Introduction
Bladder cancer is the 9th most common and the 13th
most fatal type of cancer worldwide (1). Approximately 75% of patients with
bladder cancer have non-muscle invasive bladder cancer (NMIBC) and
~25% have muscle invasive bladder cancer (MIBC) or metastatic
disease at the time of initial diagnosis (2,3). Different
molecular subtypes of bladder cancer have unique clinical
characteristics and therapeutic responses. For instance, patients
with ‘basal’ subtype of MIBC are known to have shorter
disease-specific survival and overall survival rate (4,5). MIBC is a
highly lethal disease and its overall five-year survival rate is
30–50%, whereas that for metastatic bladder cancer is 5% (3,6).
The tumor microenvironment (TME) consists
predominantly of tumor cells, stroma and infiltrating immune cells.
Immune cells in the TME may exert either tumor-suppressive or
tumor-promoting effects. CD8+ T cells and natural killer
cells (NKs) mediate antitumor functions, whereas tumor-associated
macrophages (TAMs) and regulatory T cells (Tregs) serves as
tumor-promoting agents (7,8). As an important component of the innate
and adaptive immunity, macrophages may polarize into different
functional phenotypes, such as the classically activated M1 and the
alternatively activated M2 macrophages, depending on the external
stimulus (9). Increasing evidence
suggests that M2 macrophages could perform immunosuppressive
functions and promote tumor progression and metastasis (7,10–12). It is therefore speculated that the M2
macrophages may have potential as immunotherapy targets (7).
Recently, immunotherapy based on immune checkpoint
inhibitors has achieved satisfactory results in the treatment of
bladder cancer. However, this outcome was limited to only a subset
of patients, since others failed to respond to this therapy
(13). Hence, it is important to
determine the essential immune components and relevant mechanisms
that affect tumor progression in order to improve the immunotherapy
of bladder cancer.
The majority of previous studies on the TME
associated with bladder cancer have mainly focused on a few types
of immune cells and were performed on small numbers of samples
(14,15). There is no large-scale data based on
big numbers of samples to identify important immune components in
bladder cancer. To the best of our knowledge, the molecular
mechanisms of immune activities in the bladder cancer
microenvironment have not been sufficiently researched.
The present study aimed to explore the predominant
tumor-infiltrating immune cell subsets in the bladder cancer
microenvironment and the signals recruiting them. Bioinformatics
methods were employed to analyze the immune infiltration profile
and revealed that M2 macrophage infiltration levels were associated
with the histologic grade, pathologic stage, and worse survival in
bladder cancer. Bladder cancer-specific genomic alterations, such
as gene mutation and copy number alterations, may be important
drivers of M2 macrophage infiltration.
Materials and methods
Gene expression datasets
The gene expression dataset (426 samples) and
microRNA (miRNA) mature strand expression dataset (429 samples),
based on RNA sequencing (RNA-seq) data from The Cancer Genome Atlas
(TCGA) bladder urothelial carcinoma cohort, were downloaded from
the University of California Santa Cruz (UCSC) xena browser
(https://xenabrowser.net/datapages/).
The gene expression data were presented as log2(x+1)-transformed
RNA-seq by expectation maximization (RSEM)-normalized count values
derived from TCGA level 3 data, and miRNA mature strand expression
data were presented as log2(RPM+1)-transformed reads-per-million
(RPM) values derived from TCGA level 3 data. The gene expression
validation datasets were downloaded from Gene Expression Omnibus
(GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and European
Bioinformatics Institute (EBI) ArrayExpress (https://www.ebi.ac.uk/arrayexpress/). The expression
dataset of 224 samples in GSE32894, expression dataset of 142
samples in GSE48277, and dataset of 85 samples in E-MTAB-1803 were
analyzed to confirm the results derived from the analyses of TCGA
dataset in the present study. All expression data was provided as
normalized data, after performing log2 conversion on GSE32894 and
E-MTAB-1803 datasets.
Clinical data
Clinical data of TCGA dataset were downloaded from
the UCSC Xena browser, including histologic grade, pathologic stage
and survival information for patients with bladder cancer. The
molecular subtype information of TCGA samples was obtained from the
supplementary file of the MIBC Molecular Characterization reported
by Robertson et al (3).
Clinical data for GSE32894, GSE48277 and E-MTAB-1803 datasets were
obtained from GEO and EBI ArrayExpress. The clinical data
associated with the GSE32894 and E-MTAB-1803 datasets contained the
molecular subtype information for each sample.
Mutation and copy number alteration
data
The mutation and copy number alteration data of TCGA
samples were obtained from the supplementary file of the study
reported by Robertson et al (3). The mutation data of E-MTAB-1803 and
GSE48277 were obtained from the detailed sample information.
Data processing
Gene expression data, miRNA mature strand expression
data, clinical data, mutation and copy number alteration data were
integrated according to sample ID using Perl script. Values of
GSE48277 dataset were log2 transformed by R script. The GSE32894,
GSE48277 and E-MTAB-1803 datasets were further processed by R
script. This included matching gene symbols, probes, calculating
mean expression value for the gene symbol when several probes
corresponded to one gene symbol, and defining mean value as the
expression level of the gene symbol.
Immune infiltration analysis based on
single-sample geneset enrichment analysis (ssGSEA) scores
To investigate the immune infiltration landscape of
bladder cancer, ssGSEA was performed to assess the level of immune
infiltration (recorded as ssGSEA score) in a sample according to
the expression levels of immune cell-specific marker genes. Marker
genes for most immune cell types were obtained from the article
published by Bindea et al (16). Marker genes for M1 macrophages, M2
macrophages, myeloid-derived suppressor cells (MDSCs) and Tregs
were obtained from published studies (10,14,17–25).
The ssGSEA analysis was performed based on GenePattern environment
(26). To run ssGSEA online analysis
(https://cloud.genepattern.org), gene
expression dataset file (GCT file), immune marker gene set file
(GMT file), and other parameters were uploaded as a set. Finally,
the ssGSEA scores, representing infiltration levels of immune cells
for individual samples, were presented in the output file.
Immune infiltration analysis based on
cell type identification by estimating relative subsets of known
RNA transcripts (CIBERSORT) method
The CIBERSORT analytical tool was developed to
analyze the 22 distinct leukocyte subsets in the tumors based on
bulk transcriptome data (27).
CIBERSORT (https://cibersort.stanford.edu/) was employed to
analyze the immune landscape of bladder cancer microenvironment
based on the TCGA RNA-seq dataset. The TCGA RNA-seq dataset was
used as the gene expression input and LM22 (22 immune cell types)
was set as the signature gene file. The analysis was conducted with
1,000 permutations. The CIBERSORT values generated were defined as
immune cell infiltration fraction per sample.
RNA-seq analysis
A previous study from our group has used RNA-seq
analysis of 10 tumor samples from patients with bladder cancer to
identify novel prognostic biomarkers (28). These RNA-seq data were re-analyzed in
the present study to investigate the TME, as aforementioned.
Primary sequencing data (raw reads) were subjected to quality
control and aligned to hg19 human genome. Reads per kilobase
million (RPKM)-normalized values were computed to quantify gene
expression levels. RNA-seq analysis and data processing was
conducted by Beijing Genomics Institute (Shenzhen, China).
Subsequently, the normalized data was analyzed by ssGSEA to
quantify the immune infiltration levels in the tissue samples.
Immunohistochemical (IHC) assay
Formalin-fixed (10% formalin at room temperature for
24 h), paraffin-embedded tumor tissues were collected from 48
patients with bladder cancer (including the 10 patients enrolled in
the aforementioned RNA sequencing analysis) diagnosed at Changhai
Hospital, Shanghai, China, from December 2011 to June 2017. Written
informed consent was obtained from all patients prior to surgery.
The deparaffinized sections (4 µm thickness) were incubated in
Tris/EDTA buffer (pH 9.0) for antigen retrieval. Subsequently,
blocking of endogenous peroxidase and non-specific epitopes were
performed using the UltraSensitive IHC kit (cat. no. KIT-9710;
Fuzhou Maixin Biotech. Co., Ltd.). The slides were incubated with
primary antibodies at 4°C overnight, then with biotin-conjugated
secondary antibodies at 37°C for 30 min and with
streptavidin-peroxidase at room temperature for 10 min, using the
UltraSensitive IHC kit. Immunostaining of slides was performed with
3,3′ diaminobenzidine followed by counterstaining with hematoxylin,
dehydrating, and mounting. All mounted specimens were scanned into
high resolution digital slides through slide scanners
NanoZoomer-S60 (Hamamatsu Photonics). The primary antibodies were
as follows: Monoclonal mouse anti-CD68 antibody (cat. no. M0876;
1:100 dilution; Dako; Agilent Technologies, Inc.), monoclonal
rabbit anti-CD163 antibody (cat. no. ab182422; 1:500 dilution;
Abcam) and monoclonal rabbit anti-CD31 antibody (cat. no. ab207090;
1:2,000 dilution; Abcam).
The IHC results for CD68 and CD163 were analyzed via
a combination of the scores based on the percentage of positively
stained cells (0, negative; 1, <15%; 2, 15–50%; 3, >50%) and
the scores based on the color intensity (0, negative staining; 1+,
weak staining, light yellow; 2+, moderate staining, yellow brown;
and 3+, strong staining, brown). Samples with IHC score ≤3 were
defined as low expression, while samples with IHC score >3 were
defined as high expression. Microvessels were evaluated by CD31 IHC
staining, as described previously (29). Slides were first observed at ×100
magnification to identify hotspots with the highest density of
microvessels and each hotspot was then evaluated at ×400
magnification. Any brown-staining endothelial cell or cell cluster
was considered a countable microvessel.
Statistical analysis
For ssGSEA scores, CIBERSORT values, miRNA
expression values and microvessel counts, comparison between two
groups was conducted using the Mann-Whitney test for data with
abnormal distribution and Student's t-test for data with normal
distribution. Survival analyses were performed using Kaplan-Meier
curves and log-rank test. IHC results (scores) for CD68 and CD163
were analyzed by Chi-square test. Pearson correlation analysis was
used to estimate the consistency between ssGSEA scores and IHC
scores in assessing M2 macrophage infiltration, and between miRNA
expression values and ssGSEA scores of M2 macrophage. Statistical
analysis was performed using SPSS pack 13.0 statistical software
(SPSS, Chicago, IL, USA) and GraphPad Prism 7.0 software (GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Immune landscape related to
histopathologic characteristics of bladder cancer
To explore the influence of immune cells on the
malignant progression of bladder cancer, first, the RNA-seq data of
426 patients with bladder cancer from TCGA database were analyzed
to evaluate the immune landscape. The marker genes of the majority
of types of immune cells were evaluated between low and high
histologic grade bladder cancer samples in TCGA dataset. The
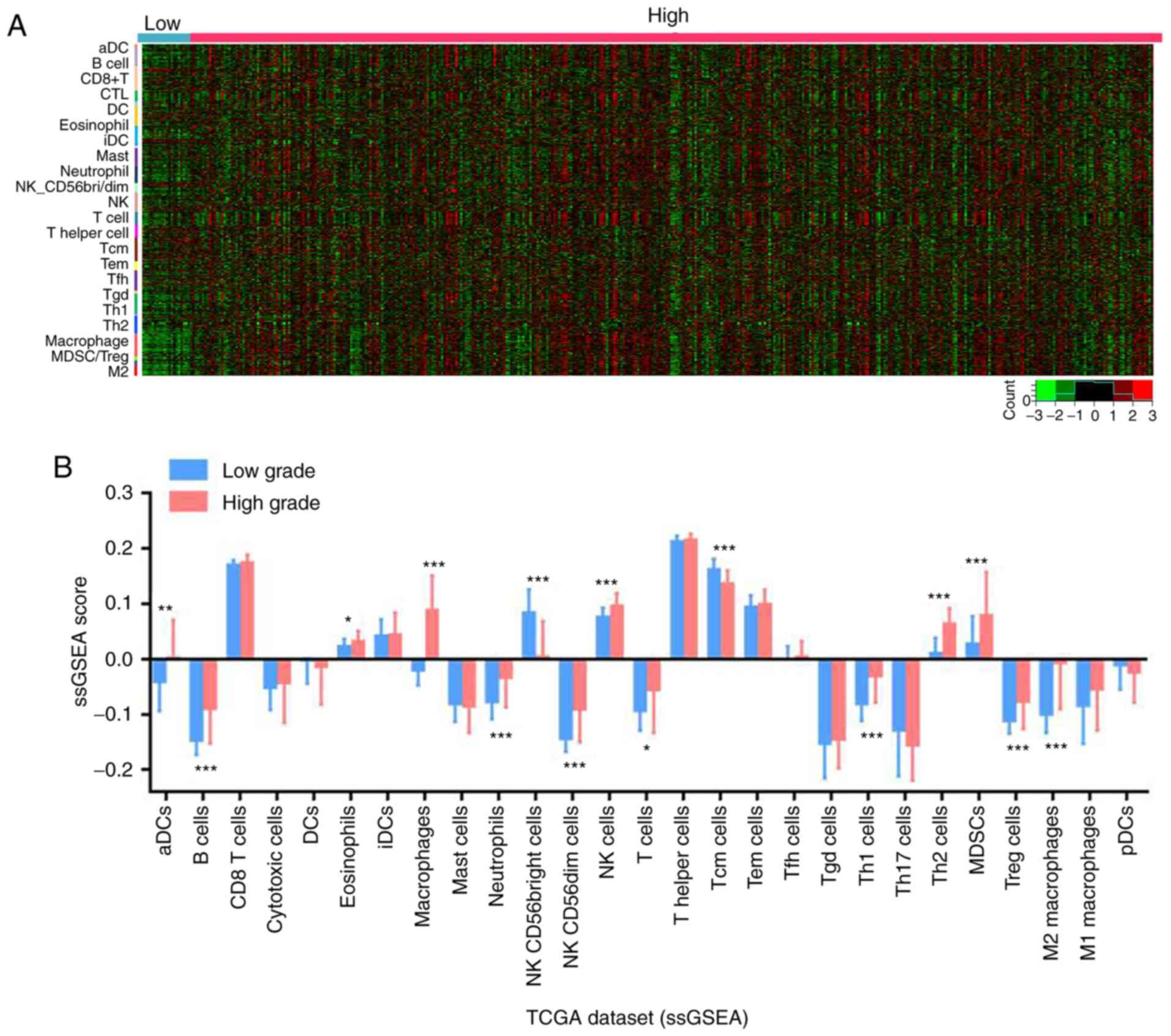
results revealed that B cells, macrophages and M2 macrophages were
enriched in high grade bladder cancer, as presented in the
corresponding heat map in Fig. 1A.
Given that ssGSEA had previously been implemented to determine the
immune landscape in clear cell renal cell carcinoma by utilizing
RNA-seq data (7), the infiltration of
each immune cell in the bladder cancer RNA-seq data was
quantitatively analyzed using this method. The results indicated
that B cells, macrophages, neutrophils, NK CD56dim cells, NK cells,
T helper (Th) 1 cells, Th2 cells, MDSCs, Tregs and M2 macrophages
were significantly enriched in high grade compared with low grade
bladder cancer tissues (Fig. 1B).
CIBERSORT was next applied to statistically estimate the relative
proportions of immune cell subsets among different grades of
bladder cancer samples in the TCGA dataset, following the method
reported by Gentles et al (27). It was demonstrated that the
infiltration levels of macrophage subsets (M0/M1/M2) and CD4 cell
subsets (memory activated/memory resting) were higher in high grade
bladder cancer. Furthermore, M2 macrophages accounted for the
largest proportion among the 22 subsets of tumor-infiltrating
immune cells, suggesting that they may have a key role in the
progression of bladder cancer (Fig.
1C). Data from the GSE32894 dataset were used to validate the
composition and proportion of tumor-infiltrating immune cell
subsets in bladder cancer. The results were consistent with those
observed in TCGA dataset, with higher levels of macrophage and M2
macrophage infiltration in the high-grade tumors compared with the
low-grade tumors (Fig. 1D).
The immune cell infiltration in different pathologic
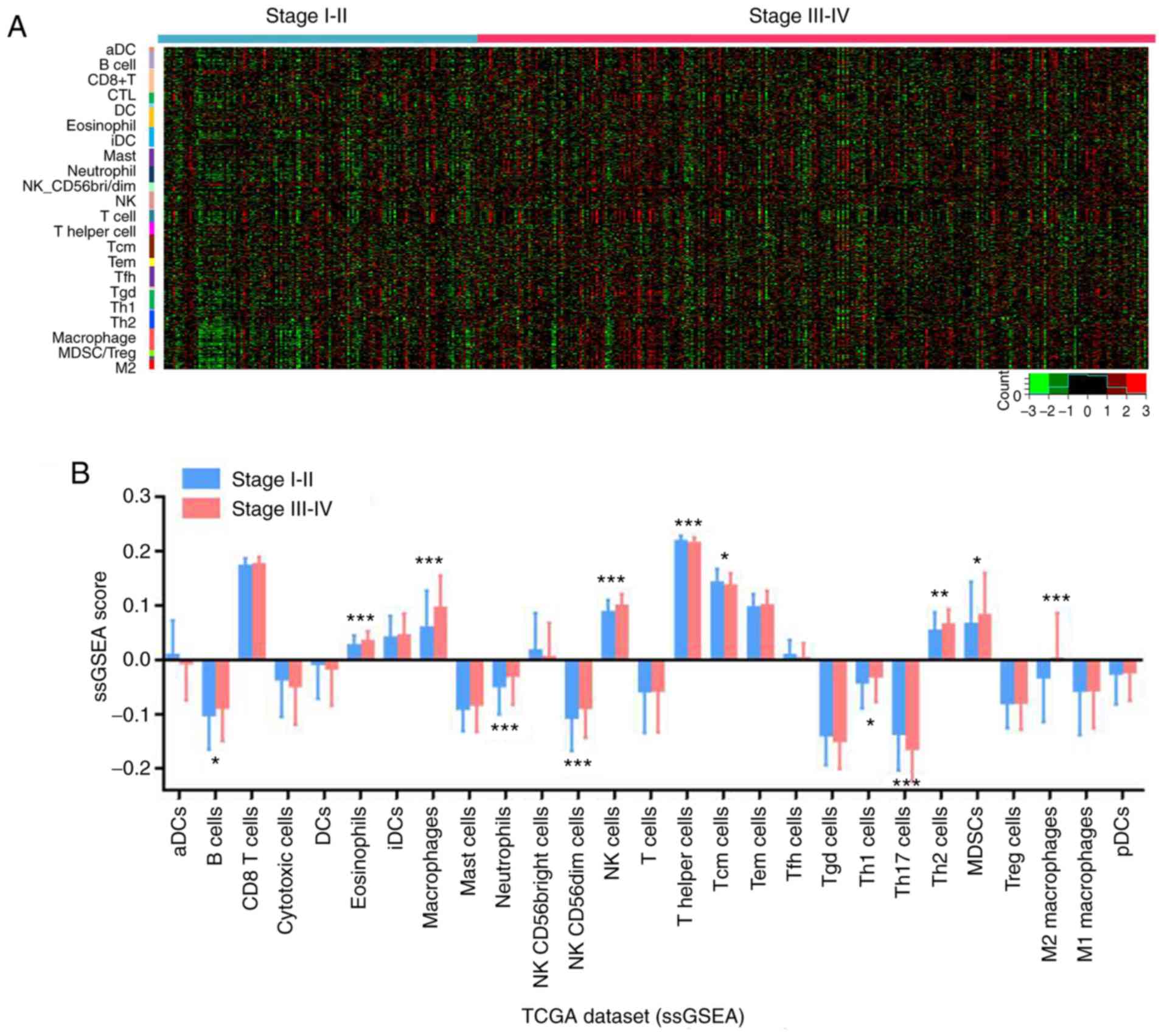
stages of bladder cancer was then explored. As presented in
Fig. 2A-C, in the TCGA dataset, B
cells, macrophages and M2 macrophages exhibited a high infiltration
level in higher compared with lower stage bladder cancer, with M2
macrophages having the highest proportion. Furthermore, the
infiltration levels of macrophages and M2 macrophages were
positively associated with the pathologic stage of bladder cancer,
as confirmed by the data analysis from the validation datasets
GSE32894 (Fig. 2D) and E-MTAB-1803
(Fig. 2E).
In summary, the present results demonstrated that
the infiltration levels or proportions of macrophages, especially
M2 macrophages, were positively associated with histologic grade
and pathologic stage of bladder cancer, suggesting that M2
macrophages may be involved in the progression of bladder
cancer.
Association of M2 macrophage
infiltration with the prognosis of patients with bladder
cancer
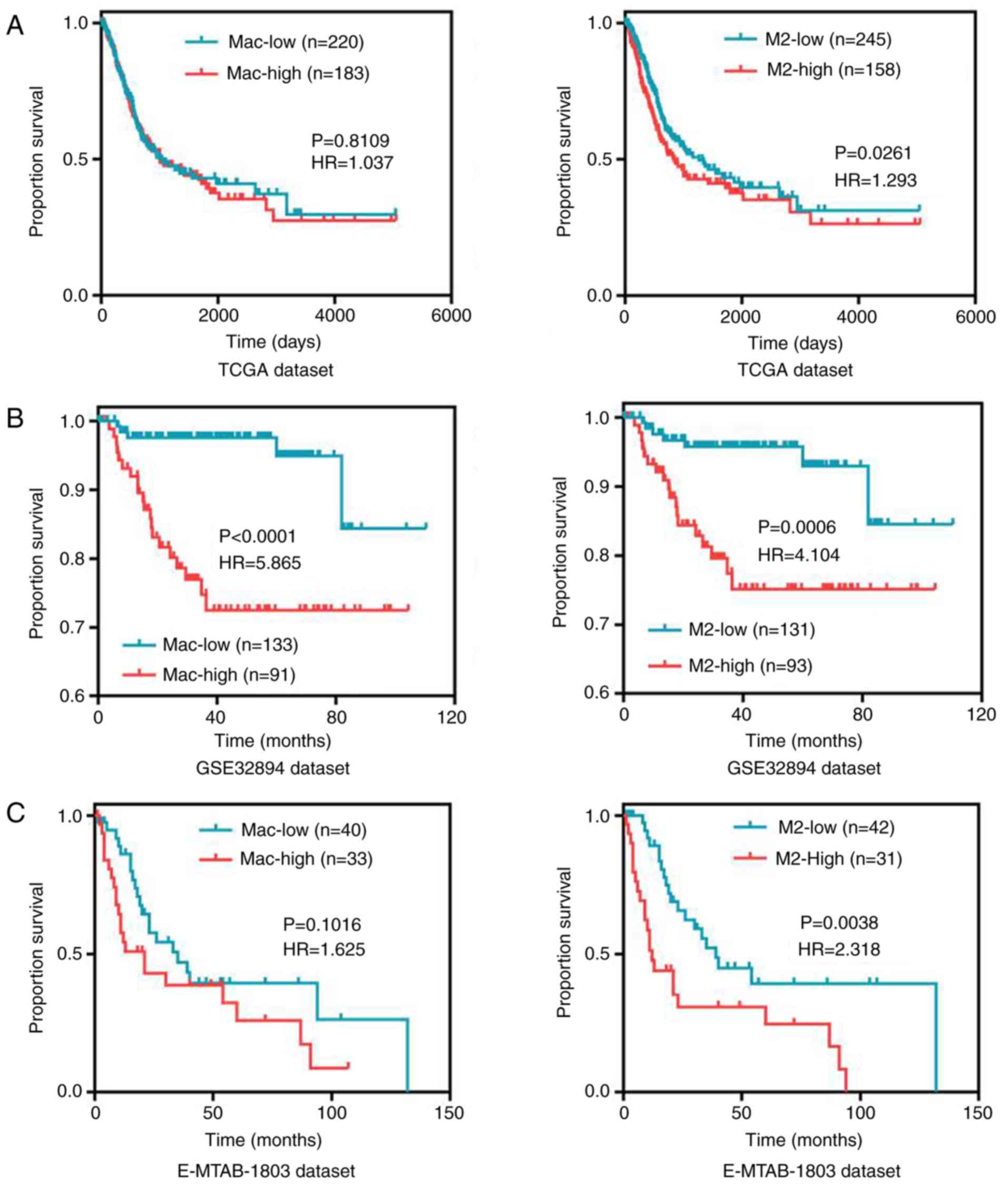
Kaplan-Meier survival curve analysis and ssGSEA
scores were used to further explore the effect of M2 macrophage
infiltration on the prognosis of patients with bladder cancer. In
TCGA dataset, it was observed that patients with high infiltration
of M2 macrophages had worse overall survival compared with patients
with low infiltration (Fig. 3A). No
significant difference was observed in patient survival with
varying total macrophage infiltration (Fig. 3A). Patients in the validation dataset
GSE32894 with high macrophage and M2 macrophage infiltration
exhibited worse disease-free survival (Fig. 3B). Finally, patients in the validation
dataset E-MTAB-1803 with high M2 macrophage infiltration (but not
total macrophage infiltration) exhibited worse overall survival
(Fig. 3C). Consequently, high
infiltration of M2 macrophages was associated with poor prognosis
in patients with bladder cancer.
Validation of macrophage and M2
macrophage infiltration in bladder cancer
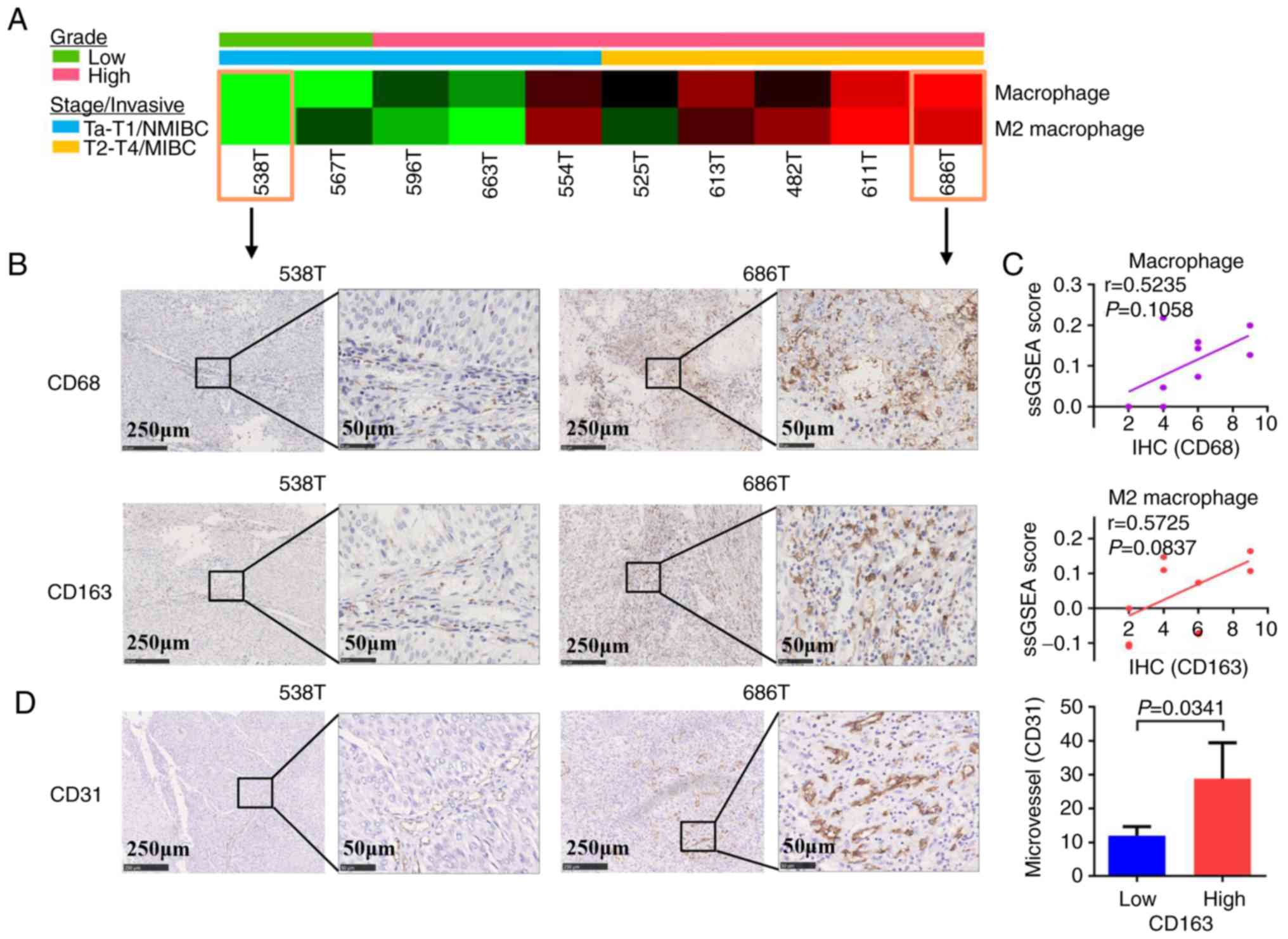
To confirm the infiltration of macrophages and M2
macrophages in tumor tissues from patients with bladder cancer in
Eastern China, the present study calculated the immune infiltration
scores for 10 newly collected tumor tissues with different
histologic grades and pathologic stages (NMIBC or MIBC) via
analyzing RNA-seq data with ssGSEA algorithm. It was demonstrated
that higher grade and stage (MIBC) bladder cancer tissues had
higher infiltration levels of macrophages and M2 macrophages,
compared with lower grade and stage (NMIBC) bladder cancer tissues
(Fig. 4A). Consequently, the
infiltration of macrophages and M2 macrophages in the 10 samples
was evaluated by IHC staining with anti-CD68 and anti-CD163
antibody, respectively. IHC scores revealed that there was high
macrophage and M2 macrophage infiltration in higher grade and stage
bladder cancer tissues (MIBC) compared with lower grade and stage
(NMIBC) bladder cancer tissues (Fig.
4B). In addition, the IHC scores were consistent with the
ssGSEA scores in the same bladder cancer patient. Pearson
correlation analysis was used to evaluate the consistency between
the IHC scores and ssGSEA scores, and the results revealed that the
two methods were positively correlated, albeit not significantly
(Fig. 4C). The reason for the lack of
statistical significance may be related to the small numbers of
samples used. Further analysis of the infiltration of macrophages
and M2 macrophages in 48 tumor tissues by IHC revealed that
macrophages and M2 macrophages were associated with the grade and
the invasiveness/stage of bladder cancer, but not with age or sex
(Table I). To investigate the
relationship between tumor-infiltrating M2 macrophages and
angiogenesis, IHC experiments were performed for the evaluation of
microvessel density via staining of CD31 in the 10 samples. The
results revealed that there were more microvessels in higher grade
and stage bladder cancer tissues and that the microvessel count was
significantly higher in the CD163-high compared with the CD163-low
tissues (Fig. 4C).
 | Table I.Macrophage and M2 macrophage
infiltration in bladder cancer samples as analyzed by
immunohistochemistry staining. |
Table I.
Macrophage and M2 macrophage
infiltration in bladder cancer samples as analyzed by
immunohistochemistry staining.
|
| CD68
expression | CD163
expression |
|---|
|
|
|
|
|---|
| Variable | High | Low | P-value | High | Low | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Male | 21 | 14 | 0.740 | 20 | 15 | 0.522 |
|
Female | 9 | 4 |
| 9 | 4 |
|
| Age (years) |
|
|
|
|
|
|
|
≥60 | 22 | 8 | 0.066 | 21 | 9 | 0.127 |
|
<60 | 8 | 10 |
| 8 | 10 |
|
| Grade |
|
|
|
|
|
|
|
Low-grade | 6 | 12 | 0.002 | 5 | 13 | 0.001 |
|
High-grade | 24 | 6 |
| 24 | 6 |
|
| Invasiveness |
|
|
|
|
|
|
|
NMIBC | 10 | 10 | 0.019 | 9 | 11 | 0.008 |
|
MIBC | 19 | 3 |
| 19 | 3 |
|
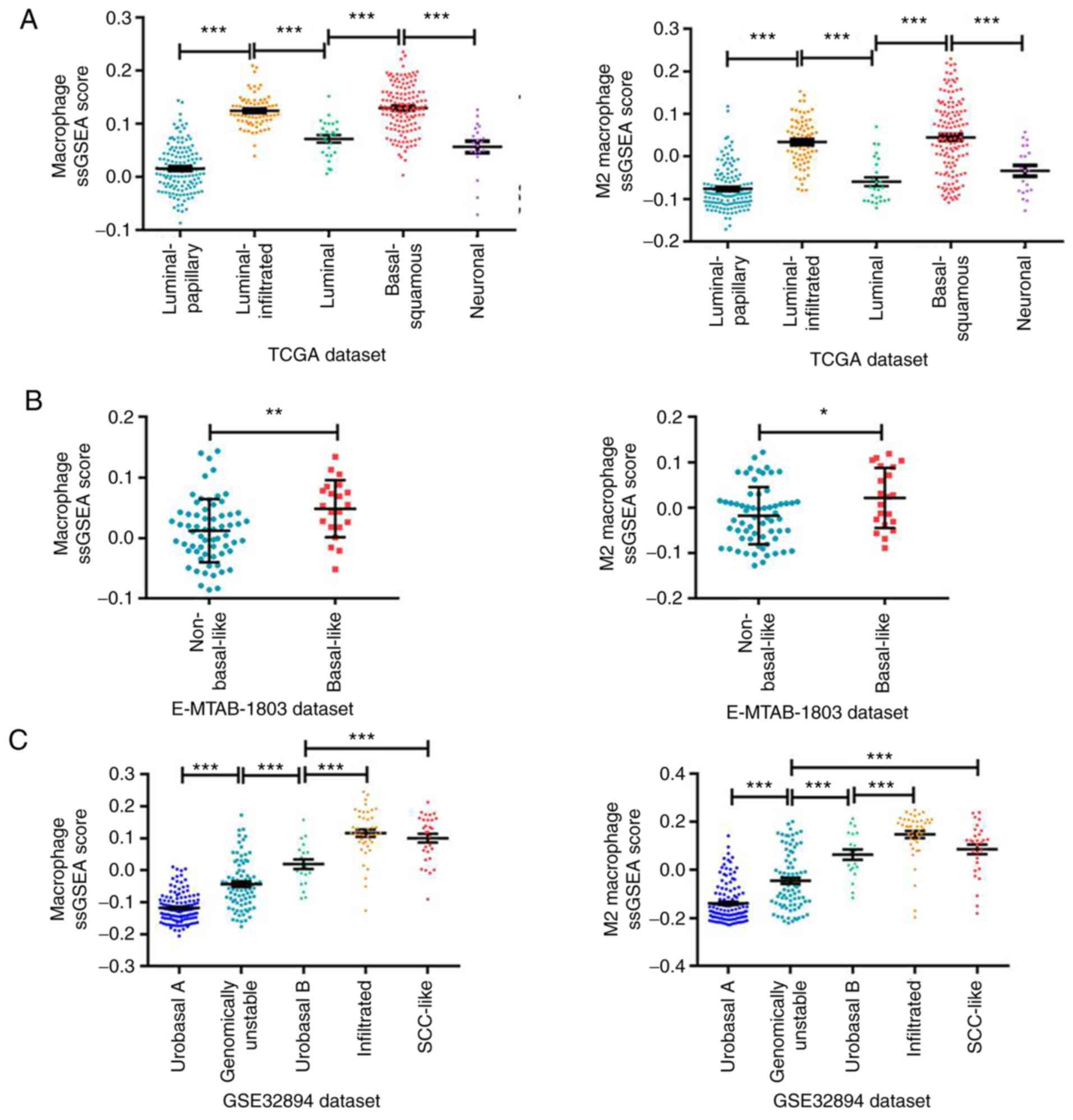
M2 macrophage infiltration varies by
molecular subtype in MIBC
Based on the high M2 macrophage infiltration in
MIBC, further investigations were conducted on whether M2
macrophage infiltration was associated with the transcriptional
molecular subtypes of MIBC. Analysis of the association of the
RNA-seq data from TCGA database with the ssGSEA scores revealed
that macrophages and M2 macrophages displayed significantly
different infiltration levels among the MIBC subtypes (Fig. 5A), with the highest infiltration
observed in the basal-squamous subtype, as defined by a previous
study using TCGA dataset (30). This
was consistent with the aforementioned results, where the
infiltration of macrophages and M2 macrophages was higher in
bladder cancer with higher grade and stage and associated with low
survival and poor prognosis of patients with basal-squamous
subtype. In addition, macrophage and M2 macrophage infiltration was
significantly higher in the basal-like subtype in the validation
dataset E-MTAB-1803 (Fig. 5B),
similar to the basal-squamous subtype in TCGA classification.
Finally, it was confirmed in the validation dataset GSE32894 that
macrophage and M2 macrophage infiltration was higher in the
infiltrated and SCC-like subtypes (5,31), which
are similar to the basal subtype. These results indicated that the
macrophage and M2 macrophage infiltration was associated with
molecular subtypes of MIBC. Additionally, M2 macrophages accounted
for the majority proportion of all subtypes of infiltrating
macrophages in bladder cancer, as presented in Figs. 1C and 2C, which indicated that the infiltration of
M2 macrophages may provide a good estimate of the overall
macrophage population in bladder cancer.
M2 macrophage infiltration is
associated with specific mutations in bladder cancer
The discovery that the infiltration of macrophages,
especially M2 macrophages, was associated with malignant
progression of bladder cancer and with the molecular subtypes of
MIBC implied that specific intrinsic genomics might affect the
infiltration of M2 macrophages. Therefore, the difference in M2
macrophage infiltration levels among different gene mutation types
was explored. Analysis of a large number of mutant genes using
ssGSEA algorithm in the TCGA dataset revealed that the M2
macrophage infiltration levels differed among multiple mutant
genes. As presented in Fig. 6A, the
M2 macrophage infiltration was higher in bladder tumor tissues with
mutant TP53, RB transcriptional corepressor 1 (RB1),
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
(PIK3CA), lysine methyltransferase 2A (KMT2A), lysine demethylase
6A (KDM6A) and apolipoprotein B mRNA editing enzyme
catalytic-polypeptide-like (APOBEC), but lower in tissues with
mutant fibroblast growth factor receptor 3 (FGFR3), E74-like ETS
transcription factor 3 (ELF3), PC4 and SFRS1 interacting protein 1
(PSIP1) and transmembrane and coiled-coil domains 4 (TMCO4). In the
validation datasets E-MTAB-1803 and GSE48277, it was confirmed that
M2 macrophage infiltration was lower in tissues with mutant FGFR3
(Fig. 6B and C); however, no
significant difference was observed for the mutation status of TP53
and RB1 (Fig. 6B and C). In summary,
M2 macrophage infiltration varied among bladder cancer mutant gene
types, suggesting that specific mutations may influence M2
macrophage infiltration.
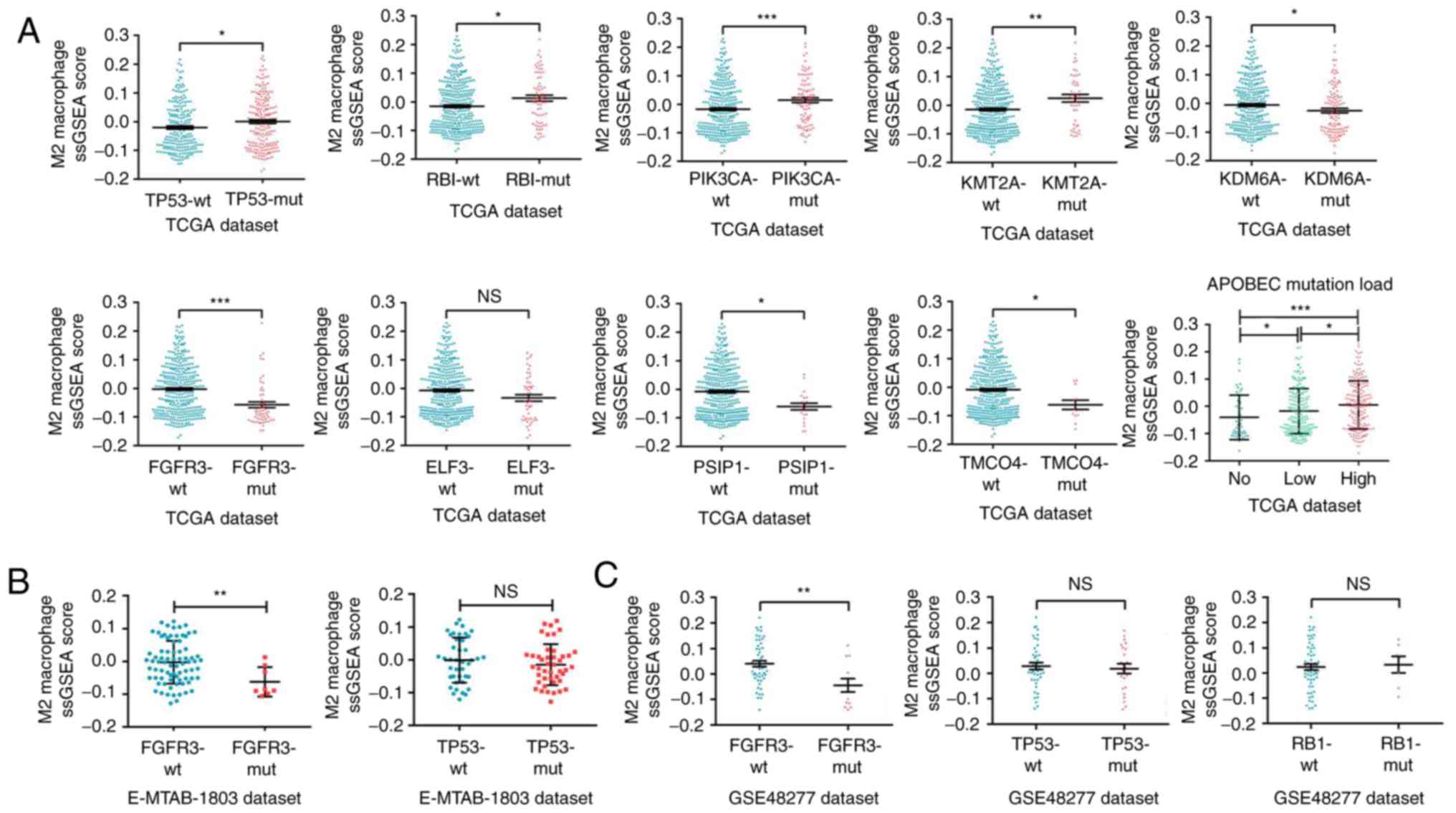 | Figure 6.M2 macrophage infiltration is
associated with specific mutations in bladder cancer. (A) Analyses
of M2 macrophage infiltration profiles in bladder cancer samples
with various mutant statuses from TCGA dataset. (B) Validation
analyses of M2 macrophage infiltration profiles in samples with
FCFR3 and TP53 mutant statuses from the E-MTAB-1803 and (C)
GSE48277 datasets. *P<0.05, **P<0.01 and ***P<0.001, with
comparisons indicated by brackets. TCGA, The Cancer Genome Atlas;
RB1, RB transcriptional corepressor 1; PIK3CA,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α;
KMT2A, lysine methyltransferase 2A; KDM6A, lysine demethylase 6A;
FGFR3, growth factor receptor 3; ELF3, E74-like ETS transcription
factor 3; PSIP, PC4 and SFRS1 interacting protein 1; TMCO4,
transmembrane and coiled-coil domains 4; APOBEC, apolipoprotein B
mRNA editing enzyme catalytic-polypeptide-like; ns, not
significant. |
M2 macrophage infiltration is
correlated with specific copy number variations in bladder
cancer
Further exploration was performed on whether there
were differences in the M2 macrophage infiltration among different
bladder cancer-specific copy number variations. It was demonstrated
that the M2 macrophage infiltration was influenced by specific
amplification types of copy number variations. The results of the
ssGSEA algorithm in TCGA dataset indicated that the infiltration in
the tissues with amplified FGFR3, erb-b2 receptor tyrosine kinase 2
(ERBB2), BCL2-like 1 (BCL2L1), telomerase reverse transcriptase
(TERT) and tyrosine-3-monooxygenase/tryptophan-5-monooxygenase
activation protein ζ (YWHAZ) and with deleted cyclin-dependent
kinase inhibitor 2A (CDKN2A), CREB binding protein (CREBBP),
AT-rich interaction domain 1A (ARID1A), fragile histidine triad
diadenosine triphosphatase (FHIT), phosphodiesterase 4D (PDE4D),
RAD51 paralog B (RAD51B), nuclear receptor corepressor 1 (NCOR1)
and protein tyrosine phosphatase receptor type D (PTPRD) was
significantly reduced (Fig. 7A and B,
respectively). Thus, the intrinsic specific copy number variations
of bladder cancer and related molecular mechanisms may influence
the M2 macrophage infiltration in the TME of bladder cancer.
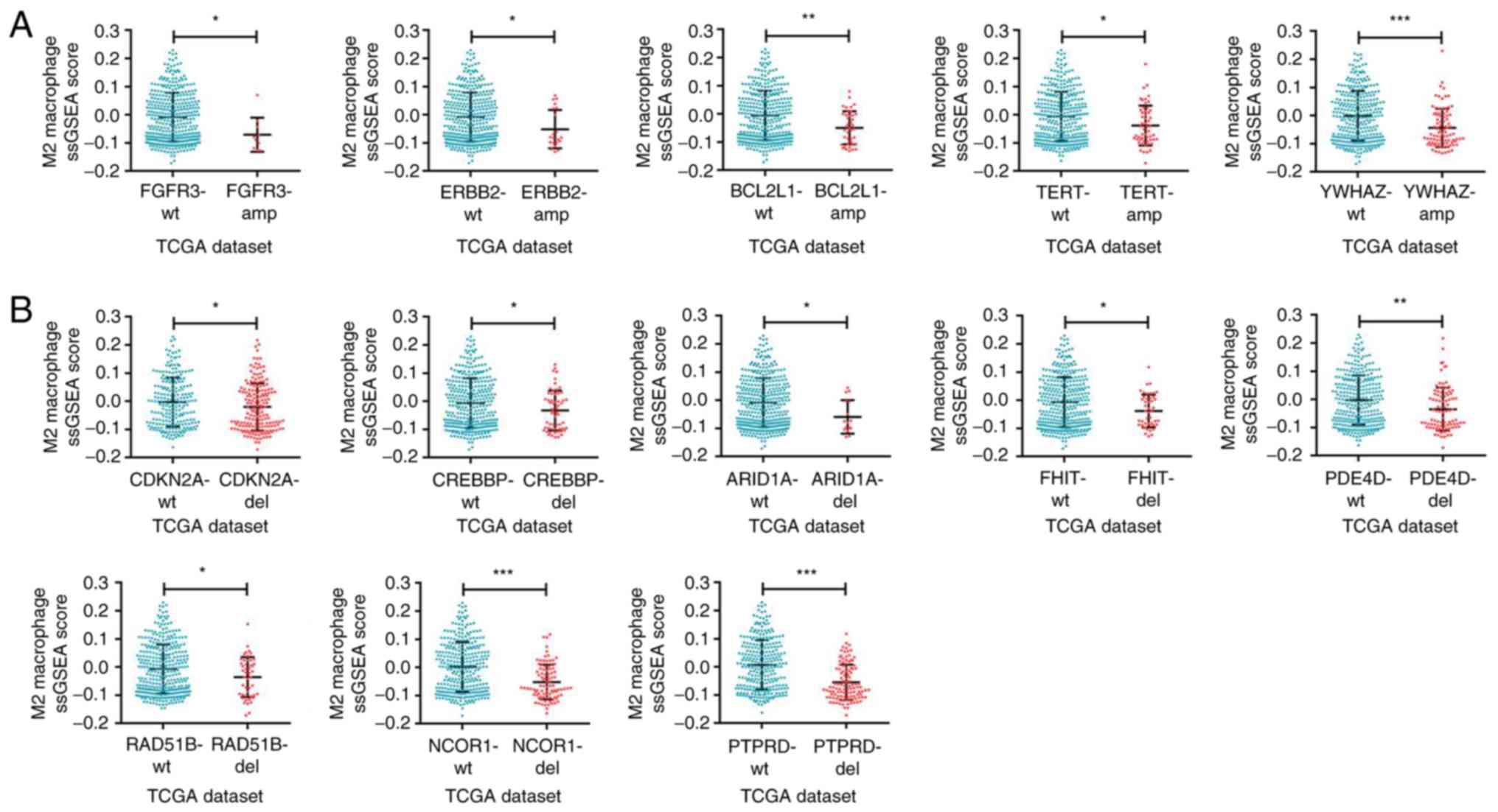 | Figure 7.M2 macrophage infiltration is
associated with specific copy number alterations in bladder cancer.
(A) Analyses of M2 macrophage infiltration profiles in bladder
cancer samples with specific amplification of gene copy number from
TCGA dataset. (B) M2 macrophage infiltration profiles in bladder
cancer samples with specific deletion of gene copy number from TCGA
dataset. *P<0.05, **P<0.01 and ***P<0.001, with
comparisons indicated by brackets. TCGA, The Cancer Genome Atlas;
FGFR3, growth factor receptor 3; ERBB2, erb-b2 receptor tyrosine
kinase 2; BCL2L1, BCL2-like 1; TERT, telomerase reverse
transcriptase; YWHAZ,
tyrosine-3-monooxygenase/tryptophan-5-monooxygenase activation
protein ζ; CDKN2A, cyclin-dependent kinase inhibitor 2A; CREBBP,
CREB binding protein; ARID1A, AT-rich interaction domain 1A; FHIT,
fragile histidine triad diadenosine triphosphatase; PDE4D,
phosphodiesterase 4D; RAD51B, RAD51 paralog B; NCOR1, nuclear
receptor corepressor 1; PTPRD, protein tyrosine phosphatase
receptor type D. |
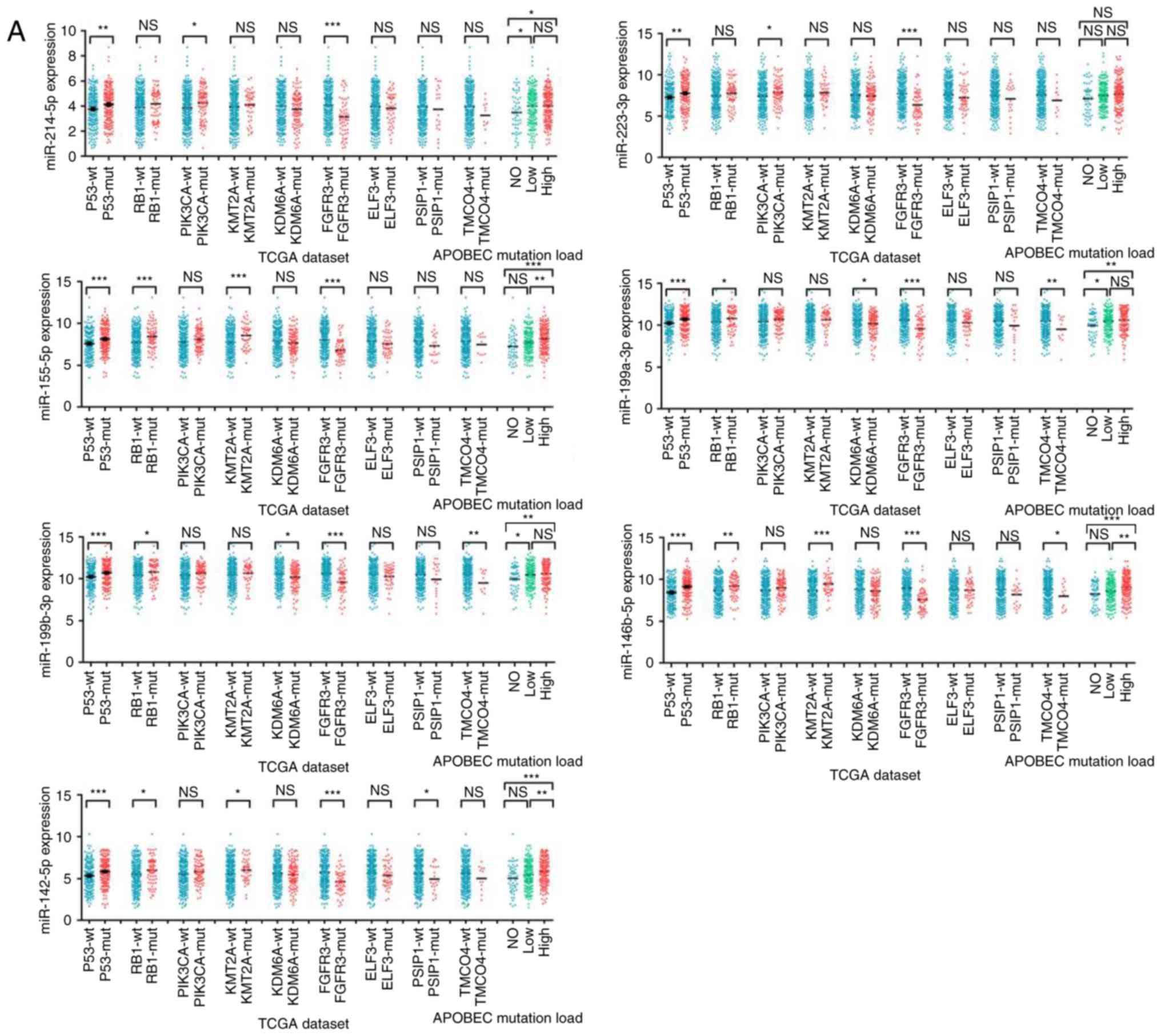
M2 macrophage infiltration is
associated with differentially expressed miRNAs in bladder
cancer
To further investigate the mechanisms of modulating
the tumor-infiltrating M2 macrophages in bladder cancer, miRNA
expression data were analyzed between tissues containing wild type
and mutant TP53, RB1, PIK3CA, KMT2A, KDM6A, FGFR3, ELF3, PSIP1,
TMCO4, and APOBEC. There was not a specific miRNA consistently
expressed differentially in all of the mutant genes. Since the
PIK3CA mutation has been previously reported to be associated with
macrophage infiltration (32), miRNAs
expressed differentially (P<0.1) between wild type and mutant
type of PIK3CA were selected. The differential expression of these
miRNAs in the remaining mutant genes were analyzed in sequence.
Seven miRNAs (miR-214-5p, miR-223-3p, miR-155-5p, miR-199a-3p,
miR-199b-3P, miR-146b-5p, miR-142-5p), which were expressed
differentially in at least three mutant genes and positively
correlated with M2 macrophage infiltration, as well as expressed
highly in high grade bladder cancer, were finally identified
(Fig. 8A-C). The differential
expression of these miRNAs in each copy number variant subtype and
stage of bladder cancer was further analyzed (Fig. S1).
Discussion
The present study demonstrated that M2 macrophages
were the predominant tumor-infiltrating immune cells in the bladder
cancer microenvironment, most abundant in the ‘basal’ subtype of
MIBC and associated with the histopathologic grade and stage, as
well as the prognosis of patients. Furthermore, miRNAs expressed
differentially due to cancer-specific genomic alterations were
identified as potential triggers of M2 macrophage recruitment in
the TME of bladder cancer.
Previous studies on the immune microenvironment of
bladder cancer focused on a few selected immune cells, such as
tumor-infiltrating dendritic cells (TIDCs), tumor-infiltrating
lymphocytes (TILs), and TAMs (detected by immunohistochemical
staining for CD68), and mostly involving a small number of samples
(14,33–35). In
the present study, ssGSEA and CIBERSORT algorithms were employed to
explore the infiltration of various immune cells in a large-scale
data and the results revealed that M2 macrophages were the main
immune components in bladder cancer and were associated with the
progression of tumors and the poor prognosis of patients.
Numerous studies have investigated the bladder
cancer-associated macrophages, focusing on the role of macrophages
in the treatment of bladder cancer with Bacillus Calmette-Guerin
(BCG), neoadjuvant chemotherapy and immunological checkpoint
inhibitors (15,35–37).
However, few studies have focused on M2 macrophages. In the present
study, high-throughput data obtained from multiple datasets were
used to demonstrate the importance of M2 macrophages in bladder
cancer progression.
Sjödahl et al (14) observed that a high CD68/CD3 ratio
reflected a poor prognosis among MIBC. Recently, Wu et al
(38) performed a meta-analysis and
demonstrated that TAMs identified with CD68 alone were not
significantly correlated with the prognosis of bladder cancer
patients, but TAMs detected with CD163 were significantly
correlated with poor relapse-free survival. Maniecki et al
(39) reported that CD163 mRNA
expression in bladder cancer biopsies was associated with advanced
tumor stages, aggressive tumor grade and poor overall survival. In
addition, high tumor macrophage infiltration (by CD163-positive
staining in IHC) was associated with advanced stage and tumor grade
(39). Aljabery et al
(40) assessed bladder cancer samples
from cystectomy by IHC and observed that M2 infiltration was not
correlated to stage or grade, but appeared to be inversely
correlated with improved cancer-specific survival; however,
patients collected in their study were all MIBC and patients with
NMIBC, which account for ~75% of all bladder cancer, were not
included. In the present study, NMIBC and MIBC samples were
included and analyzed, and M2 macrophages were demonstrated to be
associated with histologic grade and pathologic stage of bladder
cancer, as well as prognosis of patients with bladder cancer, based
on large-scale data bioinformatics analysis and biological
experimental analyses (IHC for CD163). These results suggested that
M2 macrophages may be potential targets for immunotherapy of
bladder cancer. The increased infiltration of TAMs (M2-like) is
associated with a poor response to BCG in patients with NMIBC
(35). The stromal immunotypes could
effectively predict response to adjuvant chemotherapy, recurrence
and survival in patients with MIBC (41). Nowadays, several approaches (including
depletion of TAMs, inhibition of monocyte recruitment into the
cancer, blockade of M2 macrophages, reprogramming of TAMs toward M1
macrophages) have been evaluated as strategies to target TAMs in
cancer (42). Novel immunotherapies
targeting M2 macrophages or their combination with immune
checkpoint inhibitors may present promising results in the
treatment of bladder cancer and other types of cancer (43,44).
In the present study, it was observed that
microvessel count was significantly higher in bladder cancer
tissues with highly infiltrated M2 macrophages, which indicated
that M2 macrophages may stimulate angiogenesis (45). In addition, there were more
microvessels in higher grade and stage bladder cancer tissues,
which was consistent with the hypothesis that M2 macrophages
promote neovascularization and cancer progression (46).
Tumor intrinsic genomics were found to affect the
composition of the immune microenvironment. Molecular subtypes of
glioblastoma differentially activate the immune microenvironment
and the tumor-promoting M2 macrophages exhibit a high association
with the mesenchymal subtype relative to proneural and classical
subtypes (47). It has also been
reported that TILs are associated with neurofibromin 1 and RB1
mutations, as well as epidermal growth factor receptor-amplified
and homozygous PTEN-deleted alterations in glioblastomas (48). In the present study, the levels of M2
macrophage infiltration varied among different bladder cancer
molecular subtypes, with the ‘basal’ subtype of MIBC having the
highest infiltration levels. Several gene mutations and copy number
variations were associated in the current study with M2 macrophage
infiltration in bladder cancer. Bladder cancer cells can produce
factors, such as macrophage-colony stimulating factor or monocyte
chemoattractant protein family-1, that trigger the amplification
and mobilization of macrophages in the TME (49–51).
Bladder cancer also affects macrophages by releasing and delivering
miRNAs, which may impede the M1 phenotype polarization, drive M2
phenotype polarization of macrophages, or switch phenotypes from M1
to M2 by binding to downstream targets or macrophages (9,52,53). The current results demonstrated that
there were miRNAs expressed differentially in cancer-specific
genomic alterations, related with M2 macrophage infiltration, and
highly expressed in high grade and advanced stage of bladder
cancer. Taken together, these results indicated that the bladder
cancer-specific genomic alterations may lead to differentially
expressed miRNAs, that may subsequently influence M2 macrophage
infiltration and promote bladder cancer progression. It has been
suggested that exosomes mediate the communication between tumor
cells and macrophages (54,55). Exosomes are a type of nanometric
membrane vesicles released by cells into the extracellular
environment, that regulate the biological activities of target
cells via delivering DNAs, RNAs and proteins (56). Consequently, it can be speculated that
bladder cancer with special genomic alterations may promote the
recruitment and polarization of M2 macrophages in the TME via
exosomal miRNAs. Future studies are required to validate this
hypothesis and to explore the intrinsic genomic mechanisms
affecting M2 macrophage infiltration, with the aim to provide a
potential therapeutic target of immunotherapy for bladder
cancer.
In conclusion, the present study demonstrated that
M2 macrophages were the predominant tumor-infiltrating immune cells
in bladder cancer. Their infiltration was associated with the
histologic grade, pathologic stage of bladder cancer and ‘basal’
subtype of MIBC, as well as prognosis of patients. Differentially
expressed miRNAs, associated with cancer-specific genomic
alterations, may be important drivers of M2 macrophage
infiltration, which may serve as a potential immunotherapy target
for bladder cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Fund of China (grant no. 8150101083, 81572509), the
Innovation Program of Shanghai Municipal Education Commission
(grant no. 2017-01-07-00-07-E00014), the Leading Talents Training
Program of Shanghai in 2017.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX and LT conceived the study, performed the
experiment, and wrote the manuscript. FL collected the clinical
samples and data and participated in the analysis. AL, SZ, XH and
QX participated in the experiments. LT and ZY analyzed the data. CX
and YS contributed to the initial design of the study, supervised
the bioinformatic and experimental analyses, and critically edited
the manuscript. All authors discussed the results and approved the
final manuscript.
Ethics approval and consent to
participate
The present study's protocol involving human
subjects was approved by the Ethics Committee of Changhai Hospital
of the Second Military Medical University (Shanghai, China).
Written informed consent for using tumor tissue in scientific
research was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NMIBC
|
non-muscle invasive bladder cancer
|
|
MIBC
|
muscle invasive bladder cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
TME
|
tumor microenvironment
|
|
NKs
|
natural killer cells
|
|
TAMs
|
tumor-associated macrophages
|
|
Tregs
|
regulatory T cells
|
|
GEO
|
Gene Expression Omnibus
|
|
EBI
|
European Bioinformatics Institute
|
|
ssGSEA
|
single-sample geneset enrichment
analysis
|
|
CIBERSORT
|
Cell Type Identification By Estimating
Relative Subsets Of known RNA Transcripts
|
|
RNA-seq
|
RNA sequencing
|
|
TIDCs
|
tumor-infiltrating dendritic cells
|
|
TILs
|
tumor-infiltrating lymphocytes
|
References
|
1
|
Sanli O, Dobruch J, Knowles MA, Burger M,
Alemozaffar M, Nielsen ME and Lotan Y: Bladder cancer. Nat Rev Dis
Primers. 3:170222017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Babjuk M, Böhle A, Burger M, Capoun O,
Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M,
et al: EAU guidelines on non-muscle-invasive urothelial carcinoma
of the bladder: Update 2016. Eur Urol. 71:447–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Robertson AG, Kim J, Al-Ahmadie H,
Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA,
Akbani R, et al: Comprehensive molecular characterization of
muscle-invasive bladder cancer. Cell. 171:540–556.e25. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sjödahl G, Lauss M, Lövgren K, Chebil G,
Gudjonsson S, Veerla S, Patschan O, Aine M, Fernö M, Ringnér M, et
al: A molecular taxonomy for urothelial carcinoma. Clin Cancer Res.
18:3377–3386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi W, Porten S, Kim S, Willis D, Plimack
ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al:
Identification of distinct basal and luminal subtypes of
muscle-invasive bladder cancer with different sensitivities to
frontline chemotherapy. Cancer Cell. 25:152–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Ye YL, Li MX, Ye SB, Huang WR,
Cai TT, He J, Peng JY, Duan TH, Cui J, et al: CXCL2/MIF-CXCR2
signaling promotes the recruitment of myeloid-derived suppressor
cells and is correlated with prognosis in bladder cancer. Oncogene.
36:2095–2104. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kitamura T, Qian BZ and Pollard JW: Immune
cell promotion of metastasis. Nat Rev Immunol. 15:73–86. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Şenbabaoğlu Y, Gejman RS, Winer AG, Liu M,
Van Allen EM, de Velasco G, Miao D, Ostrovnaya I, Drill E, Luna A,
et al: Tumor immune microenvironment characterization in clear cell
renal cell carcinoma identifies prognostic and
immunotherapeutically relevant messenger RNA signatures. Genome
Biol. 17:2312016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage m1-m2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Na YR, Yoon YN, Son DI and Seok SH:
Cyclooxygenase-2 inhibition blocks M2 macrophage differentiation
and suppresses metastasis in murine breast cancer model. PLoS One.
8:e634512013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruffell B and Coussens LM: Macrophages and
therapeutic resistance in cancer. Cancer Cell. 27:462–472. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Korpal M, Puyang X, Jeremy Wu Z, Seiler R,
Furman C, Oo HZ, Seiler M, Irwin S, Subramanian V, Julie Joshi J,
et al: Evasion of immunosurveillance by genomic alterations of
PPARγ/RXRα in bladder cancer. Nat Commun. 8:1032017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sjödahl G, Lövgren K, Lauss M, Chebil G,
Patschan O, Gudjonsson S, Månsson W, Fernö M, Leandersson K,
Lindgren D, et al: Infiltration of CD3+ and
CD68+ cells in bladder cancer is subtype specific and
affects the outcome of patients with muscle-invasive tumors. Urol
Oncol. 32:791–797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miyake M, Tatsumi Y, Gotoh D, Ohnishi S,
Owari T, Iida K, Ohnishi K, Hori S, Morizawa Y, Itami Y, et al:
Regulatory T cells and tumor-associated macrophages in the tumor
microenvironment in non-muscle invasive bladder cancer treated with
intravesical Bacille Calmette-Guérin: A long-term follow-up study
of a Japanese Cohort. Int J Mol Sci. 18(pii): E21862017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sanyal R, Polyak MJ, Zuccolo J, Puri M,
Deng L, Roberts L, Zuba A, Storek J, Luider JM, Sundberg EM, et al:
MS4A4A: A novel cell surface marker for M2 macrophages and plasma
cells. Immunol Cell Biol. 95:611–619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ambarus CA, Krausz S, van Eijk M, Hamann
J, Radstake TR, Reedquist KA, Tak PP and Baeten DL: Systematic
validation of specific phenotypic markers for in vitro polarized
human macrophages. J Immunol Methods. 375:196–206. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lugo-Villarino G, Troegeler A, Balboa L,
Lastrucci C, Duval C, Mercier I, Bénard A, Capilla F, Al Saati T,
Poincloux R, et al: The C-type lectin receptor DC-SIGN has an
anti-inflammatory role in human M(IL-4) macrophages in response to
Mycobacterium tuberculosis. Front Immunol. 9:11232018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi JW, Kwon MJ, Kim IH, Kim YM, Lee MK
and Nam TJ: Pyropia yezoensis glycoprotein promotes the M1 to M2
macrophage phenotypic switch via the STAT3 and STAT6 transcription
factors. Int J Mol Med. 38:666–674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu M, Yang W, Liu S, Hock D, Zhang B, Huo
RY, Tong X and Yan H: LXRα is expressed at higher levels in healthy
people compared to atherosclerosis patients and its over-expression
polarizes macrophages towards an anti-inflammatory MΦ2 phenotype.
Clin Exp Hypertens. 40:213–217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lo TH, Silveira PA, Fromm PD, Verma ND, Vu
PA, Kupresanin F, Adam R, Kato M, Cogger VC, Clark GJ and Hart DN:
Characterization of the expression and function of the C-type
lectin receptor CD302 in mice and humans reveals a role in
dendritic cell migration. J Immunol. 197:885–898. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bellora F, Castriconi R, Dondero A,
Reggiardo G, Moretta L, Mantovani A, Moretta A and Bottino C: The
interaction of human natural killer cells with either unpolarized
or polarized macrophages results in different functional outcomes.
Proc Natl Acad Sci USA. 107:21659–21664. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ornstein MC, Diaz-Montero CM, Rayman P,
Elson P, Haywood S, Finke JH, Kim JS, Pavicic PG Jr, Lamenza M,
Devonshire S, et al: Myeloid-derived suppressors cells (MDSC)
correlate with clinicopathologic factors and pathologic complete
response (pCR) in patients with urothelial carcinoma (UC)
undergoing cystectomy. Urol Oncol. 36:405–412. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gielen PR, Schulte BM, Kers-Rebel ED,
Verrijp K, Petersen-Baltussen HM, ter Laan M, Wesseling P and Adema
GJ: Increase in both CD14-positive and CD15-positive
myeloid-derived suppressor cell subpopulations in the blood of
patients with glioma but predominance of CD15-positive
myeloid-derived suppressor cells in glioma tissue. J Neuropathol
Exp Neurol. 74:390–400. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Reich M, Liefeld T, Gould J, Lerner J,
Tamayo P and Mesirov JP: GenePattern 2.0. Nat Genet. 38:500–501.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al: The
prognostic landscape of genes and infiltrating immune cells across
human cancers. Nat Med. 21:938–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeng S, Yu X, Ma C, Song R, Zhang Z, Zi X,
Chen X, Wang Y, Yu Y, Zhao J, et al: Transcriptome sequencing
identifies ANLN as a promising prognostic biomarker in bladder
urothelial carcinoma. Sci Rep. 7:31512017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chai CY, Chen WT, Hung WC, Kang WY, Huang
YC, Su YC and Yang CH: Hypoxia-inducible factor-1alpha expression
correlates with focal macrophage infiltration, angiogenesis and
unfavourable prognosis in urothelial carcinoma. J Clin Pathol.
61:658–664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of urothelial bladder
carcinoma. Nature. 507:315–322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
An Y, Adams JR, Hollern DP, Zhao A, Chang
SG, Gams MS, Chung PED, He X, Jangra R, Shah JS, et al: Cdh1 and
Pik3ca mutations cooperate to induce immune-related invasive
lobular carcinoma of the breast. Cell Rep. 25:702–714.e6. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanada T, Nakagawa M, Emoto A, Nomura T,
Nasu N and Nomura Y: Prognostic value of tumor-associated
macrophage count in human bladder cancer. Int J Urol. 7:263–269.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ayari C, LaRue H, Hovington H, Decobert M,
Harel F, Bergeron A, Têtu B, Lacombe L and Fradet Y: Bladder tumor
infiltrating mature dendritic cells and macrophages as predictors
of response to bacillus Calmette-Guérin immunotherapy. Eur Urol.
55:1386–1395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takayama H, Nishimura K, Tsujimura A,
Nakai Y, Nakayama M, Aozasa K, Okuyama A and Nonomura N: Increased
infiltration of tumor associated macrophages is associated with
poor prognosis of bladder carcinoma in situ after intravesical
bacillus Calmette-Guerin instillation. J Urol. 181:1894–1900. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tervahartiala M, Taimen P, Mirtti T,
Koskinen I, Ecke T, Jalkanen S and Boström PJ: Immunological tumor
status may predict response to neoadjuvant chemotherapy and outcome
after radical cystectomy in bladder cancer. Sci Rep. 7:126822017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang X, Ni S, Chen Q, Ma L, Jiao Z, Wang C
and Jia G: Bladder cancer cells induce immunosuppression of T cells
by supporting PD-L1 expression in tumour macrophages partially
through interleukin 10. Cell Biol Int. 41:177–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu SQ, Xu R, Li XF, Zhao XK and Qian BZ:
Prognostic roles of tumor associated macrophages in bladder cancer:
A system review and meta-analysis. Oncotarget. 9:25294–25303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maniecki MB, Etzerodt A, Ulhøi BP,
Steiniche T, Borre M, Dyrskjøt L, Orntoft TF, Moestrup SK and
Møller HJ: Tumor-promoting macrophages induce the expression of the
macrophage-specific receptor CD163 in malignant cells. Int J
Cancer. 131:2320–2331. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Aljabery F, Olsson H, Gimm O, Jahnson S
and Shabo I: M2-macrophage infiltration and macrophage traits of
tumor cells in urinary bladder cancer. Urol Oncol.
36:159.e19–159.e26. 2018. View Article : Google Scholar
|
|
41
|
Fu H, Zhu Y, Wang Y, Liu Z, Zhang J, Xie
H, Fu Q, Dai B, Ye D and Xu J: Identification and validation of
stromal immunotype predict survival and benefit from adjuvant
chemotherapy in patients with muscle-invasive bladder cancer. Clin
Cancer Res. 24:3069–3078. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rubio C, Munera-Maravilla E, Lodewijk I,
Suarez-Cabrera C, Karaivanova V, Ruiz-Palomares R, Paramio JM and
Dueñas M: Macrophage polarization as a novel weapon in conditioning
tumor microenvironment for bladder cancer: Can we turn demons into
gods? Clin Transl Oncol. 21:391–403. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pyonteck SM, Akkari L, Schuhmacher AJ,
Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT,
Teijeiro V, et al: CSF-1R inhibition alters macrophage polarization
and blocks glioma progression. Nat Med. 19:1264–1272. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhu Y, Knolhoff BL, Meyer MA, Nywening TM,
West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC and
DeNardo DG: CSF1/CSF1R blockade reprograms tumor-infiltrating
macrophages and improves response to T-cell checkpoint
immunotherapy in pancreatic cancer models. Cancer Res.
74:5057–5069. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bronte V and Murray PJ: Understanding
local macrophage phenotypes in disease: Modulating macrophage
function to treat cancer. Nat Med. 21:117–119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Q, Hu B, Hu X, Kim H, Squatrito M,
Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, et al: Tumor
evolution of glioma-intrinsic gene expression subtypes associates
with immunological changes in the microenvironment. Cancer Cell.
32:42–56.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rutledge WC, Kong J, Gao J, Gutman DA,
Cooper LA, Appin C, Park Y, Scarpace L, Mikkelsen T, Cohen ML, et
al: Tumor-infiltrating lymphocytes in glioblastoma are associated
with specific genomic alterations and related to transcriptional
class. Clin Cancer Res. 19:4951–4960. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lin EY, Nguyen AV, Russell RG and Pollard
JW: Colony-stimulating factor 1 promotes progression of mammary
tumors to malignancy. J Exp Med. 193:727–740. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qian BZ, Li J, Zhang H, Kitamura T, Zhang
J, Campion LR, Kaiser EA, Snyder LA and Pollard JW: CCL2 recruits
inflammatory monocytes to facilitate breast-tumour metastasis.
Nature. 475:222–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cortez-Retamozo V, Etzrodt M, Newton A,
Ryan R, Pucci F, Sio SW, Kuswanto W, Rauch PJ, Chudnovskiy A,
Iwamoto Y, et al: Angiotensin II drives the production of
tumor-promoting macrophages. Immunity. 38:296–308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ying W, Tseng A, Chang RC, Morin A, Brehm
T, Triff K, Nair V, Zhuang G, Song H, Kanameni S, et al:
MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative
macrophage activation. J Clin Invest. 125:4149–4159. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He X, Tang R, Sun Y, Wang YG, Zhen KY,
Zhang DM and Pan WQ: MicroR-146 blocks the activation of M1
macrophage by targeting signal transducer and activator of
transcription 1 in hepatic schistosomiasis. EBioMedicine.
13:339–347. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu Y, Gu Y and Cao X: The exosomes in
tumor immunity. Oncoimmunology. 4:e10274722015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cooks T, Pateras IS, Jenkins LM, Patel KM,
Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG and Harris
CC: Mutant p53 cancers reprogram macrophages to tumor supporting
macrophages via exosomal miR-1246. Nat Commun. 9:7712018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Junker K, Heinzelmann J, Beckham C, Ochiya
T and Jenster G: Extracellular vesicles and their role in urologic
malignancies. Eur Urol. 70:323–331. 2016. View Article : Google Scholar : PubMed/NCBI
|