Eb AJ and Noteborn MH:
Apoptin, a protein encoded by chicken anemia virus, induces cell
death in various human hematologic malignant cells in vitro.
Leukemia. 9 (Suppl 1):S118–S120. 1995.PubMed/NCBI
|
|
8
|
Greider CW: Telomere length regulation.
Annu Rev Biochem. 65:337–365. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shay JW and Bacchetti S: A survey of
telomerase activity in human cancer. Eur J Cancer. 33:787–791.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takakura M, Kyo S, Kanaya T, Hirano H,
Takeda J, Yutsudo M and Inoue M: Cloning of human telomerase
catalytic subunit (hTERT) gene promoter and identification of
proximal core promoter sequences essential for transcriptional
activation in immortalized and cancer cells. Cancer Res.
59:551–557. 1999.PubMed/NCBI
|
|
11
|
Kyo S, Takakura M, Taira T, Kanaya T, Itoh
H, Yutsudo M, Ariga H and Inoue M: Sp1 cooperates with c-Myc to
activate transcription of the human telomerase reverse
transcriptase gene (hTERT). Nucleic Acids Res. 28:669–677.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Liu Y, Wen Z, Li C, Lu H, Tian M,
Jin K, Sun L, Gao P, Yang E, et al: Potent anti-tumor effects of a
dual specific oncolytic adenovirus expressing apoptin in vitro and
in vivo. Mol Cancer. 9:102010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu L, Wu W, Zhu G, Liu L, Guan G, Li X,
Jin N and Chi B: Therapeutic efficacy of an hTERT promoter-driven
oncolytic adenovirus that expresses apoptin in gastric carcinoma.
Int J Mol Med. 30:747–754. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Wang J, Li C, Hu N, Wang K, Ji H,
He D, Quan C, Li X, Jin N, et al: Potent growth-inhibitory effect
of a dual cancer-specific oncolytic adenovirus expressing apoptin
on prostate carcinoma. Int J Oncol. 42:1052–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qi Y, Guo H, Hu N, He D, Zhang S, Chu Y,
Huang Y, Li X, Sun L and Jin N: Preclinical pharmacology and
toxicology study of Ad-hTERT-E1a-Apoptin, a novel dual
cancer-specific oncolytic adenovirus. Toxicol Appl Pharmacol.
280:362–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang G, Meng X, Sun L, Hu N, Jiang S,
Sheng Y, Chen Z, Zhou Y, Chen D, Li X, et al: Antitumor effects of
a dual cancer-specific oncolytic adenovirus on colorectal cancer in
vitro and in vivo. Exp Ther Med. 9:327–334. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Watanabe E, Muenzer JT, Hawkins WG, Davis
CG, Dixon DJ, McDunn JE, Brackett DJ, Lerner MR, Swanson PE and
Hotchkiss RS: Sepsis induces extensive autophagic vacuolization in
hepatocytes: A clinical and laboratory-based study. Lab Invest.
89:549–561. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mukhopadhyay S, Panda PK, Sinha N, Das DN
and Bhutia SK: Autophagy and apoptosis: Where do they meet?
Apoptosis. 19:555–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kubisch J, Türei D, Földvári-Nagy L, Dunai
ZA, Zsákai L, Varga M, Vellai T, Csermely P and Korcsmáros T:
Complex regulation of autophagy in cancer-integrated approaches to
discover the networks that hold a double-edged sword. Semin Cancer
Biol. 23:252–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim Y, Jeong IG, You D, Song SH, Suh N,
Jang SW, Kim S, Hwang JJ and Kim CS: Sodium meta-arsenite induces
reactive oxygen species-dependent apoptosis, necrosis, and
autophagy in both androgen-sensitive and androgen-insensitive
prostate cancer cells. Anticancer Drugs. 25:53–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen S, Kepp O and Kroemer G: The end of
autophagic cell death? Autophagy. 8:1–3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Denton D, Xu T and Kumar S: Autophagy as a
pro-death pathway. Immunol Cell Biol. 93:35–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berry DL and Baehrecke EH: Autophagy
functions in program-med cell death. Autophagy. 4:359–360. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and self-killing: Crosstalk between
autophagy and apoptosis. Nat Rev Mol Cell Biol. 8:741–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen HM and Codogno P: Autophagic cell
death: Loch Ness monster or endangered species? Autophagy.
7:457–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lum JJ, Bauer DE, Kong M, Harris MH, Li C,
Lindsten T and Thompson CB: Growth factor regulation of autophagy
and cell survival in the absence of apoptosis. Cell. 120:237–248.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amaravadi RK and Thompson CB: The roles of
therapy-induced autophagy and necrosis in cancer treatment. Clin
Cancer Res. 13:7271–7279. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu YT, Tan HL, Huang Q, Kim YS, Pan N, Ong
WY, Liu ZG, Ong CN and Shen HM: Autophagy plays a protective role
during zVAD-induced necrotic cell death. Autophagy. 4:457–466.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo JY, Chen HY, Mathew R, Fan J,
Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM,
Karantza V, et al: Activated Ras requires autophagy to maintain
oxidative metabolism and tumorigenesis. Genes Dev. 25:460–470.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shimizu S, Kanaseki T, Mizushima N, Mizuta
T, Arakawa-Kobayashi S, Thompson CB and Tsujimoto Y: Role of Bcl-2
family proteins in a non-apoptotic programmed cell death dependent
on autophagy genes. Nat Cell Biol. 6:1221–1228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharifi MN, Mowers EE, Drake LE, Collier
C, Chen H, Zamora M, Mui S and Macleod KF: Autophagy promotes focal
adhesion disassembly and cell motility of metastatic tumor cells
through the direct interaction of paxillin with LC3. Cell Rep.
15:1660–1672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Biederbick A, Kern HF and Elsässer HP:
Monodansylcadaverine (MDC) is a specific in vivo marker for
autophagic vacuoles. Eur J Cell Biol. 66:3–14. 1995.PubMed/NCBI
|
|
35
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Saiki S, Sasazawa Y, Imamichi Y, Kawajiri
S, Fujimaki T, Tanida I, Kobayashi H, Sato F, Sato S, Ishikawa K,
et al: Caffeine induces apoptosis by enhancement of autophagy via
PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 7:176–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perkins ND: The diverse and complex roles
of NF-κB subunits in cancer. Nat Rev Cancer. 12:121–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Janku F, McConkey DJ, Hong DS and Kurzrock
R: Autophagy as a target for anticancer therapy. Nat Rev Clin
Oncol. 8:528–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blessing AM, Rajapakshe K, Reddy Bollu L,
Shi Y, White MA, Pham AH, Lin C, Jonsson P, Cortes CJ, Cheung E, et
al: Transcriptional regulation of core autophagy and lysosomal
genes by the androgen receptor promotes prostate cancer
progression. Autophagy. 13:506–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Poon IK, Oro C, Dias MM, Zhang JP and Jans
DA: A tumor cell-specific nuclear targeting signal within chicken
anemia virus VP3/apoptin. J Virol. 79:1339–1341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen Ni L, Allaudin ZN, Mohd Lila MA,
Othman AM and Othman FB: Selective apoptosis induction in MCF-7
cell line by truncated minimal functional region of Apoptin. BMC
Cancer. 13:4882013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kroemer G and Levine B: Autophagic cell
death: The story of a misnomer. Nat Rev Mol Cell Biol. 9:1004–1010.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pradhan AK, Talukdar S, Bhoopathi P, Shen
XN, Emdad L, Das SK, Sarkar D and Fisher PB: mda-7/IL-24
mediates cancer cell-specific death via regulation of miR-221 and
the beclin-1 axis. Cancer Res. 77:949–959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mizushima N, Yoshimori T and Ohsumi Y: The
role of Atg proteins in autophagosome formation. Annu Rev Cell Dev
Biol. 27:107–132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kabeya Y, Mizushima N, Yamamoto A,
Oshitani-Okamoto S, Ohsumi Y and Yoshimori T: LC3, GABARAP and
GATE16 localize to autophagosomal membrane depending on form-II
formation. J Cell Sci. 117:2805–2812. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tanida I, Ueno T and Kominami E: Human
light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121
to expose Gly120 for lipidation and targeting to autophagosomal
membranes. J Biol Chem. 279:47704–47710. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Klionsky DJ, Cuervo AM and Seglen PO:
Methods for monitoring autophagy from yeast to human. Autophagy.
3:181–206. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mariño G, Uría JA, Puente XS, Quesada V,
Bordallo J and López-Otín C: Human autophagins, a family of
cysteine proteinases potentially implicated in cell degradation by
autophagy. J Biol Chem. 278:3671–3678. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Akin D, Wang SK, Habibzadegah-Tari P, Law
B, Ostrov D, Li M, Yin XM, Kim JS, Horenstein N and Dunn WA Jr: A
novel ATG4B antagonist inhibits autophagy and has a negative impact
on osteosarcoma tumors. Autophagy. 10:2021–2035. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Thongchot S, Vidoni C, Ferraresi A,
Loilome W, Yongvanit P, Namwat N and Isidoro C: Dihydroartemisinin
induces apoptosis and autophagy-dependent cell death in
cholangiocarcinoma through a DAPK1-BECLIN1 pathway. Mol Carcinog.
57:1735–1750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Song L, Ma L, Zhang X, Jiang Z, Liu H and
Jiang C: Effect of tunicamycin combined with cisplatin on
proliferation and apoptosis of human nasopharyngeal carcinoma cells
in vitro. Nan Fang Yi Ke Da Xue Xue Bao. 32:766–771. 2012.(In
Chinese). PubMed/NCBI
|