Introduction
Breakpoint cluster region (BCR)-Abelson murine
leukemia (ABL)1+ acute B-lymphoblastic leukemia (B-ALL)
is characterized by the presence of the Philadelphia chromosome
arising from the t(9;22)(q34;q11.2) translocation (1). The fusion gene constitutively encodes
BCR-ABL1 tyrosine kinase, which activates its downstream signaling
molecules/pathways (2). The
accentuation of these pathways leads to the arrest of B-cell
differentiation at an immature stage (progenitor or precursor),
followed by uncontrolled proliferation and invasion (3). The use of tyrosine kinase inhibitors
(TKIs) is the most effective method for suppressing BCR-ABL1
tyrosine kinase activity (4,5). However, single-agent TKI has not
produced sustained responses in BCR-ABL1+ B-ALL, as this
specific subtype of B-ALL is likely to develop significant
involvement of the CNS, which acts as a ‘sanctuary’ site for
leukemic cells to escape anticancer treatments (6–8).
Therefore, understanding the molecular mechanisms of
BCR-ABL1+ B-ALL associated with CNS metastasis is a
critical step to improve therapeutics.
Previous studies have reported that leukemia follows
a blood-vessel track to enter the nervous system, indicating that
lymphocytes and leukemic cells may share similar mechanisms of
dissemination into tissues (9,10). These
cells leave the blood circulation and enter extramedullary tissues
by extravasation (9). Following
selectin-mediated rolling, cells adhere to endothelial vascular
walls by chemokine- and integrin-mediated adhesion to the
endothelium (11). Subsequently,
lymphocytes and leukemic cells cross the endothelial barrier via
transendothelial migration (12,13). The
morphological changes and mechanical force required to cross the
endothelial barrier rely on integrins and adhesion molecules
expressed on lymphocytes or leukemic cells (7,10). These
phenomena imply that BCR-ABL1+ leukemic cells also have
the ability to enter and disseminate into the CNS. The BCR-ABL1
retroviral bone marrow transduction/transplantation model and the
human xenograft NOD/severe combined immunodeficiency (SCID) mouse
model are widely used to investigate the mechanisms of
BCR-ABL1+ B-ALL transformation (14,15).
However, the characteristics of BCR-ABL1+ B-ALL with CNS
metastasis have not been previously investigated, at least to the
best of our knowledge.
In the present study, the experimental procedure of
BCR-ABL1 retroviral transduction was employed to establish a murine
model of BCR-ABL1+ B-ALL with CNS metastasis. The
BCR-ABL1+ leukemic cells have an immature B-cell
phenotype, including pro-B and pre-B populations with a high
expression of integrin subunit alpha 6 (Itga6) and L-selectin
adhesion molecules. BCR-ABL1+ leukemic B cells were
observed to predominantly invade the meninges, medulla oblongata,
cerebrum, cerebellum and lumbar spinal cord. The present results
demonstrate that BCR-ABL1+ B-ALL with CNS metastasis is
dependent on the intrinsic infiltration ability of
BCR-ABL1+ cells, but is independent on increased
vascular permeability and structural damage. The present study
provides a suitable murine model for studying BCR-ABL1-dependent
leukemogenesis and may aid in the development of novel therapeutic
strategies for B-ALL disorders.
Materials and methods
Retroviral construction
Murine stem cell virus (MSCV) vector co-expressing
human BCR-ABL1 (p210) and green fluorescence protein (GFP;
MSCV-BCR-ABL1-IRES-GFP, MIG-p210) and MSCV vector expressing GFP
(MIG) have been previously described (16). K562 cells were cultured at 37°C with
5% CO2 in RPMI-1640 (HyClone; Thermo Fisher Scientific,
Inc.) medium supplemented with 10% fetal calf serum (Gibco; Thermo
Fisher Scientific, Inc.). 293T cells were cultured at 37°C with 5%
CO2 in DMEM (HyClone; Thermo Fisher Scientific, Inc.)
medium supplemented with 10% fetal calf serum. 293T cells were
transfected with PKAT2 packaging vector and MIG-p210 or MIG using
X-tremeGENE HP DNA Transfection reagent (Roche), respectively. At
48 h following transfection, viral supernatants were collected,
filtered and stored at −80°C. K562 and 293T cells were kindly
provided by Professor Yanmin Zhang (Xi'an Jiaotong University
Health Science Centre).
Bone marrow
transduction/transplantation
As previously described (14,15),
experimental C57BL/6 mice (age, 7–10 weeks; weight, ~18 g) were
purchased from the Experimental Animal Center of Xi'an Jiaotong
University and allowed to acclimatize to the environment under
specific pathogen-free conditions. The donors (males) and
recipients (females) were of the same mouse strain. All animal
experiments were performed according to the guidelines of and were
approved by the Institutional Animal Care and Use Committee of
Xi'an Jiaotong University. In brief, in order to induce ALL, femurs
and tibias collected from the donor mice were placed in cold PBS,
clipped of the end of the bone, and bone marrow cells were flushed
out with PBS. Cells were spun down at 200 × g for 10 min at 4°C and
the supernatant was removed. Bone marrow cells from 2 donor C57BL/6
mice without 5-fluorouracil treatment were subjected to a single
round of co-sedimentation with MIG-p210 retroviral supernatants
supplemented with 5% WEHI-3B-conditioned medium, 10 ng/ml
interleukin (IL)-7 (PeproTech, Inc.) and 20 µg/ml polybrene
(Beijing Solarbio Science and Technology Co., Ltd.). The cells were
collected 2 h later, washed once in HBSS (Beijing Solarbio Science
and Technology Co., Ltd.) and transplanted into lethally irradiated
(2×460 cGy) syngeneic 6 female recipient mice (1×106
cells each) or subjected to culture in RPMI-1640 medium (HyClone,
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal calf
serum (Gibco; Thermo Fisher Scientific, Inc.), 200 µmol/l
L-glutamine, 50 µmol/l 2-mercaptoethanol (Amersco, Inc., Cleveland,
OH, USA) and penicillin/streptomycin (HyClone, Thermo Fisher
Scientific, Inc.) for 48 h. For the control group, bone marrow
cells from 1 donor C57BL/6 mouse were subjected to co-sedimentation
with MIG retroviral supernatants and transplanted into 3 recipient
mice (details were the same as mentioned above). A GFP+
cell population of >10% was considered to indicate successful
transduction. When the transplanted recipient mice exhibited
clinical signs of leukemia, which included weight loss, failure to
thrive, limb paralysis and splenomegaly, they were euthanized. For
secondary transplantation, 1×106, 1×105 and
1×104 bone marrow cells from primary
BCR-ABL1+ ALL mice were transplanted into non-irradiated
recipient mice, respectively. The secondary transplant recipients
were monitored for clinical signs of leukemia as mentioned above
for euthanization.
Flow cytometry
The bone marrow and spleen cells were collected from
leukemic mice. Red blood cells (RBC) were lysed with
NH4Cl RBC lysis buffer for 5 min at room temperature.
The nucleated cells were then washed with cold PBS and stained with
anti-Gr1-APC (1:20 dilution; cat. no. 553129; BD Biosciences),
anti-CD3-PE (1:20 dilution; cat. no. 561824; BD Biosciences) and
anti-CD19-PerCP-CyTM5.5 (1:20 dilution; cat. no. 551001;
BD Biosciences) for cell lineages, and incubated with anti-CD43-PE
(1:20 dilution; cat. no. 553271; BD Biosciences) and anti-B220-APC
(1:20 dilution; cat. no. 553092; BD Biosciences) antibodies for
B-cell development. The cells were incubated with the antibodies
for 20 min at 4°C. Analysis was performed using CytoFLEX flow
cytometer and the data were analyzed with CytExpert 2.1 (Beckman
Coulter, lnc.).
Histology
Mice were anesthetised with chloral hydrate (500
mg/kg) by intraperitoneal injection and the mice were monitored for
the absence of tail, foot and ear reflexes, as well as a reduced
respiratory rate, which are all indications of effective anesthesia
(17). The mice were firstly perfused
with 0.9% saline and then with 4% paraformaldehyde in 0.1 M
phosphate buffer (pH 7.4). The brain and spinal cord tissues were
harvested and post-fixed in 4% paraformaldehyde for 4–6 h at 4°C.
The tissues were then dehydrated in 30% sucrose. All the tissues
were embedded in Tek Optimal Cutting Temperature compound (Sakura
Finetek Japan Co., Ltd.), sectioned with a semiconductor freezing
microtome and stained with hematoxylin and eosin (H&E). For
H&E staining (cat. no. G1005; Wuhan Servicebio Co., Ltd.) the
slides were washed in PBS and stained with Harris's hematoxylin
(10%) for 3–8 min and eosin (70%) for 1–3 min at room temperature.
Subsequently, the sections were dehydrated through an increasing
concentration of ethanol and xylene. Images of H&E staining
were captured using a Zeiss Axio Scope. A1 microscope (Carl Zeiss
Microscopy GmbH).
Wright-Giemsa staining
A 10 µl aliquot of peripheral blood or bone marrow
cell flushed out with PBS was spread on a slide to produce a smear
and dried at room temperature for 30 min. The smear was fixed with
methanol for 15 min and dried at room temperature. The smear was
stained with Wright-Giemsa Stain (Heart Biological Technology Co.,
Ltd) for 8 min at room temperature and was then rinsed with water
until the edges turned a pinkish-red color. After air-drying, the
smear was inspected under a Zeiss Axio Scope. A1 microscope (Carl
Zeiss Microscopy GmbH).
Analysis of CNS metastasis of leukemia
by intravascular labeling
Individual BCR-ABL1+ B-ALL mice were
intravenously injected with 3 mg
anti-CD19-PerCP-CyTM5.5, at 4 min prior to
CO2 euthanasia (flow rate of CO2 used for the
euthanasia of the mice was displaced 20% of the chamber volume per
min). Blood was then harvested by cardiocentesis, and mice were
subsequently perfused with saline to eliminate residual blood in
vessels within tissues (18). The
cerebrum, cerebellum, medulla oblongata, meninges, thoracic spinal
cord and lumbar spinal cord were dissected and mechanically
processed to obtain a single-cell suspension. The samples were
examined for GFP and CD19 expression by flow cytometry.
BCR-ABL1+ B-ALL cells in the brain tissues were
identified as GFP+CD19− cells, since the
anti-CD19 antibody had been removed by saline in the experimental
procedure and the B-ALL cells remaining in the vasculature were
GFP+CD19+.
Western blot analysis
Cell pellet was collected and lysed in RIPA buffer
[50 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 1% Triton X-100, 0.5% NaDoc,
0.1% sodium dodecyl sulphide (SDS), 1 mM ethylenediamine
tetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid
(EGTA), 1 µM phenylmethane sulphonyl fluoride (PMSF) (Amersco,
Inc.) and 1 µg/ml Pepstatin A (Sigma Chemical Co.) protease
inhibitors]. Cell lysates were centrifuged for 15 min at 17,000 g
at 4°C and the protein supernatant was collected. Protein
concentration was determined via bicinchoninic acid protein assay
using a quantification kit (cat. no. BCA02; Beijing Dingguo
Changsheng Biotechnology Co., Ltd.). Proteins (40 µg) were run on a
7.5% (w/v) Tris-HCl sodium dodecyl sulphate-poly-acrylamide gel
electrophoresis (SDS-PAGE) for electrophoresis, transferred to
polyvinylidene difluoride (PVDF) membranes (Millipore Corp,
Billerica, MA, USA) and blotted, and then probed with c-ABL
antibody (1:1,000 dilution; cat. no. 2862; Cell Signaling
Technology, Inc.) overnight at 4°C. GAPDH (1:25,000 dilution; cat.
no. ab9485; Abcam Plc.) was incubated overnight at 4°C and used as
a loading control. The signal was further detected using the
secondary antibody of goat anti-rabbit IgG conjugated with
horseradish peroxidase (1:5,000 dilution; cat. no. 35560; Thermo
Fisher Scientific, Inc.) at room temperature for 1 h. The band
signal was visualized by Immobilon™ Western Chemiluminescent HRP
substrate (Millipore Corp.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, lnc.). Complementary
DNA (cDNA) was then generated using a PrimeScript™ RT reagent kit
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
protocol. qPCR was performed with FAST SYBR®− Green
Master Mix (Applied Biosystems) using an Mx3000P (Agilent
Technologies, Inc.) under following conditions: Initial
denaturation at 95°C for 30 sec; amplified for 40 cycles of 15 sec
at 95°C, 30 sec at 60°C and 30 sec at 72°C; dissociation for 1
cycle of 1 min at 95°C, 30 sec at 55°C and 30 sec at 95°C. The
relative mRNA levels of genes were calculated using the following
equation: 2−∆∆Cq=2−(Cq target gene-Cq GAPDH) target
sample-(Cq target gene-Cq GAPDH) control sample (19). The following the forward and reverse
primers were used: Itga6 5′-GATCCCGGGAGCCTCTTC-3′ and
5′-GATGTCACAGCTGTACAGGC-3′; L-selectin, 5′-CTTACTGGGGCTCGAGGAAC-3′
and 5′-TCTCTCTTGTTTTGTATGGCGAC-3′; GAPDH,
5′-CGTCCCGTAGACAAAATGGT-3′ and 5′-AGGTCAATGAAGGGGTCGTT-3′.
Statistical analysis
Statistical analysis was performed using SPSS 20.0
(IBM Corp.) and GraphPad Prism 6.0 (GraphPad Software). For
continuous variables, descriptive results were presented as the
means ± standard error of the mean (SEM). Levene's test was used
for equality of variances. The independent-samples t-test, one-way
analyses of variance (ANOVA) with post-hoc Fisher's LSD test and
log-rank Mantel-Cox analysis were performed for 2-group
comparisons, multiple groups comparisons and survival comparisons,
respectively. Kaplan-Meier survival curves wereused to describe the
changes of survival rate with time. P<0.05 was considered to
indicate a statistically significant difference.
Results
Establishment of a murine model of
BCR-ABL1+ ALL with extramedullary hematopoietic site
infiltration
To generate a murine model of BCR-ABL1+
ALL with metastasis, 1×106 bone marrow cells harvested
from donor mice were subjected to co-sedimentation with a
retrovirus co-expressing BCR-ABL1 and GFP, and then transplanted
into syngeneic lethally irradiated recipient mice. Of the primary
recipients, 100% exhibited symptoms of clinical leukemia with hind
limb paralysis to various degrees or other observable CNS symptoms,
weight loss, respiratory or other types of distress, or extreme
lethargy. The leukemic mice were euthanized from 40 to 83 days
following transplantation (Fig.
1A-C). As a result of cachexia, the maximum percentage of body
weight loss observed in the leukemic mice was 15.23% (Fig. 1C). The peripheral blood contained
lymphoblastic cells with 32.00% GFP+ cells (Fig. 1D). The bone marrow cells appeared to
be of a lymphoblastic phenotype with 43.49% GFP+ cells
(Fig. 1E). The leukemic mice
developed splenomegaly with 54.21% GFP+ cells (Fig. 1F) and lymphadenopathy with 86.83%
GFP+ cells (Fig. 1G).
These results indicate that the transplanted mice developed
classical ALL. Furthermore, BCR-ABL1+ leukemic cells
were observed to metastasize to the subcutaneous tissues, lung and
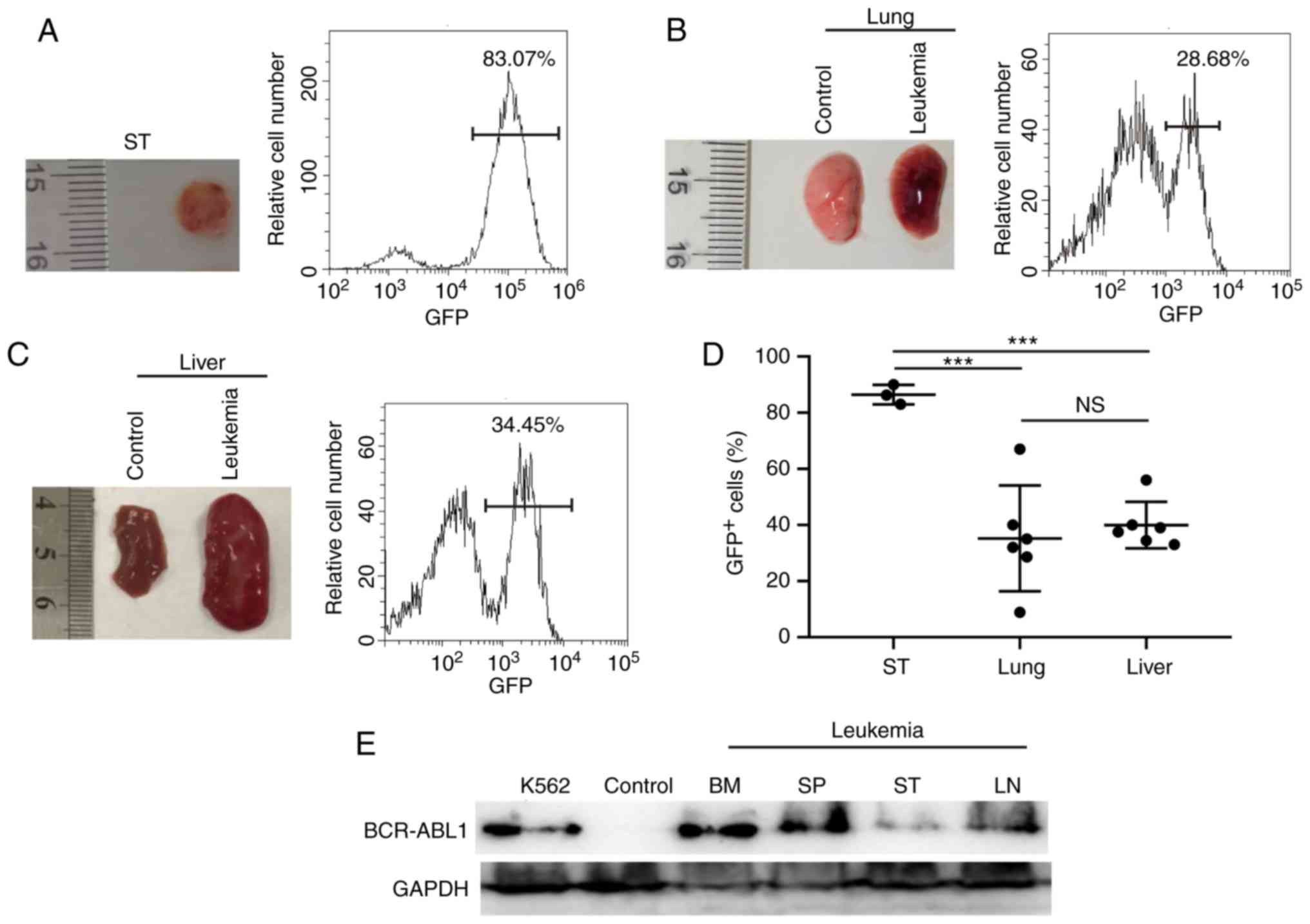
liver with varying amounts of GFP+ cells (Fig. 2A-C and Table
I). Of note, the degree of infiltration into the lymph nodes
and subcutaneous tissues was significantly higher compared with
that into other tissues in the BCR-ABL1+ ALL mice
(Figs. 1H and 2D). BCR-ABL1 expression were detected in the
bone marrow, spleen and subcutaneous tissues (Fig. 2E). Taken together, these results
demonstrate that the mouse model of BCR-ABL1+ ALL with
infiltration into non-hematopoietic tissues was successfully
established by using the BCR-ABL1 retroviral transduction
system.
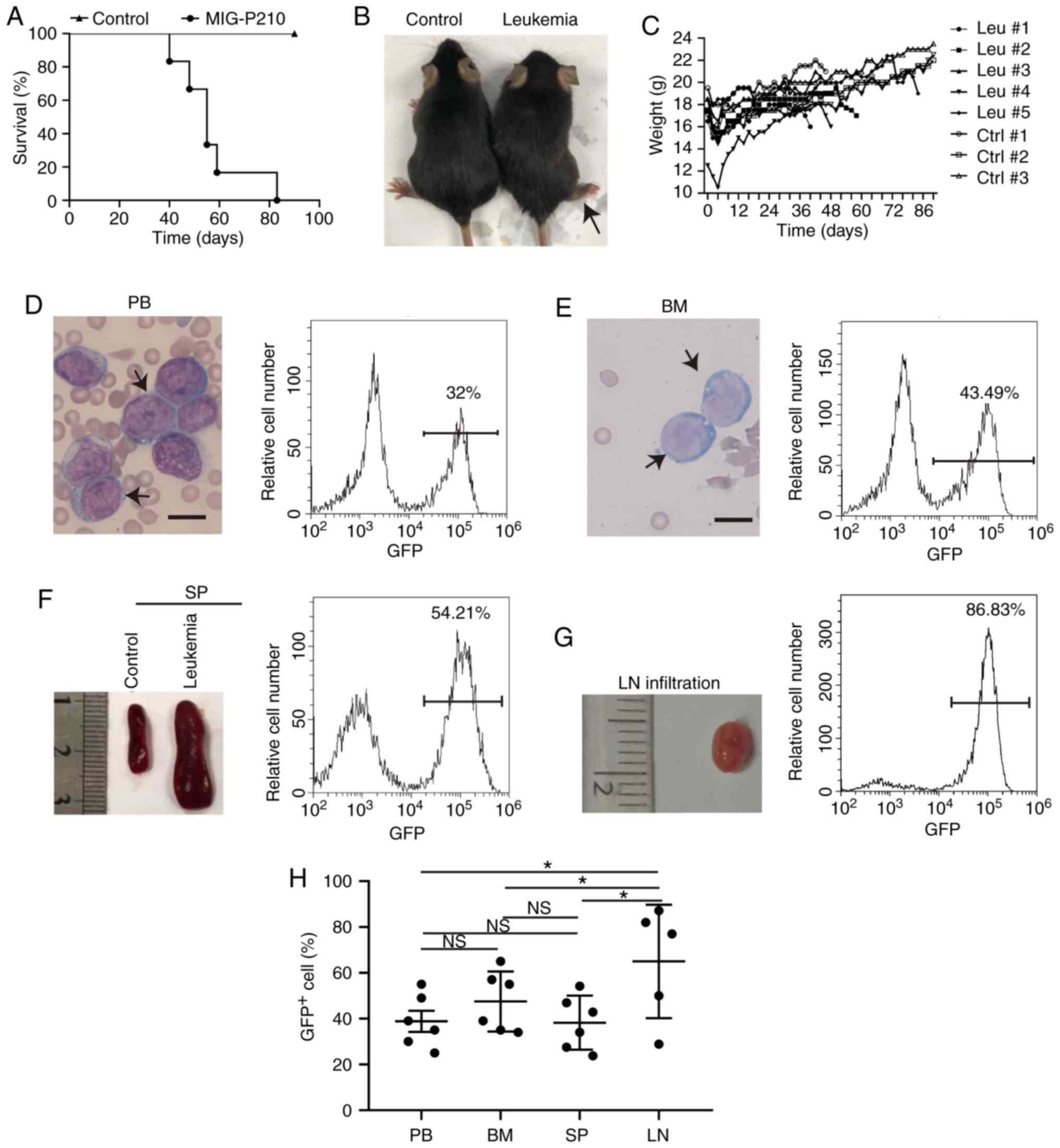 | Figure 1.Establishment of mouse
BCR-ABL1+ ALL models. (A) Kaplan-Meier survival curves
of syngeneic lethally irradiated recipient mice transplanted with
donor bone marrow cells transduced by the MIG-p210 (n=6) or MIG
(Control, n=3) retroviral supernatants, respectively. (B)
Representative leukemic mouse showing hind leg paralysis, as
indicated by the black arrow. (C) Monitoring of body weights of
leukemic and control mice until they were euthanized. Leukemic mice
(Leu, n=6) and control mice (Ctrl, n=3). (D) Peripheral blood (PB)
lymphoblastic cells were stained by Wright-Giemsa indicated with
the black arrows and the PB GFP+ cell percentage was
detected by flow cytometry in a leukemia mouse. Scale bar, 10 µm.
(E) Bone marrow (BM) lymphoblastic cells were stained by
Wright-Giemsa (indicated by black arrows) and the BM
GFP+ cell percentage was examined by flow cytometry in
one leukemic mouse. Scale bar, 10 µm. (F) Gross appearance of the
spleen (SP) and the percentage of GFP+ cells of SP in
one leukemic mouse. (G) Gross appearance of the lymph node (LN) and
the percentage of GFP+ cells of LN in one leukemic
mouse. (H) Scatter plot showing the percentages of GFP+
cells in BCR-ABL1+ ALL mice. PB (n=6), BM (n=6), SP
(n=6) and LN (n=5). Data indicate the means of independent mouse
data with the error bars representing the SEM. *P<0.05; NS, not
significant, P>0.05 (PB vs. BM, P=0.350; PB vs. SP, P=0.947; PB
vs. LN, P=0.012; BM vs. SP, P=0.318; BM vs. LN, P=0.049; SP vs. LN,
P=0.011). |
 | Table I.Summary of metastasis in primary
transplantation. |
Table I.
Summary of metastasis in primary
transplantation.
| Mouse ID | Hind limb
paralysis | Spleen
metastasis | Subcutaneous
metastasis | Lymph node
metastasis | Lungmetastasis | Liver
metastasis |
|---|
| 1# | + | + | + | – | + | + |
| 2# | + | + | – | – | ND | ND |
| 3# | + | + | – | – | ND | ND |
| 4# | + | + | + | + | + | + |
| 5# | + | + | – | + | ND | ND |
| 6# | + | + | + | – | + | + |
BCR-ABL1+ leukemic cells
have a B-lymphoid lineage-specific immunophenotype
To precisely define the BCR-ABL1+
lymphoblastic cells with respect to their phenotype and lineage
origin, it was examined whether BCR-ABL1+ leukemic cells
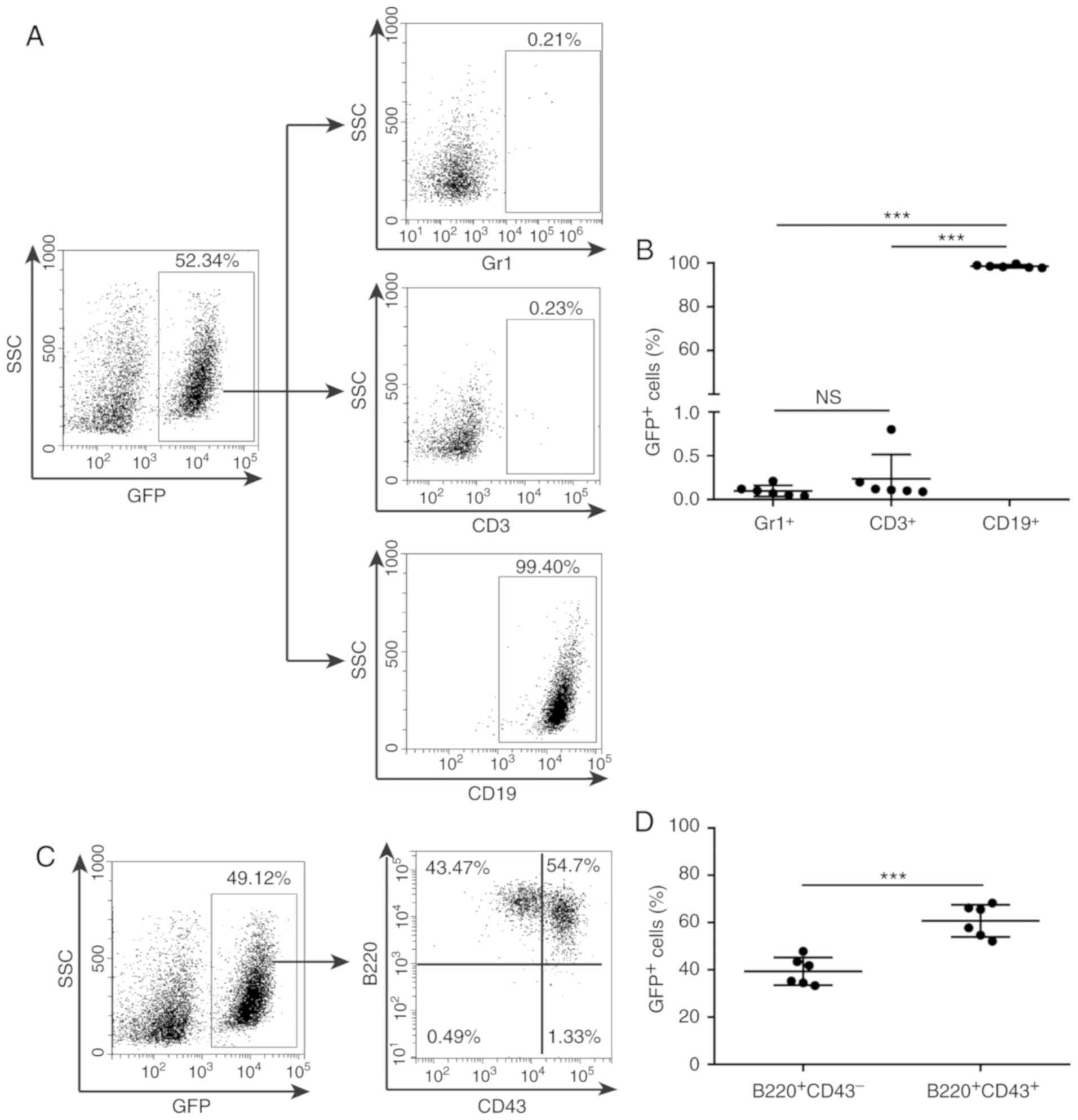
were lineage-specific. The bone marrow or spleen cells from
leukemic mice were incubated with antibodies against Gr1 for
myeloid cells, CD3 for T-lymphoid cells and CD19 for B-lymphoid
cells, respectively, and analyzed by flow cytometry. The results
indicated that GFP+ cells derived from individual
leukemic mice expressed the B lymphoid-specific marker CD19, but
not CD3 or Gr1 (Fig. 3A and B, and
Fig. S1A and B), suggesting that the
BCR-ABL1+ leukemic cells were of the B-cell lineage. To
evaluate the B-cell development stage of the leukemic
GFP+CD19+ population, the CD43- and
B220-expressing cells among GFP+ cells were gated. The
results indicated that GFP+ cells contained a variable
proportion of CD43 and B220 populations (Fig. 3C and D, and Fig. S1C and D), suggesting that the
BCR-ABL1+ leukemic cells remained fixed at the pro-B to
pre-B stage of B-cell development. The results demonstrated that
almost all BCR-ABL1+ leukemic cells exhibited a
pro-B/pre-B cell immunophenotype and that the BCR-ABL1+
leukaemia murine model generated in the present study may be
classified as BCR-ABL1+ B-ALL.
BCR-ABL1+ B-ALL leukemic
cells invade the CNS
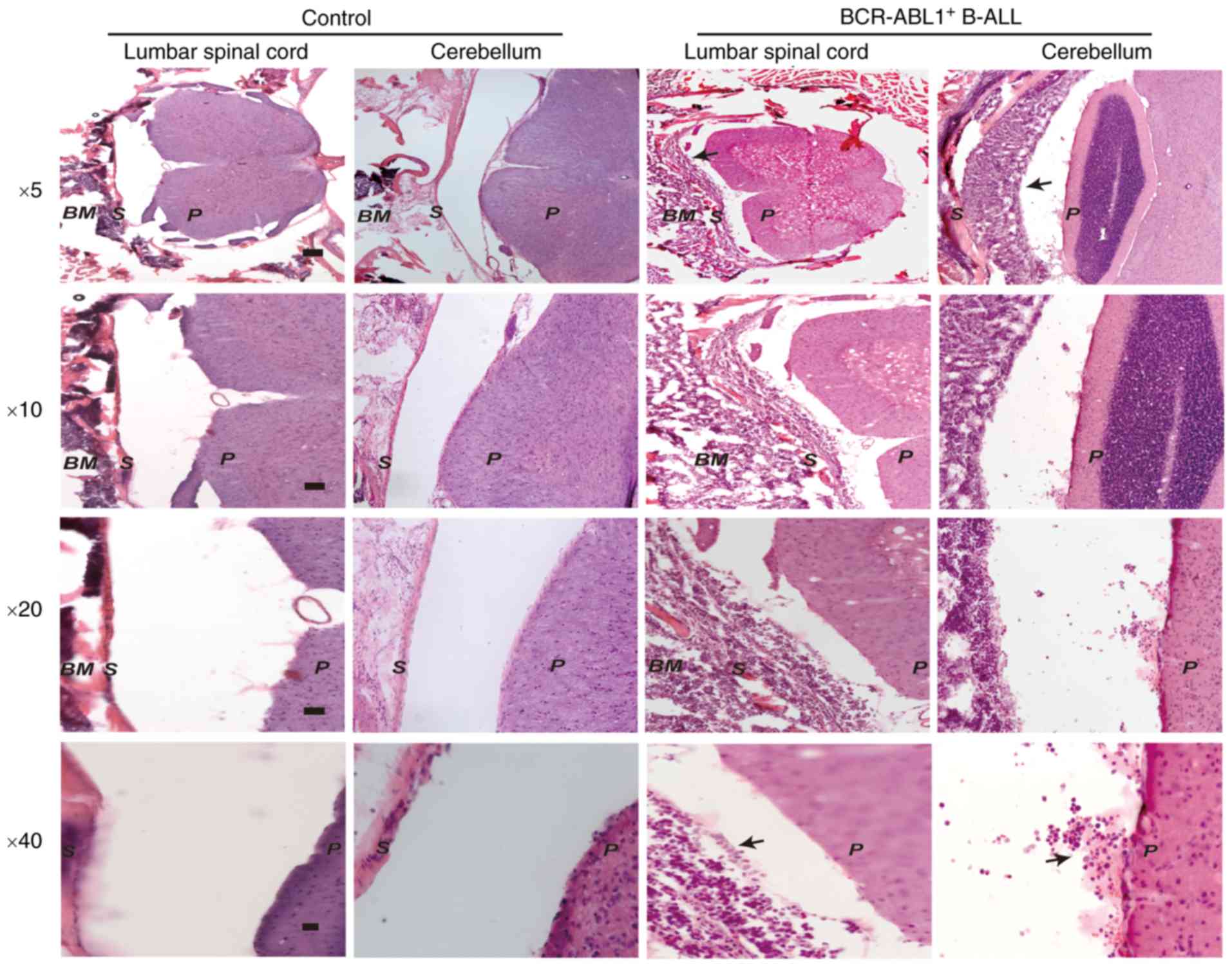
Since the BCR-ABL1+ B-ALL mice exhibited
severe CNS symptoms, the lumbar spinal cord and cerebellar tissues
were first dissected for H&E staining in the
BCR-ABL1+ B-ALL mice and healthy control mice,
respectively. The histological sections revealed a significant
number of leukemic cells in the lumbar spinal cord and meninges of
the BCR-ABL1+ B-ALL mice compared to the healthy
controls (Fig. 4), suggesting that
the BCR-ABL1+ cells had already invaded into the CNS
when the leukemic mice developed clinical symptoms of limb
paralysis.
Subsequently, to identify the exact proportions of
leukemic infiltration in the nervous system, an intravascular
staining approach was pursued. First, the anti-CD19 antibody was
injected into the vasculature of BCR-ABL1+ B-ALL mice
and the blood was then harvested immediately. The leukemic mouse
was perfused with saline to remove residual blood cells and
anti-CD19 antibody after euthanasia. The single-cell suspension was
collected from the blood, brain parenchyma, meninges and spinal
cord, and further analyzed for GFP and CD19 expression. The
BCR-ABL1+ B-ALL leukemic cells in the vasculature
revealed a GFP+CD19+ population, as the cells
were stained by anti-CD19 antibody (Fig.
5A). The leukemic cells that had infiltrated into the CNS had
no opportunity to be stained by anti-CD19 antibody and were only
GFP+, and their proportion varied between the meninges
(72.70%), lumbar spinal cord (8.84%), medulla oblongata (4.25%),
cerebrum (4.14%) and cerebellum (3.25%; Fig. 5B and C), which was in agreement with
the observation from H&E staining that massive leukemic cell
infiltration had occurred in the meninges. In addition, no
infiltration in the thoracic spinal cord was observed, indicating
that the leukemic cells in the lumbar spinal cord probably did not
originate from the spreading of CNS residual leukemia by extension
along spinal nerves. These results suggest that the
BCR-ABL1+ B-ALL cells have the ability to enter and
disseminate into the CNS.
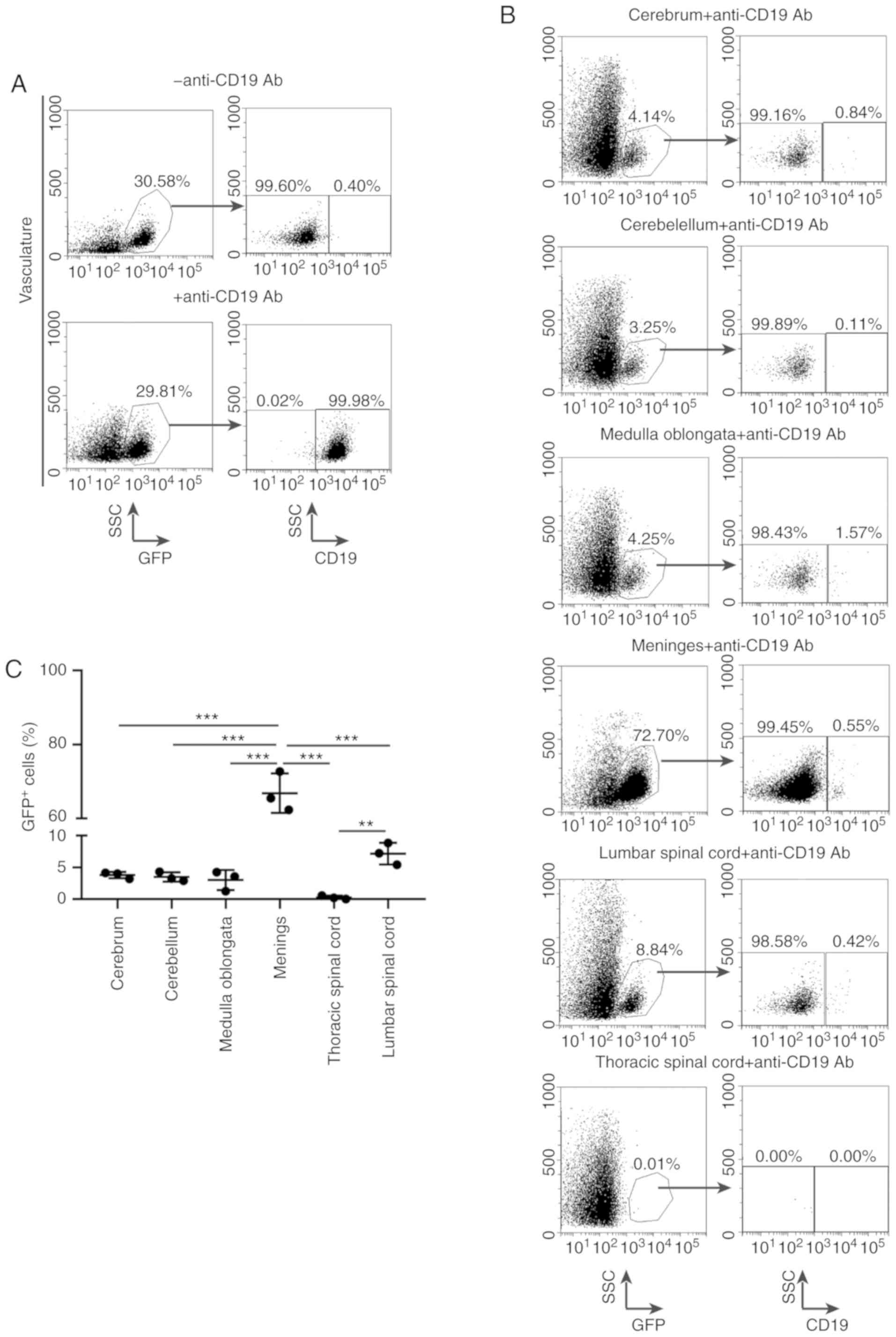 | Figure 5.Evaluation of BCR-ABL1+
B-ALL leukemic cell accumulation in various central nervous system
(CNS) tissues. Representative flow cytometric analysis of leukemic
cells metastasis into the nervous system. (A) The
BCR-ABL1+ B-ALL mouse was intravenously injected with
CD19-PerCP-CyTM5.5 antibodies then quickly euthanized.
Living cells were gated by forward- and side-scatter (data not
shown), and then gated on side-scatter and GFP (left panels).
GFP+ cells arrested in the vasculature were labelled by
CD19+ gate (right panels). (B) Living cells from various
CNS tissues were gated by forward- and side-scatter (data not
shown), followed by gating on side-scatter and GFP (left).
GFP+ cells were gated on CD19 to distinguish
CD19+ and CD19− population depending on
whether brain parenchyma GFP+ cells had either already
metastasized before injection with CD19-PerCP-CyTM5.5
antibodies or had been contaminated by vessel leukemic cells during
dissection, respectively (right panels). (C) Scatter plot showing
the percentages of GFP+ cells in CNS derived from
BCR-ABL1+ ALL mice. cerebrum (n=3), cerebellum (n=3),
medulla oblongata (n=3), meninges (n=3), lumbar spinal cord (n=3),
thoracic spinal cord (n=3). Data indicate the means of independent
mouse data with the error bars representing the SEM. **P<0.01
and ***P<0.001 (cerebrum vs. meninges, P<0.001; cerebellum
vs. meninges, P<0.001; medulla oblongata vs. meninges,
P<0.001; thoracic spinal cord vs. meninges, P<0.001; lumbar
spinal cord vs. meninges, P<0.001; lumbar spinal cord vs.
thoracic spinal cord, P=0.004; P-values between other groups were
no significance). |
BCR-ABL1+ B-ALL leukemic
cell invasion into the CNS is dependent on their own intrinsic
properties
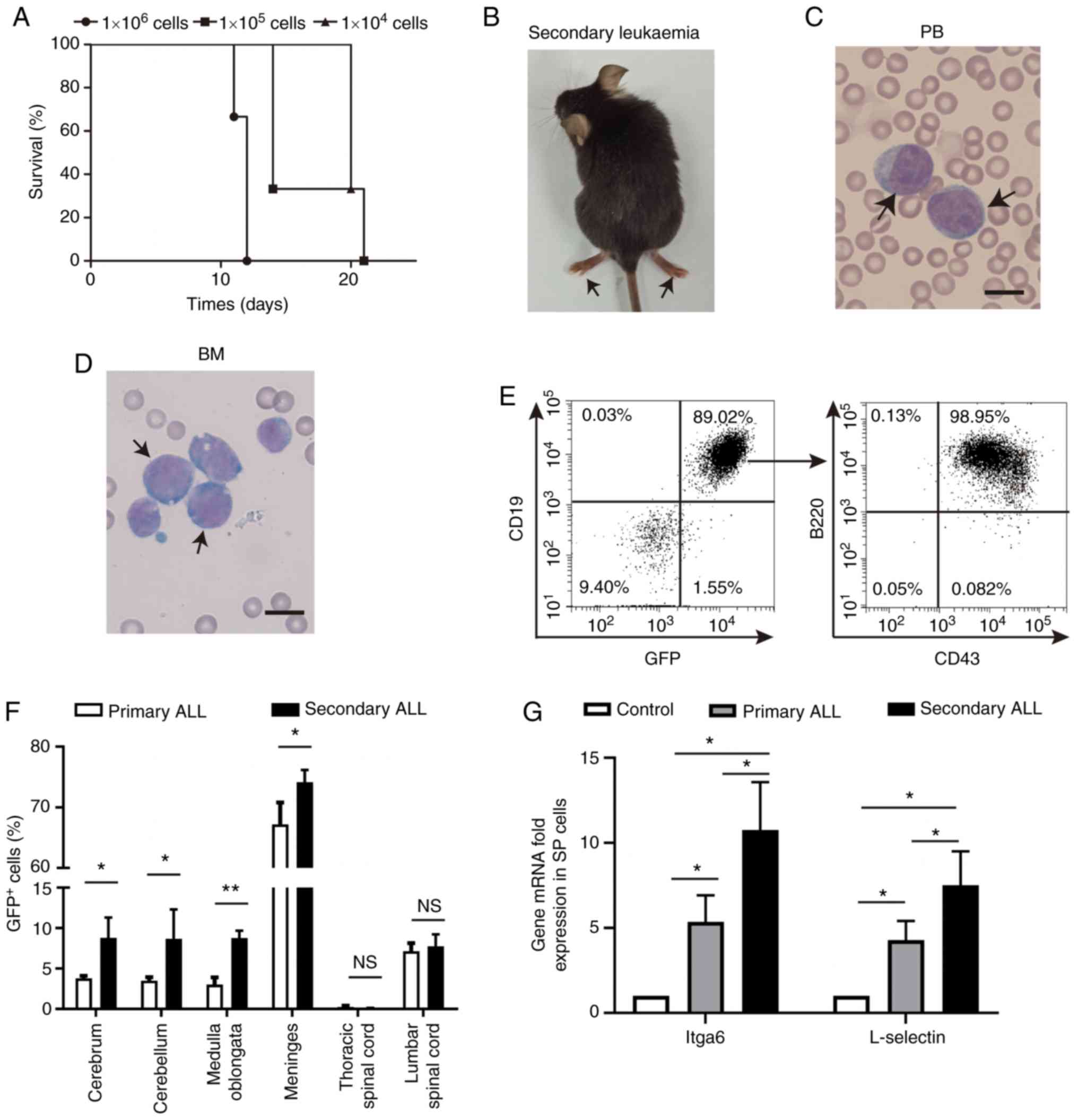
To further confirm whether the BCR-ABL1+
B-ALL with CNS metastasis was driven by the ability of leukemic
cells to enter and disseminate into the CNS, experiments were
performed using immunocompetent recipient mice injected with
1×106, 1×105 or 1×104 primary
BCR-ABL1+ leukemic cells. Irradiation-induced CNS damage
was avoided through this injection. It was observed that the
secondary recipients developed BCR-ABL1+ leukemia with
severe paralysis of their hind limbs within 25 days following
transplantation (Fig. 6A and B). The
onset of secondary leukemia was shorter in comparison with that of
primary leukemia (Fig. 1A). The
morphological lymphoblastic cells were enriched in the peripheral
blood and bone marrow (Fig. 6C and
D). The immunophenotype of GFP+ cells displayed a
B-cell lineage with block at pro-B to pre-B stages of B-cell
development (Fig. 6E). Of note, the
percentage of GFP+ cells in the CNS of secondary
BCR-ABL1+ B-ALL was significantly increased when
compared to that in primary B-ALL (Fig.
6F). These results re-emphasize that BCR-ABL1+
leukemogenic cells have their own intrinsic properties to induce
B-ALL with a high incidence of CNS infiltration.
It was hypothesized that the molecules associated
with adhesion are deregulated in BCR-ABL1+ B-ALL mice
(20). The expression of cell
adhesion molecule Itga6 and L-selectin was detected in splenic
GFP+CD19+ cells sorted from primary and
secondary BCR-ABL1+ B-ALL mice. The splenic
CD19+ cells derived from healthy mice were used as
controls. The results indicated that the transcripts of Itga6 and
L-selectin were upregulated in primary and secondary
BCR-ABL1+ B-ALL compared with those in healthy control
mice, respectively (Fig. 6G and
Fig. S2). Furthermore, the
expression levels of Itga6 and L-selectin were also significantly
increased in the secondary BCR-ABL1+ B-ALL mice compared
with those in the primary ones. These results may imply that in
BCR-ABL1+ B-ALL mice, the invasion of
BCR-ABL1+ leukemic cells into the CNS probably depends
on abnormal gene expression associated with metastasis.
Discussion
BCR-ABL1+ B-ALL has a poor prognosis and
is associated with refractoriness and CNS damage (21–24).
Current disease models fail to precisely resemble
BCR-ABL1+ B-ALL with CNS infiltration, posing a
limitation for in vivo studies aiming at elucidating the
pathogenesis of the disease and the associated mechanisms. In the
present study, a mouse model of BCR-ABL1+ B-ALL with CNS
metastasis was established. The majority of the
BCR-ABL1+ cell population were immature B cells with a
pro-B/pre-B phenotype. BCR-ABL1+ B-ALL cells were found
in CNS tissues at various proportions, with a specific accumulation
in the meninges. BCR-ABL1+ B-ALL cells have the ability
to propagate, maintain their phenotypes, and upregulate Itga6 and
L-selectin expression, which, together, are associated with
migration or metastasis. With these characteristics, the newly
established model of BCR-ABL1+ B-ALL with CNS metastasis
may provide further insight into the pathogenesis of
BCR-ABL-mediated leukemogenesis.
Previous studies have been reported that
approximately 30–50% of patients with BCR-ABL1+
ALL harbor the p210 fusion protein, with the remaining 50–70% being
characterized by the p190 fusion protein (25). The BCR-ABL1 fusion oncogene
exists in two principal forms (p190 and p210) that arise from
distinct breakpoints in the BCR gene on chromosome 22,
resulting in translocation of BCR exon 1 or exons 1–12/13,
respectively, to the c-ABL1 gene on chromosome 9. p210 and
p190 contain the same portion of the c-ABL1 tyrosine kinase and
acquire uncontrolled tyrosine kinase activity, both of which
constantly turn on the downstream signaling molecules/pathways, and
promoting proliferation of leukemia cells (16). Previous studies have reported that
chronic myeloid leukemia (CML) can be reproduced in mice by the
retroviral transduction of p190 or p210 into hematopoietic stem
cell (HSC)-enriched bone marrow (BM) cells in the presence of
myeloid cytokines, followed by transplantation into irradiated
recipients (14,26), which is a myeloproliferative disorder
characterized by the increased proliferation of granulocytic cells
without the loss of their capability to differentiate (27–30).
In the present study, this approach was utilized by
donor bone marrow cells pre-stimulated with the lymphoid cytokine
IL-7, which provides critical signals for B-cell proliferation and
survival (31,32). As a result, the mouse p210+
B-ALL mouse model was not only successfully generated, but it was
also observed that prominent infiltration in non-hematopoietic
tissues with clinical severe paralysis of CNS was achieved. The
present results indicated that the majority of the leukemic cells
may be characterized as immature pro-B
(CD43+B220+) and pre-B
(CD43−B220+) populations (33–36). In
addition to this observation, rare GFP+ cells, either
from the bone marrow or spleen, lacked expression of
lineage-specific antigens (data not shown), implying that the
BCR-ABL1+ B-ALL cells contain leukemic stem and
progenitor cells and propagate leukemia to the next generation. It
was also demonstrated that the transplantation of primary
BCR-ABL1+ B-ALL cells is capable of generating secondary
B-ALL in non-irradiated mice. This murine model resembles human
BCR-ABL1+ B-ALL and may be used to study the prognosis
of human B-ALL.
The model established in the present study has an
attractive feature of enabling leukemic cells to expand into
various types of non-hematopoietic tissue, including the CNS,
subcutaneous tissue, lung and liver. H&E staining and
intravascular staining technology were applied to precisely assess
the exact portions of leukemic cells dissemination into CNS. The
results suggest that leukemic cells are predominantly deposited in
the meninges, medulla oblongata, cerebrum, lumbar spinal cord and
cerebellum, which is consistent with the results of previous
studies (37–39). A previous study detected small vessels
transiting between the bone marrow and subarachnoid space in
leukemic mice, and it was indicated that numerous openings in the
vertebral cortical bone were filled with ALL cells, which appeared
to be in transit between the bone marrow involved and the
subarachnoid space (13,40). Therefore, BCR-ABL1+
leukemic cells may directly metastasize into the lumbar spinal
cord, but not into the thoracic spinal cord.
Laminin is localized in the extracellular matrix of
meninges, the choroid plexus and peripheral nerve sheaths in the
nervous system (41,42). Metastasis to the CNS has been
indicated to be mediated through the interaction of molecules
associated with cell motility and adhesion (43). This information supports the present
result that highly expressed Itga6 and L-selectin may allow
BCR-ABL1+ B-ALL leukemic cells to interact with laminin
located in the ECM of meninges. This specific interaction hijacks
and recruits leukemic cells into the meninges, but fails to enter
the brain parenchyma (7,12,42,43). These
results support the phenomenon that a large number of leukemic
cells accumulate in the meninges.
In conclusion, in the present study, a murine model
of BCR-ABL1+ B-ALL with CNS metastasis was generated.
The infiltrated BCR-ABL1+ immature B cells have the
ability to metastasize in various proportions into the CNS, with
significant accumulation in the meninges, which may be associated
with the upregulation of numerous adhesion molecules, including
Itga6 and L-selectin. These results encourage further studies to
address whether TKIs combined with the targeting of proteins
associated with cell migration and adhesion may offer a therapeutic
approach for the CNS metastasis/relapse of BCR-ABL1+
B-ALL.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Shaoguang
Li from the Division of Hematology/Oncology, University of
Massachusetts Medical School, for providing the MIG-p210 construct.
The authors would also like to thank Mr. Huixun Ren (Xi'an Jiaotong
University Health Science Centre) for assisting the with mouse
colony maintenance and Mr. Xiaofei Wang (Xi'an Jiaotong University
Health Science Centre) for providing expert technical assistance
with cell sorting.
Funding
This study was supported by grants (no. 31170821,
no. 31370874 and no. 81670157) from the National Natural Scientific
Foundation of China and by a grant (no. 2016JZ030) from the Natural
Scientific Foundation of Shaanxi.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article and its supplementary
information files.
Authors' contributions
XY, HZ and MZ performed the experiments and
participated in designing some experiments. MY, PZ and YW
participated in the experiment of viral supernatants and article
revision. YW, ZL and WOO performed the flow cytometry experiments,
and CLi, CLiu, YJ and YM performed some of the in vivo
experiments. XY and YJ wrote the manuscript. All authors have read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All animal procedures were approved by the
Institutional Animal Care and Use Committee of Xi'an Jiaotong
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BCR-ABL1+ B-ALL
|
Breakpoint cluster region-Abelson
murine leukemia acute B-lymphoblastic leukemia
|
|
CNS
|
central nervous system
|
|
TKIs
|
tyrosine kinase inhibitors
|
|
SCID
|
severe combined immunodeficiency
|
|
CML
|
chronic myeloid leukemia
|
|
MIG-p210 vector
|
MSCV-BCR-ABL1-IRES-GFP
|
|
MIG vector
|
MSCV-GFP
|
|
SPF
|
specific pathogen-free
|
|
H&E
|
hematoxylin and eosin
|
|
PB
|
peripheral blood
|
|
SP
|
spleen
|
|
LN
|
lymph node
|
|
BM
|
bone marrow
|
|
ST
|
subcutaneous tumor
|
References
|
1
|
Bernt KM and Hunger SP: Current concepts
in pediatric Philadelphia chromosome-positive acute lymphoblastic
leukemia. Front Oncol. 4:542014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quintás-Cardama A and Cortes J: Molecular
biology of bcr-abl1-positive chronic myeloid leukemia. Blood.
113:1619–1630. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schjerven H, Ayongaba EF, Aghajanirefah A,
McLaughlin J, Cheng D, Geng H, Boyd JR, Eggesbø LM, Lindeman I,
Heath JL, et al: Genetic analysis of Ikaros target genes and tumor
suppressor function in BCR-ABL1+ pre-B ALL. J Exp Med.
214:793–814. 2017.PubMed/NCBI
|
|
4
|
Pfeifer H, Wassmann B, Hofmann WK, Komor
M, Scheuring U, Brück P, Binckebanck A, Schleyer E, Gökbuget N,
Wolff T, et al: Risk and prognosis of central nervous system
leukemia in patients with Philadelphia chromosome-positive acute
leukemias treated with imatinib mesylate. Clin Cancer Res.
9:4674–4681. 2003.PubMed/NCBI
|
|
5
|
Mullighan CG: The molecular genetic makeup
of acute lymphoblastic leukemia. Hematology Am Soc Hematol Educ
Program. 2012:389–396. 2012.PubMed/NCBI
|
|
6
|
Hamdi A, Mawad R, Bassett R, di SA, Ferro
R, Afrough A, Ram R, Dabaja B, Rondon G, Champlin R, et al: Central
nervous system relapse in adults with acute lymphoblastic leukemia
after allogeneic hematopoietic stem cell transplantation. Biol
Blood Marrow Transplant. 20:1767–1771. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trendowski M: The inherent metastasis of
leukaemia and its exploitation by sonodynamic therapy. Crit Rev
Oncol Hematol. 94:149–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jabbour E, Thomas D, Cortes J, Kantarjian
HM and O'Brien S: Central nervous system prophylaxis in adults with
acute lymphoblastic leukemia: Current and emerging therapies.
Cancer. 116:2290–2300. 2010.PubMed/NCBI
|
|
9
|
Arbonés ML, Ord DC, Ley K, Ratech H,
Maynard-Curry C, Otten G, Capon DJ and Tedder TF: Lymphocyte homing
and leukocyte rolling and migration are impaired in
L-selectin-deficient mice. Immunity. 1:247–260. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Campbell JJ, Hedrick J, Zlotnik A, Siani
MA, Thompson DA and Butcher EC: Chemokines and the arrest of
lymphocytes rolling under flow conditions. Science. 279:381–384.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ley K, Laudanna C, Cybulsky MI and
Nourshargh S: Getting to the site of inflammation: The leukocyte
adhesion cascade updated. Nat Rev Immunol. 7:678–689. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nourshargh S, Hordijk PL and Sixt M:
Breaching multiple barriers: Leukocyte motility through venular
walls and the interstitium. Nat Rev Mol Cell Biol. 11:366–378.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yao H, Price TT, Cantelli G, Ngo B, Warner
MJ, Olivere L, Ridge SM, Jablonski EM, Therrien J, Tannheimer S, et
al: Leukaemia hijacks a neural mechanism to invade the central
nervous system. Nature. 560:55–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peng C and Li S: CML mouse model in
translational research. Methods Mol Biol. 602:253–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roumiantsev S, de Aos IE, Varticovski L,
Ilaria RL and Van Etten RA: The src homology 2 domain of Bcr/Abl is
required for efficient induction of chronic myeloid leukemia-like
disease in mice but not for lymphoid leukemogenesis or activation
of phosphatidylinositol 3-kinase. Blood. 97:4–13. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li S, Ilaria RL, Million RP Jr, Daley GQ
and Van Etten RA: The P190, P210, and P230 forms of the BCR/ABL
oncogene induce a similar chronic myeloid leukemia-like syndrome in
mice but have different lymphoid leukemogenic activity. J Exp Med.
189:1399–1412. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maheras KJ and Gow A: Increased anesthesia
time using 2,2,2-tribromoethanol-chloral hydrate with low impact on
mouse psychoacoustics. J Neurosci Method. 219:61–69. 2013.
View Article : Google Scholar
|
|
18
|
Anderson KG, Mayer-Barber K, Sung H, Beura
L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL
and Masopust D: Intravascular staining for discrimination of
vascular and tissue leukocytes. Nat Protoc. 9:209–222. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krause DS, Katherine L, Lewis JB, Andrian
UH and Van Etten RA: Selectins and their ligands are required for
homing and engraftment of BCR-ABL1+ leukemic stem cells
in the bone marrow niche. Blood. 123:1361–1371. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Levinsen M, Taskinen M, Abrahamsson J,
Forestier E, Frandsen TL, Harila-Saari A, Heyman M, Jonsson OG,
Lähteenmäki PM, Lausen B, et al: Clinical features and early
treatment response of central nervous system involvement in
childhood acute lymphoblastic leukemia. Pediatr Blood Cancer.
61:1416–1421. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Camicia R, Winkler HC and Hassa PO: Novel
drug targets for personalized precision medicine in
relapsed/refractory diffuse large B-cell lymphoma: A comprehensive
review. Mol Cancer. 14:2072015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Krause S, Pfeiffer C, Strube S, Alsadeq A,
Fedders H, Vokuhl C, Loges S, Waizenegger J, Ben-Batalla I, Cario
G, et al: Mer tyrosine kinase promotes the survival of
t(1;19)-positive acute lymphoblastic leukemia (ALL) in the central
nervous system (CNS). Blood. 125:820–830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Buchner M, Swaminathan S, Chen Z and
Müschen M: Mechanisms of pre-B-cell receptor checkpoint control and
its oncogenic subversion in acute lymphoblastic leukemia. Immunol
Rev. 263:192–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Score J, Calasanz MJ, Ottman O, Pane F,
Yeh RF, Sobrinho-Simões MA, Kreil S, Ward D, Hidalgo-Curtis C, Melo
JV, et al: Analysis of genomic breakpoints in p190 and p210 BCR-ABL
indicate distinct mechanisms of formation. Leukemia. 24:1742–1750.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gu S, Sayad A, Chan G, Yang W, Lu Z,
Virtanen C, Van Etten RA and Neel BG: SHP2 is required for
BCR-ABL1-induced hematologic neoplasia. Leukemia. 32:203–213. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong Y, Liu F, Wu C, Li S, Zhao X, Zhang
P, Jiao J, Yu X, Ji Y and Zhang M: Illegitimate RAG-mediated
recombination events are involved in IKZF1 Δ3–6 deletion in
BCR-ABL1 lymphoblastic leukaemia. Clin Exp Immunol. 185:320–331.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morotti A, Panuzzo C, Crivellaro S,
Pergolizzi B, Familiari U, Berger AH, Saglio G and Pandolfi PP:
BCR-ABL disrupts PTEN nuclear-cytoplasmic shuttling through
phosphorylation-dependent activation of HAUSP. Leukemia.
28:1326–1333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dash AB, Williams IR, Kutok JL, Tomasson
MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J,
Housman D, et al: A murine model of CML blast crisis induced by
cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci
USA. 99:7622–7627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang H, Peng C, Hu Y, Li H, Sheng Z, Chen
Y, Sullivan C, Cerny J, Hutchinson L, Higgins A, et al: The Blk
pathway functions as a tumor suppressor in chronic myeloid leukemia
stem cells. Nat Genet. 44:861–871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vegh P, Winckler J and Melchers F:
Long-term ‘in vitro’ proliferating mouse hematopoietic progenitor
cell lines. Immunol Lett. 130:32–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holl TM, Haynes BF and Kelsoe G: Stromal
cell independent B cell development in vitro: Generation and
recovery of autoreactive clones. J Immunol Methods. 354:53–67.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ji Y, Resch W, Corbett E, Yamane A,
Casellas R and Schatz DG: The in vivo pattern of binding of RAG1
and RAG2 to antigen receptor loci. Cell. 141:419–431. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dong Y, Wu C, Zhao X, Zhang P, Zhang H,
Zheng M, Li S, Jiao J, Yu X, Lv Z and Ji Y: Epigenetic
modifications of the VH region after DJH recombination in Pro-B
cells. Immunology. 152:218–231. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hardy RR and Hayakawa K: B cell
development pathways. Annu Rev Immunol. 19:595–621. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu C, Dong Y, Zhao X, Zhang P, Zheng M,
Zhang H, Li S, Jin Y, Ma Y, Ren H and Ji Y: RAG2 involves the Igκ
locus demethylation during B cell development. Mol Immunol.
88:125–134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pippard MJ, Callender ST and Sheldon PW:
Infiltration of central nervous system in adult acute myeloid
leukaemia. Br Med J. 1:227–229. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prieto C, López-Millán B, Roca-Ho H, Stam
RW, Romero-Moya D, Rodríguez-Baena FJ, Sanjuan-Pla A, Ayllón V,
Ramírez M, Bardini M, et al: NG2 antigen is involved in leukemia
invasiveness and central nervous system infiltration in
MLL-rearranged infant B-ALL. Leukemia. 32:633–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roddie P, Collie D and Johnson P:
Myelomatous involvement of the dura mater: A rare complication of
multiple myeloma. J Clin Pathol. 53:398–399. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheung LC, Tickner J, Hughes AM, Skut P,
Howlett M, Foley B, Oommen J, Wells JE, He B, Singh S, et al: New
therapeutic opportunities from dissecting the pre-B leukemia bone
marrow microenvironment. Leukemia. 32:2326–2338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Colognato H, ffrench-Constant C and Feltri
ML: Human diseases reveal novel roles for neural laminins. Trends
Neurosci. 28:480–486. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang L, Dong Z, Zhang Y and Miao J: The
roles of integrin β4 in vascular endothelial cells. J Cell Physiol.
227:474–478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Salomão DR, Pulido JS, Johnston PB,
Canal-Fontcuberta I and Feldman AL: Vitreoretinal presentation of
secondary large B-cell lymphoma in patients with systemic lymphoma.
JAMA Ophthalmol. 131:1151–1158. 2013. View Article : Google Scholar : PubMed/NCBI
|