Introduction
Intrahepatic cholangiocarcinoma (ICC) is the second
most common primary hepatobiliary malignancy following
hepatocellular carcinoma (1). ICC
accounts for approximately 5–10% of all cases of
cholangiocarcinoma, and both its incidence and mortality rate have
been increasing in recent decades, especially in Southeast Asia
(2,3).
Despite advances in the diagnosis and treatment of patients with
ICC, the prognosis of this disease is still poor, with a 5-year
survival of only 25–35% (4). This
poor survival rate is due to patients being diagnosed at the
advanced stage, when cancer cell invasion into the blood and
lymphatic vessels has already led to metastatic spread (5). Therefore, the prognosis of ICC patients
may be improved by identifying novel and effective therapeutic
targets.
Chromodomain helicase/ATPase DNA-binding protein
1-like gene (CHD1L), also named amplified in liver cancer 1
gene (ALC1), was first isolated from 1q21 by Ma et al
(6) in 2008, and it was identified as
a target oncogene in hepatocellular carcinoma (HCC) (7). CHD1L belongs to the sucrose
non-fermenting 2 (SNF2)-like subfamily of the SNF2
family consisting of a helicase superfamily c-terminal
(HELICc) and a Macro domain (7). Hence, CHD1L has also been
hypothesized to play important roles in transcriptional regulation,
maintenance of chromosome integrity and DNA repair, similar to the
SNF2 family members (7).
CHD1L was first found to play a vital role in the
development and progression of HCC (8). More interestingly, a number of studies
have found that amplification of CHD1L is extremely common
in many solid tumors, including breast (9), gastric (10) and nasopharyngeal carcinoma (11). Recently, He et al reported that
CHD1L protein is overexpressed in human ovarian carcinomas
and is a novel predictive biomarker for patient survival (12). However, the expression of CHD1L
and its significance in ICC is far from clear; even less is known
about its function and how CHD1L contributes to cancer
development and progression.
In the present study, CHD1L expression levels
were detected in ICC tissues and cell lines. The relationship
between CHD1L and clinical characteristics of ICC patients
was analyzed, and its oncogene function was examined further in
vitro and in vivo. Our results suggest that CHD1L
is markedly upregulated and promotes the proliferation and
metastasis of ICC cells. CHD1L acts as an oncogene and may
be a prognostic factor or therapeutic target for patients with
ICC.
Materials and methods
Patients and tissue samples
Eighty ICC tissue and thirty hepatolithiasis tissue
sections used for paraffin embedding were collected from ICC
patients who underwent curative surgery without prior radiotherapy
or chemotherapy between January 2007 and January 2012 at the
Department of Hepatobiliary Surgery, Jiangxi Provincial People's
Hospital (Nanchang, China) and were confirmed by a pathologist. The
present study was approved by the Ethics Committee of Jiangxi
Provincial People's Hospital, and all patients provided informed
consent. The tumor stage was classified according to the 7th
tumor-node-metastasis (TNM) classification of the International
Union against Cancer (UICC) (13).
Among the 80 ICC patients, there were 49 males and 31 females with
ages ranging from 42 to 73 years (mean age, 55 years). Information
concerning the clinical characteristics and survival prognosis was
extracted from medical records and follow-ups. Fresh ICC tissues
and paired non-tumor tissue samples were obtained from 34 ICC
patients, and these samples were frozen and stored at −80°C. Paired
non-tumor tissues were dissected at least 2 cm away from the cancer
border and were verified to lack cancer cells by microscopy.
RNA extraction and RT-qPCR
Total RNA was extracted from fresh tissues and
cultured cells using TRIzol reagent (TransGen Biotech Co., Ltd.,
Beijing, China) according to the manufacturer's protocol. cDNA was
synthesized from 2 µg of total RNA using PrimeScript™ RT Master Mix
(Takara Bio, Inc., Shiga, Japan). RNA expression was measured by
RT-qPCR using the SYBR-Green Fast qPCR Mix in an Applied
Biosystems® 7500 Real-Time PCR Systems (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. The 2−ΔΔCq
method (14) was used to calculate
the expression level (defined as the fold change) of CHD1L
compared with GAPDH expression. Primer sequences are listed
in Table I.
 | Table I.Primer sequences of CHD1L and
GAPDH. |
Table I.
Primer sequences of CHD1L and
GAPDH.
| Gene promoter | Sequence
(5′-3′) |
|---|
| CHD1L | F:
5′-GGGAAGACCTGCCAGATTTGCT-3′ |
|
| R:
5′-ACGTGACTCCTGTTTCAGGTCTTG-3′ |
| GAPDH | F:
5′-GTTGGAGGTCGGAGTCAACGGA-3′ |
|
| R:
5′-GAGGGATCTCGCTCCTGGAGGA-3′ |
Immunohistochemistry (IHC)
IHC staining of sections was performed using a
standard streptavidin-peroxidase staining method. Paraffin-embedded
samples of CHD1L expression were cut into 5-µm-thick
sections and processed for IHC method, as previously described by
Renshaw (15). Tissue sections with
antigen retrieval by microwave treatment in citrate buffer (pH 6.0)
were then incubated at 4°C overnight with primary antibodies for
anti-E-cadherin (dilution 1:250; cat. no. sc-71008; Santa Cruz
Biotechnology, Santa Cruz, CA, USA), anti-N-cadherin (dilution
1:250; cat. no. sc-53488; Santa Cruz Biotechnology) and
anti-CHD1L (dilution 1:500; cat. no. ab-197019; Abcam,
Cambridge, MA, USA). Immunostaining was performed using Mayer's
hematoxylin (Beyotime Biotechnology Institute of Biotechnology,
Shanghai, China) and images were captured and assessed using a
light microscope (Olympus Corp., Tokyo, Japan). The degree of
immunostaining of sections was reviewed and estimated independently
by two observers in a blinded manner, based on both the staining
intensity and the proportion of positive tumor cells (16). The staining intensity was
semi-quantitatively classified as 0 (negative), 1 (weak), 2
(intermediate), or 3 (strong). Additionally, the proportion of
positive tumor cells was scored as follows: 0, 0–5%, no positive
tumor cells; 1, >5-25%, positive tumor cells; 2, >25-50%,
positive tumor cells; and 3, >50%, positive tumor cells. The
staining index=the staining intensity × proportion of positive
tumor cells; the final immunoreaction score was defined as negative
(0–1), weak (2–3), moderate (4–6) and strong
(6–9)
staining. For statistical purposes, the staining index score was
graded as negative (negative and weak) or positive (moderate and
strong) expression.
Cell lines and cell culture
The human ICC cell lines RBE and HCCC9810 were
purchased from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China), and cell lines QBC939 and HuCCT1 were obtained
from the American Type Culture Collection (ATCC, Manassas, VA,
USA). Human intrahepatic biliary epithelial cells (HiBECs) were
stored at the Key Molecular Medical Laboratory of Jiangxi Province
(Nanchang, China). Cells were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Thermo Fisher Scientific, Inc.). Cells were incubated
for 48 or 96 h in a humidified incubator supplied with 5%
CO2 at 37°C.
Antibodies and western blotting
A rabbit anti-CHD1L antibody was purchased from
Abcam (dilution 1:5,000; cat. no. ab197019). Mouse anti-N-cadherin
(dilution 1:500; cat. no. sc-53488), anti-E-cadherin (dilution
1:500; cat. no. sc-71008), anti-vimentin (dilution 1:600; cat. no.
sc-80975), anti-p53 (dilution 1:600; cat. no. sc-47698),
anti-cyclin D1 (dilution 1:750; cat. no. sc-8396) and anti-Cdk2
(dilution 1:750; cat. no. sc-128295; all were from Santa Cruz
Biotechnology) antibodies were purchased from Santa Cruz
Biotechnology and rabbit anti-GAPDH antibody was obtained from
ProteinTech Group, Inc. (Chicago, IL, USA; diluted at 1:10,000).
Briefly, equal quantities of cellular proteins were resolved by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis,
transferred onto polyvinylidene difluoride (PVDF) membranes, and
immunoblotted with primary antibodies against CHD1L, N-cadherin,
E-cadherin, vimentin, p53, cyclin D1, Cdk2 and GAPDH. After
incubation for 1 h at room temperature with horseradish peroxidase
(HRP)-conjugated secondary antibody (dilution 1:10,000; cat. no.
HS101-01; TransGen Biotech Co., Ltd.), western blot analyses were
visualized by enhanced chemiluminescence (EMD Millipore, Billerica,
MA, USA). GAPDH was used as the loading control.
Lentivirus-mediated RNA interference
and construction of plasmids
Silencing of CHD1L was carried out using
short hairpin RNAs (shRNAs), which were synthesized and inserted
into the lentivirus vector containing a cytomegalovirus-driven
enhanced green fluorescent protein (EGFP) gene. Vectors expressing
CHD1L-shRNA (shCHD1L) or negative control shRNA
(shNC) were designed by Shanghai GeneChem Co., Ltd. (Shanghai,
China). The full-length CHD1L cDNA was cloned into the GV362
expression vector (Shanghai GeneChem Co., Ltd.) and empty
vector-transfected cells (MOCK) were used as control. The detailed
sequences are listed in Table II.
RBE and HUCCT1 cells were transfection with Lv-shRNA in serum-free
medium using concentrated virus and replaced with complete culture
medium after 24 h. Similarly, the overexpression CHD1L and
MOCK plasmids were transfected into HCCC9810 cells using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
Stably transfected cells were selected for 1–2 weeks by using 500
µg/ml puromycin (TransGen Biotech Co., Ltd.); CHD1L
expression in surviving cells was validated by western blotting and
RT-qPCR analysis after transfection for 72 h.
 | Table II.Sequences of the CHD1L-RNAi. |
Table II.
Sequences of the CHD1L-RNAi.
| CHD1L-RNAi | Sequence
(5′-3′) |
|---|
| CHD1L-RNAi1 | F:
5′-CGTATTGGACATGCCACGAAA-3′ |
|
| R:
5′-TTTCGTGGCATGTCCAATACG-3′ |
| CHD1L-RNAi2 | F:
5′-GCCAAGAGAAGGAGACTCATA-3′ |
|
| R:
5′-TATGAGTCTCCTTCTCTTGGC-3′ |
| CHD1L-RNAi3 | F:
5′-GCACAAACTCTTGCAGCCATT-3′ |
|
| R:
5′-AATGGCTGCAAGAGTTTGTGC-3′ |
| NC | F:
5′-TTCTCCGAACGTGTCACGT-3′ |
|
| R:
5′-GUGACACGUUCGGAGAAT-3′ |
Cell proliferation assay
Cells after transfection for 24 h were planted at a
density of 4×104 cells/well in a 96-well plate and
cultured at 37°C with 5% CO2 in an incubator. Ten
microliters of CCK-8 reagent (TransGen Biotech Co., Ltd.) was added
to each well at 0, 24, 48, 72 and 96 h and incubated for 2 h.
Finally, OD values at 450 nm were measured with a microplate
reader, and the growth curve was plotted. Anchorage-independent
growth was assessed by a colony formation assay. Briefly, 1,000
cells were seeded in 6-well plates. The cells were cultured for
~1–2 weeks, changed into fresh medium after 2–3 days to see visible
clones. Afterwards, cells were fixed with 4% paraformaldehyde for
20 min and stained with 0.1% crystal violet for 30 min. The total
number of colonies containing >50 cells and ranging in size from
0.3–1.0 mm was counted, and the images were photographed at ×100
magnification under a light microscope.
Flow cytometry
ICC cells (1.5×105 cells) were seeded
into 100-mm culture dishes. Twenty-four hours after seeding, the
cells were treated with 0.1 or 0.3 mg/ml vehicle (0.1% DMSO) for 24
h. After treatment, the cells at 70–80% confluence were digested
into a single-cell suspension, fixed in 70% ethanol, stained with
propidium iodide (PI), and analyzed by flow cytometry. In addition,
the percentages of cells within each phase of the cell cycle were
analyzed with ModFit version 4.0 (Verity Software House, Inc.,
Topsham, ME, USA) and CellQuest version 5.1 (Thermo Fisher
Scientific, Inc.).
Cell migration and invasion
assays
For the wound healing assay, cells were planted at a
density of 5×106 cells/ml in a 6-well plate and
incubated at 37°C overnight. A cell-free area of the culture medium
was wounded by scratching with a 200-µl pipette tip. Cell migration
into the wound area was viewed and photographed at 0 and 24 h after
scratching. Cell migration rate was calculated as follows:
(original gap distance -current gap distance)/original gap distance
×100%. Transwell migration and invasion assays were examined using
24-well chambers (8 µm Transwell filters per chamber) (Corning
Inc., Corning, MA, USA). Then, 3×104 cells in 200 µl
serum-free medium were added to the upper chamber containing an
uncoated or Matrigel-coated (BD Biosciences, San Jose, CA, USA)
membrane. The lower chamber contained 600 µl culture medium
supplemented with 20% FBS. After being cultured for 24 h in an
incubator, cells on the upper surface of the microporous membrane
were wiped off with a cotton swab, fixed with 4% paraformaldehyde
for 20 min, and stained with 0.1% crystal violet for 30 min.
Migrated or invaded cells were counted in five randomly chosen
fields in each chamber. Imaging and counting were performed at ×200
magnification under a light microscope. The experiments were
executed in triplicate.
Subcutaneous and peritoneal xenograft
tumor models
BALB/c-nu mice, 4–6 weeks old, were purchased from
Hunan SJA Laboratory Animal Company (Changsha, China). All animal
protocols were approved by the Ethics Committee of Jiangxi Province
People's Hospital. Five mice were divided randomly into each group.
To explore the effects of CHD1L on tumor growth and
metastasis in vivo, 1×107 cells were injected
into the left axilla and enterocoelia of the mice (17,18) (5
mice/group). Tumor growth was observed every week and measured in
two dimensions. The tumor volume (V) was calculated using the
following formula: V=4π/3 × (width/2)2 × (length/2).
After 4 weeks, the mice were sacrificed under cervical dislocation
and the tumors were dissected out and weighed. Then, metastatic
tumors were fixed with formalin and embedded in paraffin. Finally,
the expression levels of CHD1L, E-cadherin and N-cadherin
were evaluated by IHC.
Statistical analysis
GraphPad Prism 6.0 software (GraphPad Software,
Inc., Chicago, IL. USA) was used to process data and images. Each
experimental value was expressed as the mean ± standard deviation
(SD). The paired Student's t-test mainly applies to CHD1L
protein levels in tumor and adjacent non-tumor tissues. The
Chi-square (χ2) test was used to analyze the association
of CHD1L expression with the clinicopathological
characteristics of ICC. Kaplan-Meier plots and log-rank tests were
used for survival analysis. Univariate and multivariate Cox
proportional hazard regression models were used to analyze
independent prognostic factors. P<0.05 was considered
statistically significant, and all assays were performed in
triplicate independent experiments.
Results
Expression of CHD1L is upregulated and
is correlated with a poor prognosis in ICC
To assess the potential role of CHD1L in ICC,
we used RT-qPCR and western blotting to measure the expression of
CHD1L in 34 fresh clinical ICC tissues and their paired
adjacent non-tumor tissues. The relative expression level of
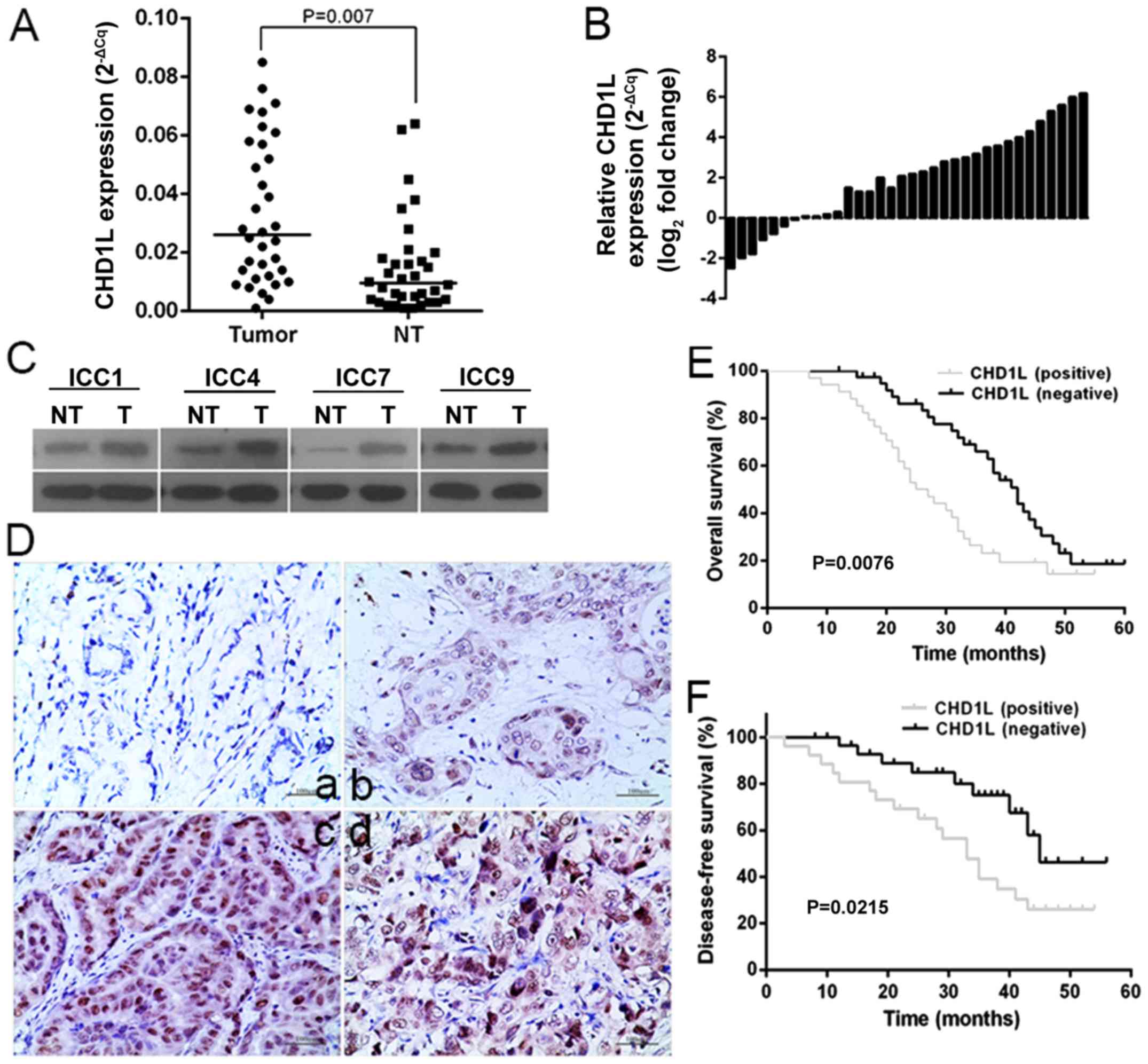
CHD1L in ICC tissues was significantly higher than that
noted in the paired adjacent non-tumor tissues (P=0.007; Fig. 1A and B). Consistent with the RT-qPCR
results, significantly increased CHD1L protein levels were
observed in the ICC tissues, when compared with the matched
adjacent non-cancerous tissues (P<0.01; Fig. 1C). Furthermore, protein expression
levels of CHD1L were measured in 80 samples of stored
paraffin-embedded ICC tissues and 30 hepatolithiasis tissues by
immunohistochemistry (Fig. 1D).
CHD1L was overexpressed in tumor tissues compared with that
noted in hepatolithiasis tissues (P=0.03, Table III). Then, the clinical significance
of CHD1L overexpression in the ICC cohort was investigated
by statistical analysis. We found that CHD1L overexpression
was closely related to histological differentiation (P=0.011),
vascular invasion (P=0.002), lymph node metastasis (P=0.008) and
TNM stage (P=0.001), suggesting that CHD1L may play roles in
ICC metastasis (Table IV).
Kaplan-Meier analysis showed that ICC patients with positive
CHD1L expression had reduced overall and disease-free
survival than those with negative CHD1L expression (log
rank, 7.117; P=0.0076; Fig. 1E) (log
rank, 5.285; P=0.0215; Fig. 1F). Cox
regression statistical analysis was used to test the effects of
CHD1L on the independent prognostic value of ICC patients.
Overexpression of the CHD1L protein, as well as other
clinicopathological variables (histological differentiation,
vascular invasion, lymph node metastasis and TNM stage) that showed
significance by univariate analysis (all P<0.05; Table V), were included in the multivariate
analysis (hazard ratio, 5.875; 95% confident interval: 3.243–8.023,
P<0.001; Table V). The
CHD1L protein was identified as an independent prognostic
factor for poor overall and disease-free survival in ICC patients.
Taken together, these results revealed that CHD1L was
aberrantly overexpressed in ICC tissues, and this overexpression
was associated with ICC progression.
 | Table III.Immunohistochemical analysis of CHD1L
protein expression in the specimens. |
Table III.
Immunohistochemical analysis of CHD1L
protein expression in the specimens.
|
|
| CHD1L
expression |
|
|---|
|
|
|
|
|
|---|
| Groups | N | Negative (0–1) | Weak (2–3) | Moderate (4–6) | Strong (6–9) | P-value |
|---|
| Intrahepatic
cholangiocarcinoma | 80 | 16 | 28 | 15 | 21 | 0.03 |
|
Hepatolithiasis | 30 | 15 | 8 | 5 | 2 |
|
 | Table IV.Correlation between CHD1L expression
and the clinicopathological features of 80 patients with ICC. |
Table IV.
Correlation between CHD1L expression
and the clinicopathological features of 80 patients with ICC.
|
|
| CHD1L |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | No. | High n (%) | Low n (%) | χ2
value | P-value |
|---|
| Total cases | 80 | 36 (45.0) | 44 (55.0) |
|
|
| Sex |
|
|
| 0.522 | 0.47 |
|
Male | 49 | 23 (46.9) | 26 (53.1) |
|
|
|
Female | 31 | 12 (38.7) | 19 (61.3) |
|
|
| Age (years) |
|
|
| 1.204 | 0.273 |
|
≥55 | 46 | 20 (43.5) | 26 (56.5) |
|
|
|
<55 | 34 | 19 (55.9) | 15 (44.1) |
|
|
| Histological
differentiation |
|
|
| 8.973 | 0.011a |
|
Well | 23 | 7
(30.4) | 16 (69.6) |
|
|
|
Moderate | 42 | 20 (47.6) | 22 (52.4) |
|
|
|
Poor | 15 | 12 (80.0) | 3 (20) |
|
|
| Tumor size
(cm) |
|
|
| 2.161 | 0.142 |
|
≥4.0 | 52 | 33 (63.5) | 19 (36.5) |
|
|
|
<4.0 | 28 | 13 (46.4) | 15 (53.6) |
|
|
| Vascular
invasion |
|
Present | 36 | 23 (63.9) | 13 (36.1) | 9.436 | 0.002a |
|
Absent | 44 | 13 (29.5) | 31 (70.5) |
|
|
| CA19-9 (U/ml) |
|
|
| 0.671 | 0.413 |
|
≥35 | 45 | 26 (57.8) | 19 (42.2) |
|
|
|
<35 | 35 | 17 (48.6) | 18 (51.4) |
|
|
| Lymphatic node
metastasis |
|
|
| 7.026 | 0.008a |
|
Present | 48 | 31 (64.6) | 17 (35.4) |
|
|
|
Absent | 32 | 11 (34.4) | 21 (35.6) |
|
|
| TNM stage
(AJCC) |
|
|
| 14.831 | 0.001a |
|
I–II | 30 | 6
(20.0) | 24 (80.0) |
|
|
|
III | 36 | 19 (52.8) | 17 (47.2) |
|
|
| IV | 14 | 11 (78.6) | 3
(21.4) |
|
|
 | Table V.Univariate and multivariate analyses
of the different prognostic variables in the ICC patients. |
Table V.
Univariate and multivariate analyses
of the different prognostic variables in the ICC patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| CHD1L | 4.852 | 1.758–8.758 |
<0.001a | 5.875 | 3.243–8.023 |
<0.001a |
| Sex | 1.356 | 0.475–2.354 | 0.123 |
|
|
|
| Age | 0.875 | 0.652–1.102 | 0.763 |
|
|
|
| Histologic
differentiation | 3.142 | 1.246–5.863 |
<0.001a |
|
|
|
| Tumor size | 0.968 | 0.389–1.821 | 0.369 |
|
|
|
| Vascular
invasion | 8.257 | 3.012–13.562 |
<0.001a |
|
|
|
| CA-199 | 1.102 | 0.526–1.956 | 0.425 |
|
|
|
| Lymphatic node
metastasis | 4.232 | 1.897–8.014 | 0.0035a | 3.653 | 1.536–8.524 | 0.016a |
| TNM stage
(AJCC) | 4.452 | 1.356–7.485 |
<0.001a | 4.265 | 1.875–9.231 | 0.002a |
CHD1L is overexpressed in ICC cell
lines
To further explore the role of CHD1L in the
progression of ICC, we examined the expression of CHD1L in
one normal bile duct epithelial cell line (HiBECs) and four ICC
cell lines (HCCC9810, RBE, QBC939 and HuCCT1) by western blotting
and RT-qPCR. The results showed that the protein levels of
CHD1L were relatively higher in ICC cell lines when compared
to HiBECs, especially in RBE and HuCCT1 (Fig. 2A). Consistent mRNA levels were
observed in RT-qPCR (Fig. 2B). We
chose RBE and HUCCT1 cell lines for stable transfection with
CHD1L-shRNA lentivirus vectors, and we chose HCCC9810 cell
lines for stable transfection with CHD1L-overexpression
plasmid vector. We detected the transduction efficiency by EGFP
expression under a fluorescence microscope at 36–48 h after
transduction. The efficiency of lentiviral transduction in both RBE
and HUCCT1 cell lines was higher than 85%. The transfection
efficiency was further confirmed by western blotting (Fig. 2C) and RT-qPCR (Fig. 2D). Our results showed that the effect
of shRNA transduction on the expression of CHD1L was
examined using western blotting and RT-qPCR analysis with the most
efficient knockdowns by shCHD1L-1 in RBE and HUCCT1 cell
lines compared with those of the other two vectors
(shCHD1L-2 and shCHD1L-3), and the expression of
CHD1L in the control group (CTRL) and the shNC groups were
significantly higher than in the CHD1L-silenced group. In
addition, the expression of CHD1L with
CHD1L-expression plasmid vector in HCCC9810 cell lines was
significantly higher than that in the empty vector-transfected cell
(MOCK) group and the control (CRTL).
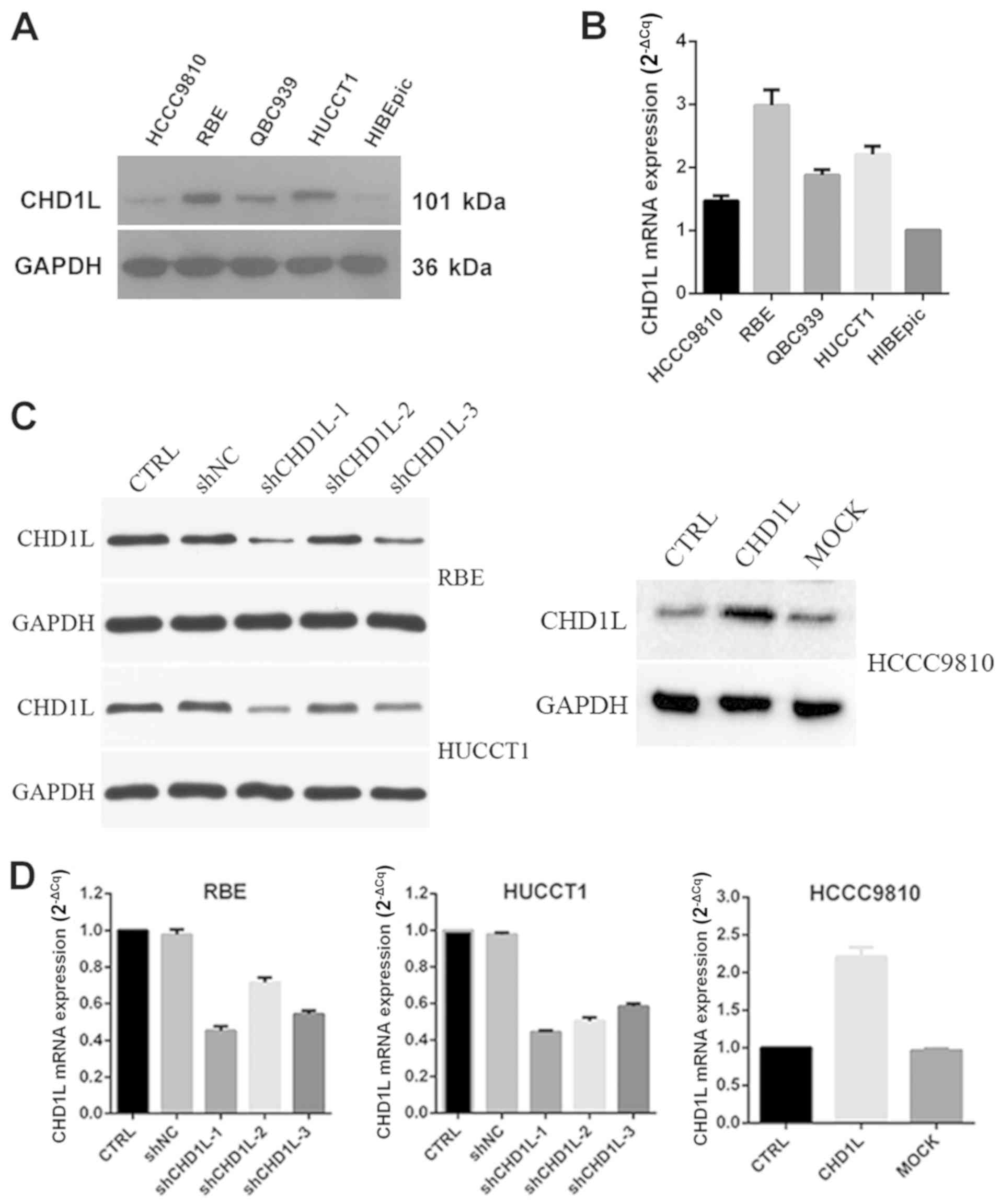 | Figure 2.CHD1L expression in ICC cell
lines. (A and B) Protein and mRNA expression of CHD1L in
normal epithelial cells, HiBECs, and ICC cell lines HCCC9810, RBE,
QBC939 and HuCCT1. (C and D) Transduction efficiency of
CHD1L expression in CHD1L-silenced RBE and HUCCT1
cells (shCHD1L−1, −2 and −3) and CHD1L-overexpressing
HCCC9810 cells (CHD1L) was examined by western blotting and
RT-qPCR. GAPDH was used as the loading control.
CHD1L, chromodomain helicase/ATPase DNA-binding protein
1-like gene; ICC, intrahepatic cholangiocarcinoma; CTRL, control;
MOCK, empty vector-transfected cells; shNC, negative control. |
CHD1L has strong tumorigenic
function
As previously reported, CHD1L may show
pro-cancer effects. Therefore, the tumorigenic ability of
CHD1L was evaluated by CCK-8 and plate colony formation
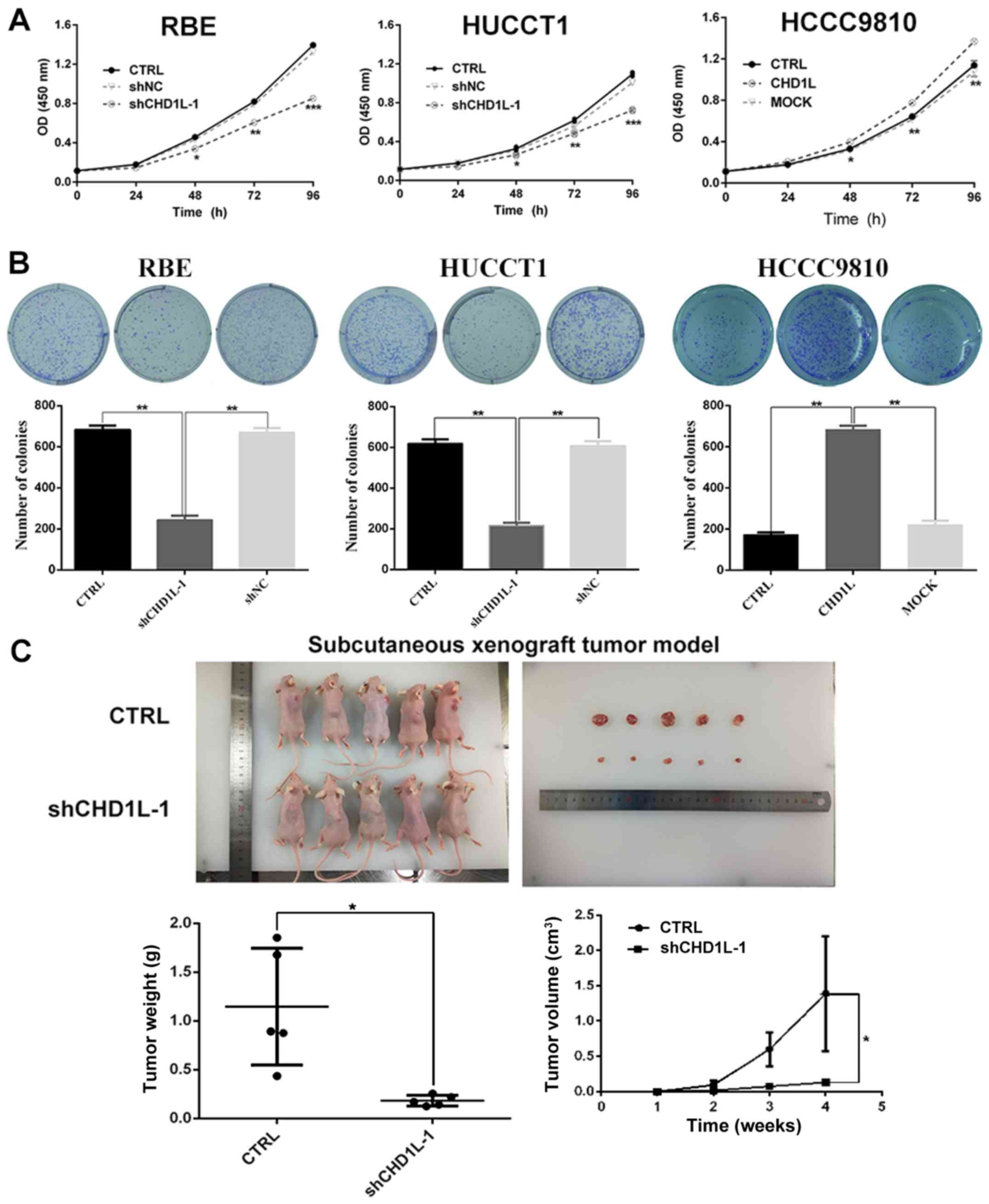
assays in vitro. A CCK-8 assay showed that the proliferative
capacity of the shCHD1L-1 transfected group was clearly
gradually inhibited compared with the control group at 48, 72 and
96 h in the RBE and HUCCT1 ICC cell lines (Fig. 3A). Additionally, as compared with the
shNC and CTRL groups, plate colony formation assays demonstrated
that the number of colonies formed by RBE and HUCCT1 ICC cell lines
was significantly reduced by CHD1L depletion (Fig. 3B). Moreover, to assess the
tumorigenicity of CHD1L in vivo, tumor formation in nude
mice was performed by injecting CHD1L-knockdown or control
RBE cells into the right back of 5 nude mice, respectively. Then,
the tumor volume was calculated. The results showed that the tumors
formed in the CHD1L-silencing xenografts were significantly
smaller than that noted in the control (CTRL) cells (Fig. 3C). In contrast, compared with the MOCK
and CRTL, CHD1L-transfected HCCC9810 cells showed a stronger
proliferation rate (Fig. 3A) and
increased numbers of colony forming units (Fig. 3B). In total, these results indicated
that overexpression of CHD1L had strong tumorigenic
ability.
Overexpression of CHD1L promotes G1/S
phase transition
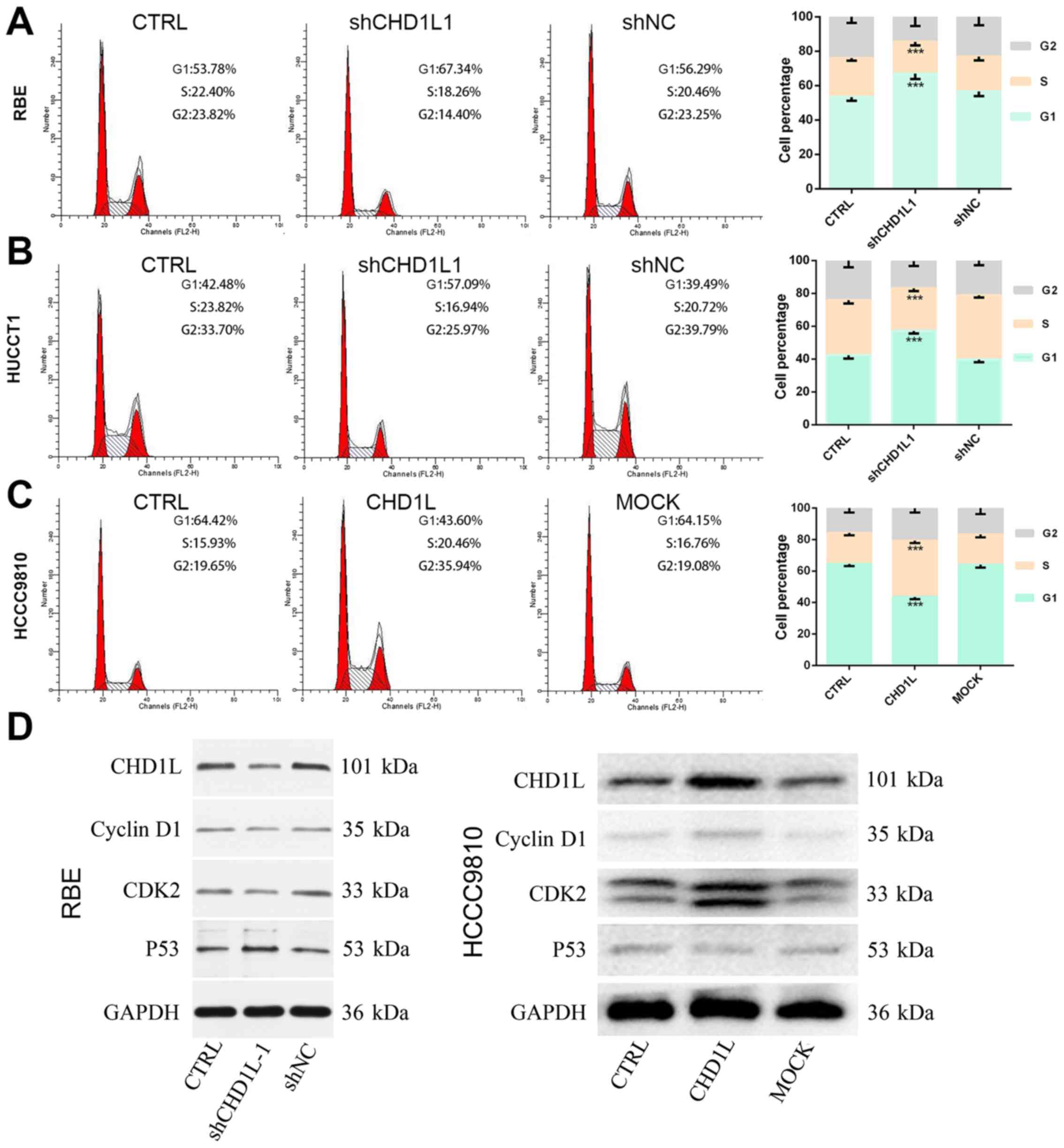
To investigate the effect of CHD1L on cell
cycle distribution, we evaluated alterations in the cell cycle
phase after treatment with the CHD1L-knockdown lentivirus by
flow cytometry. As shown in the results, cells in the G1 phase
increased, while cells in the S phase decreased in the
shCHD1L-1 cells, compared with control cells and shNC
(P<0.05; Fig. 4A and B). This
finding indicated that inhibition of CHD1L could obstruct
G1/S cell cycle transition. To further confirm the results,
CHD1L-overexpression vector was transfected into HCCC9810
cells. The MOCK and CRTL groups were used as control. The results
showed that the percentages of cells in the G1 and S phases were
significantly decreased and increased, respectively, in the
CHD1L-overexpression cells, compared with cells in the MOCK
and CRTL groups (P<0.05; Fig. 4C).
To further explore the mechanisms of CHD1L in promoting G1/S
phase transition, the effects of CHD1L on cell cycle
regulators, namely, P53, cyclin D1 and CDK2, were
examined by western blotting. As shown in the results, P53
expression was upregulated while CDK2 and cyclin D1
were downregulated in the shCHD1L-1 group in RBE cells
whereas P53 expression was downregulated and CDK2 and
cyclin D1 were upregulated in the
CHD1L-overexpressing HCCC9810 cells (Fig. 4D). This finding indicated that
CHD1L may promote G1/S transition by affecting critical cell
cycle proteins.
CHD1L increases ICC cell migration and
invasion by inducing EMT
To study the role of CHD1L in ICC cell
migration and invasion, we applied wound healing, Transwell
migration and Matrigel invasion assays in vitro. The wound
healing assay showed that the rate of motility was significantly
decreased in RBE and HUCCT1 cells by CHD1L-depletion
compared to the CTRL and shNC groups, while overexpression of
CHD1L in HCCC9810 cells showed the opposite effect
(P<0.001; Fig. 5A). Similarly,
Transwell migration and Matrigel invasion assays demonstrated that
the cells number of migration and invasion of the CTRL and shNC
groups were significantly more than the treated group. Conversely,
the invasiveness of the CHD1L-expressing cells was
significantly higher than CTRL and MOCK cells (P<0.0001;
Fig. 5B). Therefore, these results
suggest that CHD1L increases ICC cell migration and
invasion. To explore whether CHD1L promotes the invasiveness
of ICC through EMT, we examined EMT-related biomarkers by western
blotting (Fig. 5C). The result showed
that RBE and HUCCT1 cells transfected with shCHD1L expressed
high levels of E-cadherin, which is characteristic of epithelial
cells. However, in ICC cell lines transfected with shCHD1L,
expression levels of proteins related to the mesenchymal phenotype
(N-cadherin and vimentin) were downregulated. Overexpression of
CHD1L could reverse this phenotype in HCCC9810 cells. These
results indicated that CHD1L increased ICC cell migration
and invasion by inducing EMT.
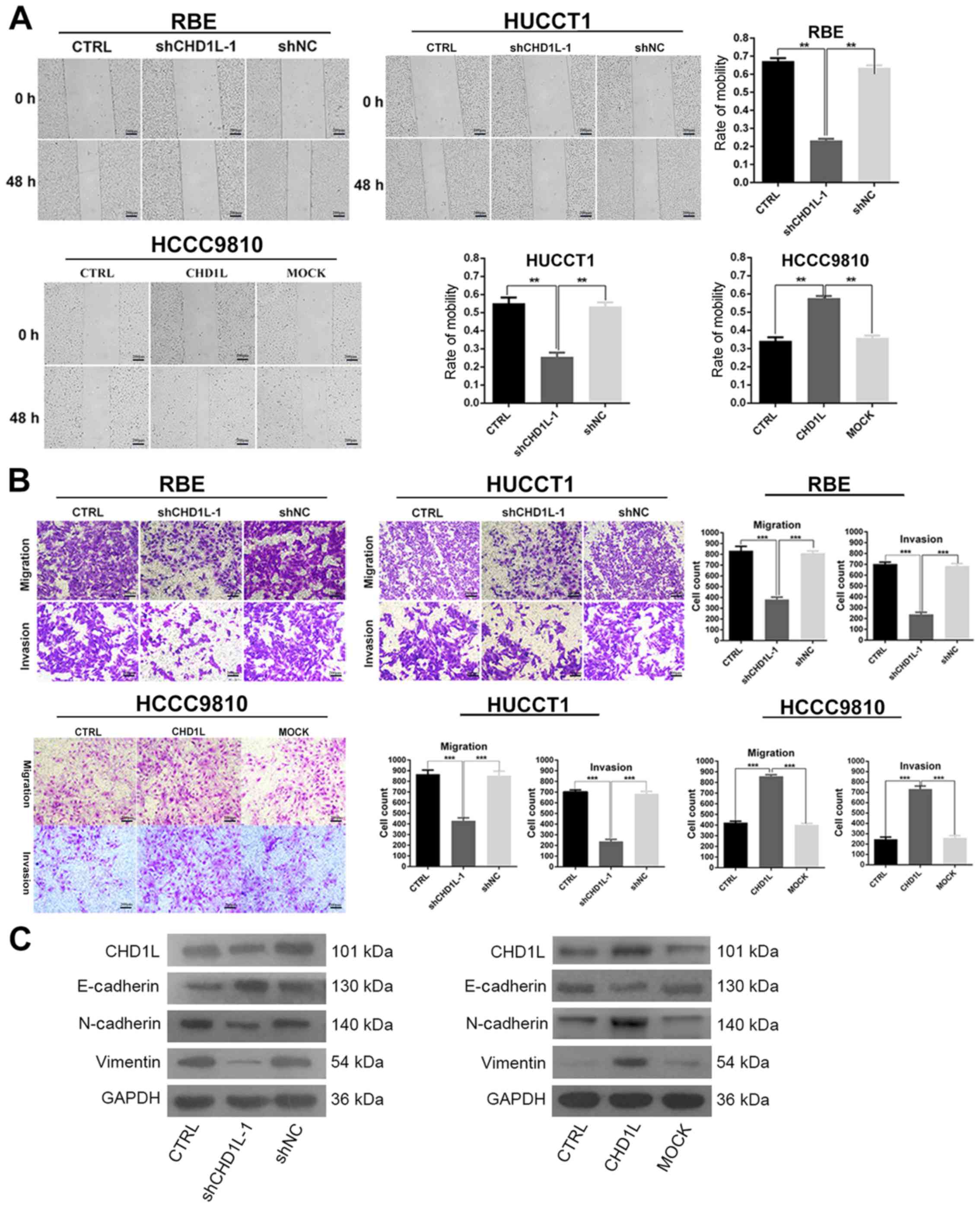 | Figure 5.CHD1L increases ICC cell
migration and invasion by inducing EMT. (A) Cell migration ability
in RBE, HUCCT1 and HCCC9810 cells in the various groups was
detected at 48 h by wound healing assay. The rate of mobility was
calculated and is depicted in the bar chart (**P<0.01). Scale
bar, 200 µm. (B) Transwell migration and invasion assays
demonstrated that the number of migrated and invasive cells in the
CHD1L-knockdown (shCHD1L-1) groups were significantly
reduced compared with the other two groups in RBE and HUCCT1 cells.
Overexpression of CHD1L in the HCCC9810 cells had the
opposite effects (***P<0.001). Scale bar, 200 µm. (C) The
protein expression of EMT-related biomarkers (vimentin, N-cadherin
and E-cadherin) in the indicated cells was examined by western
blotting. CHD1L, chromodomain helicase/ATPase DNA-binding
protein 1-like gene; ICC, intrahepatic cholangiocarcinoma; EMT,
epithelial-mesenchymal transition; CTRL, control; MOCK, empty
vector-transfected cells; shNC, negative control. |
CHD1L promotes tumor metastasis in
BALB/c-nu mice via mesenchymal-epithelial transition (MET)
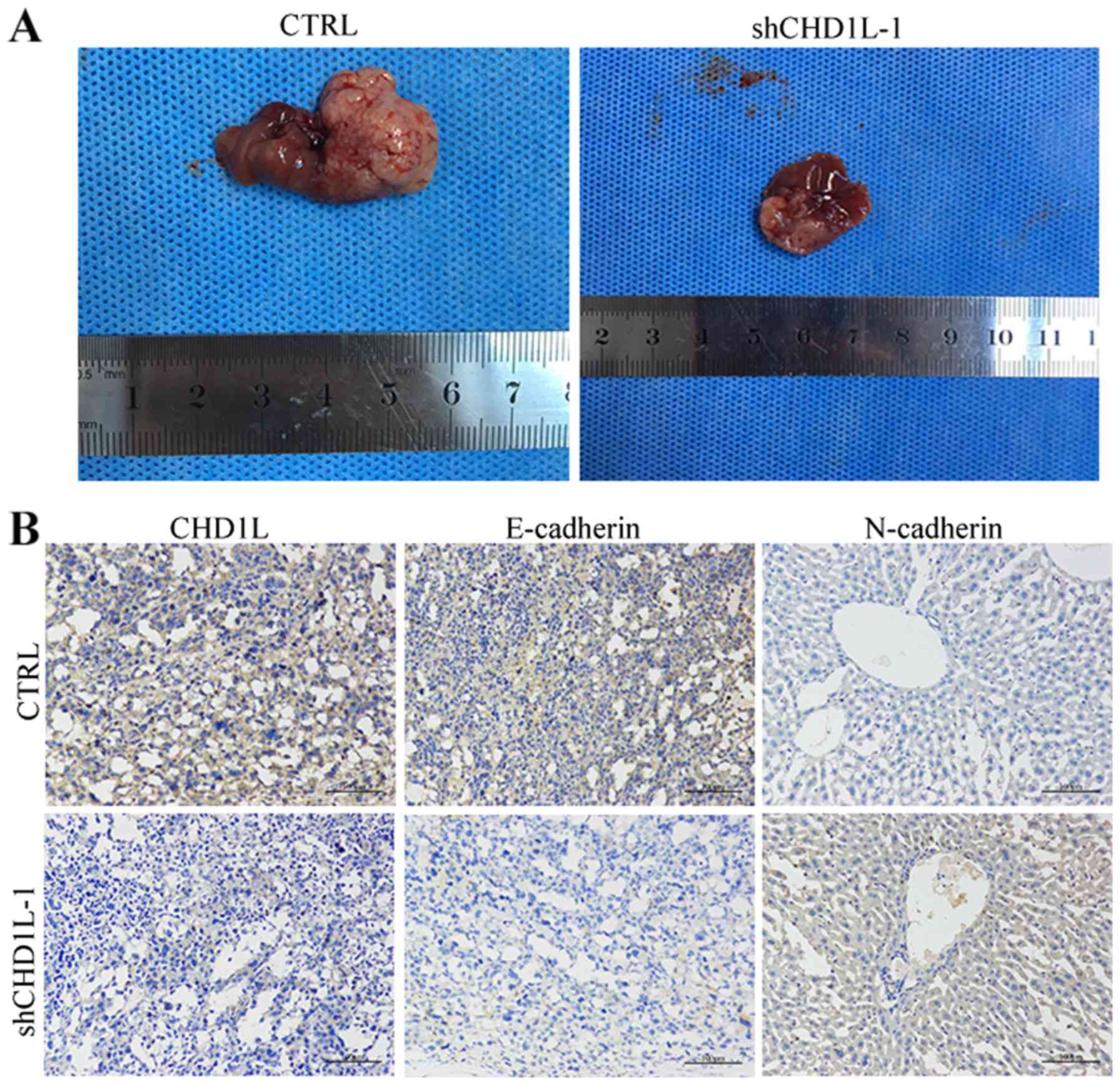
To confirm the in vivo effects of
CHD1L on metastasis, we performed a liver metastasis model
with BALB/c-nu mice in vivo after inoculation with
shCHD1L-1 or CTRL-transfected cells. The RBE cell suspension
was injected into the abdominal cavity of nude mice. After the
peritoneal metastatic tumors grew, the mice were euthanized and the
livers were harvested (Fig. 6A).
Metastatic liver tumors were observed in 2 of 5 and 4 of 5
shCHD1L-1- and CTRL-injected nude mice, respectively
(Table VI). To determine whether MET
plays a role in tumor metastasis induced by CHD1L in the nude
mouse, IHC with antibodies against E-cadherin and N-cadherin was
performed on serial sections of each tumor (Fig. 6B). The IHC results showed that
decreased expression of E-cadherin and increased expression of
N-cadherin were observed in tumors induced by shCHD1L-1
cells compared with tumors induced by CTRL cells. This suggests
that the CHD1L promotes ICC metastasis in vivo by
MET.
 | Table VI.Tumor incidence rate during the
4-week observation period between CTRL and shCHD1L-1. |
Table VI.
Tumor incidence rate during the
4-week observation period between CTRL and shCHD1L-1.
|
| Tumor incidence
rate (n/total) for week |
|---|
|
|
|
|---|
| Groups | 1 | 2 | 3 | 4 |
|---|
| CTRL | 0/5 | 1/5 | 3/5 | 4/5 |
| shCHD1L-1 | 0/5 | 0/5 | 1/5 | 2/5 |
Discussion
In a previous study, a candidate oncogene
chromodomain helicase/ATPase DNA-binding protein 1-like gene
(CHD1L) was identified in hepatocellular carcinoma (HCC)
(6). It is overexpressed in many
tissues, including the bladder (19)
and colorectal cancer (20), glioma
(21), myeloma (22) and lung adenocarcinoma (23). Recently, CHD1L expression was
found to increase tumor progression in pancreatic cancer (24). Although CHD1L is reported to be
overexpressed in several other types of carcinoma, it has not been
linked to intrahepatic cholangiocarcinoma (ICC). In the present
study, we aimed to detect CHD1L expression in tumor tissues
and evaluate its prognostic significance in ICC. Our results showed
that CHD1L was overexpressed in ICC tissues compared with
the adjacent non-tumor tissues, indicating that CHD1L may
have an effect on ICC development. Moreover, IHC results
demonstrated that overexpression of CHD1L was significantly
related to histological differentiation, vascular invasion, lymph
node metastasis, TNM stage and a shorter overall and disease-free
survival time of ICC patients. In addition, Cox regression
statistical analysis further indicated that high CHD1L
expression was an independent predictor for poor prognosis in ICC
patients. Therefore, CHD1L overexpression in ICC may serve
as a biomarker for early diagnosis and precise prognoses.
Additionally, we evaluated the expression of
CHD1L in ICC cell lines by western blotting and RT-qPCR. The
expression level of CHD1L was relatively higher in ICC cell
lines than that in HiBECs and the normal bile duct epithelial cell
line, which suggested that CHD1L may have oncogenic ability.
Meanwhile, the highest CHD1L levels were found in highly
aggressive cell lines RBE (25) and
HUCCT1 (26) and the lowest
CHD1L levels were found in ICC cell line HCCC9810 (27). Hence, we chose RBE and HUCCT1 cell
lines for stable transfection with CHD1L-shRNA lentivirus
vectors, and HCCC9810 cell lines for stable transfection with the
CHD1L-overexpression plasmid vector. Our results
demonstrated the effect of shRNA transduction on the expression of
CHD1L and the most efficient knockdown by shCHD1L-1
was noted when compared with those of the other two vectors
(shCHD1L-2 and shCHD1L-3).
In addition, promotion of cell proliferation is a
major molecular mechanism of an oncogene in cancer development. In
the present study, we demonstrated that CHD1L had strong
tumorigenic ability in vitro and in vivo functional
studies. Inhibition of CHD1L obstructed G1/S cell cycle
transition and the effect was reversed by CHD1L
overexpression. P53 is crucial for effective tumor
suppression in humans (28) and can
upregulate the expression of P21, which in turn functions as
a CDK2 inhibitor to control S phase entry via the
inactivation of the cyclin D1-CDK2 complex (29,30).
Consistent with this theory, we found that CHD1L reduced
P53 expression and increased S-phase-specific protein
expression, including CDK2 and cyclin D1, after
CHD1L-overexpression transfection. This evidence suggested
that the dysregulation of the P53/cyclin D1/CDK2 pathway
maybe involved in CHD1L-induced G1/S transition in ICC.
Finally, we found that CHD1L promoted ICC
cell migration and invasion, suggesting that CHD1L may
promote metastasis-related genetic alterations in ICC cells. It has
been reported that metastasis is the most common cause of death
from malignant neoplasms (31).
Metastasis is a multistep cellular process by which tumor cells
disseminate from their primary site and form secondary tumors at a
distant site. The pathophysiological course of metastasis is
mediated by the dynamic plasticity of cancer cells, which enables
them to shift between epithelial and mesenchymal phenotypes through
a transcriptionally regulated program termed EMT and its reverse
process MET (32). EMT includes loss
of cell-cell adhesion and activation of mesenchymal markers, as
well as increased motility of tumor cells (33). It has been reported that CHD1L
promotes HCC progression and metastasis by induction of EMT
(34). In the present study, we found
that inhibition of the expression of CHD1L induced EMT by
upregulation of expression of the epithelial marker, E-cadherin,
and downregulation of expression of the mesenchymal markers,
N-cadherin and vimentin. Furthermore, silencing of CHD1L
expression inhibited MET phenotype, which involved decreased
expression of E-cadherin and increased expression of N-cadherin in
nude mice by IHC. Our findings indicate that CHD1L may drive
EMT and MET in cancer cells, resulting in metastasis.
In summary, we confirmed that the overexpression of
CHD1L was associated significantly with histological
differentiation, vascular invasion, lymph node metastasis, TNM
stage, and the shorter overall and disease-free survival time of
ICC patients. CHD1L promotes ICC cell proliferation and
metastasis both in vitro and in vivo. We hypothesize
that the dysregulation of the P53/cyclin D1/CDK2 pathway
maybe involved in CHD1L-induced G1/S transition and that
CHD1L may drive EMT and MET in ICC cells, resulting in
metastasis. Taken together, this study offers new insights that
CHD1L may serve as an oncogene in ICC pathogenesis. However,
our research has some shortcomings. First, further research is
still needed to validate the more reasonable approach to silence or
overexpress CHD1L. Second, a better understanding of the
oncogenic mechanisms of CHD1L during ICC initiation and
progression may have implications for future patient treatment.
Acknowledgements
The authors thank the Key Molecular Medical
Laboratory of Jiangxi Province (Nanchang, China) for providing the
technical assistance, and offering human intrahepatic biliary
epithelial cells (HiBECs).
Funding
The present study was supported by the Natural
Science Foundation of Jiangxi Province, China (grant no.
20151512070090), the Science and Technology Program of Health and
Planning Commission in Jiangxi Province (grant no. 20161048) and
the Science and Technology Research of Education Department in
Jiangxi Province (grant no. GJJ150049).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SL and YC were responsible for the experimental
design. SL contributed to the execution of the experiments, data
statistical analysis and the manuscript content. YC and YD
participated in performing the experiments and in the manuscript
mapping and submission. TY participated in the writing of the
Discussion and data interpretation. CW conceived the study and
revised the manuscript. CH was responsible for the funding
application, the supervision and management of the project and
offered technical instruction and assistance. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All procedures in studies were in accordance with
the ethical standards of the Human Biomedical Research and Animal
Research Ethics Review Board of the People's Hospital of Jiangxi
Provincial (Nanchang, China). Informed consent was obtained from
all individual participants included in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CHD1L
|
chromodomain helicase/ATPase
DNA-binding protein 1-like gene
|
|
ICC
|
intrahepatic cholangiocarcinoma
|
|
HCC
|
hepatocellular carcinoma
|
|
SNF2
|
sucrose non-fermenting 2
|
|
EMT
|
epithelial-mesenchymal transition
|
|
MET
|
mesenchymal-epithelial transition
|
|
CTRL
|
control
|
|
LV-shNC
|
lentivirus-shorthairpin negative
control
|
|
RT-qPCR
|
real-time quantitative polymerase
chain reaction
|
|
IHC
|
immunohistochemistry
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EGFP
|
enhanced green fluorescent
protein
|
|
MOCK
|
empty vector-transfected
|
|
HR
|
hazard ratio
|
|
CI
|
confident interval
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Li J, Xia Y, Gong R, Wang K, Yan
Z, Wan X, Liu G, Wu D, Shi L, et al: Prognostic nomogram for
intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin
Oncol. 31:1188–1195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Haga H and Patel T: Molecular diagnosis of
intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Sci.
22:114–123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bridgewater J, Galle PR, Khan SA, Llovet
JM, Park JW, Patel T, Pawlik TM and Gores GJ: Guidelines for the
diagnosis and management of intrahepatic cholangiocarcinoma. J
Hepatol. 60:1268–1289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okumura S, Kaido T, Hamaguchi Y, Kobayashi
A, Shirai H, Fujimoto Y, Iida T, Yagi S, Taura K, Hatano E, et al:
Impact of skeletal muscle mass, muscle quality, and visceral
adiposity on outcomes following resection of intrahepatic
cholangiocarcinoma. Ann Surg Oncol. 24:1037–1045. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma NF, Hu L, Fung JM, Xie D, Zheng BJ,
Chen L, Tang DJ, Fu L, Wu Z, Chen M, et al: Isolation and
characterization of a novel oncogene, amplified in liver cancer 1,
within a commonly amplified region at 1q21 in hepatocellular
carcinoma. Hepatology. 47:503–510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng W, Su Y and Xu F: CHD1L: A novel
oncogene. Mol Cancer. 12:1702013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen L, Yuan YF, Li Y, Chan TH, Zheng BJ,
Huang J and Guan XY: Clinical significance of CHD1L in
hepatocellular carcinoma and therapeutic potentials of
virus-mediated CHD1L depletion. Gut. 60:534–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu J, Zong Y, Fei X, Chen X, Huang O, He
J, Chen W, Li Y, Shen K and Zhu L: Presence of CHD1L
over-expression is associated with aggressive tumor biology and is
a novel prognostic biomarker for patient survival in human breast
cancer. PLoS One. 9:e986732014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Su Z, Zhao J, Xian G, Geng W, Rong Z, Wu Y
and Qin C: CHD1L is a novel independent prognostic factor for
gastric cancer. Clin Transl Oncol. 16:702–707. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su FR, Ding JH, Bo L and Liu XG:
Chromodomain helicase/ATPase DNA binding protein 1-like protein
expression predicts poor prognosis in nasopharyngeal carcinoma. Exp
Ther Med. 8:1745–1750. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He WP, Zhou J, Cai MY, Xiao XS, Liao YJ,
Kung HF, Guan XY, Xie D and Yang GF: CHD1L protein is overexpressed
in human ovarian carcinomas and is a novel predictive biomarker for
patients survival. BMC Cancer. 12:4372012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Farges O, Fuks D, Le Treut YP, Azoulay D,
Laurent A, Bachellier P, Nuzzo G, Belghiti J, Pruvot FR and
Regimbeau JM: AJCC 7th edition of TNM staging accurately
discriminates outcomes of patients with resectable intrahepatic
cholangiocarcinoma. Cancer. 117:2170–2177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Renshaw S: Immunohistochemistry and
immunocytochemistry: Essential methods. John Wiley Sons.
6:2482017.
|
|
16
|
Li J, Guan HY, Gong LY, Song LB, Zhang N,
Wu J, Yuan J, Zheng YJ, Huang ZS and Li M: Clinical significance of
sphingosine kinase-1 expression in human astrocytomas progression
and overall patient survival. Clin Cancer Res. 14:6996–7003. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koya Y, Kajiyama H, Liu W, Shibata K,
Senga T and Kikkawa F: Murine experimental model of original tumor
development and peritoneal metastasis via orthotopic inoculation
with ovarian carcinoma cells. J Vis Exp. 9:543532016.
|
|
18
|
Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao
Y, Wang XA, Zhang F, Xiang SS, Li HF, et al: SPOCK1 as a potential
cancer prognostic marker promotes the proliferation and metastasis
of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol
Cancer. 14:122015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tian F, Xu F, Zhang ZY, Ge JP, Wei ZF, Xu
XF and Cheng W: Expression of CHD1L in bladder cancer and its
influence on prognosis and survival. Tumor Biol. 34:3687–3690.
2013. View Article : Google Scholar
|
|
20
|
Ji X, Li J, Zhu L, Cai J, Zhang J, Qu Y,
Zhang H, Liu B, Zhao R and Zhu Z: CHD1L promotes tumor progression
and predicts survival in colorectal carcinoma. J Surg Res.
185:84–91. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhong D, He G, Zhao S, Li J, Lang Y, Ye W,
Li Y, Jiang C and Li X: LRG1 modulates invasion and migration of
glioma cell lines through TGF-β signaling pathway. Acta Histochem.
117:551–558. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu X, He Y, Miao X, Wu Y, Han J, Wang Q,
Liu J, Zhong F, Ou Y, Wang Y and He S: Cell adhesion induces
overexpression of chromodomain helicase/ATPase DNA binding protein
1-like gene (CHD1L) and contributes to cell adhesion-mediated drug
resistance (CAM-DR) in multiple myeloma cells. Leuk Res. 47:54–62.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He LR, Ma NF, Chen JW, Li BK, Guan XY, Liu
MZ and Xie D: Overexpression of CHD1L is positively associated with
metastasis of lung adenocarcinoma and predicts patients poor
survival. Oncotarget. 6:31181–31190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu C, Fu X, Zhong Z, Zhang J, Mou H, Wu
Q, Sheng T, Huang B and Zou Y: CHD1L expression increases tumor
progression and acts as a predictive biomarker for poor prognosis
in pancreatic cancer. Dig Dis Sci. 62:2376–2385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He Q, Cai L, Shuai L, Li D, Wang C, Liu Y,
Li X, Li Z and Wang S: Ars2 is overexpressed in human
cholangiocarcinomas and its depletion increases PTEN and PDCD4 by
decreasing microRNA-21. Mol Carcinog. 52:286–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang XQ, Xu YF, Guo S, Liu Y, Ning SL, Lu
XF, Yang H and Chen YX: Clinical significance of nerve growth
factor and tropomyosin-receptor-kinase signaling pathway in
intrahepatic cholangiocarcinoma. World J Gastroenterol.
20:4076–4084. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shen G, Lin Y, Yang X, Zhang J, Xu Z and
Jia H: MicroRNA-26b inhibits epithelial-mesenchymal transition in
hepatocellular carcinoma by targeting USP9X. BMC Cancer.
14:3932014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Green DR and Chipuk JE: p53 and
metabolism: Inside the TIGAR. Cell. 126:30–32. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zolota V, Sirinian C, Melachrinou M,
Symeonidis A and Bonikos DS: Expression of the regulatory cell
cycle proteins p21, p27, p14, p16, p53, mdm2, and cyclin E in bone
marrow biopsies with acute myeloid leukemia. Correlation with
patients' survival. Pathol Res Pract. 203:199–207. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen M, Huang JD, Hu L, Zheng BJ, Chen L,
Tsang SL and Guan XY: Transgenic CHD1L expression in mouse induces
spontaneous tumors. PLoS One. 4:e67272009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
López-Soto A, Gonzalez S, Smyth MJ and
Galluzzi L: Control of metastasis by NK cells. Cancer Cell.
32:135–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:eaak95572017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen L, Chan TH, Yuan YF, Hu L, Huang J,
Ma S, Wang J, Dong SS, Tang KH, Xie D, et al: CHD1L promotes
hepatocellular carcinoma progression and metastasis in mice and is
associated with these processes in human patients. J Clin Invest.
120:1178–1191. 2010. View Article : Google Scholar : PubMed/NCBI
|