Introduction
Colorectal cancer (CRC) is one of the most common
malignancy types globally (1). In
the USA, from 2000–2013, although the morbidity and mortality rates
of CRC have decreased in adults ≥50 years of age, they have
increased significantly in adults <50 years of age (2). According to the latest figures, there
was an estimated 18.1 million new cases and 9.6 million
cancer-associated mortalities globally in 2018. However, the global
incidence (6.1%) and mortality (9.2%) rates of CRC in 2018 are the
third and second highest, respectively, of all cancer types
(3). The transition from normal
epithelium to development of CRC is a process involving multiple
genes, including the activation of pro-oncogenes and the
inactivation of tumor suppressor genes (4). Therefore, identification of novel
tumor markers and underlying molecular mechanisms may contribute to
the diagnosis, treatment and prognosis of CRC.
The leucine zipper downregulated in cancer 1 (LDOC1)
is a differentially-expressed gene identified by Nagasaki using the
RNA differential display technique in cancer cells (5). It encodes a protein that has the
leucine zipper-like motif and the SH3-binding domain that can
regulate gene transcription and intracellular signal transduction
(6). Previous studies indicated
that LDOC1 expression is decreased in numerous cancer types,
including papillary thyroid carcinoma, liver cancer and prostate
cancer (6–11). As a tumor suppressor gene, it has
been demonstrated to be involved in the regulation of the nuclear
factor-κB (NF-κB) signaling pathway in numerous cancer types,
including papillary thyroid carcinoma, cervical cancer and
pancreatic cancer, thereby promoting apoptosis and inhibiting
proliferation of cancer cells (6,12–13).
The decreased expression of LDOC1 is also associated with
methylation in ovarian and cervical cancer types (14,15).
Additionally, LDOC1 can regulate the release of inflammatory
mediators and thus affect inflammation (11); however, the significance of LDOC1
expression for cancer metastasis and progression is rarely
reported. Furthermore, only one publication has reported that LDOC1
may regulate the metastasis of osteosarcoma through the Wnt5a
signaling pathway (16). Studies
demonstrated that there is an indirect association between the
Wnt5a and Wnt/β-catenin signaling pathways (17,18).
It is well known that the Wnt/β-catenin signaling pathway serves a
crucial role in the development of numerous cancer types, including
cervical, ovarian and lung cancer, particularly in invasion,
migration and epithelial-mesenchymal transition (EMT) (19–21). A
number of studies demonstrated that some genes, including PLAG1
like zinc finger 2, G protein nucleolar 3 and deleted in bladder
cancer protein 1, that regulate the Wnt/β-catenin signaling pathway
affect invasion, migration and EMT in CRC (22–24).
However, the association between LDOC1 and the occurrence and
development of CRC has not been reported, and the potential
mechanisms of LDOC1 action in CRC have not been elucidated.
Therefore, the aim of the present study was to investigate the
LDOC1 expression and elucidate its molecular mechanism in CRC, thus
providing a novel potential biomarker for CRC.
Materials and methods
Tissue samples and cell culture
A total of 14 fresh samples of CRC and paired
adjacent normal colorectal tissue were collected from the Operation
Room of the First Affiliated Hospital of Chongqing Medical
University (Chongqing, China; June, 2017 to December, 2017)
(Table I). Subsequently, the RNA
was immediately extracted, which would be detected via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
according to subsequent protocols. However, another 53 paraffin
sections of CRC and normal tissue, which were obtained from the
Department of Pathology of the First Affiliated Hospital of
Chongqing Medical University (June, 2018 to September, 2018), were
used for detecting the levels of proteins, via the subsequent
immunohistochemistry protocols (Table
II). Therefore, there is no overlap between the two groups of
patients. All patients provided informed consent, and the present
study was approved by the Ethics Committee of the First Affiliated
Hospital of Chongqing Medical University. CRC cell lines (SW480,
Caco2, HCT116 and HT29) were purchased from the American Type
Culture Collection (Manassas, VA, USA). Cells were cultured in
RPMI-1640 (HyClone; GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% fetal bovine serum (cat. no. FSP500; ExCell
Biology Inc., Shanghai, China) for 3 days in 5% CO2 at
37°C. The Wnt/β-catenin pathway activator IM-12 (Selleck Chemicals,
Houston, TX, USA) was added to culture medium (1 µg/ml). After 24 h
post treatment at 37°C, the total cell lysates were collected and
protein level detected by western blotting, according to the
subsequent protocol.
 | Table I.Clinicopathological features of the
study cohort used for quantitative PCR. |
Table I.
Clinicopathological features of the
study cohort used for quantitative PCR.
|
Characteristics | Number |
|---|
| Number of
patients | 14 |
| Age (mean ± SD),
years | 61.36±10.60 |
| Sex
(male/female) | 8/6 |
| Cancer types (colon
cancer/rectal cancer) | 12/2 |
| Lymph node
metastasis (absent/present) | 9/5 |
| Distant metastasis
(absent/present) | 14/0 |
| TNM stage
(I/II/III/IV)a | 1/8/5/0 |
 | Table II.Clinicopathological features of the
study cohort used for immunohistochemistry analysis. |
Table II.
Clinicopathological features of the
study cohort used for immunohistochemistry analysis.
|
Characteristics | Number |
|---|
| Number of
patients | 53 |
| Age (mean ± SD),
years | 63.68±13.96 |
| Sex
(male/female) | 31/22 |
| Cancers types
(colon cancer) | 53 |
| Tumor size
(T1/T2) | 15/38 |
| Lymph node
metastasis | 28/25 |
| Distant
metastasis | 41/12 |
| TNM stage
(I/II/III/IV)a | 13/15/13/12 |
Cell transfection and puromycin
screening
Human LDOC1 cDNA was amplified and inserted into a
lentiviral vector (lentivirus titer, 2.1×108 TU/ml). The
recombinant lentiviral vector expressing LDOC1 (Lv-LDOC1) and the
empty vector (Lv-NC) were purchased from GeneCopoeia, Inc.
(Rockville, MD, USA). With the assistance of Polybrene (5 µg/ml;
GeneCopoeia, Inc.), HCT-116 and Caco2 cells were transfected with
Lv-LDOC1 or Lv-NC vectors, according to the manufacturer's
protocol. Puromycin resistance markers were included in the vector;
therefore, stable cell lines were selected by adding 1 µg/ml
puromycin (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) to
the cell culture medium at 37°C for 3–5 days after 48 h of
transfection.
Reverse transcription-polymerase chain
reaction (RT-PCR) and RT-qPCR
Total RNA was extracted from fresh CRC tissue
samples and CRC cells with TRIzol® reagent (Takara
Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocols. For RT-PCR, RNA was reverse-transcribed
into cDNA using PrimeScript™ RT reagent kit (Takara Biotechnology
Co., Ltd.). RT-PCR primers were purchased from Takara Biotechnology
Co., Ltd., and the sequences were as follows: LDOC1, forward,
5′-CTATGCTGCCACTTCACATCC-3′ and reverse,
5′-GTGAGCTGTCCAAATCAATGTC-3′ (126 bp); and β-actin, forward,
5′-ACTCTTCCAGCCTTCCTTCCT-3′ and reverse,
5′-ACTCGTCATACTCCTGCTTGCT-3′ (314 bp). RT-PCR was performed with Ex
Taq® DNA polymerase (Takara Biotechnology Co., Ltd.)
under the following thermal conditions: 95°C for 2 min, 95°C for 30
sec, 55°C for 30 sec, 72°C for 30 sec and a final extension at 72°C
for 3 min. The RT-PCR duration for LDOC1 was 33 cycles, and for
β-actin it was 23 cycles. The products of RT-PCR were
electrophoresed in 2% agarose gels, and subjected to densitometric
analysis with a gel imaging system (Quantity One 1-D Analysis
Software version 4.6.2; Bio-Rad Laboratories, Inc., Hercules, CA,
USA). For RT-qPCR, total RNA was used for first-strand cDNA
synthesis using a PrimeScript™ RT reagent kit with gDNA Eraser
(Takara Biotechnology Co., Ltd.). Primers were designed by
Genscript (Nanjing, China), and the sequences were as follows:
LDOC1, forward, 5′-GCATTCCTAATCAGCCTCCTCA-3′ and reverse,
5′-CAAAGCACTGTTTCATCTCATCG-3′ (121 bp); and β-actin, forward,
5′-CCACGAAACTACCTTCAACTCC-3′ and reverse,
5′-GTGATCTCCTTCTGCATCCTGT-3′ (132 bp). qPCR was performed with TB
Green™ Premix Ex Taq (Takara Biotechnology Co., Ltd.) in
triplicate. The amplification conditions were as follows: 95°C for
30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 1 min.
All samples were run in triplicate. The relative expression levels
of LDOC1 were measured using the 2−∆∆Cq method (25).
Cell proliferation assay
Cell Counting Kit-8 (cat. no. E1CK-000208-5; EnoGene
Biotechnology, Nanjing, China) was used to detect cell
proliferation. The stably transfected cell lines (HCT-116-LDOC1,
HCT-116-NC, Caco2-LDOC1 and Caco2-NC) were seeded in 96-well plates
at a density of 3,500 cells/well. CCK8 reagent was added into the
wells, and cell proliferation was evaluated at 0, 24, 48 and 72 h
by measuring the 450 nm absorbance with a microplate reader
(Bio-Rad Laboratories, Inc.).
Colony formation assay
The stably transfected cell lines were seeded in
6-well plates at a density of 500 cells/well for 2 weeks at 37°C,
and RPMI-1640 supplemented with 10% fetal bovine serum was replaced
every 3 days. At room temperature, the clones were fixed with 4%
paraformaldehyde for 15 min and stained with 0.1% crystal violet
for 15 min. The numbers of clones (>50 cells per colony) were
counted under a fluorescent microscope (×200 magnification, Leica
Microsystems GmbH, Wetzlar, Germany).
Cell cycle and apoptosis assays
For the cell cycle assay, HCT-116 and Caco2 cells
were transfected for 48 h and then digested with 0.25% EDTA-free
trypsin. The cells were washed with pre-chilled PBS twice and fixed
in 70% pre-chilled ethanol for 48 h at 4°C. RNase A (10 mg/ml;
Sigma-Aldrich; Merck KGaA) was added to cells and incubated for 30
min at 37°C in an atmosphere containing 5% CO2.
Subsequently, the cells were stained with propidium iodide (PI;
Beyotime Institute of Biotechnology, Shanghai, China) under the
same conditions. The cell-cycle distributions were examined by a
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and
analyzed by FlowJo version 7.6 software (FlowJo LLC, Ashland, OR,
USA). For cell apoptosis, the transfected cells (HCT-116 and Caco2)
were also digested with 0.25% EDTA-free trypsin and suspended in
PBS buffer. The cells were stained using an Annexin V-fluorescein
isothiocyanate (FITC)/PI kit (BD Pharmingen; BD Biosciences) in the
dark, and apoptosis was examined by a flow cytometer (BD
Biosciences) and analyzed by FlowJo version 7.6 software (FlowJo
LLC, Ashland, OR, USA).
Cell migration and invasion
assays
Cell migration and invasion assays were detected
using Transwell chambers (Corning Inc., Corning, NY, USA). For the
migration assays, the stably transfected cell lines (HCT-116 and
Caco2) were digested with trypsin and suspended (2.5×105
cells/ml) with RPMI-1640 medium. Cell suspension (200 µl) was added
to the upper chamber, and 700 µl RPMI-1640 medium containing 10%
FBS was added to the lower chamber. After 46 h, the migrated cells
were fixed with 4% paraformaldehyde and stained with 0.1% crystal
violet for 15 min each at room temperature. Subsequently, the cells
from five random fields were counted under a microscope (×200
magnification; Leica Microsystems GmbH, Wetzlar, Germany). For the
invasion assays, Matrigel (BD Biosciences) was diluted with
RPMI-1640 medium at a 1:8 ratio. A 100 µl aliquot of the mixture
was added to the upper chamber and incubated at 37°C for 5 h to
solidify. A 100 µl RPMI-1640 aliquot containing 1×105
cells was added to the upper chamber; the subsequent steps were
identical to the migration assay.
Immunohistochemical (IHC)
staining
The colorectal cancer tissue samples (n=53) were
fixed with 10% formalin at room temperature for 48 h and
paraffin-embedded to make paraffin sections with thickness of 5 µm.
The paraffin sections were incubated at 60°C for 2 h, successively
placed into xylene and graded ethanol (100% ethanol for 5 min, 95%
ethanol for 5 min and 70% ethanol for 5 min) for dewaxing and
hydration, and then washed with PBS thrice. For antigen repair, the
sections were placed into citric acid buffer (10 mmol/l, pH6.0;
Sigma-Aldrich; Merck KGaA) for 25 min at 95°C, and then cooled to
room temperature and washed with PBS thrice. Subsequently, the
sections were incubated with 3% hydrogen peroxide (cat. no.
SP-0023; Beijing Biosynthesis Biotechnology Co., Ltd., Beijing,
China) for 20 min at room temperature, followed by three washes
with PBS. These sections were blocked by incubating with 5% goat
serum (cat. no. SP-0023; Beijing Biosynthesis Biotechnology Co.,
Ltd.) for 20 min at room temperature, and incubated with a rabbit
polyclonal antibody (cat. no. bs-6543R; anti-LDOC1 antibody; 1:200
dilution; Beijing Biosynthesis Biotechnology Co., Ltd.) at 4°C
overnight. Subsequently, the sections were washed with PBS thrice
and horseradish peroxidase-conjugated goat anti-rabbit IgG (cat.
no. SP-0023; 1:500 dilution; Beijing Biosynthesis Biotechnology
Co., Ltd.) was added as a secondary antibody and incubated at room
temperature for 20 min. The slices were stained with
diaminobenzidine (DAB kit; OriGene Technologies, Inc., Beijing,
China) at room temperature for 2 min. The nuclei were
counterstained with hematoxylin at room temperature for 1 min and
washed immediately with tap water for 10 min. The slices were
successively placed into graded ethanol (70% ethanol for 5 min, 95%
ethanol for 5 min and 100% ethanol for 5 min) and xylene for
dehydration and sealed with neutral gum pieces. Image was captured
using a Leica microscope image system (×100 and ×400 magnification;
Leica Microsystems GmbH, Wetzlar, Germany). IHC scores were
determined according to the intensity of immunostaining (0, no
staining reaction; 1, mild reaction; 2, moderate reaction; and 3,
intense reaction) and the percentage of positive cells (0, no
positive cells; 1, ≤10% positive cells; 2, 11–50% positive cells;
3, 51–80% positive cells; and 4, >80% positive cells). An
overall score was derived by multiplying the intensity and
percentage scores (26).
Immunofluorescence assay
The HCT-116 and Caco2 cells were transfected for 48
h and seeded in 24-well plates with glass coverslips at 37°C
overnight. The cells were fixed with 4% paraformaldehyde for 20 min
at room temperature and washed with PBS thrice. The samples were
soaked in 0.5% Triton X-100 (Beyotime Institute of Biotechnology)
at room temperature for 20 min. Subsequently, the cells were
blocked with 1% bovine serum albumin (Beyotime Institute of
Biotechnology) at room temperature for 1 h and incubated with
primary antibody (cat. no. 8480; anti-β-catenin antibody; 1:100
dilution; Cell Signaling Technology, Inc., Danvers, MA, USA) at 4°C
overnight. Subsequently, the samples were incubated with Alexa
Fluor® 555-conjugated (cat. no. 4413; 1:500 dilution;
Cell Signaling Technology, Inc.) secondary antibody against rabbit
IgG for 1 h in the dark at 37°C, and washed with PBS thrice. The
cell nuclei were stained with DAPI (Sigma-Aldrich; Merck KGaA) at
room temperature for 5 min in the dark. The samples were sealed
with the sealing liquid, and then the images were observed under a
fluorescence microscope (×400 magnification; Leica Microsystems
GmbH).
Western blotting
Proteins were extracted from Caco2 and HCT-116 cells
at 48 h post-transfection using radioimmunoprecipitation assay
lysis buffer and phenylmethanesulfonyl fluoride (both from Beyotime
Institute of Biotechnology). These protein concentrations were
quantified with a Bicinchoninic Acid kit (Beyotime Institute of
Biotechnology). Total proteins (20–40 mg) were separated by 10%
SDS-PAGE and then transferred onto polyvinylidene fluoride
membranes. The membrane was blocked in 5% skim milk at room
temperature for 2–3 h and incubated with the primary antibodies at
4°C overnight. Subsequently, the membrane was washed with PBS
thrice and incubated with horseradish peroxidase-conjugated goat
anti-rabbit IgG (cat. no. bs-0295G-HRP; 1:5,000 dilution; Beijing
Biosynthesis Biotechnology Co., Ltd.) at room temperature for 1–2
h. Chemiluminescent reagents (BeyoECL Moon; Beyotime Institute of
Biotechnology) were used to detect protein levels. Western blot
bands were analyzed with ImageJ software (version 6.0; National
Institutes of Health, Bethesda, MD, USA). The primary antibodies
used included: Rabbit anti-p65 (cat. no. 8242; 1:1,000),
anti-phospho-IκBα (cat. no. 2859; 1:1,000), anti-B-cell lymphoma-2
(Bcl-2)-associated X (Bax; cat. no. 5023; 1:1,000), anti-Bcl-2
(cat. no. 3498; 1:1,000), anti-E-cadherin (cat. no. 3195; 1:1,000),
anti-N-cadherin (cat. no. 13116; 1:1,000), anti-vimentin (cat. no.
5741; 1:1,000), anti-β-catenin (cat. no. 8480; 1:1,000),
anti-glycogen synthase kinase-3β (GSK-3β; cat. no. 9315; 1:1,000)
and anti-c-myc (cat. no. 5605; 1:1,000) from Cell Signaling
Technology, Inc.; anti-cleaved caspase-3 (cat. no. WL01992; 1:500),
anti-Bcl-extra large (Bcl-xl; cat. no. WL01558; 1:500) and
anti-matrix metallopeptidase 2 (MMP2; cat. no. WL1579; 1:300) from
Wanleibio Co., Ltd. (Shanghai, China); and anti-GAPDH (cat. no.
bs-2188R; 1:5,000) from Beijing Biosynthesis Biotechnology Co.,
Ltd.
Statistical analysis
Statistical analyses were performed with GraphPad
Prism software version 5.0 (GraphPad Software, Inc., La Jolla, CA,
USA). Data are presented as the mean ± standard deviation. The
differences between two groups were analyzed using Student's
t-test. Any statistical differences between three groups were
evaluated using a one-way of variance with Dunnett's multiple
comparison post-hoc tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of LDOC1 in CRC tissues and
cell lines
mRNA and protein expression levels of LDOC1 in CRC
and normal colorectal tissues were detected by RT-qPCR and IHC.
mRNA levels of LDOC1 in 14 CRC tissues were significantly
downregulated, compared with the levels in paired normal colorectal
tissues (P<0.05; Fig. 1A). The
IHC results demonstrated that protein expression levels of LDOC1 in
CRC tissues (n=53) were significantly decreased, compared with
normal tissues (n=18; P<0.0001; Fig.
1B). Furthermore, the expression levels of LDOC1 in four CRC
cell lines were measured by RT-PCR analysis. LDOC1 expression was
notably repressed in the four CRC cell lines, compared with normal
colorectal tissues, particularly in HCT-116 and Caco2 cells
(Fig. 1C). Therefore, these two
cell lines were selected for subsequent experiments.
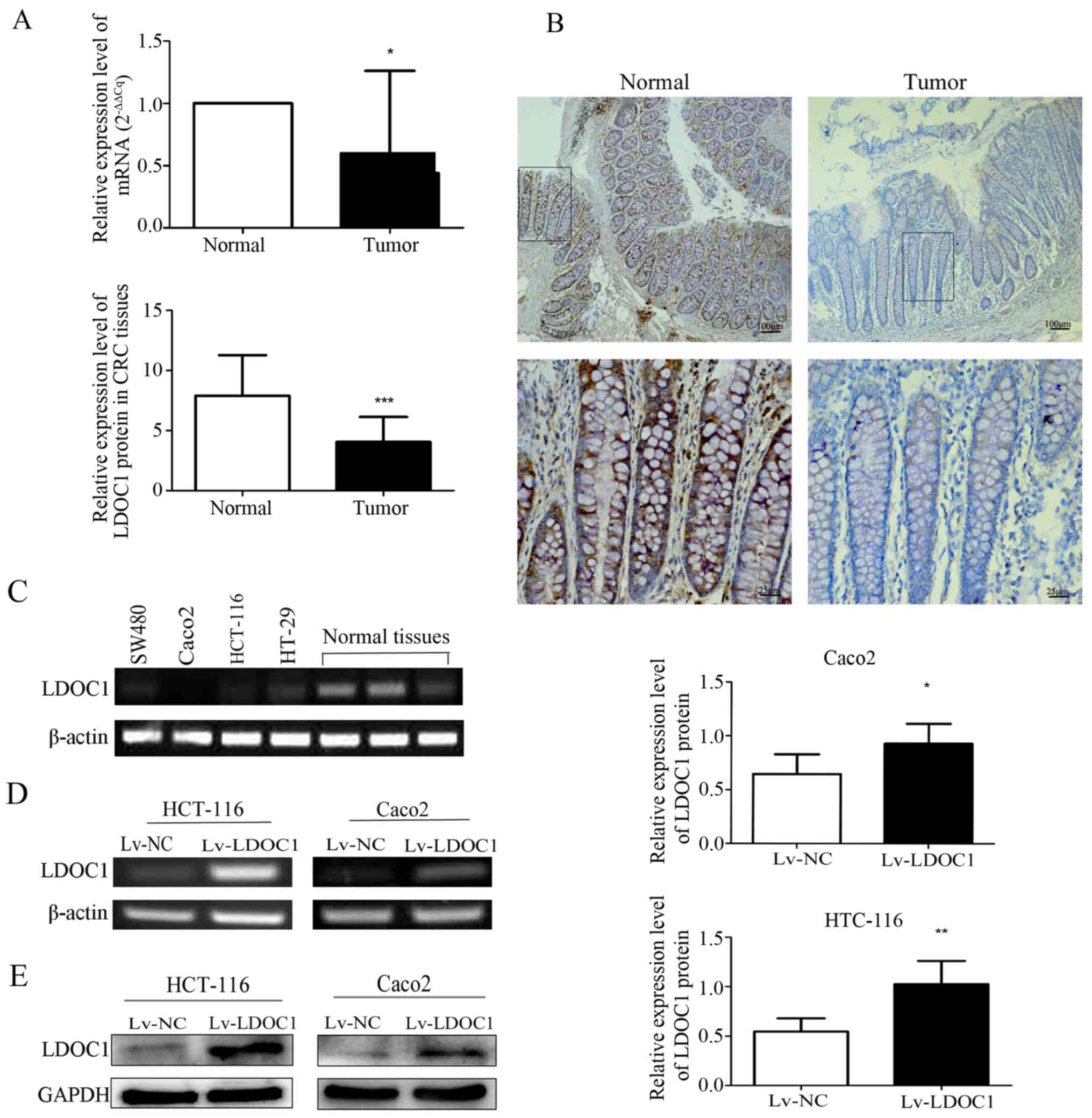 | Figure 1.Expression of LDOC1 in colorectal
cancer tissues, cell lines, and Caco2 and HCT-116 cells transfected
with lentiviral expression vector. (A) mRNA expression level of
LDOC1 in 14 paired CRC tissues and normal adjacent tissues was
detected by reverse transcription-quantitative polymerase chain
reaction with β-actin as a control. According to the quantitative
analysis, LDOC1 was expressed at significantly reduced levels in
CRC tissues, compared with normal adjacent tissues. (B)
Representative images of immunohistochemical staining for LDOC1
protein in normal colorectal tissues and colorectal cancer tissues
(×100 and ×400). LDOC1 was decreased significantly in CRC tissues,
compared with normal adjacent tissues. (C) RT-PCR was used to
detect mRNA expression of LDOC1 in four colorectal cancer cell
lines and normal colorectal tissues. LDOC1 expression level was
significantly reduced in four CRC cell lines, compared with normal
colorectal tissues. (D) mRNA expression level of LDOC1 in the
transfected cells was detected by RT-PCR, LDOC1 was notably
increased in Lv-LDOC1, compared with Lv-NC. (E) Protein expression
level of LDOC1 in the transfected cells was detected by western
blot analysis, LDOC1 was notably increased in Lv-LDOC1, compared
with Lv-NC. *P<0.05, **P<0.01 and ***P<0.001. LDOC1,
leucine zipper downregulated in cancer 1; CRC, colorectal cancer;
RT-PCR, reverse transcription-polymerase chain reaction; Lv-LDOC1,
LDOC1-overexpressing cells; Lv-NC, negative control cells. |
Verification of the LDCO1
overexpression following transfection
HCT-116 and Caco2 cells were transfected with
Lv-LDOC1 or Lv-NC. The mRNA and protein levels of LDOC1 were
detected by RT-PCR and western blot analysis in the HTC-116 and
Caco2 cell lines. The results demonstrated that the mRNA and
protein expression levels were significantly upregulated in the
HCT-116 (Lv-LDOC1; P<0.01) and Caco2 (Lv-LDOC1; P<0.05) cell
lines (Fig. 1D and E).
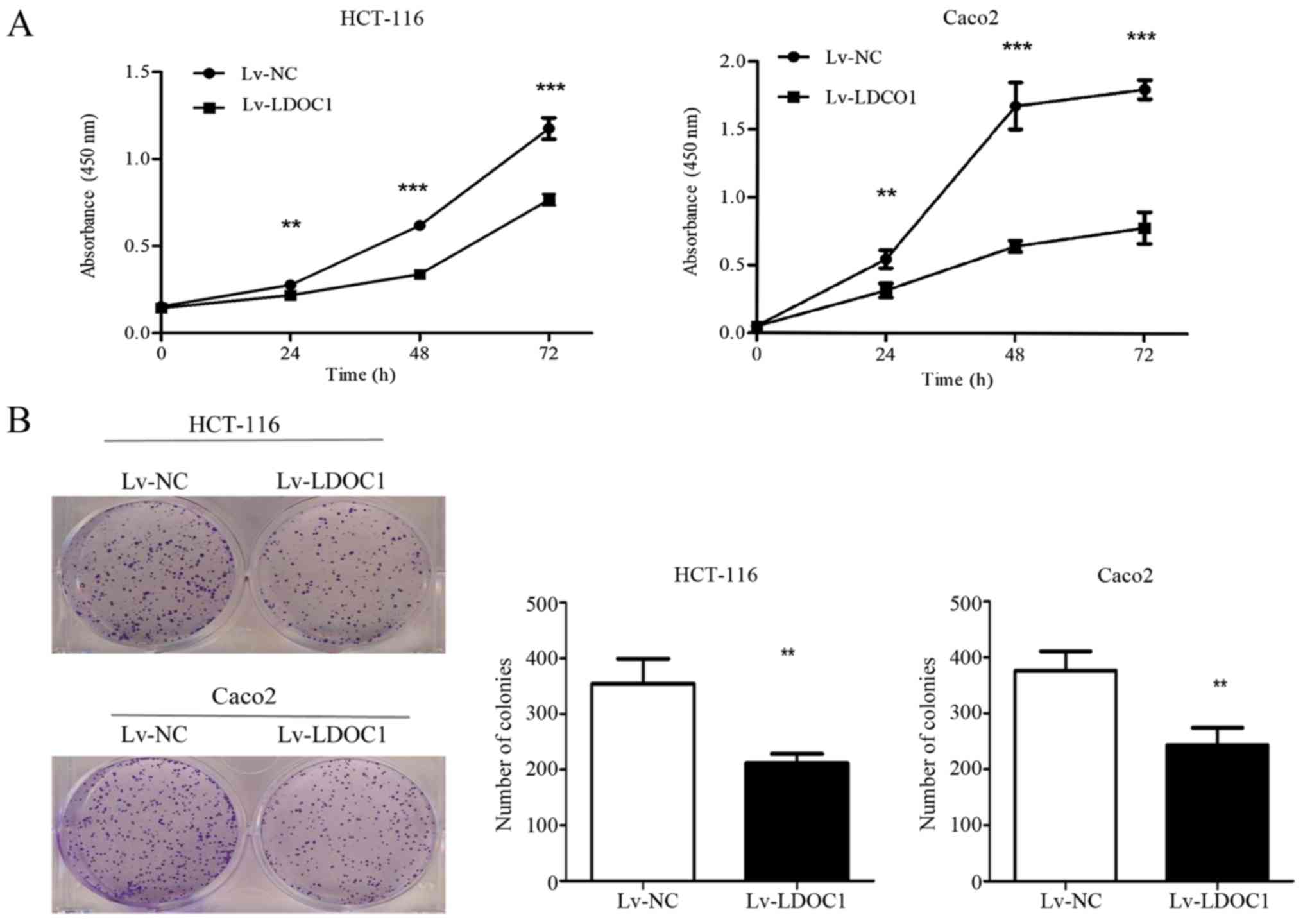
LDOC1 inhibits CRC cell proliferation
and colony formation
CCK8 and colony formation experiments were used to
examine the effect of LDOC1 on the proliferation capacity of CRC
cells. The CCK8 results demonstrated that LDOC1 overexpression
significantly inhibits the proliferation of CRC cells (HTC-116 and
Caco2), compared with the negative control at 24, 28 and 72 h
(P<0.01; Fig. 2A). Additionally,
the colony formation assay demonstrated that Lv-LDOC1 cells formed
significantly reduced colonies, compared with Lv-NC cells
(P<0.01; Fig. 2B). However, the
aforementioned results indicated that the effect of LDOC1 on Caco2
cells in the colony formation experiment is not as notable as that
in the CCK8 experiment. It is probable that the period of the CCK8
experiment is significantly shorter, compared with the colony
formation experiment. In the early stage, LDOC1 strongly inhibited
the proliferation of Caco2 cells, while in the later stage, Caco2
cells grew relatively faster, compared with the earlier stage, due
to LDOC1 having a weak effect on promoting apoptosis and cycle
arrest. However, these results demonstrated that LDOC1 could
suppress the proliferation of CRC cells.
LDOC1 induces cell-cycle arrest and
apoptosis in CRC cells
The effects of LDOC1 on cell cycle and apoptosis of
CRC cells were measured by flow cytometry. The results demonstrated
that the HCT116 (Lv-LDOC1) cells were significantly arrested in the
G0/G1 phase (P<0.0001), and Caco2 (Lv-LDCO1) cells were
significantly arrested in the G2/M phase (P<0.001) (Fig. 3A). The aforementioned results may be
due to LDOC1 regulating the expression of cyclin D1, cyclin E and
cyclin dependent kinase 2 (CDK2) in HCT-116 cells, resulting in
G0/G1 phase block, while LDOC1 regulates the expression of cyclin B
and CDK1 in Caco2 cells, resulting in G2/M phase block (27,28).
Annexin V-FITC/PI staining demonstrated that the rate of apoptosis
of the HTC116 (Lv-LDCO1; P<0.0001) and Caco2 (Lv-LDCO1;
P<0.0001) cells were significantly increased, compared with the
empty lentiviral vector cells (Fig.
3B). Additionally, LDOC1 has been indicated to regulate the
NF-κB signaling pathway, thereby affecting apoptosis of cancer
cells in a number of cancer types, including papillary thyroid
carcinoma, cervical cancer and pancreatic cancer (6,12,13).
Therefore, the critical factors of the NF-κB signaling pathway and
apoptosis-associated factors were detected by western blot
analysis. As depicted in Fig. 3,
expression of the apoptosis-associated factors cleaved caspase-3
and Bax increased, whereas expression of Bcl-2 and Bcl-xl, as well
as the critical factors of NF-κB signaling pathway, including p65
and p-IκBα, decreased in Lv-LDOC1 cells, compared with the
expression in Lv-NC cells (Fig.
3C). These results indicated that LDOC1 arrests cell cycle and
promotes apoptosis.
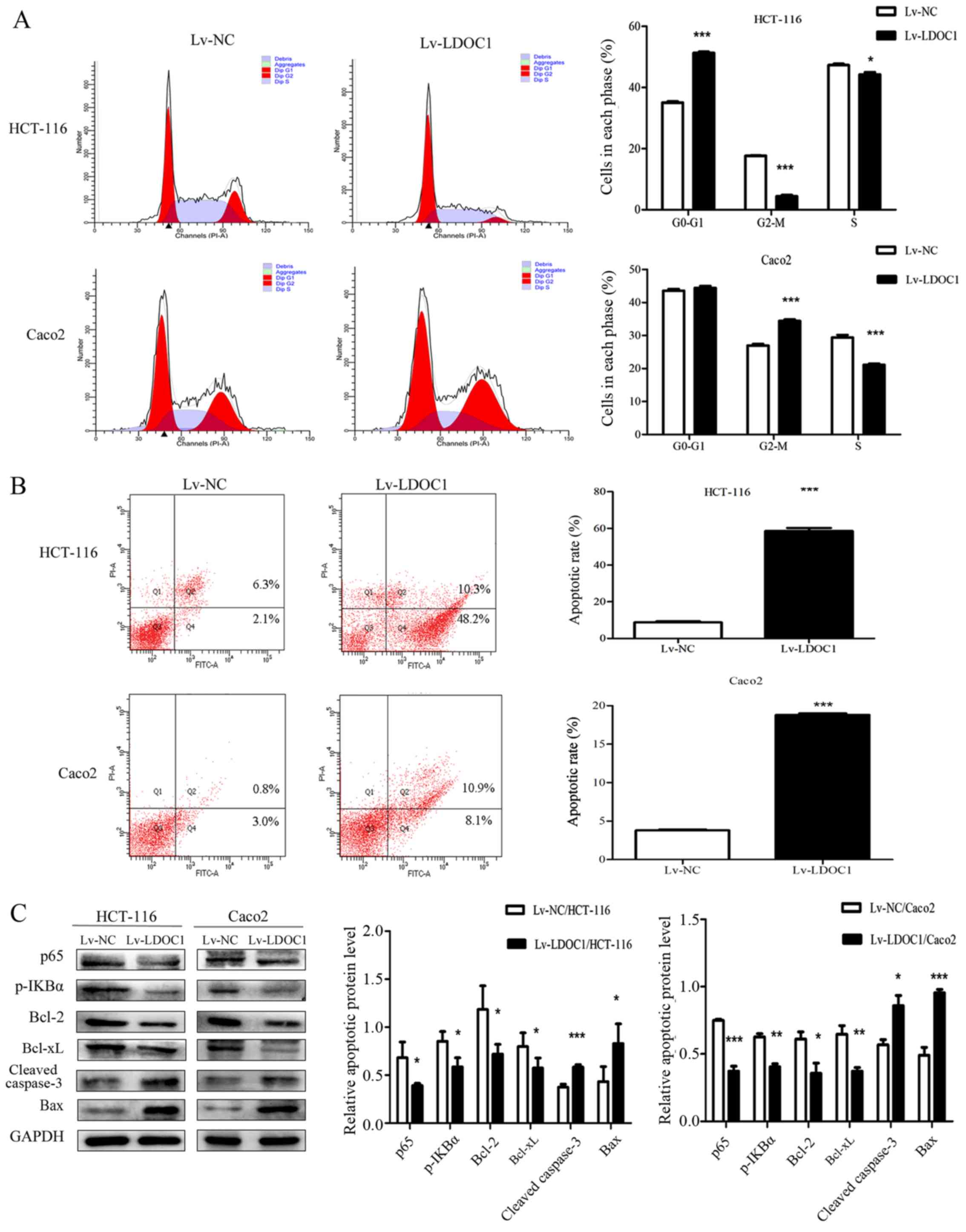 | Figure 3.Apoptosis rate and cell cycle phase
distribution in the transfected cells were detected by flow
cytometry, and the expression of apoptosis-associated proteins was
analyzed by western blotting. (A) From the cell-cycle
distributions, HCT116 (Lv-LDOC1) cells were significantly arrested
in the G0/G1 phase and Caco2 (Lv-LDOC1) cells were significantly
arrested in the G2/M phase. (B) The apoptosis rates of Lv-LDOC1
were increased, compared with Lv-NC. (C) Lv-LDOC1 increased cleaved
caspase-3 and Bax levels and reduced p65, p-IKBα, Bcl-2 and Bcl-xl
levels, compared with Lv-NC. *P<0.05, **P<0.01 and
***P<0.001 vs. Lv-NC. Bcl-2, B-cell lymphoma-2; Bcl-xl, B-cell
lymphoma extra-large; Bax, Bcl-2-associated X protein; p-IKBα:
phosphorylated inhibitor-κ-Bα; Lv-LDOC1, LDOC1-overexpressing
cells; Lv-NC, negative control cells. |
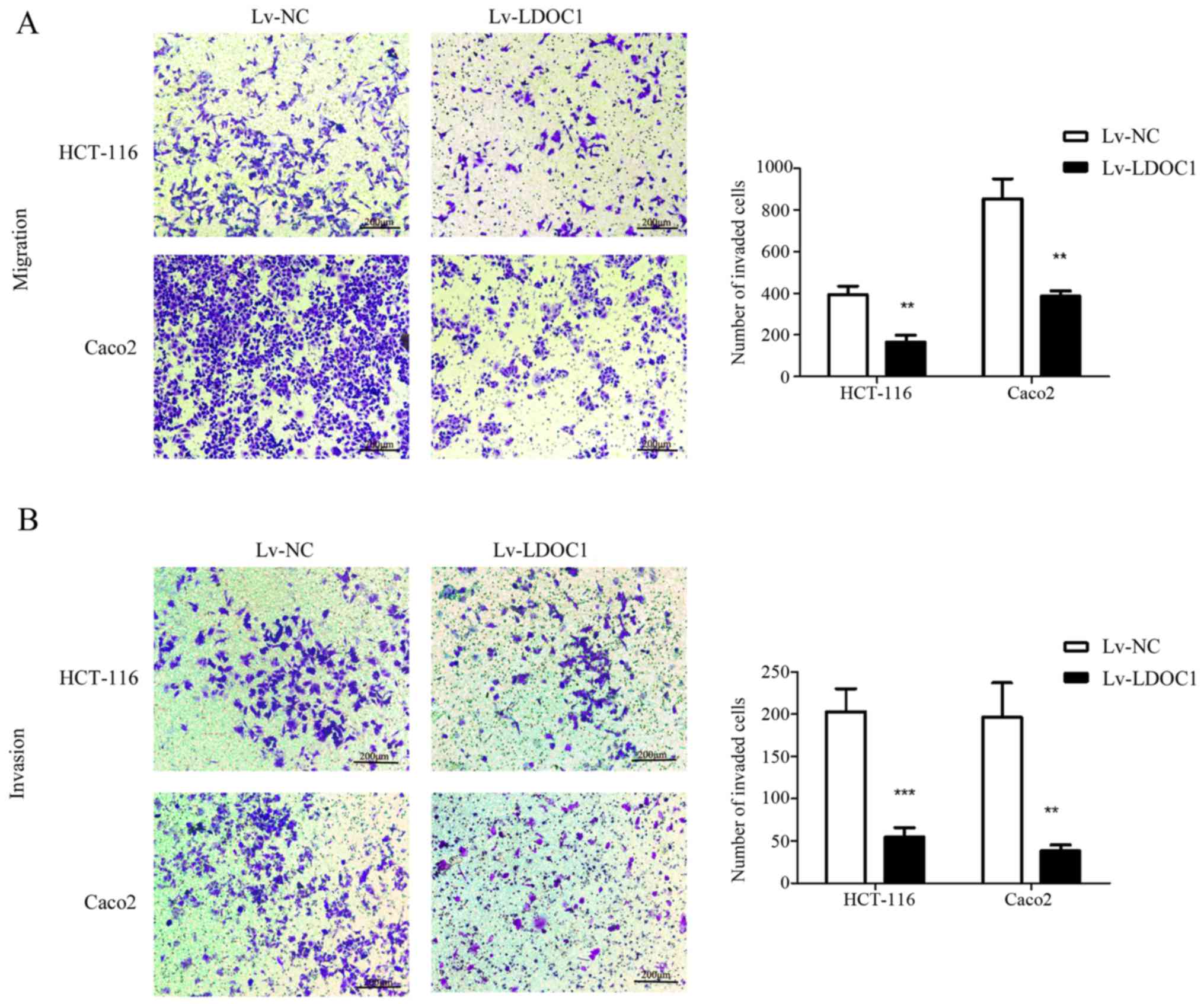
LDOC1 inhibits CRC cell migration and
invasion
To investigate the effect of LDOC1 on migration and
invasion of colon cancer cells, Transwell and Matrigel assays were
conducted. According to the Transwell and Matrigel assay results,
the number of migrating and invasive cells in the LDOC1
overexpression group was significantly reduced, compared with the
control group (P<0.01; Fig. 4A and
B). Therefore, LDOC1 inhibits the migration and invasion of CRC
cells.
LDOC1 inhibits migration, invasion and
EMT by downregulation of the Wnt/β-catenin pathway in CRC
cells
The Wnt/β-catenin signaling pathway serves a crucial
role in the development of numerous cancer types, including
cervical, ovarian and lung cancer, particularly in invasion,
migration and EMT (19–21). Therefore, western blot analysis was
used to investigate whether overexpression of LDOC1 had any effect
on the Wnt/β-catenin pathway, migration, invasion and EMT. As the
results demonstrated, the levels of the core protein β-catenin and
the downstream target gene c-myc of the Wnt/β-catenin pathway were
decreased in LDOC1-overexpressing CRC cells, compared with their
expression in control CRC cells. However, expression of GSK-3β, a
critical upstream enzyme of the Wnt/β-catenin pathway, was
increased in LDOC1-overexpressing cells, compared with the NC
cells. Furthermore, the levels of EMT-associated proteins,
including N-cadherin, vimentin and MMP2 were decreased, while
E-cadherin levels were increased in LDOC1-overexpressing cells,
compared with NC cells (Fig. 5A).
According to the results of cellular immunofluorescence,
LDOC1-overexpressing cells reduced the levels of total β-catenin
and nuclear β-catenin, compared with NC cells (P<0.05; Fig. 5B).
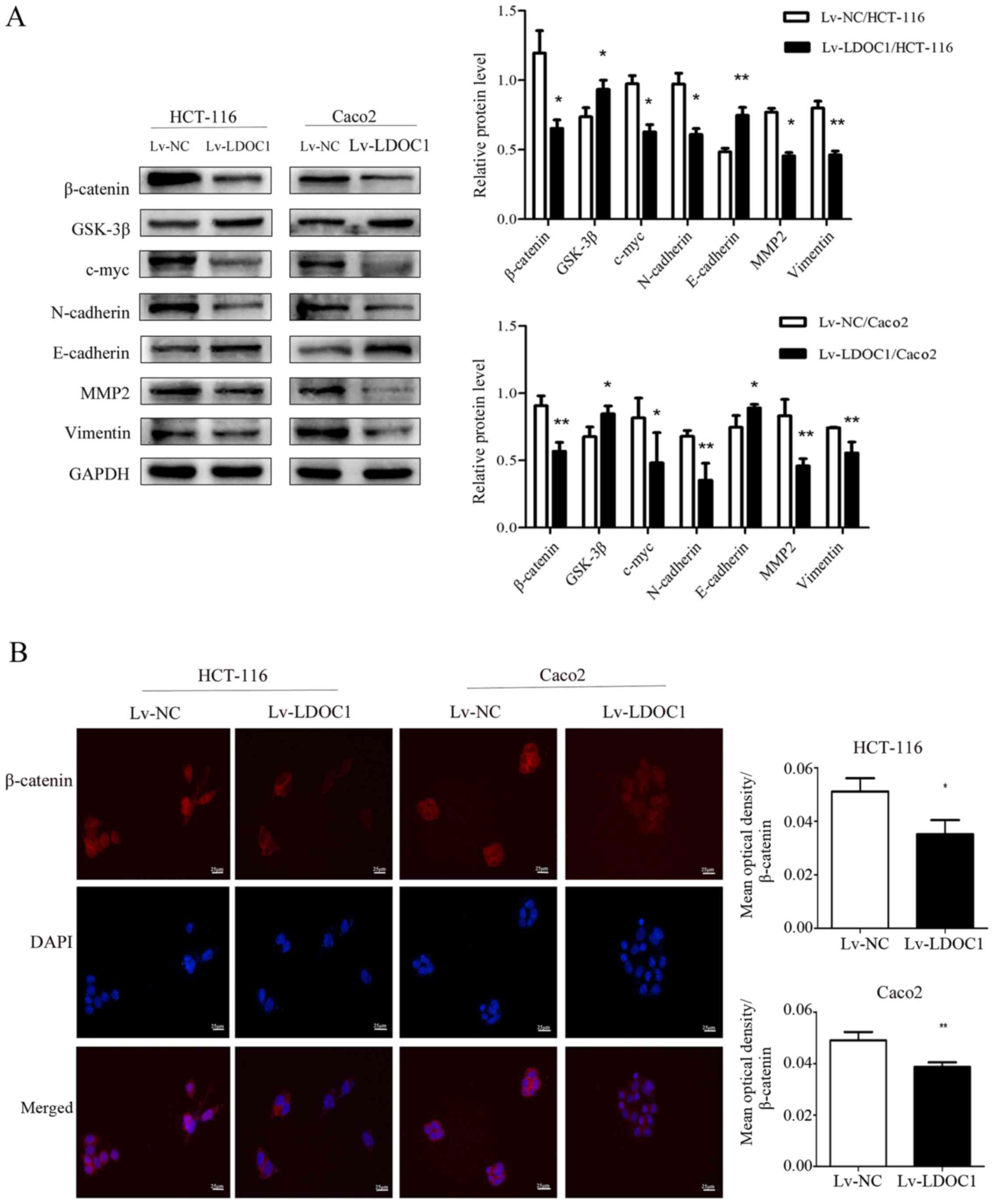 | Figure 5.Effects of LDOC1 overexpression on
proteins associated with the Wnt/β-catenin signaling pathway,
metastasis and epithelial-mesenchymal transition. (A) Western blot
analysis demonstrated that Lv-LDOC1 increased GSK-3β and E-cadherin
levels, and decreased β-catenin, c-myc, N-cadherin, MMP2 and
vimentin levels in HCT-116 and Caco2 cells. (B) Immunofluorescence
analysis for β-catenin in the transfected cells (×400). The
β-catenin protein is labelled in red, and cell nuclei are labelled
in blue by DAPI staining. Lv-LDOC1 decreased β-catenin levels,
compared with Lv-NC in HCT-116 and Caco2 cells. *P<0.05 and
**P<0.01 vs. Lv-NC. LDOC1, leucine zipper downregulated in
cancer 1; MMP2, matrix metalloproteinase 2; GSK-3β, glycogen
synthase kinase-3β; Lv-LDOC1, LDOC1-overexpressing cells; Lv-NC,
negative control cells. |
To further verify whether LDOC1 inhibits CRC cell
migration and invasion by regulating the Wnt/β-catenin pathway, the
Wnt/β-catenin pathway activator IM-12 was used. IM-12 has been
demonstrated to inhibit the expression level of GSK-3β, thereby
reducing phosphorylation of β-catenin and promoting the expression
of β-catenin as a core protein of the Wnt/β-catenin pathway
(29,30). Protein expression in Lv-NC cells,
Lv-NC cells treated with IM-12 (Lv-NC+IM-12 group) and Lv-LDOC1
cells treated with IM-12 (Lv-LDOC1+IM-12 group), was evaluated with
a western blot assay. The western blot assay results demonstrated
that GSK-3β was notably decreased, and β-catenin, MMP2, and
N-cadherin were notably increased in Lv-NC cells treated with
IM-12, compared with their levels in Lv-NC cells. This indicates
that the Wnt/β-catenin pathway is activated, and promotes migration
and invasion following IM-12 addition to CRC cells. It was also
determined that GSK-3β was notably increased and β-catenin, MMP2,
and N-cadherin were significantly decreased in Lv-LDOC1 cells
treated with IM-12, compared with their levels in Lv-NC cells
treated with IM-12 (Fig. 6). It was
further verified that LDOC1 inhibited migration, invasion and EMT
of CRC cells via downregulation of the Wnt/β-catenin pathway.
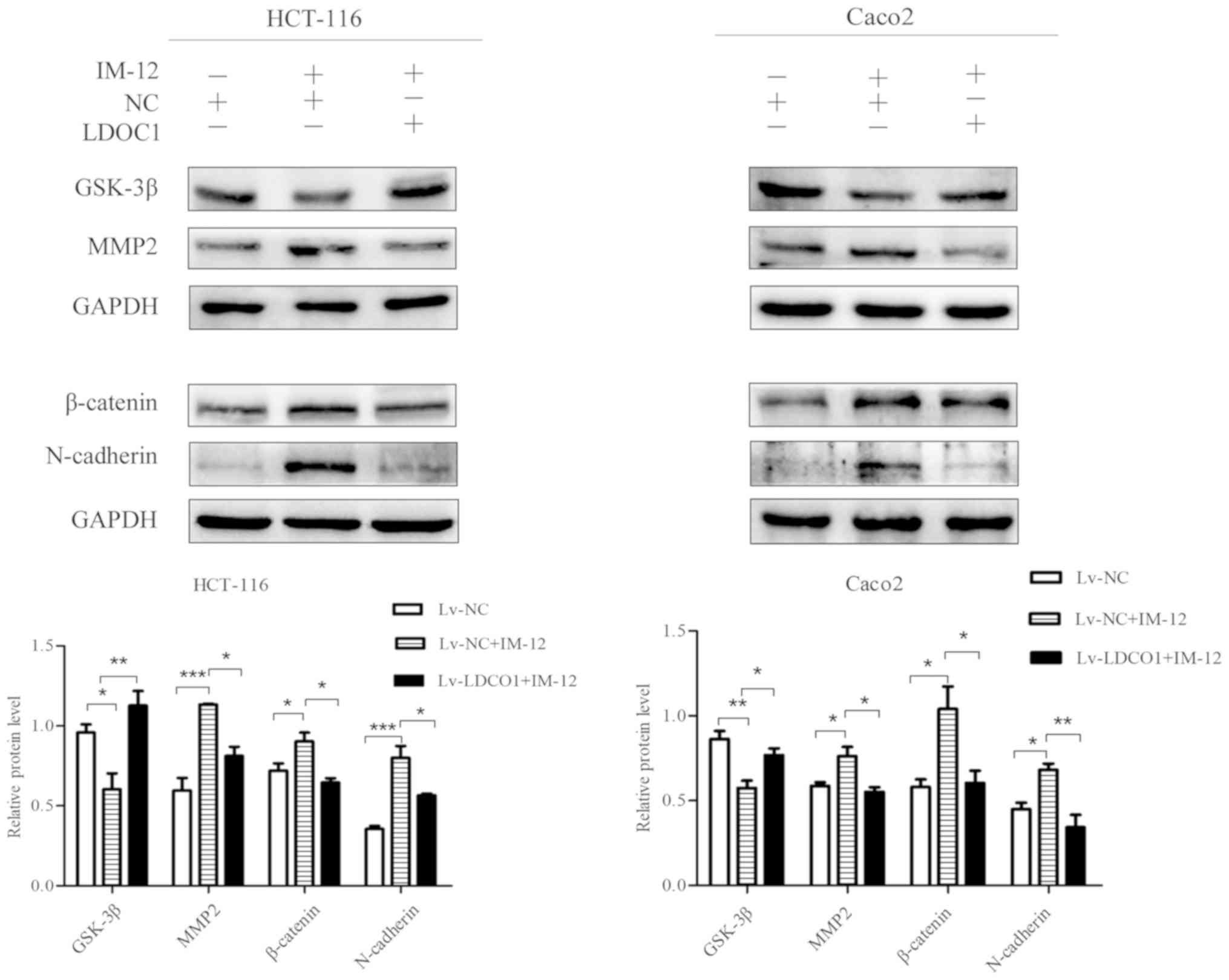 | Figure 6.Effects of IM-12 and LDOC1
overexpression on proteins associated with the Wnt/β-catenin
signaling pathway and metastasis in colon cancer cells. Western
blot analysis demonstrated that IM-12 decreased GSK-3β expression,
and increased expression of β-catenin, MMP2 and N-cadherin in Lv-NC
treated with IM-12, compared with their levels in Lv-NC. Lv-LDOC1
increased GSK-3β expression, and decreased expression of β-catenin,
MMP2 and N-cadherin in Lv-LDOC1 treated with IM-12, compared with
the levels in Lv-NC treated with IM-12. *P<0.05, **P<0.01 and
***P<0.001, with comparisons shown by brackets. LDOC1, leucine
zipper downregulated in cancer 1; GSK-3β: glycogen synthase
kinase-3β; MMP2: matrix metalloproteinase 2; Lv-LDOC1,
LDOC1-overexpressing cells; Lv-NC, negative control cells. |
Discussion
The occurrence and development of tumors result from
interactions of biological and environmental factors (31). However, in terms of the molecular
mechanism, the progression from normal to cancer cell is the result
of gene mutation, which includes activation of pro-oncogenes and
inactivation of tumor suppressor genes (4). Therefore, the identification of novel
tumor markers, their biological functions and molecular mechanisms
in CRC may contribute to the diagnosis, treatment and prognosis of
CRC.
The decreased expression of LDOC1 has been observed
in certain malignancy types, including papillary thyroid carcinoma,
liver cancer and prostate cancer (6–11).
Studies demonstrated that LDOC1 inhibits cell proliferation and
promotes apoptosis via downregulation of the NF-κB signaling
pathway in a number of cancer types, papillary thyroid carcinoma,
cervical cancer and pancreatic cancer (6,12–13).
However, the effect of LDOC1 expression on the migration and
invasion of cancer cells is rarely reported. In the present study,
using RT-qPCR, IHC and RT-PCR, it was determined, for the first
time, that LDOC1 expression is decreased in CRC tissues and cells.
Subsequently, it was indicated that LDOC1 overexpression inhibits
cell proliferation and cell cycle arrest, and promotes cell
apoptosis. Subsequently, the associated proteins were examined and
it was determined that the expression levels of p65, phosphorylated
inhibitor-κ-Bα (p-IKBα), Bcl-2 and Bcl-xl are decreased, while
expression levels of cleaved caspase-3 and Bax are increased. In
the NF-κB signaling pathway, p50/p65 binds to IκBα to form an
inactive trimer. Following phosphorylation of IκBα, p65 activates
and enters the nucleus to regulate the expression of
apoptosis-associated target genes, including anti-apoptotic Bcl-2
family, pro-apoptotic Bax and apoptosis executioner caspase-3
(6,32–36).
LDOC1 may promote apoptosis by inhibiting the NF-κB pathway in CRC,
but the regulatory mechanism involved requires further
elaboration.
Additionally, LDOC1 overexpression inhibited
migration and invasion of CRC cells, which was confirmed with
Transwell and Matrigel assays. In osteosarcoma, LDOC1 has been
demonstrated to inhibit cell metastasis by downregulating the Wnt5a
pathway (16). However, there is an
indirect association between the Wnt5a and Wnt/β-catenin signaling
pathways (17,18). The Wnt/β-catenin signaling pathway
serves an important role in regulating cell metastasis and EMT of
CRC (19–21). Therefore, the protein expression
levels of the core factors of the Wnt/β-catenin pathway and
metastasis-associated genes were detected. Western blot analysis
results indicated that LDOC1 overexpression reduced β-catenin and
c-myc levels, and increased GSK-3β levels, compared with control
cells. Immunofluorescence results also demonstrated that total and
nuclear β-catenin has reduced levels in the LDOC1 overexpression
group, compared with the control group. Particularly in HCT-116
cells, the localization of β-catenin in the control group was
notably changed from nucleus to cytoplasm following LDOC1
overexpression. As a core protein of the Wnt/β-catenin signaling
pathway, β-catenin accumulates in the cytoplasm and enters the
nucleus when the pathway is activated, which promotes the
transcription of multiple target genes and thus promotes
proliferation and metastasis (37,38).
Therefore, β-catenin can indirectly promote cell metastasis, while
reducing the expression of β-catenin in the nucleus can inhibit
cell metastasis. Additionally, the E-cadherin expression increased,
while MMP2, vimentin and N-cadherin protein levels decreased in
LDOC1-overexpressing cells, which were associated with EMT. In the
process of EMT, E-cadherin is reduced as a marker of epithelial
cells, resulting in the loss of epithelial cell polarity and the
reduction of intercellular adhesion (39). N-cadherin and vimentin increased as
markers of mesenchymal cells, which, together with MMP2, enhance
cell movement ability. This indicated that LDOC1 inhibited
migration, invasion and EMT of colon cancer cells by downregulating
the Wnt/β-catenin signaling pathway. Furthermore, IM-12 was used to
inhibit the expression of GSK-3β and reduce phosphorylation of
β-catenin to activate the Wnt/β-catenin signaling pathway. The
western blot assay results demonstrated that GSK-3β level is
notably increased, and β-catenin, MMP2 and N-cadherin levels are
significantly decreased in Lv-LDOC1 cells treated with IM-12,
compared with the levels in Lv-NC cells treated with IM-12. It was
further verified that LDOC1 inhibits metastasis of colon cancer
cells by downregulating the Wnt/β-catenin signaling pathway.
However, it is notable that there are differences in cell cycle
arrest and immunofluorescence between HCT-116 cells and Caco2
cells, which may be associated with the different types of tumor
cells and different degrees of malignancy. Although these
experiments have fully verified the effects of LDOC1 on
proliferation, metastasis and apoptosis of CRC cells in
vitro, they lack further validation in vivo. In the
following experiments, to further validate the hypothesis in
vivo, the use of another xenograft model to detect the effects
of LDOC1 on tumorigenesis and metastasis in nude mice should be
conducted.
In conclusion, as a tumor suppressor gene, LDOC1
inhibits cell proliferation and promotes cell apoptosis in CRC;
furthermore, it was determined that LDOC1 could inhibit metastasis
of CRC cells via downregulation of the Wnt/β-catenin signaling
pathway. These observations provide the novel direction for
molecular targeted therapy and prognosis of CRC. However, the
present study has some deficiencies, including the lack of the
expression levels of LDOC1 in normal colorectal cells, which will
be compensated by further study.
Acknowledgements
The authors would like to thank the Chongqing Key
Laboratory of Molecular Oncology and Epigenetics (Chongqing, China)
for technical guidance.
Funding
No funding was received.
Availability of data and materials
All the data used and analyzed during study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JJ, YL and ZJ made substantial contributions to the
conception and design of the study. JJ performed the experiments
and analyzed the data. YL participated in the collection of samples
and the collation of data. JJ and YL were involved in the drafting
of the manuscript. ZJ revised the study critically for important
intellectual content. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All patients signed informed consent forms, and the
present study was approved by the Ethics Committee of the First
Affiliated Hospital of Chongqing Medical University (Chongqing,
China).
Patient consent for publication
Patients consented for publication.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
CCK8
|
Cell Counting Kit-8
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RG, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo Y, Tsuchiya KD, Il Park D, Fausel R,
Kanngurn S, Welcsh P, Dzieciatkowski S, Wang J and Grady WM: RET is
a potential tumor suppressor gene in colorectal cancer. Oncogene.
32:2037–2047. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagasaki K, Manabe T, Hanzawa H, Maass N,
Tsukada T and Yamaguchi K: Identification of a novel gene, LDOC1,
down-regulated in cancer cell lines. Cancer Lett. 140:227–234.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao S, Wang Q, Li Z, Ma X, Wu L, Ji H and
Qin G: LDOC1 inhibits proliferation and promotes apoptosis by
repressing NF-κB activation in papillary thyroid carcinoma. J Exp
Clin Cancer Res. 34:1462015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee CH, Pan KL, Tang YC, Tsai MH, Cheng
AJ, Shen MY, Cheng YM, Huang TT and Lin P: LDOC1 silenced by
cigarette exposure and involved in oral neoplastic transformation.
Oncotarget. 6:25188–25201. 2015.PubMed/NCBI
|
|
8
|
Riordan JD and Dupuy AJ: Domesticated
transposable element gene products in human cancer. Mob Genet
Elements. 3:e266932013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Camões MJ, Paulo P, Ribeiro FR,
Barros-Silva JD, Almeida M, Costa VL, Cerveira N, Skotheim RI,
Lothe RA, Henrique R, et al: Potential downstream target genes of
aberrant ETS transcription factors are differentially affected in
Ewing's sarcoma and prostate carcinoma. PLoS One. 7:e498192012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Inoue M, Takahashi K, Niide O, Shibata M,
Fukuzawa M and Ra C: LDOC1, a novel MZF-1-interacting protein,
induces apoptosis. FEBS Lett. 579:604–608. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Griesinger AM, Witt DA, Grob ST, Georgio
Westover SR, Donson AM, Sanford B, Mulcahy Levy JM, Wong R, Moreira
DC, DeSisto JA, et al: NF-κB upregulation through epigenetic
silencing of LDOC1 drives tumor biology and specific
immunophenotype in Group A ependymoma. Neuro Oncol. 19:1350–1360.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thoompumkal IJ, Rehna K, Anbarasu K and
Mahalingam S: Leucine zipper down-regulated in cancer-1 (LDOC1)
interacts with guanine nucleotide binding protein-like 3-like
(GNL3L) to modulate nuclear factor-kappa B (NF-κB) signaling during
cell proliferation. Cell Cycle. 15:3251–3267. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagasaki K, Schem C, von Kaisenberg C,
Biallek M, Rösel F, Jonat W and Maass N: Leucine-zipper protein,
LDOC1, inhibits NF-kappaB activation and sensitizes pancreatic
cancer cells to apoptosis. Int J Cancer. 105:454–458. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Buchholtz ML, Brüning A, Mylonas I and
Jückstock J: Epigenetic silencing of the LDOC1 tumor suppressor
gene in ovarian cancer cells. Arch Gynecol Obstet. 290:149–154.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Buchholtz ML, Jückstock J, Weber E,
Mylonas I, Dian D and Brüning A: Loss of LDOC1 expression by
promoter methylation in cervical cancer cells. Cancer Invest.
31:571–577. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yong BC, Lu JC, Xie XB, Su Q, Tan PX, Tang
QL, Wang J, Huang G, Han J, Xu HW, et al: LDOC1 regulates Wnt5a
expression and osteosarcoma cell metastasis and is correlated with
the survival of osteosarcoma patients. Tumour Biol. Feb
1–2017.(Epub ahead of print). doi.org/10.1177/1010428317691188.
PubMed/NCBI
|
|
17
|
Chen X, Jia C, Jia C, Jin X and Gu X:
MicroRNA-374a inhibits aggressive tumor biological behavior in
bladder carcinoma by suppressing Wnt/β-catenin signaling. Cell
Physiol Biochem. 48:815–826. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park HW, Kim YC, Yu B, Moroishi T, Mo JS,
Plouffe SW, Meng Z, Lin KC, Yu FX, Alexander CM, et al: Alternative
Wnt signaling activates YAP/TAZ. Cell. 162:780–794. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang LZ, Huang LY, Huang AL, Liu JX and
Yang F: CRIP1 promotes cell migration, invasion and
epithelial-mesenchymal transition of cervical cancer by activating
the Wnt/β catenin signaling pathway. Life Sci. 207:420–427. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu R, Cai L, Chi Y, Ding X and Wu X:
miR-377 targets CUL4A and regulates metastatic capability in
ovarian cancer. Int J Mol Med. 41:3147–3156. 2018.PubMed/NCBI
|
|
21
|
Ling DJ, Chen ZS, Zhang YD, Liao QD, Feng
JX, Zhang XY and Shi TS: MicroRNA-145 inhibits lung cancer cell
metastasis. Mol Med Rep. 11:3108–3114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang YP, Guo PT, Zhu Z, Zhang H, Xu Y,
Chen YZ, Liu F and Ma SP: Pleomorphic adenoma gene like-2 induces
epithelial-mesenchymal transition via Wnt/β-catenin signaling
pathway in human colorectal adenocarcinoma. Oncol Rep.
37:1961–1970. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang X, Zha L, Li H, Liao G, Huang Z, Peng
X and Wang Z: Upregulation of GNL3 expression promotes colon cancer
cell proliferation, migration, invasion and epithelial-mesenchymal
transition via the Wnt/β-catenin signaling pathway. Oncol Rep.
38:2023–2032. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim HJ, Moon SJ, Kim SH, Heo K and Kim JH:
DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key
regulator of cancer progression, in colon cancer. Cell Death Dis.
9:8312018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tao Y, Tao T, Gross N, Peng X, Li Y, Huang
Z, Liu L, Li G, Chen X and Yang J: Combined effect of IL-12Rβ2 and
IL-23R expression on prognosis of patients with laryngeal cancer.
Cell Physiol Biochem. 50:1041–1054. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen J: The cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patil M, Pabla N and Dong Z: Checkpoint
kinase 1 in DNA damage response and cell cycle regulation. Cell Mol
Life Sci. 70:4009–4021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Zhu Y, Sun C, Wang T, Shen Y, Cai
W, Sun J, Chi L, Wang H, Song N, et al: Feedback activation of
basic fibroblast growth factor signaling via the Wnt/β-catenin
pathway in skin fibroblasts. Front Pharmacol. 8:322017.PubMed/NCBI
|
|
30
|
Cai T, Sun D, Duan Y, Wen P, Dai C, Yang J
and He W: WNT/β-catenin signaling promotes VSMCs to osteogenic
transdifferentiation and calcification through directly modulating
Runx2 gene expression. Exp Cell Res. 345:206–217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Johnson IT and Belshaw NJ: Environment,
diet and CpG island methylation: Epigenetic signals in
gastrointestinal neoplasia. Food Chem Toxicol. 46:1346–1359. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayden MS and Ghosh S: Regulation of NF-κB
by TNF family cytokines. Semin Immunol. 26:253–266. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu L, Li L, Medeiros LJ and Young KH:
NF-κB signaling pathway and its potential as a target for therapy
in lymphoid neoplasms. Blood Rev. 31:77–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shukla S, Shankar E, Fu P, MacLennan GT
and Gupta S: Suppression of NF-κB and NF-κB-regulated gene
expression by apigenin through IκBα and IKK pathway in TRAMP mice.
PLoS One. 10:e01387102015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Prasad S, Ravindran J and Aggarwal BB:
NF-kappaB and cancer: How intimate is this relationship. Mol Cell
Biochem. 336:25–37. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brentnall M, Rodriguez-Menocal L, De
Guevara RL, Cepero E and Boise LH: Caspase-9, caspase-3 and
caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell
Biol. 14:322013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dai G, Zheng D, Wang Q, Yang J, Liu G,
Song Q, Sun X, Tao C, Hu Q, Gao T, et al: Baicalein inhibits
progression of osteosarcoma cells through inactivation of the
Wnt/β-catenin signaling pathway. Oncotarget. 8:86098–86116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kwon YJ, Baek HS, Ye DJ, Shin S, Kim D and
Chun YJ: CYP1B1 enhances cell proliferation and metastasis through
induction of EMT and activation of Wnt/β-catenin signaling via Sp1
upregulation. PLoS One. 11:e01515982016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|