Introduction
Dysfunction of cell cycle progression plays a key
role in tumor formation and progression, and abnormally regulated
cell cycle progression often leads to tumorigenesis and progression
(1,2). As one of the many genes regulating
cell cycle progression, UbcH10 overexpression has been found in
tumor tissues from a variety of sources and its overexpression has
been revealed to be correlated with tumor histological grade
(3,4), cell proliferation (5) and poor patient prognosis (6,7). In
recent years, we focused on the function of UbcH10 in NSCLC. Based
on the available data, the expression of UbcH10 in NSCLC was
similar to the level in other types of tumors. A significant
positive correlation with tumor pathological grade and tumor cell
proliferation was noted. The data revealed that UbcH10 may play a
key role in the pathogenesis of NSCLC. Our previous study revealed
that UbcH10 controls excessive proliferation of NSCLC cells through
the regulation of tumor protein p53 (P53) and Ki-67 and other
proliferation-related genes, while UbcH10 knockdown can
significantly inhibit the proliferation of NSCLC cells, which
causes cell cycle arrest and promotes apoptosis. On the other hand,
high expression of UbcH10 was found to reduce the sensitivity of
NSCLC cells to chemotherapeutic drugs by promoting the expression
of multi-drug resistance (MDR1) gene, while the targeted knockdown
of the UbcH10 gene could effectively enhance chemotherapy
sensitivity of SK-MES-1 and A549 cell lines to gemcitabine and
paclitaxel (8).
The fundamental purpose of gene function research is
to systematically elucidate the upstream regulatory mechanisms. By
following the study on the regulatory mechanism, we can effectively
prevent NSCLC from a macro perspective, and develop effective
personalized treatment plans for patients (9–11). In
essence, the root of cancer development involves epigenetics.
Epigenetics can control gene expression through regulation of DNA
methylation, histone modification, chromatin remodeling and
non-coded RNA, and run through the whole process of cancer
occurrence and development (12).
Therefore, the in-depth study of cancer epigenetics has important
significance for the clinical diagnosis, treatment and prevention
of cancer. In the present study, we focused on the mechanism of the
abnormal expression of UbcH10 in NSCLC. We found that the direct
cause of the abnormal post-transcriptional regulatory mechanism of
UbcH10 was the decrease in hsa-miR661-3p expression. Investigation
of epigenetic research data was not the primary goal of present
study. In the future, we are planning to explain the cause of the
aberrant expression of hsa-miR661-3p in NSCLC from the focus of
epigenetics. Therefore, we demonstrated the mode of action and
functional specificity of hsa-miR661-3p/UbcH10 in the regulatory
pathway of lung cancer through a complete and logical experimental
design. On this basis, we carried out a preliminary validation of
in vitro and in vivo inhibition experiments for
non-small cell lung cancer by the aberrant expression of
hsa-miR661-3p. The present study has great theoretical value for
the complete interpretation of UbcH10 in the regulatory mechanism
of NSCLC.
Materials and methods
Cell culture
Human NSCLC cells (A549, SK-MES-1 and NCI-H266) and
immortalized human normal lung cells (BEAS-2B), were purchased from
the Cell Bank of the Chinese Academy of Sciences (Shanghai, China).
The lentivirus-producing cell line, 293TN, was purchased from
System Biosciences (Palo Alto, CA, USA). All the cell lines were
maintained in Dulbecco's minimum essential medium (DMEM;
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.). Cells were passaged by 0.25% trypsin
digestion (Invitrogen; Thermo Fisher Scientific, Inc.) and
incubated in an atmosphere of 5% CO2 at 37°C.
Tumor tissues
Twelve pairs of NSCLC and adjacent normal tissues
were obtained from the Shanghai Pulmonary Hospital between October
2016 and December 2017 (Table I).
Written informed consents were received from the patients and the
study was approved by the Ethics Committee of the Shanghai
Pulmonary Hospital. Samples were rinsed with saline and transferred
to 2-ml microtubes, labeled and maintained in liquid nitrogen
subsequently. Total RNA and protein were extracted and used for
determination of hsa-miR-661-3p and UbcH10 protein,
respectively.
 | Table I.Clinicopathological features of 12
patients with NSCLC. |
Table I.
Clinicopathological features of 12
patients with NSCLC.
| No. | Sex | Age (years) | Pathological | Clinical stage |
|---|
| 1 | M | 54 | Squamous
carcinoma | II |
| 2 | M | 60 | Squamous
carcinoma | III |
| 3 | F | 64 | Adenocarcinoma | IV |
| 4 | M | 58 | Adenocarcinoma | IV |
| 5 | F | 52 | Adenocarcinoma | III |
| 6 | F | 68 | Squamous
carcinoma | IV |
| 7 | M | 70 | Squamous
carcinoma | II |
| 8 | M | 72 | Squamous
carcinoma | IV |
| 9 | F | 70 | Adenocarcinoma | III |
| 10 | F | 69 | Adenocarcinoma | II |
| 11 | F | 53 | Squamous
carcinoma | III |
| 12 | M | 73 | Adenocarcinoma | II |
Construction of the vectors
Construction of the cDNA expression
vector
The CDS sequence of human UbcH10 (NM_007019.2) was
amplified by using the primers
5′-GGAATTCGCCACCATGGCTTCCCAAAACCGCG-3′ and
5′-CGGGATCCTCAGGGCTCCTGGCTGGTG-3′, which contain an EcoRI
cutting site and Kozak sequence and a BamHI cutting site,
respectively, with the cDNA prepared by reverse transcription of
RNA isolated from 293T cells. The PCR product was digested and
cloned into the pcDH1 lentiviral-expressing vector; the recombinant
vector was named pcDH1-UbcH10.
Construction of the hsa-miR-661-3p
expression vector
Human genomic DNA was extracted from 293 cells and
used for amplification of the template of precursor sequence of
hsa-miR-661-3p. The primers used were:
5′-GGAATTCTGGCATGCCATAGCAGCGCAG-3′ and
5′-CGGGATCCCTCCCATCTAAGCTTCCCAAAGTGT-3′. The PCR product was
digested using EcoRI and BamHI and ligated into the
linear pcDH1 vector (System Biosciences) and transformed into DH5α
competent cells. The obtained vector was called the pcDH1-miR-661
vector. The products of the vectors were confirmed by DNA
sequencing. Endotoxin-free DNA was prepared in all cases.
Construction of the luciferase
reporter vector
The 3′-untranslated region (3′-UTR, 259 bp) of human
UbcH10 was amplified from cDNA obtained through the reverse
transcription of total RNA of 293 cells, with the following
primers: 5′-GCTCTAGAGAAACCTACTCAAAGCAG-3′ and
5′-GCTCTAGAACCACAGCTCAAGATAAA-3′. The amplification parameters were
as follows: 32 cycles of denaturation at 95°C for 10 sec, annealing
at 58°C for 30 sec and extension at 72°C for 30 sec. The product
was digested with XbaI and inserted into the pGL3-promotor
vector (Promega, Madison, WI, USA). The seed region was mutated
from 5′-CCCAGGC-3′ to 5′-CGCCACG-3 by point mutation, and the
resultant vectors were called pGL-wt-UbcH10 and pGL-mt-UbcH10,
respectively.
Lentivirus packaging
One day before the transfection, 293TN cells were
seeded into 10-cm dishes. A total of 2 µg of each shRNA or
expression vector and 10 µg of pPACK Packaging Plasmid Mix (System
Biosciences) were co-transfected using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol. The medium was replaced with DMEM plus 1%
FBS. After 48 h, the supernatant was harvested and cleared by
centrifugation at 5,000 × g at 4°C for 5 min and then passed
through a 0.45-µm polyvinylidene difluoride (PVDF) membrane (EMD
Millipore, Billerica, MA, USA). The titer of the virus was
determined by gradient dilution. The packaged lentiviruses were
named Lv-miR-661 and Lv-UbcH10.
Assessment of hsa-miR-661 and UbcH10
protein level in NSCLC specimens and NSCLC cell lines
Twelve pairs of NSCLC and para-carcinoma tissues, as
well as A549, SK-MES-1 and NCI-H266 and BEAS-2B cells
(5×106 each), were collected, followed by total RNA
extraction and real-time PCR for the measurement of hsa-miR-661-3p.
Total protein extraction and western blotting was performed for the
UbcH10 protein.
Effect of the expression of
hsa-miR-661-3p on UbcH10
A549 and SK-MES-1 in logarithmic phase were seeded
in 6-well plates at 5×105 cells/well. One day later, a
viral solution (Lv-miR-661 or Lv-UbcH10) was added at an MOI of 10.
The infection efficiency was evaluated by observing and analyzing
the fluorescence 72 h after infection. The total RNA was isolated
from the cells and subjected to real-time PCR or western blotting
for the level of hsa-miR-661-3p and UbcH10 protein,
respectively.
Experiment of luciferase assay
We used Target Scan 7.1 (Whitehead Institute,
Cambridge, MI, USA; http://www.targetscan.org/) to predict whether an
hsa-miR661-3p binding site exists within the 3′-UTR of human UbcH10
mRNA. The results revealed that a seven-base hsa-miR-661-3p seed
sequence is present in the 3′-UTR of UbcH10 mRNA. A suspension of
293T cells in logarithmic phase growth was prepared and the number
of viable cells was counted using a hemocytometer in conjunction
with trypan blue staining. The cells were seeded into 6-well plates
at a concentration of 2×105 cells/well and maintained in
Dulbecco's modified Eagle's medium supplemented with 10% FBS at
37°C for 24 h in a 5% CO2 atmosphere. The transfection
of plasmid DNA and RNA was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Transfection of cells
with pGL-TK (100 ng) served as a reference for luciferase
detection. Luciferase activity was assessed using the
Dual-Luciferase reporter assay system (Promega) 48 h after
transfection.
Cellular proliferation assay
We assessed whether regulation of has-miR-661-3p
expression could inhibit the proliferation of NSCLC cell lines.
A549 cells were infected with recombinant lentiviruses for 72 h,
trypsinized and seeded into 96-well plates at a density of
1×104 cells/well. The cells were cultured under normal
conditions and cell viability was examined using CCK-8 (Cell
Counting Kit-8; Dojindo Laboratories, Kumamoto, Japan) at 24, 48
and 72-h time-points. Briefly, 10 µl of CCK-8 solution was added,
and then the cells were cultured under normal conditions for an
additional 4 h before measurement of the absorbance at 450 nm. The
experimental groups were as follows: i) Cell; ii) Cell+Lv-NC; iii)
Cell+Lv-miR-661; and iv) Cell+Lv-miR-661+Lv-UbcH10.
Cell invasion assay
Cell invasion experiments were performed using the
QCM™ 24-well Fluorimetric Cell Invasion Assay kit
(ECM554; Chemicon International, Temecula, CA, USA) according to
the manufacturer's instructions. The kit used an insert
polycarbonate membrane with an 8-µm pore size. The insert was
coated with a thin layer of EC Matrix™ that occluded the
membrane pores and blocked the migration of non-invasive cells.
Culture medium (500 µl) supplemented with 10% FBS was used as a
chemoattractant. Cells that migrated and invaded the underside of
the membrane were fixed in 4% paraformaldehyde. The invading cells
were stained with crystal violet staining solution (0.1%), and the
number was then determined by fluorescence and reported as the
relative fluorescence units (RFUs). SK-MES-1 cells were used and
the grouping was the same as in the proliferation assay.
Effect of hsa-miR-661-3p on the
expression of functional proteins of SAC
UbcH10, cyclin B, BubR1 and Mad2 protein levels in
the A549 cells were assessed using western blotting 72 h after
infection with Lv-NC, Lv-miR-661 or Lv-miR-661combined with
Lv-UbcH0.
Cell cycle analysis
NCI-H266 cells were infected with Lv-miR-661-3p or
Lv-UbcH10 as described above. The cells at logarithmic growth phase
were typsinized, washed with phosphate-buffered saline (PBS) twice,
and then fixed with 70% ethanol at 4°C overnight. The fixed cells
were washed with PBS twice, resuspended in 100 µl PBS [(containing
100 µg/ml ribonuclease A and 50 µg/ml propidium iodide (PI)], and
incubated at room temperature for 30 min. The cell suspensions were
detected by a BD FACSCalibur flow cytometer (BD Biosciences,
Franklin Lakes, NJ, USA).
Real-time PCR
Total RNA (2 µg) was used for cDNA preparation using
M-MLV reverse transcription kit and specific primers: U6 snRNA
(NM_001101.3), 5′-TACCTTGCGAAGTGCTTAAAC-3′ and miR-661,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGATCGCG-3′. RNA
contents were detected using PCR of fluorescent dye (Takara Bio
Group, Beijing, China) in accordance with the manufacturer's
instructions. The following primers were used for quantification of
human U6 snRNA and miR-661: U6 snRNA, 5′-GTGCTCGCTTCGGCAGCACAT-3′
and 5′-TACCTTGCGAAGTGCTTAAAC-3′, producing a segment of 112 bp; and
miRNA631, 5′-GCCGGCGCCCGAGCTCTGGCTC-3′ and
5′-TGCCTGGGTCTCTGGCCTGCGCGT-3′, producing a segment of 72 bp. The
PCR system included: Takara SYBR Premix Ex Taq 10 µl, forward and
reverse primers (20 µM) 0.2 µl each, and cDNA 2 µl, added with
dH2O to 20 µl. Cycling parameters were as follows: 40
cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 20
sec and extension at 72°C for 20 sec. The levels of hsa-miR-661-3p
were normalized using the ΔΔCt method; U6 snRNA was used as a
reference. Each RNA sample was run in triplicate.
Detection of protein contents in the
cells or tissues
The total protein was extracted from the cells using
M-PER mammalian protein extraction reagent (Pierce, Rockford, IL,
USA) or from tissues using T-PER tissue protein extraction reagent
(Pierce). Equal amounts of protein (20 µg per lane) estimated by
the bicinchoninic acid (BCA) protein assay were loaded onto (11%)
SDS-PAGE gels and transferred onto nitrocellulose membranes. Blots
were blocked in Tris-buffered saline (TBS) containing Tween-20
(TBST) containing 5% non-fat milk at room temperature for 2 h and
incubated with primary antibodies against human UbcH10 (dilution
1:200; cat. no. ab12290), cyclin B (dilution 1:500; cat. no.
ab32053), BubR1 (dilution 1:600; cat. no. ab183496), Mad2 (dilution
1:250; cat. no. ab61591) and β-actin (dilution 1:1,200; cat. no.
ab227387) (Abcam, Cambridge, UK), followed by the secondary
HRP-conjugated anti-rabbit antibody (cat. no. ab97051; Abcam).
Enhanced chemiluminescence (ECL) substrates (Pierce) and X-ray film
were used to detect the bands, and the relative optical densities
were analyzed using software Total Lab v1.10 (Total Lab Ltd.,
Newcastle, UK). β-actin was used as an endogenous reference for
normalization.
Animal xenografts
Six- to eight-week old nude mice (20±2 g) were
purchased from Shanghai Slack Experimental Animal Co., Ltd.
(Shanghai, China) and housed at the Second Military Medical
University Animal Experiment Center, where the implantation
experiment was performed. All mice were bred and maintained in a
specific pathogen-free facility and were used in accordance with
the institutional guidelines for animal care and all the protocols
were previously approved by the Tongji University Experiment Animal
Ethics Committee. A549 cells (1×105) were suspended in
200 µl medium, and injected subcutaneously into the flank regions
of 48 female athymic nude mice. Two weeks after the inoculation,
visible subcutaneous tumors were detected, and the tumors were ~2.5
mm in diameter 3 weeks after inoculation. All animals were randomly
divided into 4 groups (12 mice per group): The Model group, the
Lv-control group, the miR-661 expression group and the
miR-661-expression+UbcH10 overexpression group. For the
intervention groups, each animal received 50 µl recombinant
lentivirus (5×107 IFU) twice a week (on Monday and
Thursday) from the second week for 4 weeks, while the model group
received the same volume of saline instead. The tumor diameter was
assessed weekly from the second week, and the data were used to
plot the tumor growth curves. The formula for calculating the tumor
volume (V) was: V=0.5×axbxb, where a and b are the long and short
diameters of the tumor, respectively.
Statistical analysis
All data are expressed as the mean ± SD and analyzed
by Student's t-test or one-way analysis of variance (ANOVA) test.
Least significant difference (LSD) was used for multiple
comparisons between any two means. P<0.05 was considered to
indicate a statistically significant result. All statistical
analysis was performed using SPSS 18.0 software (SPSS, Inc.,
Chicago, IL, USA).
Results
hsa-miR-661-3p is downregulated and
UbcH10 is enhanced in NSCLC tumors and cell lines
The results of the hsa-miR-661-3p quantification in
NSCLC showed that the expression of hsa-miR-661-3p was lower in
NSCLC tumors than that in adjacent tissues (P<0.01), and the
data of the UbcH10 protein evaluation indicated that the level of
UbcH10 protein was higher in NSCLC tumors than these levels in
adjacent tissues (P<0.01) (Fig.
1A). The examination in BEAS-2B, A549, SK-MES-1 and NCI-H226
cells revealed that hsa-miR-661-3p and UbcH10 protein were elevated
in these cell lines compared with BEAS-2B, which is a normal lung
cell line (P<0.01) (Fig. 1B).
These results collectively suggested that the expression of
hsa-miR-661-3p was negatively associated with UbcH10.
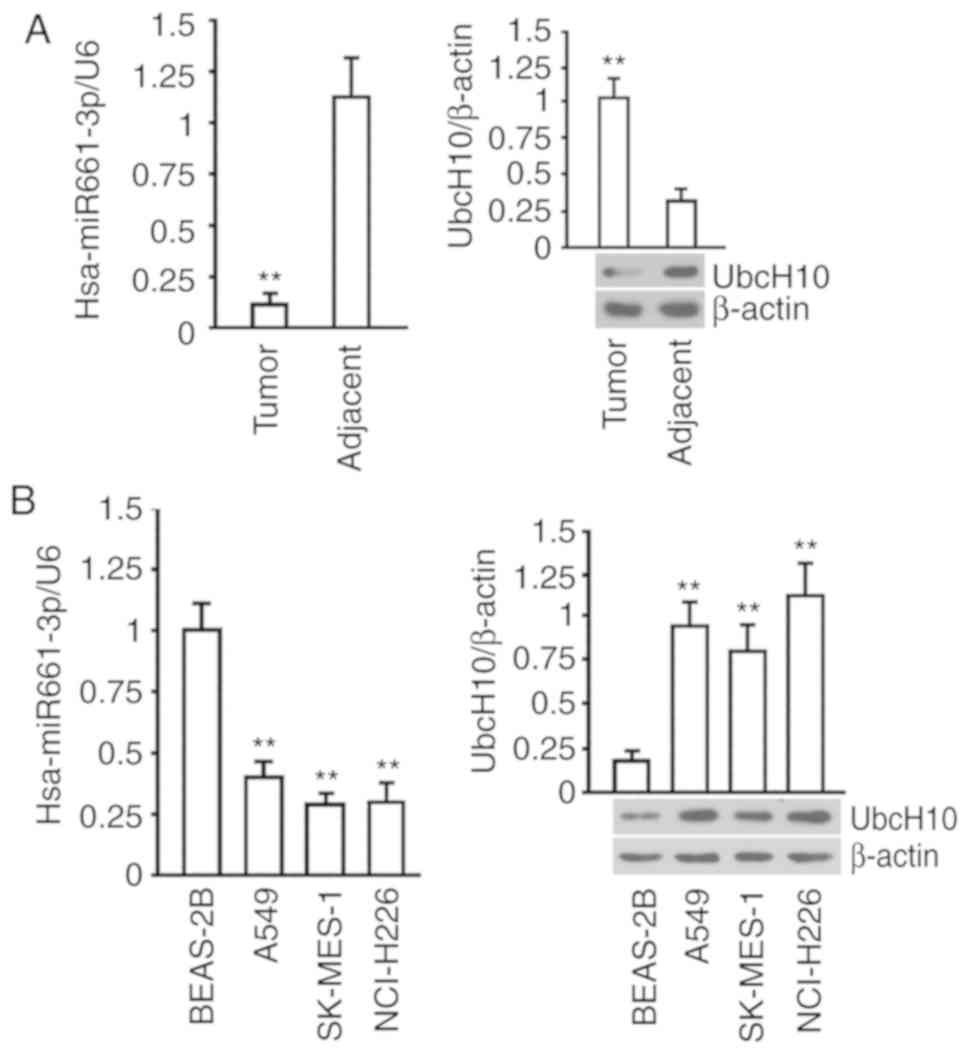 | Figure 1.Levels of hsa-miR-661-3p and UbcH10
protein in NSCLC tissue and cells. (A) Determination of
hsa-miR-661-3p and UbcH10 protein levels in 12 pairs of NSCLC
tissues and adjacent tissues. U6 served as an internal reference
for the determination of hsa-miR-661-3p, and the relative
hsa-miR-6613p expression in NSCLC tissue was used for normalization
in the comparison between groups (left image); for the
determination of UbcH10 protein expression, 12 pairs of samples
were pooled, and tested by western blotting, β-actin served as an
internal reference, and the relative UbcH10 expression in the NSCLC
samples were used for normalization in comparison between the
groups (right image). **P<0.01 vs. adjacent tissue. (B)
Determination of hsa-miR-661-3p (left) and UbcH10 protein (right
image) in A549, SK-MES-1, NCI-H226 and BEAS-2B cells. U6 served as
an internal reference for the determination of hsa-miR-661-3p, and
the relative expression of hsa-miR-661-3p in cells was used for
normalization in comparison among the groups; β-actin served as an
internal reference for determination of the UbcH10 protein, and the
relative expression of the UbcH10 protein in cells was used for
normalization in comparison among the groups. **P<0.01 vs.
BEAS-2B. The tests were carried out on three biological
triplicates, and the data are expressed as the mean ± SD. |
hsa-miR-661-3p inhibits UbcH10
expression by interacting with the 3′UTR of UbcH10 mRNA
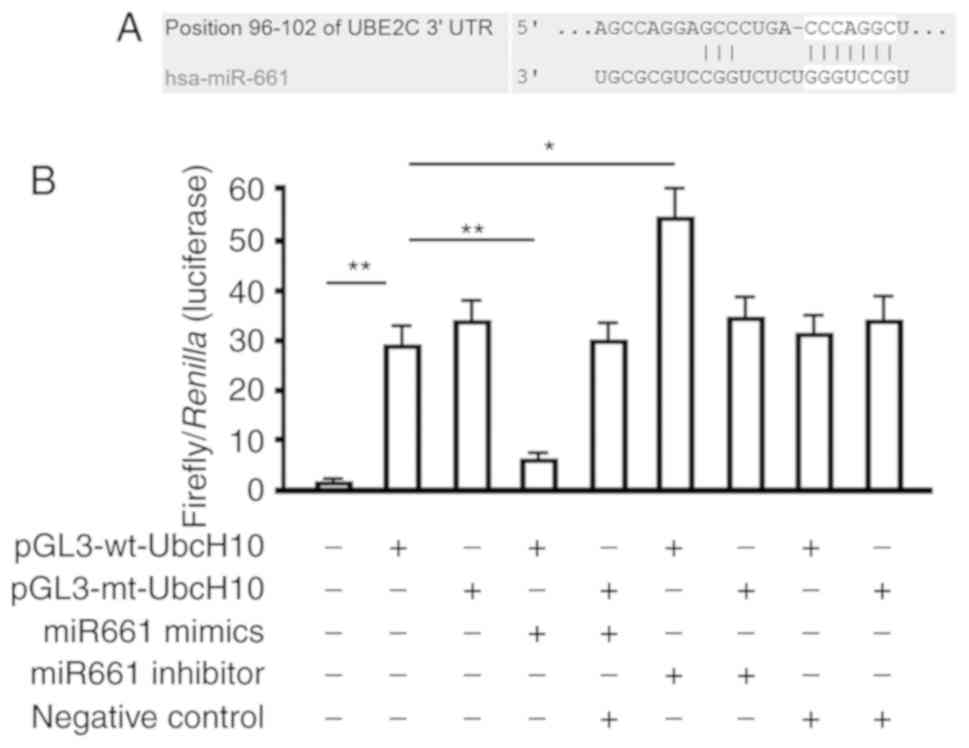
Our bioinformatic analysis identified a seven-base
hsa-miR-661-3p seed sequence in the 3′UTR of UbcH10 mRNA (Fig. 2A). Therefore, we constructed
luciferase reporter vectors to verify whether this site represents
a valid hsa-miR-661-3p target. Reporter vectors were generated that
contained the wild-type UbcH10 3′-UTR or a variant in which the
hsa-miR-661-3p target site within the 3′UTR had been mutated. Both
reporter constructs expressed luciferase at a high level (Fig. 2A). However, the miR-661-3p mimic
significantly inhibited luciferase activity in cells transfected
with the reporter vector encoding the wild-type 3′UTR (27.21±3.61
vs. 6.83±1.12; P<0.01), while the miRNA-661-3p inhibitor
significantly increased luciferase activity in these cells
(27.21±3.61 vs. 52.67±9.04; P<0.05). Conversely, in cells
transfected with the reporter vector encoding the mutated
hsa-miR-661-3p target site, neither the miR-661-3p mimic nor the
miR-661-3p inhibitor had any observable effect on luciferase
activity (P>0.05). Co-transfection of miR-661-3p-NC
(non-targeting control) had no effect on the luciferase activity of
either of the vectors (P>0.05). These results verified the
presence of a hsa-miR-661-3p target site in the 3′UTR of UbcH10
mRNA and demonstrated that binding of hsa-miR-661-3p to this target
site downregulated UbcH10 expression.
Effect of the expression of
hsa-miR-661-3p and UbcH10 via lentiviral approach NSCLC cells
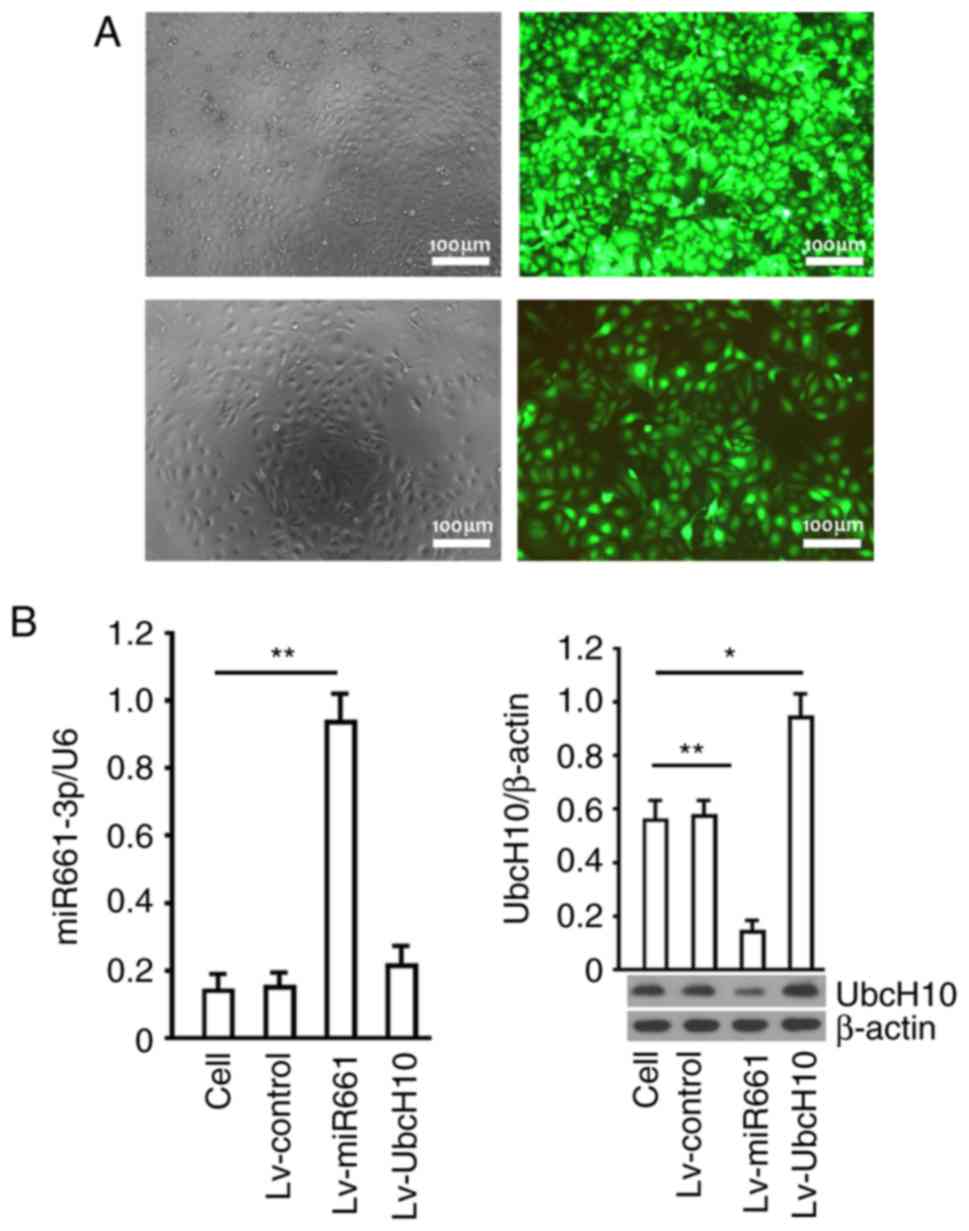
Recombinant lentiviruses, Lv-control, Lv-miR-661 and
Lv-UbcH10, were used to infect A549 cells. GFP (Green fluorescent
protein) was detected in most of the cells 72 h after infection,
while the proportion of GFP-expressing cells suggested that the
gene delivery efficiency was higher than 95% in the A549 (Fig. 3A). Hsa-miR-661-3p was significantly
increased by Lv-miR-661 (P<0.01), and no change was observed in
cells infected with Lv-UbcH10 (P>0.05). The protein level of
UbcH10 was significantly increased by Lv-UbcH10 and decreased by
Lv-miR-661 (P<0.01) (Fig. 3B).
The same data were obtained in SK-MES-1 cells (data not shown).
These findings indicated that decrease of hsa-miR-661-3p
upregulated UbcH10 expression in A549 cells, and overexpression of
UbcH10 had no obvious effect on hsa-miR-661-3p, thus, there is an
obvious upstream and downstream relationship between hsa-miR-661-3p
and UbcH10, that is to say, hsa-miR-661-3p is located upstream of
the UbcH10.
Overexpression of hsa-miR-661-3p
inhibits cellular proliferation and invasion and induces cell cycle
arrest in A549 cells
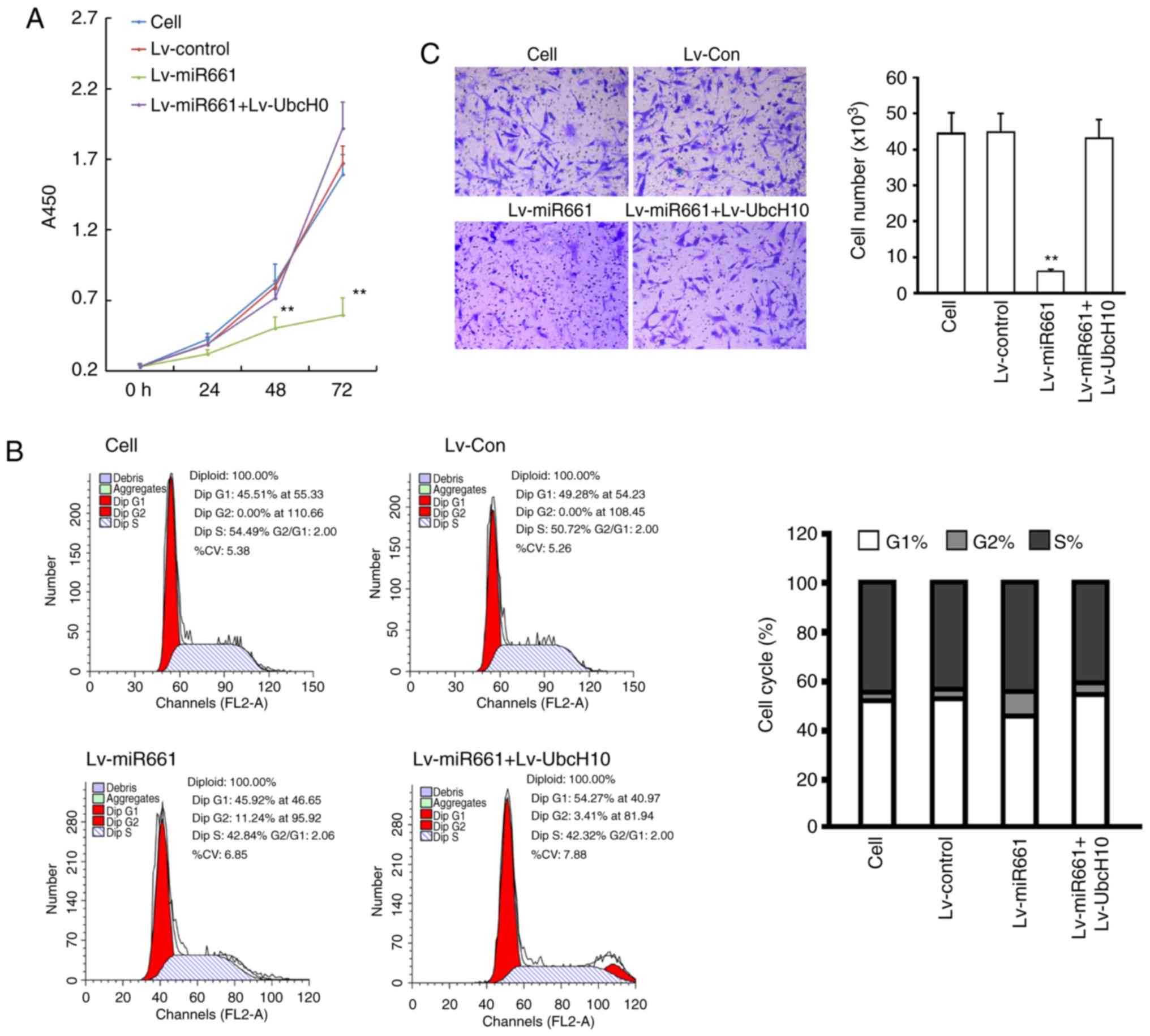
A549 cells infected with Lv-miR-661-3p or Lv-UbcH10
were subjected to the proliferation assays. The results indicated
that exogenous expression of hsa-miR-661-3p inhibited the
proliferation of A549 cells after 48 or 72 h of incubation
(P<0.05, vs. the cell group or NC group). Cells infected with
Lv-UbcH10 combined Lv-miR-661-3p have no significant difference on
the proliferation compared to cell group or NC group (P>0.05,
vs. the cell group or NC group) (Fig.
4A). To investigate whether the inhibition of hsa-miR-661-3p on
cell proliferation of SK-MES-1 was mediated by cell cycle
alteration, cell cycle distribution was determined. The results
revealed that in comparison with the cell group, hsa-miR-661-3p
expression significantly increased the cell population at the G2
phase, from 0.92±0.07 to 14.02±1.24% (P<0.01) indicating that
overexpression of hsa-miR-661-3p caused a cell cycle G2 arrest in
A549 cells (Fig. 4B). Furthermore,
cell invasion assay results demonstrated that hsa-miR-661-3p
expression inhibited invasion in NCI-H226 cells (P<0.01), and
UbcH10 overexpression could reverse the invasion inhibition caused
by hsa-miR-661-3p expression (P<0.05) (Fig. 4C).
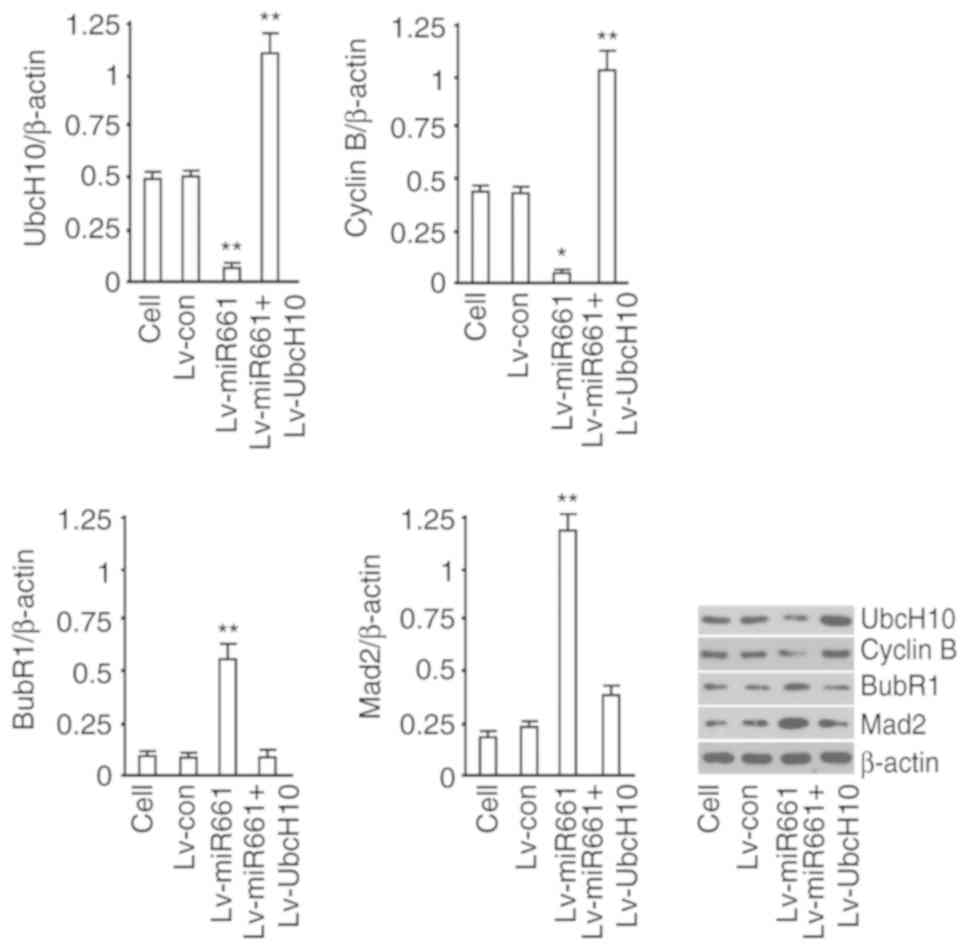
Effect of hsa-miR-661-3p on the
expression of functional proteins of SAC
We assessed UbcH10, cyclin B, BubR1 and Mad2 in the
hsa-miR-661-3p-overexpressed A549 cells. The results revealed that
UbcH10 and cyclin B were decreased, while BubR1 and Mad2 were
increased by hsa-miR-661-3p expression but not in the Lv-Control
group (Fig. 5). These results
indicated that hsa-miR-661-3p expression could regulate the
expression of functional proteins of SAC by decreasing the
expression of UbcH10 protein.
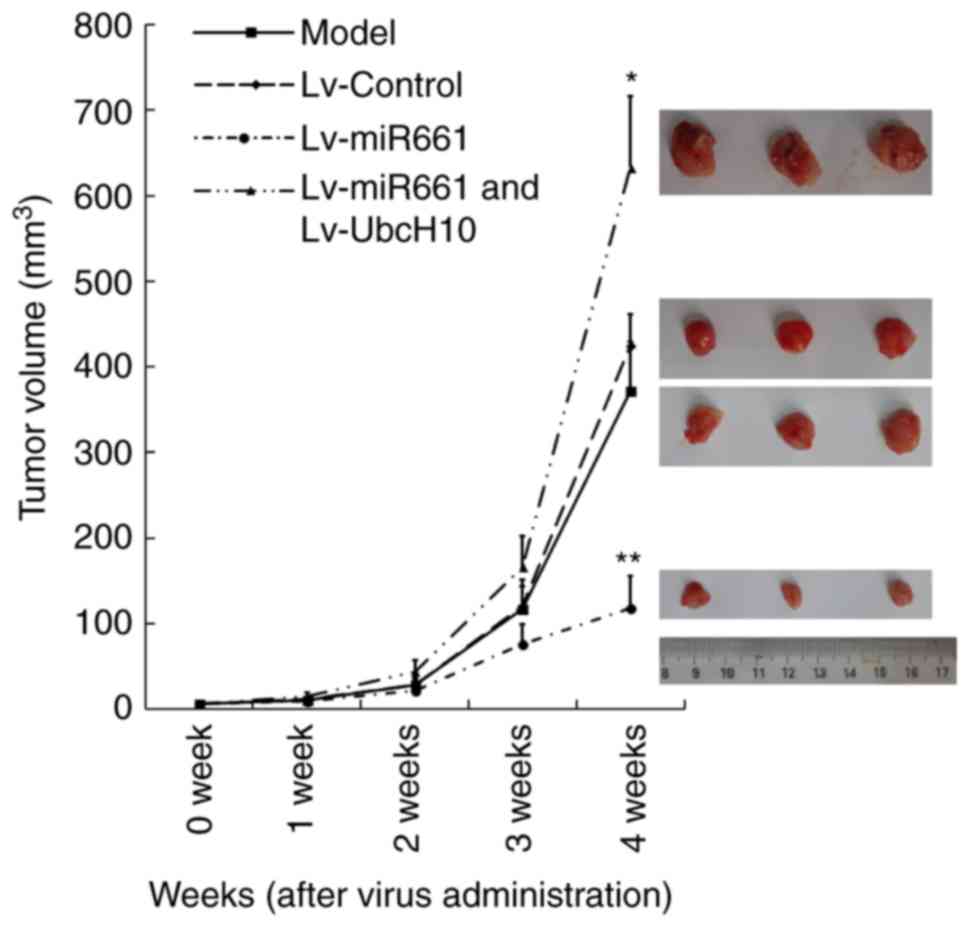
In vivo tumor suppression
An in vivo experiment revealed that 4
consecutive weeks of hsa-miR-661-3p expression significantly
decreased the tumor volume. However, UbcH10 overexpression could
significantly reverse the inhibition caused by hsa-miR-661-3p
deficiency. After administration for four weeks, the tumor volume
of the model group was 372.43±61.26 mm3, the Lv-control
group was 424.94±56.59 mm3, the hsa-miR-661-3p
expression group was 118.66±37.12 mm3, and the
hsa-miR-661-3p and UbcH10 expression group was 633.31±83.02
mm3. The tumor inhibition rates in the control group,
UbcH10 expression group and hsa-miR-661-3p and UbcH10 expression
group were 0, 68.12 and 0%, respectively, with a statistically
significant difference between the miR-661-3p expression group and
the other three groups (P<0.01) (Fig. 6).
Discussion
Since the 1980s, lung cancer has become the most
malignant cancer with the highest morbidity and mortality rates
worldwide. Statistics have revealed that the five-year survival
rate of lung cancer patients in the United States is only 15%, but
less than 8.9% in developing countries (13). In China, more than 700,000 new cases
of lung cancer are diagnosed each year, among them, non-small cell
lung cancer (NSCLC) accounts for approximately 85% of the total
cases (14). Therefore, a thorough
understanding of the mechanism of pathogenesis of NSCLC is critical
importance.
By comparing data of chromosomal status analysis in
NSCLC patients, the proportion of chromosomal abnormalities could
reach more than 40% compared with control subjects (14). The dysfunction of spindle assembly
checkpoint (SAC) is primarily responsible for this phenomenon and
it may be an important reason for the tumorigenesis of NSCLC
(15–17). Some SAC member proteins such as
BubR1, Mad2 are aberrantly expressed with the dysfunction of SAC
and are related to lung cancer (18,19).
Unfortunately, the specific mechanism of chromosome aberration in
NSCLC patients has not been clearly elucidated.
UbcH10 has been revealed to play an important role
in the dysfunction of SAC in NSCLC cells. UbcH10 is a member of the
ubiquitin binding enzyme gene family, and is a key member of the
ubiquitin/proteasome protein degradation pathway (UPP) in the human
body (20,21). Okamoto et al firstly revealed
that UbcH10 was upregulated in lung cancer tissues and cell lines
(7). UbcH10 was a significant
proto-oncogene that could cause genomic instability and cancer
susceptibility, and UbcH10-overexpressing transgenic mice lung
cancer was easily induced (14). In
our previous study, we revealed that UbcH10 expression had a
significant correlation with the pathological type and tissue
differentiation of lung cancer. It was also significant negative
correlation with the post-surgery survival time of patients with
NSCLC (P=0.012). These suggests that similar to other cancers,
UbcH10 may be used as a prognostic factor in patients with NSCLC
(8). In addition, an in
vitro study found that inhibition of UbcH10 expression
increased the sensitivity of SK-MES-1 cells to chemotherapeutic
drugs gemcitabine and paclitaxel, and the possible mechanism may be
due to the inhibition of tumor cell proliferation and cell cycle
change (8). Therefore, it was
proposed that UbcH10 maybe play an essential role in the
dysfunction of SAC in NSCLC cells through regulated mitosis.
In this study, we first detected and analyzed the
high expression of UbcH10 in NSCLC in transcription or post
transcription. The quantitative detection of the mRNA of the UbcH10
gene in NSCLC cells and clinical specimens revealed that the mRNA
of the UbcH10 gene in NSCLC tissue and NSCLC cells was slightly
higher than that of paracancerous tissue and normal lung epithelial
cells (data not show). Concurrently, western blotting detection
data revealed that the expression of UbcH10 protein in NSCLC tissue
and NSCLC cells was significantly higher than that of the
paracancerous and BEAS-2B cells. When comparing the difference of
UbcH10 mRNA and protein expression with controls, it was clearly
demonstrated that UbcH10 was highly expressed in the occurrence and
development of NSCLC and it was inactivated by post-transcriptional
regulation.
Post-transcriptional regulation is the regulation of
gene expression after RNA transcription, which is one of the
characteristics of eukaryotic gene expression (22). As one of the classical
post-reduction regulation mechanisms, miRNA combines with the 3′UTR
region of the target gene to inhibit protein translation,
therefore, it has been a hot spot in cancer research in recent
years. At present, there are approximately 1,000 types of miRNA in
human cells (of which >400 have been confirmed), which only
account for approximately 1% of the protein encoding genes,
however, they regulate approximately 30% of gene expression.
Approximately 50% of miRNAs are located in the fragile sites or
regions of chromosomes which are amplified or missing in cancer
(23,24). Studies had confirmed that the
expression profile of miRNAs is closely related to the type,
progression, patient survival rate and prognosis of lung cancer.
miRNAs could be used as cancer suppressors or cancer promoting
factors to regulate cell proliferation, apoptosis, invasion,
metastasis and angiogenesis (25).
Yanaihara et al used a miRNA microarray
analysis method to compare the miRNA expression profiles of 104
NSCLC tissues with normal lung tissues, and the results revealed
that 43 miRNAs had significant expression differences, 15 miRNAs
were upregulated and 28 miRNAs were downregulated (26). These findings built the foundation
for the research of miRNAs in lung cancer. Studies have revealed
that 5-miRNAs (miR34c-5p, miR34a, miR25, miR191 and let-7a) could
accurately classify lung adenocarcinoma and lung squamous cell
carcinoma (27). By comparing the
expression profiles of 122 NSCLC patients (62 squamous cell
carcinomas and 60 adenocarcinomas), Lebanony et al found
that the sensitivity of miR205 to squamous cell carcinoma was 96%
and the specificity was 90% when distinguishing between lung
squamous cell carcinoma and lung adenocarcinoma, which was a highly
specific biomarker for lung squamous cell carcinoma (28). A study conducted by Lima Queiroz
et al revealed that malate dehydrogenase 1 (MdH1) was
overexpressed in NSCLC tissue and it had important prognostic
value. The authors screened miRNA for the 3′UTR region of the
target MdH1 gene and the results revealed that miRNA126-5p
expression could inhibit apoptosis by inhibiting the expression of
MdH, while the inhibition of mitochondrial respiratory disease
ultimately induced apoptosis in NSCLC cell lines (29). Karagur et al demonstrated
that the high expression of miR-200 could reduce the expression of
N-cadherin and vimentin by increasing the expression of E-cadherin
and targeting ZEB1 to inhibit the ZEB1. EMT is considered to be an
initiator of tumor malignancy and miR-200 was capable of impeding
the EMT of NSCLC A549 cell line (30). Thus, we wondered whether the
increase of aberrant expression for UbcH10 in NSCLC tissues and
cells was caused by the inactivation of miRNA regulatory
mechanisms. Fortunately, in this study we were convinced that the
high expression of UbcH10 in NSCLC tissues and cells was due to
decreased expression of hsa-miR-661-3p.
The UbcH10 gene 3′UTR region was predicted by the
biological prediction software, and the content of miRNA in the
NSCLC tissues and cells in the pre-ranked seed region was
quantitatively analyzed. The results revealed that the content of
hsa-miR661-3p in the NSCLC tissues and cells was negatively
associated with the expression of the UbcH10 protein. The
luciferase reporter gene experiment further confirmed that
hsa-miR-661-3p could negatively regulate the expression of the
target gene by the combination of the 3′UTR region of gene UbcH10.
Hsa-miR-661-3p is located on human chromosome 8. At present, there
are only few studies of this miRNA and its relationship to cancer.
A study by Ali et al revealed that the combined analysis of
hsa-miR-661-3p and ATG-4B gene content could be a marker for
clinical diagnosis and prognosis of liver cancer (31). In addition, it has been reported
that hsa-miR-661-3p played an important role in the maintenance of
redox and metabolic balance of intestinal cancer cells (32). In the present study, to the best of
our knowledge, we are the first to report that hsa-miR-661-3p could
affect the dysfunction of SAC by regulating the expression of
UbcH10 as a target gene, while the dysfunction of SAC could lead to
the separation of chromosomes, rendering cell proliferation
uncontrollable. In vitro experiments revealed that the
overexpression of hsa-miR-661-3p through a lentivirus system could
effectively inhibit the proliferation of A549 cells, which invaded
and blocked the cells in the G2 phase. The in vivo
experimental data revealed that overexpression of hsa-miR-661-3p
could effectively inhibit the proliferation activity of the
subcutaneous tumor-bearing cancer of nude mice of A549 cells. The
UbcH10 gene suppression experimental group revealed that the
inhibitory effect of hsa-mi661-3p on NSCLC was achieved through the
target gene UbcH10. The SAC member protein BubR1 (Mad3), Mad2 and
cyclin B also relied on D-box and KEN-box competitively inhibiting
the combination of Cdc20 and APC, which inhibit the activity of
APC/C as a pseudo-substrate. If BubR1, Mad2 and cyclin B did not
comply with the strict timing and APC/C complex dissociation and
that were degraded by the UbcH10-APC/C complex in advance, it could
cause the dysfunction of SAC and the termination of the checkpoint,
leading eventually to the abnormal separation of the sister
chromosomes and the cells withdrawing from mitosis in advance
(33–36).
The important significance of this study is that on
the basis of the functional research performed earlier on UbcH10 in
NSCLC, we conducted a preliminary analysis of the downstream route
of action and the upstream factors of UbcH10. The decrease of the
expression of hsa-mi-661-3p in the NSCLC tissues and cells directly
led to the increase of the expression of the UbcH10 protein. UbcH10
could promote progression of NSCLC randomly occurring through cycle
regulation, indicating that hsa-miR-661-3p may be a valuable target
for the clinical treatment of NSCLC. Detection of BubR1, Mad2 and
cyclin B also revealed that overexpression of UbcH10 in A549 cells
could affect the function of the UbcH10-APC/C complex, leading to
the early recognition and degradation of SAC member proteins BubR1
and Mad2 in the G2/M phase. This may cause the dysfunction of SAC
and lead to chromosomal segregation. We believe that these data
provided an important theoretical value for fully elucidating the
molecular mechanism of UbcH10 regulation of NSCLC.
For biological experiments, the wider applicability
of the study is important. Therefore, validating the results of
this study in expanded NSCLC cell lines and tissues will be the
first step for all following future study plans. Though this study
has systematically demonstrated that overexpression of
hsa-miR-661-3p can effectively inhibited the proliferation and
metastasis of non-small cell lung cancer by inhibiting the
expression of UbcH10, it did not elucidate the reasons for the
decrease of hsa-miR-661-3p in non-small cell lung cancer, which
required further study. In the future, the collection of a large
number of NSCLC clinical tissue specimens is planned, in order to
analyze and explain the reasons for the decrease in the expression
of hsa-miR-661-3p in NSCLC from an epigenetic point of view. The
upstream regulation of hsa-miR-6613p/UbcH10 will continue to be our
research target that helps us confirm hsa-miR-661-3p as a possible
biomarker for clinical screening and prognosis evaluation of
NSCLC.
Acknowledgements
The authors thank Dr Jun Wang for his technical
support.
Funding
The present study was supported by the National
Natural Sciences Fund Project of China (NSFC no. 81372529), the
Natural Science Fund Project of Shanghai (13ZR1414400), the
Innovation Fund Project of Education Commission in Shanghai
(14ZZ079), and the Special issue of Military Medical of the Second
Military Medical University (2012JS21).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL, XG and FL made substantial contributions to the
conception or design of the work, the acquisition, the analysis, or
the interpretation of data for the work; they also drafted the
manuscript or revised it critically for important intellectual
content. ZR substantially contributed to the conception or design
of the work, the acquisition, the analysis, or interpretation of
data for the study; ML and LZ contributed substantially to the
conception or design of the work, the acquisition, the analysis, or
the interpretation of data for the work; they drafted the
manuscript or revising it critically for important intellectual
content and performed most of the research works and wrote the
manuscript. All authors agreement to be accountable for all aspects
of the work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study and use of human samples was approved by
the Ethics Committee of Shanghai Pulmonary Hospital and written
informed consents were obtained from all patients. All the
protocols involving animal experiments were previously approved by
the Tongji University Experiment Animal Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Hoeijmakers JH: Genome maintenance
mechanisms for preventing cancer. Nature. 411:366–374. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blagosklonny MV and Pardee AB: Exploiting
cancer cell cycling for selective protection of normal cells.
Cancer Res. 61:4301–4305. 2001.PubMed/NCBI
|
|
3
|
Berlingieri MT, Pallante P, Sboner A,
Barbareschi M, Bianco M, Ferraro A, Mansueto G, Borbone E,
Guerriero E, Troncone G and Fusco A: UbcH10 is overexpressed in
malignant breast carcinomas. Eur J Cancer. 43:2729–2735. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berlingieri MT, Pallante P, Guida M, Nappi
C, Masciullo V, Scambia G, Ferraro A, Leone V, Sboner A,
Barbareschi M, et al: UbcH10 expression may be a useful tool in the
prognosis of ovarian carcinomas. Oncogene. 26:2136–2140. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Troncone G, Guerriero E, Pallante P,
Berlingieri MT, Ferraro A, Del Vecchio L, Gorrese M, Mariotti E,
Iaccarino A, et al: UbcH10 expression in human lymphomas.
Histopathology. 54:731–740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ieta K, Ojima E, Tanaka F, Nakamura Y,
Haraguchi N, Mimori K, Inoue H, Kuwano H and Mori M: Identification
of over expressed genes in hepatocellular carcinoma, with special
reference to ubiquitin-conjugating enzyme E2C gene expression. Int
J Cancer. 121:33–38. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M,
Miyazaki M and Nakagawara A: UbcH10 is the cancer-related E2
ubiquitin-conjugating enzyme. Cancer Res. 63:4167–4173.
2003.PubMed/NCBI
|
|
8
|
Zhao L, Jiang L, Wang L, He J, Yu H, Sun
G, Chen J, Xiu Q and Li B: UbcH10 expression provides a useful tool
for the prognosis and treatment of non-small cell lung cancer. J
Cancer Res Clin Oncol. 138:1951–1961. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carmichael JA, Wing-San Mak D and O'Brien
MA: Review of recent advances in the treatment of elderly and poor
performance NSCLC. Cancers (Basel). 10:E2362018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharma N and Graziano S: Overview of the
LUX-lung clinical trial program of afatinib for non-small cell lung
cancer. Cancer Treat Rev. 69:143–151. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bassanelli M, Sioletic S, Martini M,
Giacinti S, Viterbo A, Staddon A, Liberati F and Ceribelli A:
Heterogeneity of PD-L1 expression and relationship with biology of
NSCLC. Anticancer Res. 38:3789–3796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rubin H: Cancer development: The rise of
epigenetics. Eur J Cancer. 28:1–2. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tomasini R, Tsuchihara K, Tsuda C, Lau SK,
Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G
and Mak TW: TAp73 regulates the spindle assembly checkpoint by
modulating BubR1 activity. Proc Natl Acad Sci USA. 106:797–802.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Ree JH, Jeganathan KB, Malureanu L and
van Deursen JM: Overexpression of the E2 ubiquitin-conjugating
enzyme UbcH10 causes chromosome missegregation and tumor formation.
J Cell Biol. 188:83–100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Draviam VM, Xie S and Sorger PK:
Chromosome segregation and genomic stability. Curr Opin Genet Dev.
14:120–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nasmyth K, Peters JM and Uhlmann F:
Splitting the chromosome: Cutting the ties that bind sister
chromatids. Science. 288:1379–1385. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu L, Liu X, Chervona Y, Yang F, Tang MS,
Darzynkiewicz Z and Dai W: Chromium induces chromosomal
instability, which is partly due to deregulation of BubR1 and Emi1,
two APC/C inhibitors. Cell Cycle. 10:2373–2379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kato T, Daigo Y, Aragaki M, Ishikawa K,
Sato M, Kondo S and Kaji M: Overexpression of MAD2 predicts
clinical outcome in primary lung cancer patients. Lung Cancer.
74:124–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Doherty FJ, Dawson S and Mayer RJ: The
ubiquitin-proteasome pathway of intracellular proteolysis. Essays
Biochem. 38:51–63. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Summers MK, Pan B, Mukhyala K and Jackson
PK: The unique N terminus of the UbcH10 E2 enzyme controls the
threshold for APC activation and enhances checkpoint regulation of
the APC. Mol Cell. 31:544–556. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barad O, Meiri E, Avniel A, Aharonov R,
Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al:
MicroRNA expression detected by oligonucleotide microarrays: System
establishment and expression profiling in human tissues. Genome
Res. 14:2486–2494. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lai EC, Tomancak P, Williams RW and Rubin
GM: Computational identification of Drosophila microRNA genes.
Genome Biol. 4:R422003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH,
Kim NH, Dhanasekaran SM, Chinnaiyan AM and Athey BD: New class of
microRNA targets containing simultaneous 5′-UTR and 3′-UTR
interaction sites. Genome Res. 19:1175–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gilad S, Lithwick-Yanai G, Barshack I,
Benjamin S, Krivitsky I, Edmonston TB, Bibbo M, Thurm C, Horowitz
L, Huang Y, et al: Classification of the four main types of lung
cancer using a microRNA-based diagnostic assay. J Mol Diagn.
14:510–517. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Landi MT, Zhao Y, Rotunno M, Koshiol J,
Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola
FM, et al: MicroRNA expression differentiates histology and
predicts survival of lung cancer. Clin Cancer Res. 16:430–441.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lebanony D, Benjamin H, Gilad S, Ezagouri
M, Dov A, Ashkenazi K, Gefen N, Izraeli S, Rechavi G, Pass H, et
al: Diagnostic assay based on hsa-miR-205 expression distinguishes
squamous from nonsquamous non-small-cell lung carcinoma. J Clin
Oncol. 27:2030–2037. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lima Queiroz A, Zhang B, Comstock DE, Hao
Y, Eriksson M, Hydbring P, Vakifahmetoglu-Norberg H and Norberg E:
miR-126-5p targets malate dehydrogenase 1 in non-small cell lung
carcinomas. Biochem Biophys Res Commun. 499:314–320. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karagur ER, Ozay C, Mammadov R and Akca H:
Anti-invasive effect of Cyclamen pseudibericum extract on A549
non-small cell lung carcinoma cells via inhibition of ZEB1 mediated
by miR-200c. J Nat Med. 72:686–693. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ali MA, Matboli M, El-Khazragy N, Saber O,
El-Nakeep S, Abdelzaher HM, Shafei AE and Mostafa R: Investigating
miRNA-661 and ATG4-B mRNA expression as potential biomarkers for
hepatocellular carcinoma. Biomark Med. 12:245–256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gómez de Cedrón M, Acín Pérez R,
Sánchez-Martínez R, Molina S, Herranz J, Feliu J, Reglero G,
Enríquez JA and Ramírez de Molina A: MicroRNA-661 modulates redox
and metabolic homeostasis in colon cancer. Mol Oncol. 11:1768–1787.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lara-Gonzalez P, Scott MI, Diez M, Sen O
and Taylor SS: BubR1 blocks substrate recruitment to the APC/C in a
KEN-box-dependent manner. J Cell Sci. 124:4332–4345. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rape M, Reddy SK and Kirschner MW: The
processivity of multiubiquitination by the APC determines the order
of substrate degradation. Cell. 124:89–103. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Townsley FM, Aristarkhov A, Beck S,
Hershko A and Ruderman JV: Dominant-negative cyclin-selective
ubiquitin carrier protein E2-C/UBCH10 blocks cells in metaphase.
Proc Natl AcadSci USA. 94:2362–2367. 1997. View Article : Google Scholar
|
|
36
|
Patel D and McCance DJ: Compromised
spindle assembly checkpoint due to altered expression of UBCH10 and
Cdc20 in human papillomavirus type 16 E6- and E7-expressing
keratinocytes. J Virol. 84:10956–10964. 2010. View Article : Google Scholar : PubMed/NCBI
|