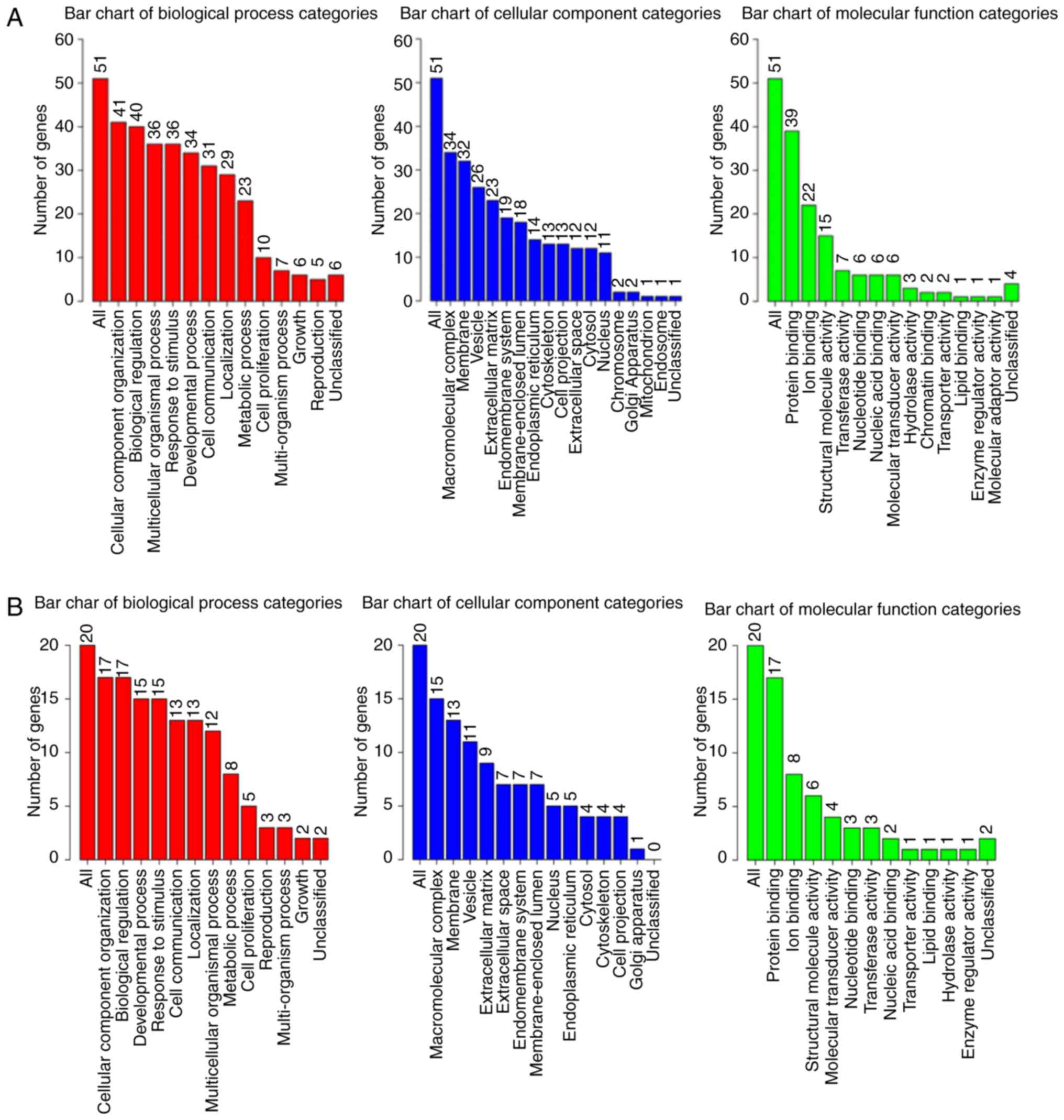
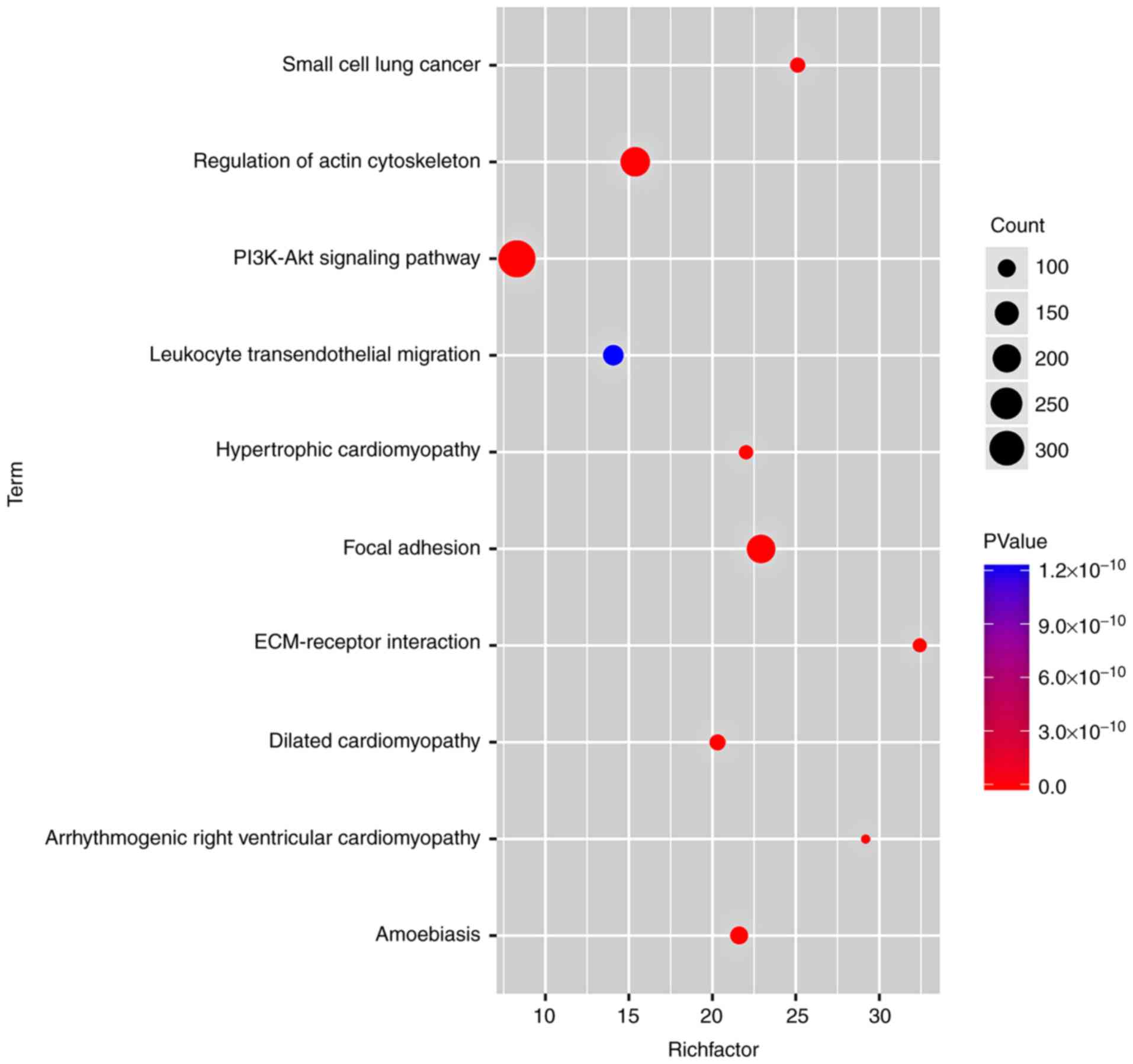
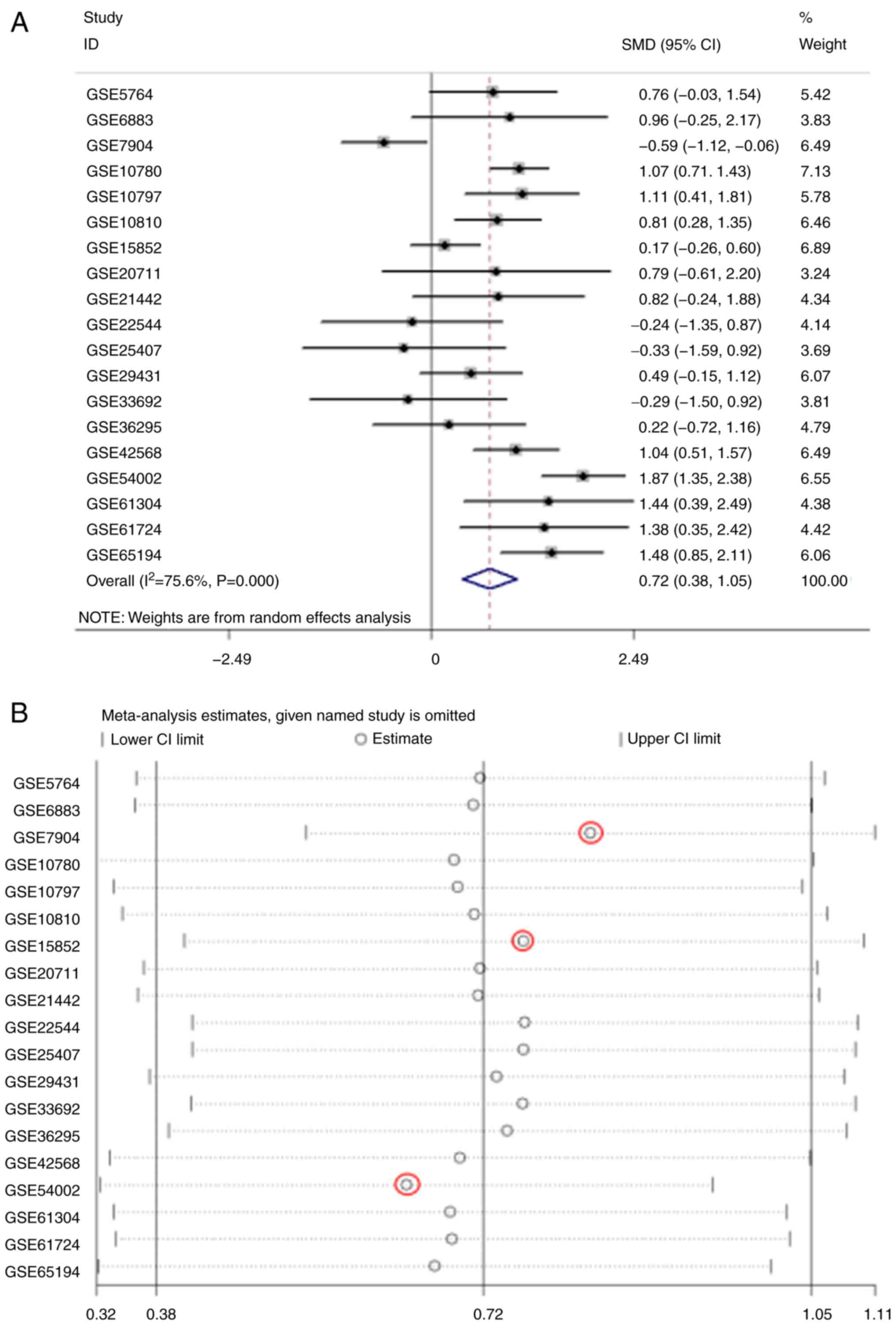
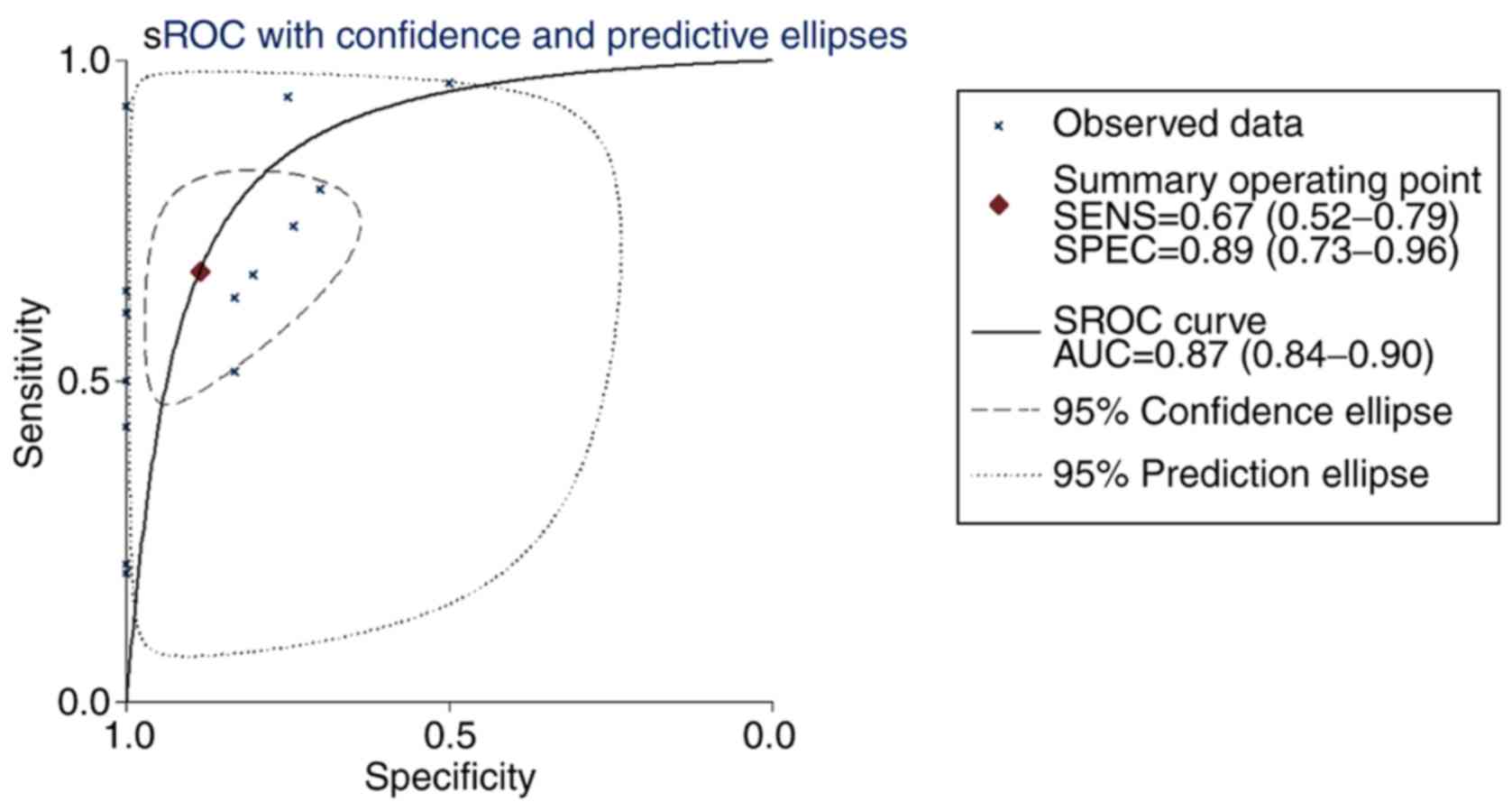
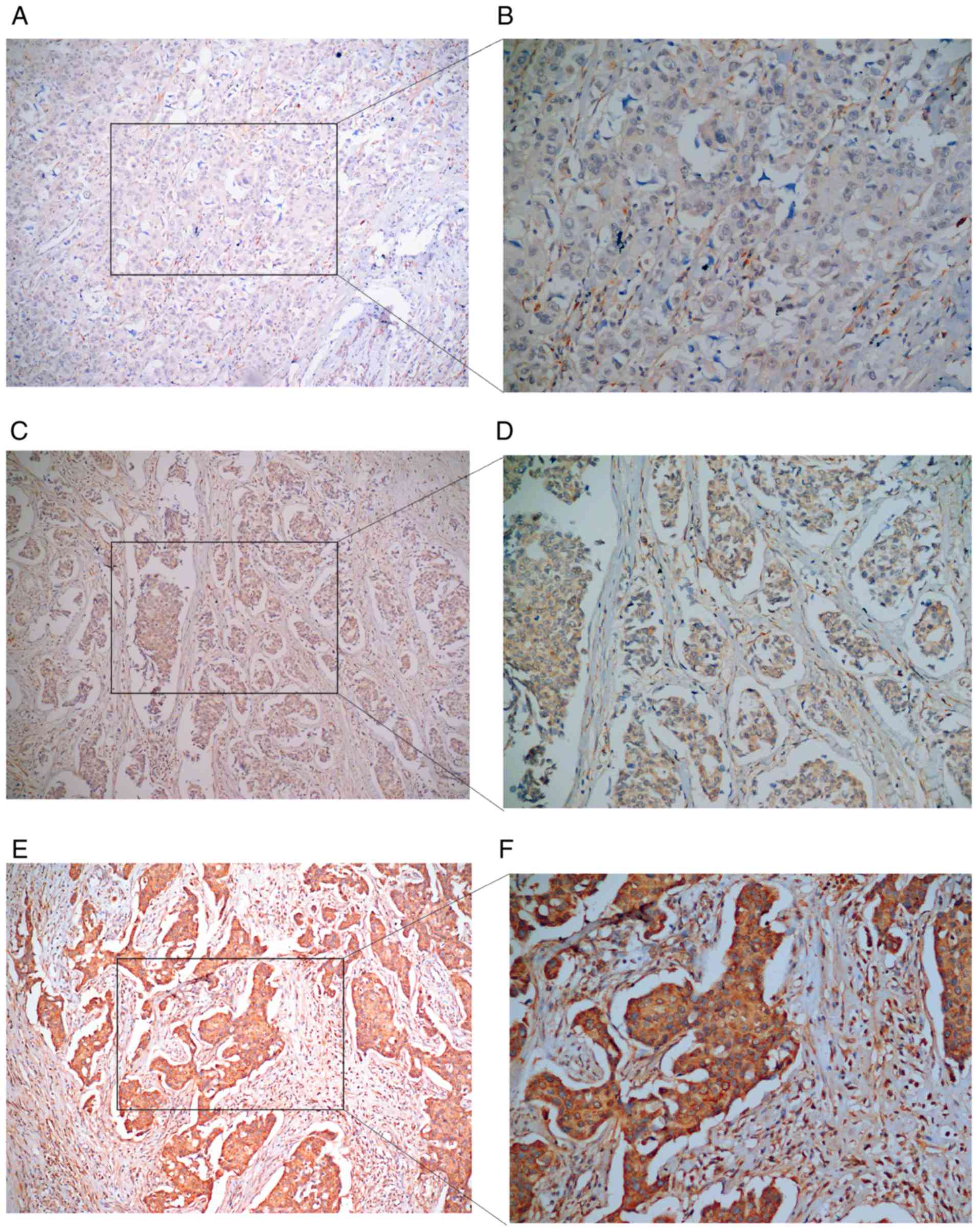
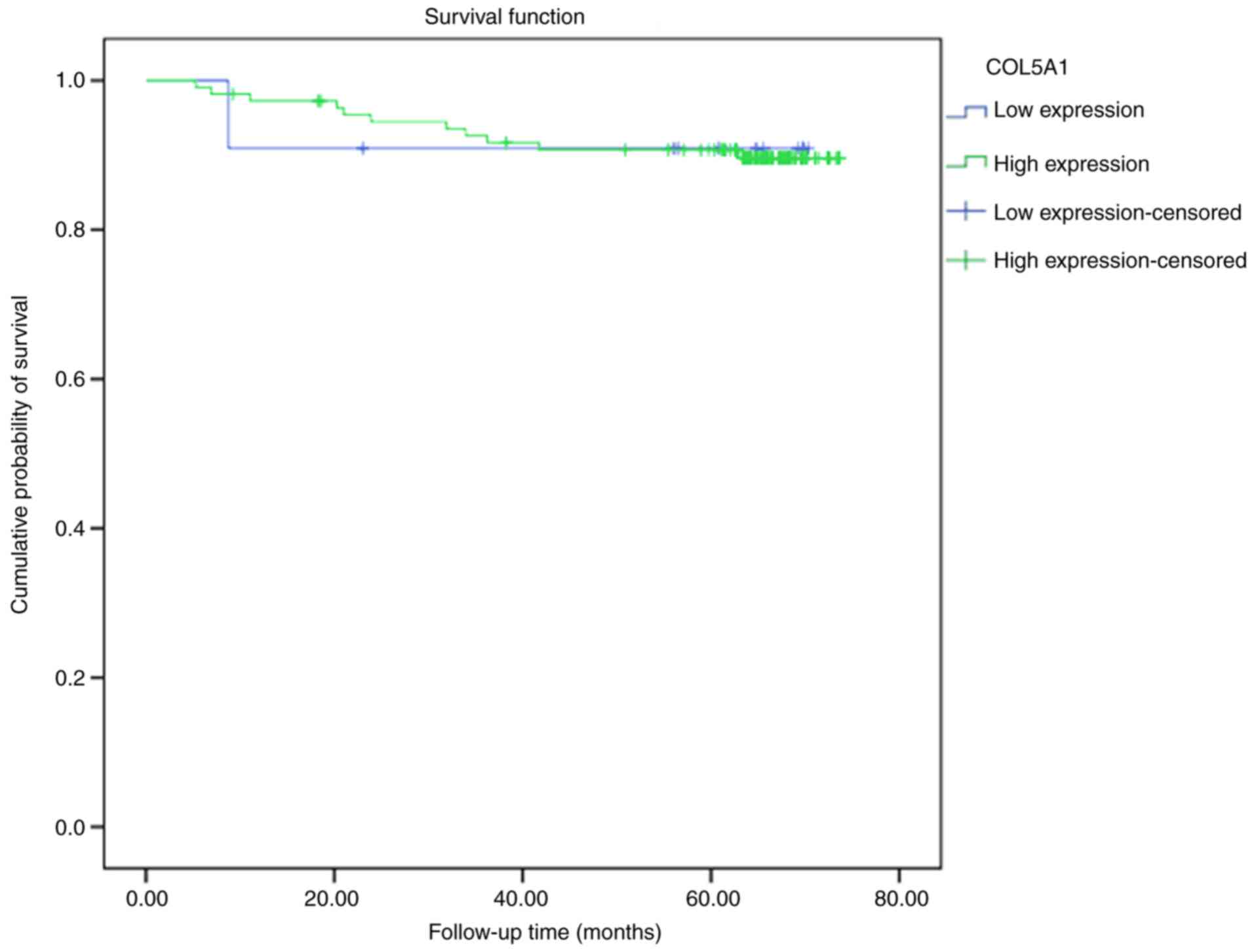
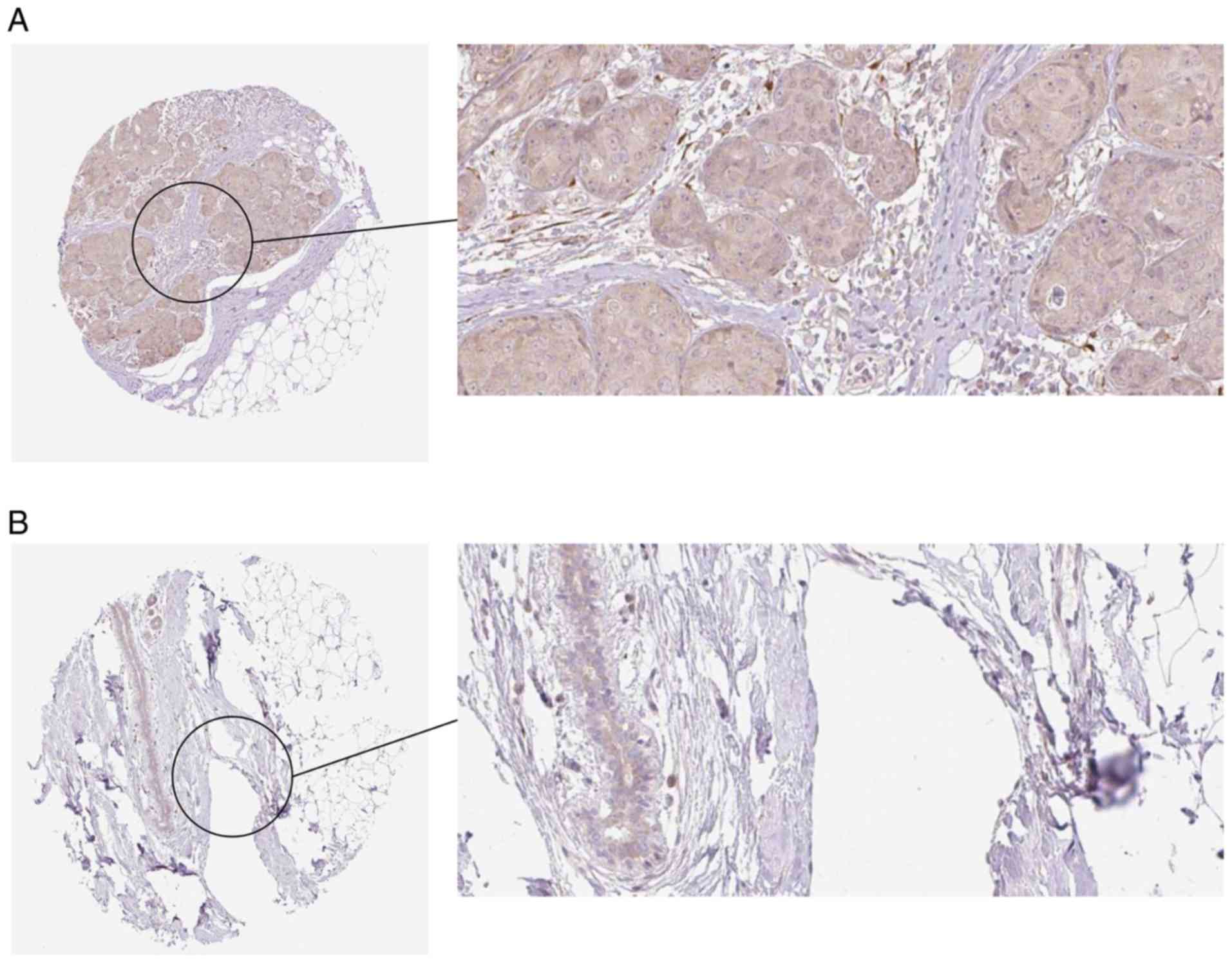
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang ZJ, Yu Y, Chi JR, Guan M, Zhao Y and
Cao XC: The combined pN stage and breast cancer subtypes in breast
cancer: A better discriminator of outcome can be used to refine the
8th AJCC staging manual. Breast Cancer. 25:315–324. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
An N, Shi Y, Ye P, Pan Z and Long X:
Association between MGMT promoter methylation and breast cancer: A
Meta-analysis. Cell Physiol Biochem. 42:2430–2440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Casey MC, Sweeney KJ, Brown JA and Kerin
MJ: Exploring circulating micro-RNA in the neoadjuvant treatment of
breast cancer. Int J Cancer. 139:12–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Savci-Heijink CD, Halfwerk H, Koster J and
Van de Vijver MJ: Association between gene expression profile of
the primary tumor and chemotherapy response of metastatic breast
cancer. BMC Cancer. 17:7552017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin YH, Hua QF, Zheng JJ, Ma XH, Chen TX,
Zhang S, Chen B, Dai Q and Zhang XH: Diagnostic value of ER, PR, FR
and HER-2-targeted molecular probes for magnetic resonance imaging
in patients with breast cancer. Cell Physiol Biochem. 49:271–281.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akram M, Iqbal M, Daniyal M and Khan AU:
Awareness and current knowledge of breast cancer. Biol Res.
50:332017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Polyak K: Breast cancer: Origins and
evolution. J Clin Invest. 117:3155–3163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ren W, Zhang Y, Zhang L, Lin Q, Zhang J
and Xu G: Overexpression of collagen type V α1 chain in human
breast invasive ductal carcinoma is mediated by TGF-β1. Int J
Oncol. Mar 15–2018.(Epub ahead of print). doi:
10.3892/ijo.2018.4317. View Article : Google Scholar
|
|
11
|
Lee S, Lee J, Sim SH, Lee Y, Moon KC, Lee
C, Park WY, Kim NK, Lee SH and Lee H: Comprehensive somatic genome
alterations of urachal carcinoma. J Med Genet. 54:572–578. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chai F, Liang Y, Zhang F, Wang M, Zhong L
and Jiang J: Systematically identify key genes in inflammatory and
non-inflammatory breast cancer. Gene. 575:600–614. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and prognosis of topoisomerase isoforms in
non-small-cell lung cancer by using Oncomine and Kaplan-Meier
plotter. PLoS One. 12:e01745152017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gyorffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang HL, Chang KK, Mei J, Zhou WJ, Liu LB,
Yao L, Meng Y, Wang MY, Ha SY, Lai ZZ, et al: Estrogen restricts
the apoptosis of endometrial stromal cells by promoting TSLP
secretion. Mol Med Rep. 18:4410–4416. 2018.PubMed/NCBI
|
|
18
|
Sas-Korczynska B, Reinfuss M, Mitus JW,
Pluta E, Patla A and Walasek T: Radiotherapy alone as a method of
treatment for sinonasal mucosal melanoma: A report based on six
cases and a review of current opinion. Rep Pract Oncol Radiother.
23:402–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res. 45:W130–W137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dai Y, Sun L and Qiang W: A new strategy
to uncover the anticancer mechanism of Chinese compound formula by
integrating systems pharmacology and bioinformatics. Evid Based
Complement Alternat Med. 2018:67078502018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Frank GA, Danilova NV, Andreeva I and
Nefedova NA: WHO classification of tumors of the breast, 2012. Arkh
Patol. 75:53–63. 2013.(In Russian). PubMed/NCBI
|
|
24
|
Gyorffy B, Pongor L, Bottai G, Li X,
Budczies J, Szabó A, Hatzis C, Pusztai L and Santarpia L: An
integrative bioinformatics approach reveals coding and non-coding
gene variants associated with gene expression profiles and outcome
in breast cancer molecular subtypes. Br J Cancer. 118:1107–1114.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yan W, Zhao Y and He J: Anti-breast cancer
activity of selected 1,3,5-triazines via modulation of EGFR-TK. Mol
Med Rep. 18:4175–4184. 2018.PubMed/NCBI
|
|
26
|
Li X, Huang X, Zhang J, Huang H, Zhao L,
Yu M, Zhang Y and Wang H: A novel peptide targets CD105 for tumour
imaging in vivo. Oncol Rep. 40:2935–2943. 2018.PubMed/NCBI
|
|
27
|
Noorlag R, van der Groep P, Leusink FK,
Leusink FK, van Hooff SR, Frank MH, Willems SM and van Es RJ: Nodal
metastasis and survival in oral cancer: Association with protein
expression of SLPI, not with LCN2, TACSTD2, or THBS2. Head Neck.
37:1130–1136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Uhlen M, Zhang C, Lee S, Sjöstedt E,
Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et
al: A pathology atlas of the human cancer transcriptome. Science.
357:eaan25072017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thul PJ, Akesson L, Wiking M, Mahdessian
D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM,
et al: A subcellular map of the human proteome. Science.
356:eaal33212017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang Y, Zhang Y, Huang Q and Li C:
Integrated bioinformatics analysis reveals key candidate genes and
pathways in breast cancer. Mol Med Rep. 17:8091–8100.
2018.PubMed/NCBI
|
|
31
|
Kim GJ, Kim DH, Min KW, Kim YH and Oh YH:
Loss of p27kip1 expression is associated with poor
prognosis in patients with taxane-treated breast cancer. Pathol Res
Pract. 214:565–571. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jimenez-Morales S, Pérez-Amado CJ, Langley
E and Hidalgo-Miranda A: Overview of mitochondrial germline
variants and mutations in human disease: Focus on breast cancer
(Review). Int J Oncol. 53:923–936. 2018.PubMed/NCBI
|
|
33
|
Do SI, Kim HS, Kim K, Lee H, Do IG, Kim
DH, Chae SW and Sohn JH: Predictive and prognostic value of
sphingosine kinase 1 expression in patients with invasive ductal
carcinoma of the breast. Am J Transl Res. 9:5684–5695.
2017.PubMed/NCBI
|
|
34
|
Kim JH, Jung ES, Kim CH, Youn H and Kim
HR: Genetic associations of body composition, flexibility and
injury risk with ACE, ACTN3 and COL5A1 polymorphisms in Korean
ballerinas. J Exerc Nutrition Biochem. 18:205–214. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lim ST, Kim CS, Kim WN and Min SK: The
COL5A1 genotype is associated with range of motion. J Exerc
Nutrition Biochem. 19:49–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Makoukji J, Makhoul NJ, Khalil M, El-Sitt
S, Aldin ES, Jabbour M, Boulos F, Gadaleta E, Sangaralingam A,
Chelala C, et al: Gene expression profiling of breast cancer in
Lebanese women. Sci Rep. 6:366392016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu G, Wu K and Sheng Y: Elucidation of
the molecular mechanisms of anaplastic thyroid carcinoma by
integrated miRNA and mRNA analysis. Oncol Rep. 36:3005–3013. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An F, Zhang Z, Xia M and Xing L: Subpath
analysis of each subtype of head and neck cancer based on the
regulatory relationship between miRNAs and biological pathways.
Oncol Rep. 34:1745–1754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumari K, Das B, Adhya AK, Rath AK and
Mishra SK: Genome-wide expression analysis reveals six contravened
targets of EZH2 associated with breast cancer patient survival. Sci
Rep. 9:19742019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Di Y, Chen D, Yu W and Yan L: Bladder
cancer stage-associated hub genes revealed by WGCNA co-expression
network analysis. Hereditas. 156:72019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abrahams Y, Laguette MJ, Prince S and
Collins M: Polymorphisms within the COL5A1 3′-UTR that alters mRNA
structure and the MIR608 gene are associated with Achilles
tendinopathy. Ann Hum Genet. 77:204–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ritelli M, Dordoni C, Venturini M,
Chiarelli N, Quinzani S, Traversa M, Zoppi N, Vascellaro A,
Wischmeijer A, Manfredini E, et al: Clinical and molecular
characterization of 40 patients with classic Ehlers-Danlos
syndrome: Identification of 18 COL5A1 and 2 COL5A2 novel mutations.
Orphanet J Rare Dis. 8:582013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Riches A, Campbell E, Borger E and Powis
S: Regulation of exosome release from mammary epithelial and breast
cancer cells-a new regulatory pathway. Eur J Cancer. 50:1025–1034.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rejon C, Al-Masri M and McCaffrey L: Cell
polarity proteins in breast cancer progression. J Cell Biochem.
117:2215–2223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
McCuaig R, Wu F, Dunn J, Rao S and
Dahlstrom JE: The biological and clinical significance of
stromal-epithelial interactions in breast cancer. Pathology.
49:133–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pizon M, Schott DS, Pachmann U and
Pachmann K: B7-H3 on circulating epithelial tumor cells correlates
with the proliferation marker, Ki-67, and may be associated with
the aggressiveness of tumors in breast cancer patients. Int J
Oncol. 53:2289–2299. 2018.PubMed/NCBI
|
|
47
|
Luo M, Clouthier SG, Deol Y, Liu S,
Nagrath S, Azizi E and Wicha MS: Breast cancer stem cells: Current
advances and clinical implications. Methods Mol Biol. 1293:1–49.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
49
|
Chen X, Pei Z, Peng H and Zheng Z:
Exploring the molecular mechanism associated with breast cancer
bone metastasis using bioinformatic analysis and microarray genetic
interaction network. Medicine (Baltimore). 97:e120322018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao H and Li H: Network-based
meta-analysis in the identification of biomarkers for papillary
thyroid cancer. Gene. 661:160–168. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li S, Chen X, Liu X, Yu Y, Pan H, Haak R,
Schmidt J, Ziebolz D and Schmalz G: Complex integrated analysis of
lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol.
73:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen M, Liu B, Xiao J, Yang Y and Zhang Y:
A novel seven-long non-coding RNA signature predicts survival in
early stage lung adenocarcinoma. Oncotarget. 8:14876–14886.
2017.PubMed/NCBI
|
|
53
|
Epstein SG, Drucker L, Pomeranz M, Fishman
A, Pasmanik-Chor M, Tartakover-Matalon S and Lishner M: First
trimester human placenta prevents breast cancer cell attachment to
the matrix: The role of extracellular matrix. Mol Carcinog.
56:62–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Giussani M, Landoni E, Merlino G, Turdo F,
Veneroni S, Paolini B, Cappelletti V, Miceli R, Orlandi R, Triulzi
T and Tagliabue E: Extracellular matrix proteins as diagnostic
markers of breast carcinoma. J Cell Physiol. 233:6280–6290. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bonnans C, Chou J and Werb Z: Remodelling
the extracellular matrix in development and disease. Nat Rev Mol
Cell Biol. 15:786–801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Boguslawska J, Kedzierska H, Poplawski P,
Rybicka B, Tanski Z and Piekielko-Witkowska A: Expression of genes
involved in cellular adhesion and extracellular matrix remodeling
correlates with poor survival of patients with renal cancer. J
Urol. 195:1892–1902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheon DJ, Tong Y, Sim MS, Dering J, Berel
D, Cui X, Lester J, Beach JA, Tighiouart M, Walts AE, et al: A
collagen-remodeling gene signature regulated by TGF-β signaling is
associated with metastasis and poor survival in serous ovarian
cancer. Clin Cancer Res. 20:711–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Alqinyah M and Hooks SB: Regulating the
regulators: Epigenetic, transcriptional, and post-translational
regulation of RGS proteins. Cell Signal. 42:77–87. 2018. View Article : Google Scholar : PubMed/NCBI
|