Introduction
Hematopoietic stem and progenitor cells (HSPCs)
maintain blood production throughout an organism's lifespan through
self-renewal and differentiation. In 1978, Schofield proposed that
HSPCs in the bone marrow require anatomically defined regions to
maintain function. These regions are composed of peripheral cells
within a milieu of local extracellular stromal cells, termed the
bone marrow microenvironment (BMM) or ‘niche’ (1). There is ample evidence to indicate that
the transformation of the BMM acts directly or indirectly on
leukemia cells, and can interfere with the development and
progression of hematopoietic malignancies (2–6). However,
contrasting reports have indicated that this interference by
leukemia cells may even promote the development of leukemia within
the BMM (3,7,8).
Regardless of the interaction, the role of the BMM is closely
related to the occurrence, development, invasion and metastasis of
leukemia.
Mesenchymal stem cells (MSCs), are a key component
of the BMM, and are essential for regulating the number of HSPCs,
as well as for maintaining normal hematopoietic function (9–11). Due to
their central role, it is conceivable that they may influence the
occurrence and development of leukemia. However, whether the
effects of MSCs are positive (12) or
negative (7,8,13) remains
uncertain. Compared with healthy donor-derived MSCs (HD-MSCs),
acute myeloid leukemia (AML)-derived MSCs (AML-MSCs) exhibit a
distinct pattern of gene expression, cytokine production,
immunophenotyped and cytogenetics (14,15), with
significant growth defects, insufficient osteogenic differentiation
and accelerated cellular senescence (4,16,17). These defects in AML-MSCs are closely
related to DNA methylation, transcriptional gene expression
(18) and chromosomal aberrations
(19). Additionally, compared with
HD-MSCs, the significant changes in cytokines, such as interleukin
(IL)-6 and IL-32 induced by AML-MSCs has been shown to be
associated with chemoresistance (20). Collectively, the BMM constructed by
AML-MSCs can be considered distinct from that of HD-MSCs.
Bone morphogenetic proteins (BMPs) are a member of
the transforming growth factor (TGF)-β family, expressed primarily
in the bone marrow stroma. They are the only known morphogenetic
factors capable of inducing the differentiation of MSCs (21–24), and
have been shown to play important roles in proliferation,
hematopoiesis and the development of leukemia (25–27). There
is increasing evidence linking BMPs to the transformation of the
BMM (26,28,29) and
this suggests a potential role of BMPs in leukemia. Along these
lines, alterations in the BMP pathway resulting in MSC
differentiation and the secretion of growth factors by MSCs has
been shown to confer a growth advantage, as well as BMM
transformation in AML (8). Moreover,
BMP4-induced NANOG expression with increased stem cell-like
characteristics has also been demonstrated in AML (30). In summary, BMP has shown to be
critical for the self-renewal and differentiation of leukemia stem
cells (LSCs) (26,28), and is considered to be a unique factor
in the development and progression of leukemia (27).
In 1991, Bradham et al found that connective
tissue growth factor (CTGF) was isolated from human endothelial
cells, and that it played an important role in cell adhesion,
migration, proliferation and chemotaxis (31). CTGF mediates adhesion mainly by
bridging extracellular matrix (ECM) components (including
fibronectin, perlecan, vitronectin and decorin) to integral cell
surface molecules, such as integrins and connexin (32,33). CTGF
protein induces the proliferation of MSCs, promotes the adhesion of
leukemia cells to MSCs, and leads to the overexpression of genes
involved in the cell cycle and ECM synthesis (34). More importantly, CTGF expression in
MSCs may even be induced via BMP/Osterix/Runx2-mediated signaling
in AML, and may enhance mouse leukemia implantation (8). Correspondingly, in a previous study, in
co-culture experiments with HSPCs co-cultured with MSCs in which
CTGF was knocked down, Smad 2/3-dependent signaling was found to be
activated, resulting in blocked cell cycle progression and
inhibited activation of HSPCs (35).
BMP-2-induced signaling and osteoblast differentiation has been
shown to be negatively regulated by CTGF (36). Therefore, the adhesion effect mediated
by CTGF may be closely related to the BMP signaling pathway.
Moreover, adhesion provides a protective BMM for leukemia cell
survival (37–39), further leading to the presence of
minimal residual disease, which becomes the source of genetic
instability and relapse (40–42).
We hypothesized that the transformation of the BMM
by AML-MSCs occurs through CTGF-mediated cell adhesion via the BMP
pathway, and ultimately contributes to the development of
chemoresistance. By performing co-culture experiments with AML-MSCs
and either sensitive K562 or chemoresistant K562-ADM cells, in this
study, we aimed to elucidate the mechanisms through which this
occurs.
Materials and methods
AML patient-derived bone marrow donor
samples
The bone marrow of patients with leukemia was
provided by the Hematology Department of the First Hospital of
Lanzhou University (January, 2015 to Novmber, 2018). All AML
patients (aged 7–82 years, male/female ratio, 34/22) met the
diagnostic criteria according to the World Health Organization
(WHO) and the French-American-British (FAB) co-operative group
(43). This study was approved by the
Institutional Ethics Committee of the First Hospital of Lanzhou
University and written informed consent was obtained from patients
and/or their legal guardians. The collection and acquisition of
MSCs was carried out according to the Declaration of Helsinki
(44). HD-MSCs was purchased from
Saiye Biotechnology Co. Ltd.
K562 and K562-ADM cells
The human ADM-resistant AML cell line, K562/ADM
cells, and the non-resistant cell line, K562 cells, were both
obtained from the Central Laboratory of the First Hospital of
Lanzhou University (YB-H1580 and YB-H1581; Yu Bo Biotech Co.,
Ltd.). The K562/ADM and K562 cells were grown in RPMI-1640 medium
supplied with 10% fetal bovine serum (ZheJiang Tianhang
Biotechnology Co., Ltd.) at 37°C in a humid atmosphere containing
5% carbon dioxide (CO2). The cells were confirmed to
have a confluence of 80–90% before being used in the
experiments.
Culture of human AML-MSCs
Bone marrow aspirates (2 ml) from patients with AML
were added to a 25 cm2 culture flask (Corning, Inc.)
containing DMEM/low glucose complete medium. The culture conditions
were the same as those described above. After 7–10 days, the
cultures were washed with PBS. The primary AML-MSCs were then
obtained. The cultured AML-MSCs were cultured in osteogenic
induction medium (500 µl vitamin C, 1 ml β-glycerophosphate and 10
µl dexamethasone in 100 ml DMEM) (45), followed by alkaline phosphatase assay
and Alizarin Red staining to verify its ability to differentiate
into osteoblasts.
For alkaline phosphatase staining, the cells were
first fixed with 4% paraformaldehyde for 10–15 min and then washed
with PBS. Subsequently, the alkaline phosphatase incubation
solution (BeiJing Solarbio Technology Co., Ltd.) was added in
dropwise manner on the cells followed by incubation for 20 min in
an incubator (37°C, 5% CO2), and washing with PBS. The
cells were counterstained with nuclear solid red staining solution
for 5 min, and washed with PBS for microscopic examination (CKX41;
Olympus).
For Alizarin Red staining, 0.2% Alizarin Red
(BeiJing Solarbio Technology Co., Ltd.) dye solution (0.1 g
Alizarin red in 50 ml PBS) was slowly added in a dropwise manner to
the surface of the cells, followed by gentle shaking until the dye
covered all the cells. The cells were then allowed to stand for 1–2
min, and then washed with PBS for microscopic examination (CKX41;
Olympus).
Co-culture of AML-MSCs and K562 or
K562-ADM cells
The primary AML-MSCs were sub-cultured by
trypsinization, and third-generation cells were selected for
testing. When cell growth reached between 80 and 90% confluence,
the AML-MSCs were added to 5×105 cells/ml of
K562/K562-ADM cells for co-culture (37°C, 5% CO2).
Morphological observations
The analysis of cell proliferation was carried out
by MTT (Sigma) assay. The formazan was dissolved in DMSO and the OD
value was measured at 490 nm. The cell proliferation curve was
plotted based on the OD value. The co-culture of AML-MSCs with
either K562 or K562-ADM cells was performed for 24, 48 or 72 h.
Morphological observations were performed using an Olympus inverted
biological microscope CKX41 (Olympus).
Peroxidase (POX) staining
Following co-culture, non-adherent cells were
discarded. The remaining adherent cells were air-dried, and
subsequently fixed with solution (3% glutaraldehyde, 60% acetone
solution) for 1 min. Benzoyl benzidine 10 mg + 5 M
Tris-hydrochloride 40 ml (pH 7.6) with 2% hydrogen peroxide at a
1:1 ratio was then added to fix the cells for 10 min. After rinsing
and drying, hematoxylin was used for counterstaining for 20 min and
the cells were then washed with water. Finally, 0.5% ammonia was
added to reverse blue coloration. Microscopic (CKX41; Olympus)
examination was carried out after complete drying.
RT-qPCR
RT-qPCR was performed on adherent spindle cells at
the end of the co-culture experiments. Cells were collected for RNA
extraction once they entered the exponential growth phase,
(approximately 5×107 cells/ml), as previously described
(46). The quality and concentration
of the RNA/total RNA were determined using a spectrophotometer (GE
Nanovue Plus). In total, <1 µg of total RNA was used for reverse
transcription. According to the manufacturer's instructions, cDNA
was synthesized using the iScript gDNA Clear cDNA Synthesis kit
(Bio-Rad). The concentration and qualitiy of all the obtained DNA
samples were determined by UV spectroscopy as described above.
BCR-ABL expression was determined using the
leukemia-associated fusion gene detection kit (Shanghai Yuanqi
Biomedical Technology Co. Ltd.). The amplification conditions were
as follows: 42°C, 30 min; 94°C, 5 min; (94°C, 15 sec; 60°C, 60 sec)
40 cycles. The reaction system was 25 µl. The fluorescence signal
was collected at the second step of the PCR cycle at 60°C and
analyzed by the standard curve method in absolute quantitative
detection. The standard reagent was a plasmid provided by the
manufacturers that had been quantified and contained fixed copies
of the BCR-ABL fusion gene (Yuanqi Biomedical Technology Co.,
Ltd.). The dilution was carried out with water. A calibration curve
of the Ct values of the standard dilution series vs. the
concentrations is calculated and used to determine the
concentrations of the unknowns, based on their Ct values.
The detection of the BMP4 and CTGF genes was carried
out by the relative quantification method. The sequences of the
primers used were as follows (Takara): BMP4 forward,
5′-AGATCCACAGCACTGGTCTTGAGTA-3′ and reverse,
5′-TCAGGGATGCTGCTGAGGTTA-3′; CTGF, forward,
5′-CTTGCGAAGCTGACCTGGAA-3′ and reverse,
5′-AGCTCAAACTTGATAGGCTTGGAGA-3′; and β-actin forward,
5′-TGGCACCCAGCACAATGAA-3′ and reverse,
5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′. The cells were fluorescently
labeled with iTaq Universal SYBR-Green Supermix (Bio-Rad). HD-MSCs
alone served as the control group. The amplification conditions
were as follows: 95°C, 5 min; (95°C, 15 sec; 60°C, 60 sec) 40
cycles; 95°C, 5 sec; 65°C, 60 sec; 40°C, 30 sec. The reaction
system was 20 µl. Detection was performed using a Roche
LightCycler® 480 Fluorescence PCR detector (Roche). The
obtained data is the Ct value. The Ct values of the repeat sample
wells were averaged and calculated by the 2−ΔΔCq method
(the reference gene was β-actin) (47).
Western blot analysis
The cells were lysed with lysis buffer (1% SDS, 10
mm Tris-HCl, pH 7.6, 20 g/ml aprotinin, 20 g/ml leupeptin and 1 mm
AEBSF). Protein concentrations were determined using the Bradford
method (48). Protein (20 µg) was
separated on 12% SDS-PAGE gels and transferred onto PVDF membranes
(Merck Millipore). After blocking with 10% skim milk, the membranes
were incubated with the primary antibodies (anti-CTGF antibody,
1:1,000, cat. no. ab6992, Abcam; anti-phospho-Smad1/5 antibody,
1:1,000, cat. no. 9516, Cell Signaling Technology) at 4°C
overnight. After washing 3 times with triethanolamine buffer
solution (Sangon Biotech Co., Ltd.), the membranes were incubated
with goat anti-rabbit IgG horseradish peroxidase-conjugated
secondary antibodies (cat. no. 31460; Invitrogen; Thermo Fisher
Scientific) (1:200 dilution in 5% skim milk) at room temperature
for 1 h. The signals were examined with the ECL kit (Elabscience
Biotechnology Co., Ltd.), using anti-β-actin antibody (1:1,000,
cat. no. 4970; Cell Signaling Technology) as an internal control.
Protein gray value detection was performed using ImageJ software
(National Institutes of Health).
Fluorescence in situ hybridization
(FISH)
The co-cultured cells as described above were placed
onto glass slides. The tightly adherent cells were washed with PBS
after 24 h. The slides were placed in hypotonic 0.075 M potassium
chloride solution at 37°C for 20 min, then placed 3 times in
fixative (methanol:glacial acetic acid, 3:1) for 1 min each time.
The slides were then placed on a roaster (Tianjin Tianli Aviation
Electro-Mechanical Co., Ltd.) at 56°C for 10–20 min, then immersed
in 2X SSC buffer (sodium chloride:sodium citrate, 2:1; pH 7.0±0.2)
twice for 5 min each time. They were then placed in 0.1 M HCl for
10 min, and again immersed in 2X SSC buffer twice for 2 min each.
Pepsin was added dropwise onto the slides followed by incubation at
37°C for 11 min. They were then washed twice in 2X SSC buffer for 5
min each. Finally, the slides were immersed in formaldehyde
fixative (1 ml formaldehyde and 0.18 g
MgCl2.6H2O in 39 ml PBS) for 10 min, and then
sequentially dehydrated through a gradient of ethanol
concentrations (70, 85 and 100%) over a period of 3 min at room
temperature.
The BCR-ABL fluorescence probe were prepared
(Jinpujia Pharmaceutical Technology Co., Ltd.) in a dark room for
in situ hybridization (75-80°C, 5 min; 42°C, overnight;
In situ hybridization apparatus, IRIS International
Inc.).
The slides for in situ hybridization were
placed into two different 2X SSC buffers (46°C, water bath for 30
min) shaken for 1–3 sec, rinsed for 15 min, and then rinsed in 0.1%
NP-40/2X SSC buffer for 15 min. Finally, the slides were soaked in
70% ethanol for 3 min at room temperature. A total of 15 µl DAPI
were added to the slides to stain the cell nuclei after which they
were observed under a fluorescence microscope (Olympus) after 10–20
min of incubation at room temperature.
Detection of IL-6 and IL-32 in the
supernatant by enzyme linked immunosorbent assay (ELISA)
Supernatants from co-culture experiments was
collected and the detection of human IL-6 and IL-32 levels by ELISA
was performed according to manufacturer's protocol (Shanghai
JiangLai Biotechnology Co., Ltd.).
Cell cycle detection
Cell suspensions containing approximately
2×105 to 1×106 cells from co-cultures at 24,
48 and 72 h were subjected to cell cycle analysis according to the
manufacturer's protocol (Cell Cycle Staining kit, Hangzhou Lianke
Biotechnology Co., Ltd.). Detection was performed on a BD FACS
Verse flow cytometer (BD Bioscienses).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7.0 software (GraphPad Software). Multigroup comparisons were
carried out by ANOVA analysis. The Bonferroni correction was
performed as a post hoc test for the data that were subjected to
two-way ANOVA analysis. P-values <0.05 were considered top
indicate statistically significant differences.
Results
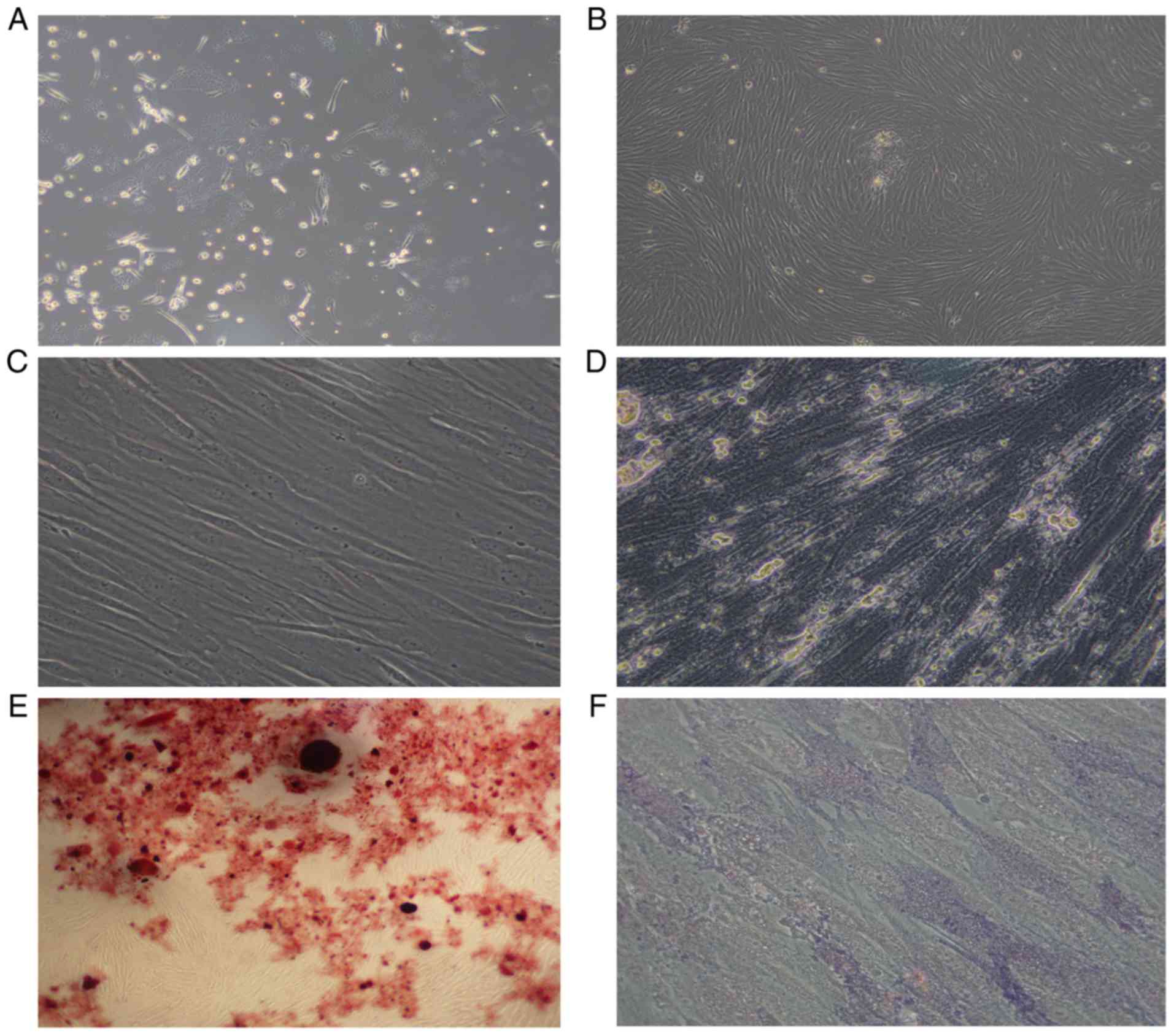
Morphology of AML-MSCs
The AML-MSCs were found to be adherent cells, with a
spindle-like shaped and to be irregularly arranged (Fig. 1A). After 14–21 days, the number of
adherent cells increased significantly, which grew in parallel or
in a spiral-like manner (Fig. 1B).
The growth rate and size of the sub-cultured AML-MSCs were also
greater than that of the primary cells (Fig. 1C). After 14 days of culture with an
osteogenic inducer (dexamethasone, vitamin C, β-sodium
glycerophosphate), the cytoplasm of the cells was filled with
granules, and calcium deposition was observed between the cells
(Fig. 1D), and red nodules were
observed following staining with Alizarin Red (Fig. 1E). Alkaline phosphatase staining
revealed a large amount of purple-brown sediment in the
extracellular matrix (Fig. 1F). Thus,
it was found that the AML-MSCs conformed to the standards of the
International Society for Cell Therapy (49).
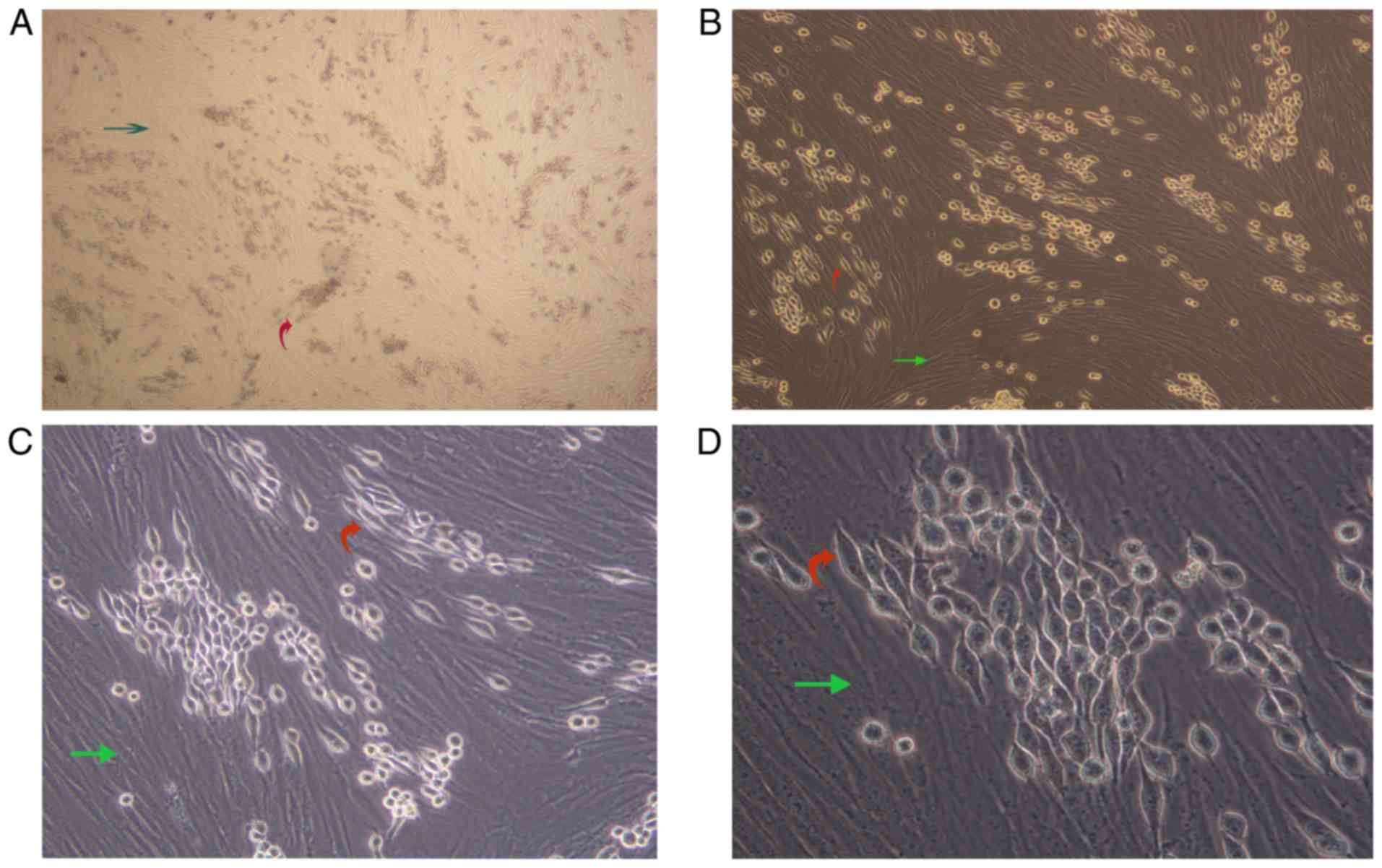
AML-MSCs promote the fusiform
transformation of K562-ADM cells
We observed a unique morphologic alteration in the
drug-resistant K562-ADM cells that was associated with the enhanced
growth advantage. There were two layers of cells in the culture
plate. The bottom layer contained AML-MSCs and the upper layer
suspended K562 or K562-ADM cells. After 24 h, the K562-ADM cells
exhibited a large amount of adhesion, which was accompanied by
fusiform transformation. Moreover, the transformed spindle cells
were larger in the middle, and the volume was smaller than that of
the AML-MSCs. The size of the transformed spindle cells was
approximately one-third that of the AML-MSCs, and the transparency
of the transformed spindle cells was better. The contents are
clearly visible (Fig. 2).
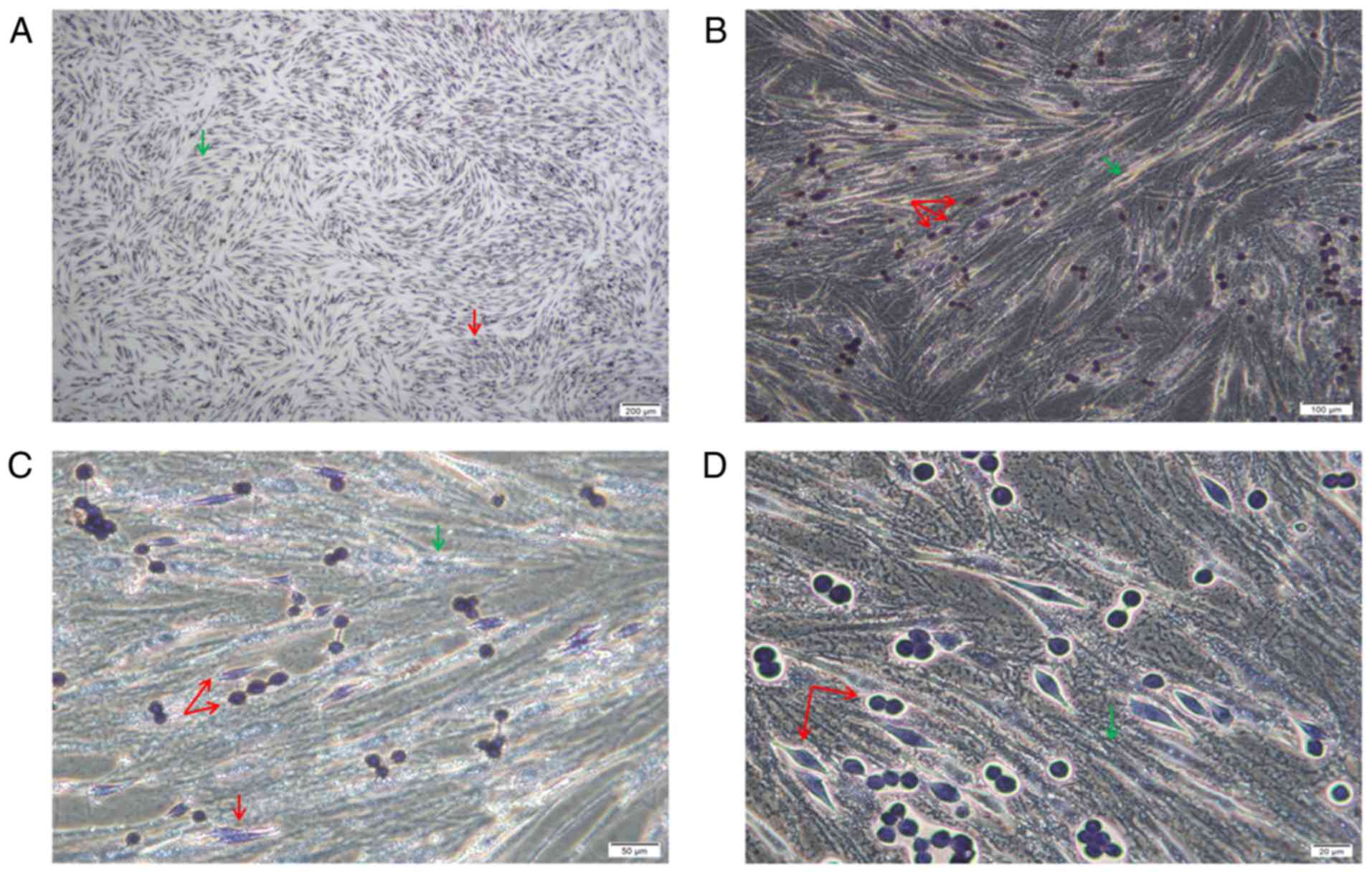
POX staining revealed that the cells were of two
origins. The bottom spindle cells were AML-MSCs and were negative
for POX, and larger. The upper adherent cells had two forms, the
cells were small and both were granulocyte sources, and were
positive or strongly positive for POX (Fig. 3).
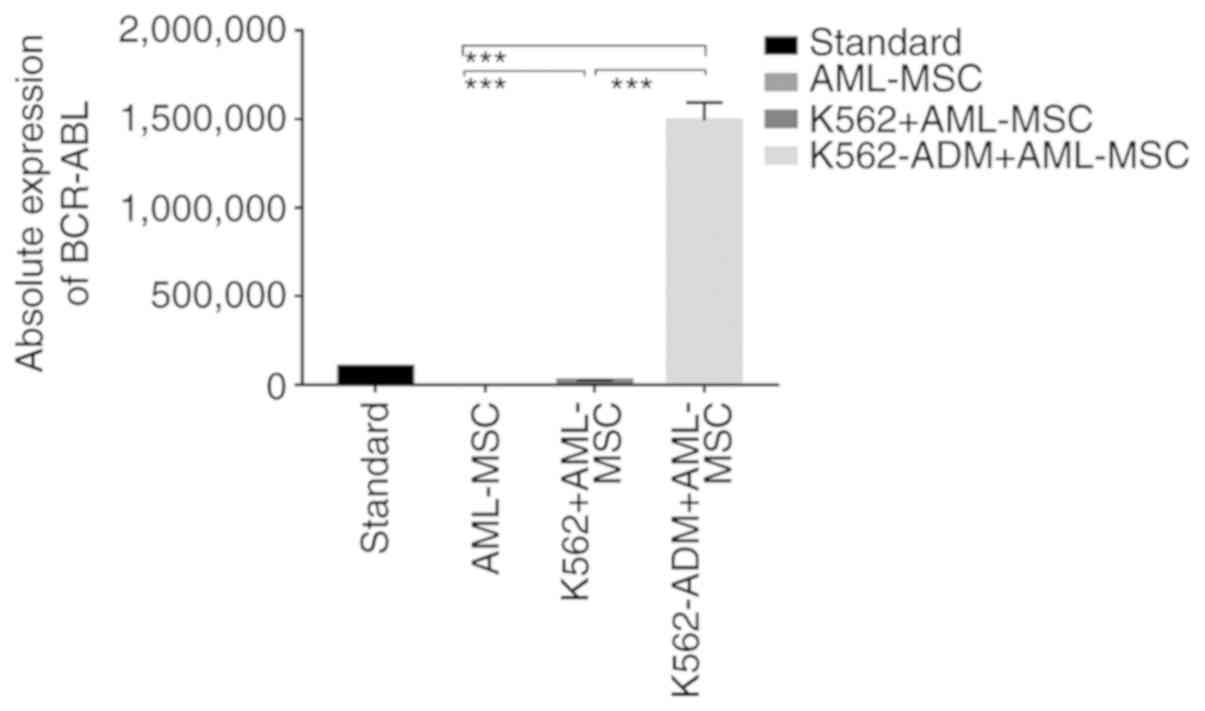
Absolute quantitative detection by RT-qPCR
demonstrated significant differences in the expression levels of
BCR-ABL between the controls (AML-MSCs) and the experimental groups
(K562 or K562-ADM-derived co-cultured cells) (P<0.001),
confirming two different populations of cells (Fig. 4). Compared with the K562 co-culture
group, the expression level of BCR-ABL in the K562-ADM co-culture
group was significantly increased (P<0.001; Fig. 4). This may be related to the
significant increase in adherent and spindle-shaped transformed
cells in the K562-ADM co-culture group (the K562 co-culture group
had fewer adherent cells and almost no fusiform transformation;
Figs. 2 and 3).
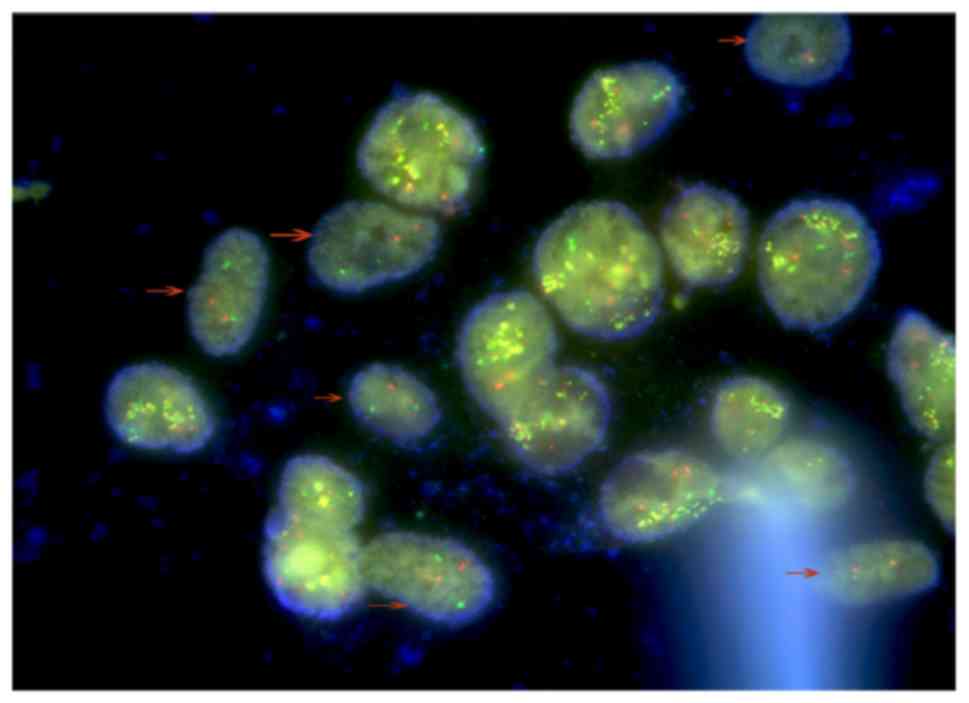
The presence of two different cell types was
furthermore confirmed by FISH, with AML-MSCs negative for BCR-ABL
indicated by arrows (Fig. 5). Due to
the polyploid genetic characteristics of the K562 or K562-ADM cell
lines, and the fact that there are multiple sets of gene loci
fusions, the results shown in Fig. 5
are in accordance with the results presented in the study by
Gribble et al (50).
These above-mentioned experimental results confirm
the morphological changes of the K562-ADM cells under co-culture
conditions with AML-MSCs. Moreover, AML-MSCs induced the
transformation of K562-ADM cells from a circular suspension cell to
an adherent spindle cell with a positive BCR/ABL expression.
Changes in BMP4 and CTGF gene
expression
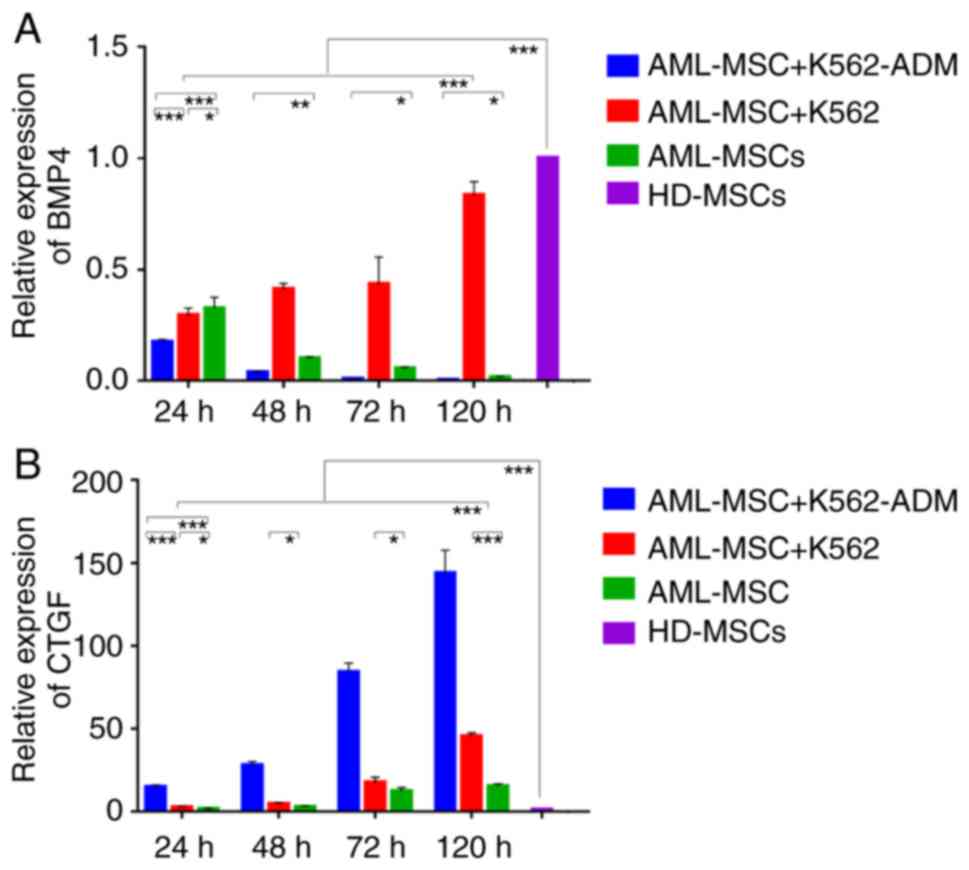
Compared with the HD-MSCs, the BMP4 levels in the
AML-MSCs decreased significantly (P<0.001; Fig. 6A). Moreover, the AML-MSCs co-cultured
with the K562-ADM cells exhibited lower BMP4 expression levels
compared to those co-cultured with the K562 cells in a time
dependent manner. Conversely, the AML-MSCs co-cultured with the
K562 cells exhibited a significant increase in BMP4 expression with
time (P<0.001; Fig. 6A).
Compared with the HD-MSCs, CTGF expression in the
AML-MSCs was upregulated significantly (P<0.001; Fig. 6B), and was found to be increased even
further following co-culture with the K562-ADM cells compared with
the K562 cells (P<0.001; (Fig.
6B). Both co-cultures exhibited a time dependent increase in
CTGF expression (P<0.001; Fig. 6B)
and this was consistent with the morphological observations.
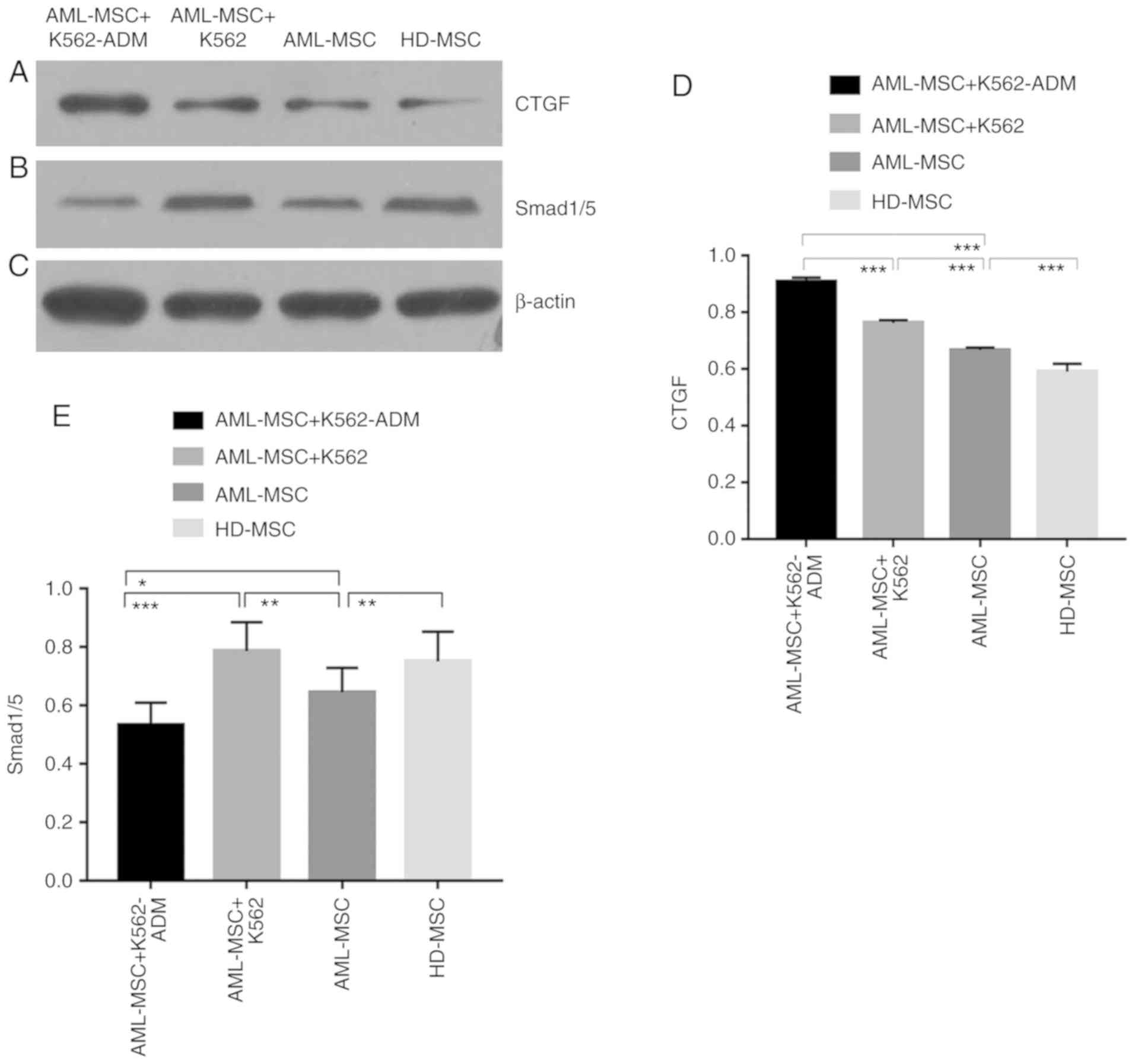
Detection of p-Smad 1/5 and CTGF
protein expression
The protein expression of CTGF was confirmed to be
present in the co-cultured cells compared to the HD-MSCs, and was
greater in the AML-MSCs with co-cultured with the AML-MSCs
co-cultured with K562-ADM cells (P<0.001; Fig. 7A and D). This was likewise consistent
with the morphological observations. Conversely, p-Smad1/5
expression was found to be lower in the K562-ADM co-culture group
compared to the K562 co-culture group (P<0.001; Fig. 7B and E).
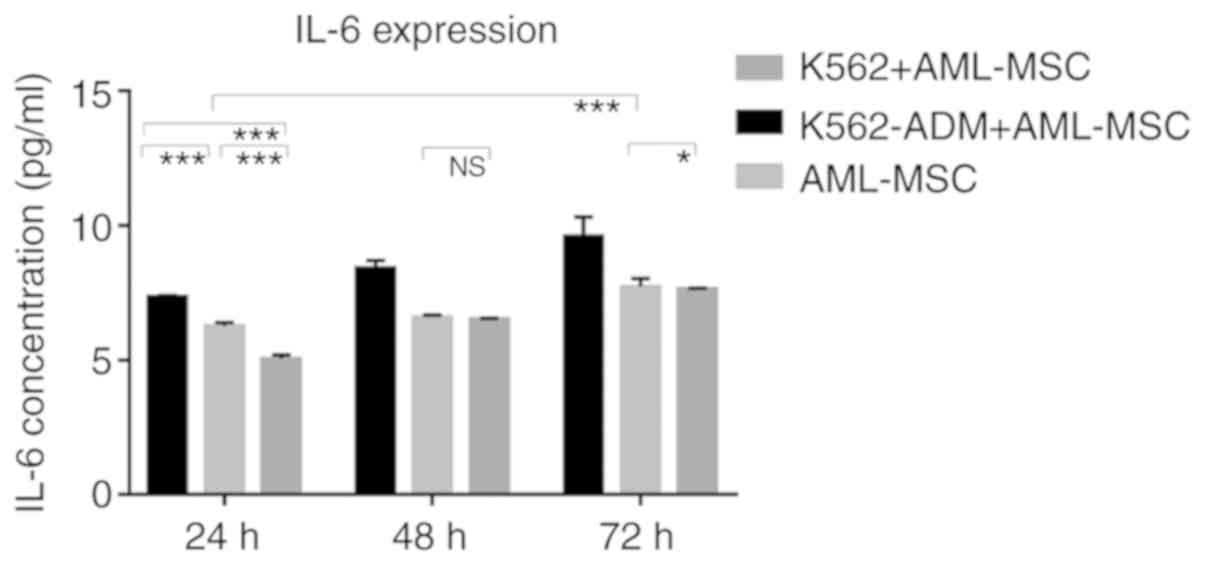
Changes in the levels of soluble
cytokines
The results of ELISA revealed that the IL-6 levels
gradually increased over time in the AML-MSC culture group. The
IL-6 levels significantly increased after the AML-MSCs were
co-cultured with the K562-ADM cells (P<0.001; Fig. 8). However, IL-6 production by the K562
cells co-cultured with the AML-MSCs was lower than that by the
AML-MSCs cultured alone (P<0.05; Fig.
8). Although these levels increased slightly with time, they
remained low, and were much lower than those in the K562-ADM
co-culture group (P<0.001; Fig.
8).
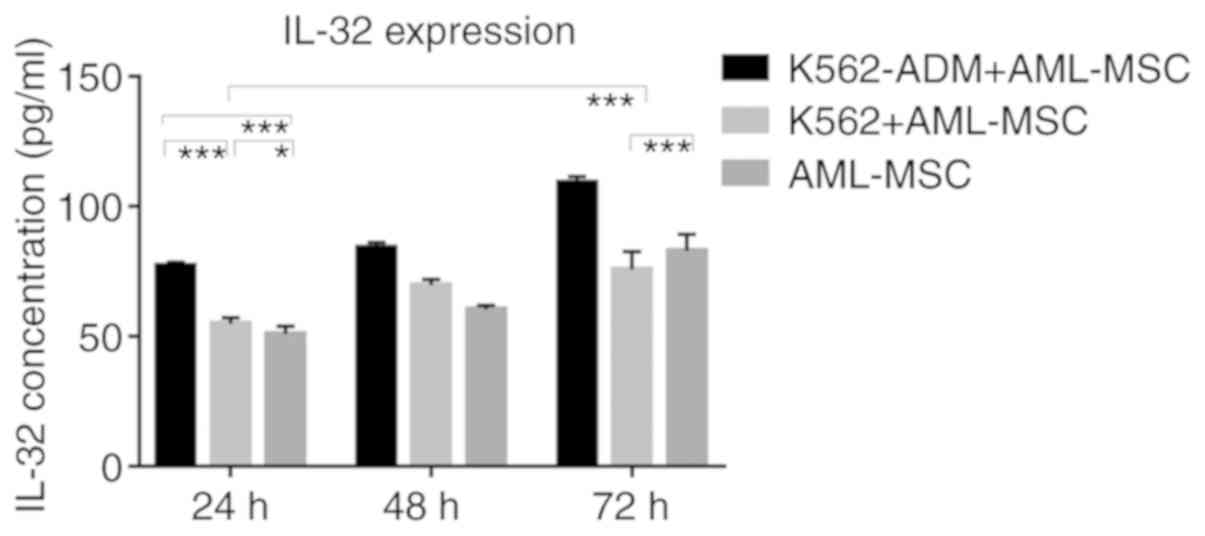
Similarly, the IL-32 levels in the K562-ADM
co-culture group were significantly higher compared with the single
culture group and the K562 co-culture group (P<0.001; Fig. 9). However, when the co-culture time
exceeded 48 h, the IL-32 levels decreased slightly in the K562
co-culture group compared to the AML-MSCs cultured alone
(P<0.001; Fig. 9).
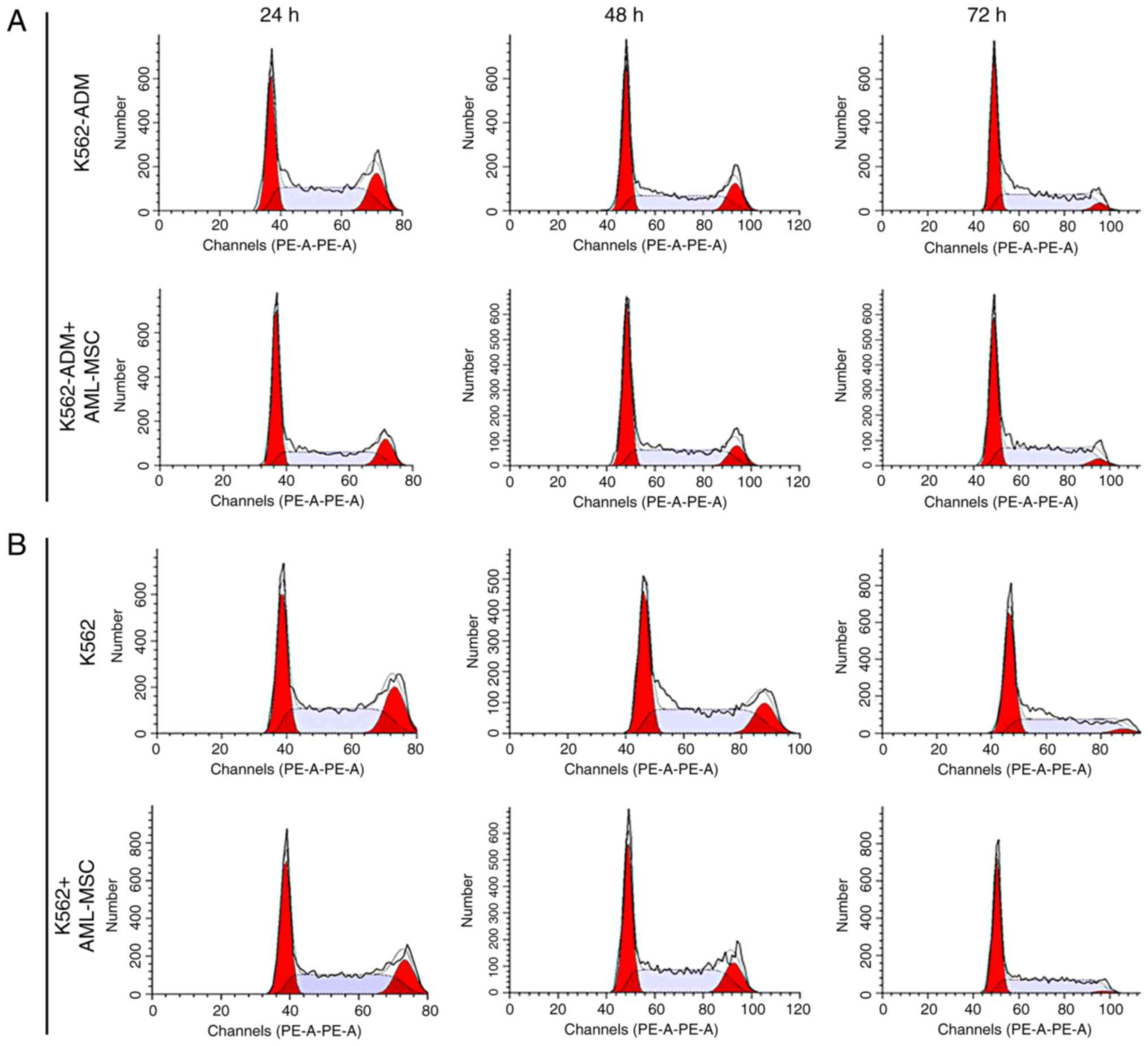
Changes in the cell cycle
At 24 h, the AML-MSCs co-cultured with the K562-ADM
cells exhibited a decreased percentage in the population of cells
in the in G2+S phases compared with the K562-ADM cells cultured
alone; however, this percentage gradually increased, with the
percentage of cells in the G2+S phase exceeding that of the
individual culture group at 72 h (Fig.
10A). The results also revealed the increased proliferation of
K562-ADM cells with time spent in co-culture with the AML-MSCs,
whereas the opposite trend was observed in the K562-ADM cells
cultured alone (P<0.001; Fig.
10C). The cell cycle analysis results of the K562 cells
co-cultured with the AML-MSCs were consistent with those of the
K562 cells cultured alone, and cell proliferation was inhibited
over time (P<0.01; Fig. 10D).
Therefore, AML-MSCs exert a promoting effect on the proliferation
of K562-ADM cells, but exert an inhibitory effect on the
proliferation of K562 cells (P<0.001; Fig. 10E).
Discussion
In our study, we observed a unique morphological
alteration in the drug-resistant K562-ADM cells. The results of
this study found that a fusiform transformation of the K562-ADM
cells occurred following co-culture with AML-MSCs (Figs. 2–5).
Since this change is necessarily accompanied by the adhesion of the
two co-cultured cells, we further examined the levels of CTGF and
its related pathway, BMP. Combined with the results of cytokine and
cell cycle analyses, we found that the AML-MSCs utilize the BMP
pathway to induce the upregulation of CTGF expression and cytokine
secretion (Figs. 6–9), ultimately promoting the proliferation of
K562-ADM cells (Fig. 10). However,
due to the differential expression of BMP4, CTGF and cytokines,
these morphological changes were not observed in the non-resistant
K562 cells. The AML-MSCs significantly inhibited cell proliferation
when co-cultured with the K562 cells (Fig. 10E). This further confirmed the
existence of an interaction between the BMM and AML cells.
Recently, there has been widespread debate over the
role of MSCs in tumor cells. Some studies have suggested that
leukemia-derived MSCs have a normal differentiation, adhesion,
expression and survival, and the ability to support hematopoiesis
(13,19,51–54).
Moreover, MSCs may inhibit the progression of leukemia cells
(13). This is in stark contrast to
the findings of other studies showing that an abnormal
differentiation, defective hematopoietic capacity, a reduced
expression of adhesion molecules, and increased an apoptosis occur
in leukemia-derived MSCs (4,14,55–58),
within the tumor microenvironment to promote the growth of leukemia
cells.
Nevertheless, finding an exact mechanism of tumor
resistance remains to be discerned. Mohammadi et al found
that AML cells co-cultured with MSCs and osteoblasts became more
resistant to drug-induced apoptosis (59). Related studies have also confirmed
that MSCs support AML cells survival and bone marrow
transplantation, thereby promoting drug resistance (60–62).
Moreover, the physical contact or adhesion of leukemia cells to
cellular components in the BMM has been found to mediate
chemoresistance (60), and K562 cells
specifically have been shown to exhibit cell adhesion and
resistance to apoptosis when exposed to BCR/ABL inhibitors AG957
and STI-571 (63). Furthermore,
related factors that cause cell adhesion promote the long-term
protection of leukemia stem cells (LSCs) by the BMM, which is an
important cause of clonal proliferation and disease recurrence
(41,42). In this study, adhesion occurred prior
to this morphological transformation. Therefore, we hypothesized
that the adhesion of K562-ADM cells mediated by AML-MSCs is the
cause of fusiform transformation and drug resistance.
As a key factor in adhesion, CTGF can induce the
formation of spherical cell aggregates, causing attached cells to
exhibit morphological changes, from spherical to flat or elongated
shapes (64). By examining CTGF
expression at the gene and protein levels in AML-MSCs, we observed
a significant upregulation following co-culture with the K562-ADM
cells (Figs. 6 and 7) vs. the non-resistant K562 cells. This was
consistent with the fusiform transformation. The continuous
overexpression of CTGF negatively regulates the BMP-2-induced
signaling pathway and osteoblast differentiation, resulting in
decreased protein levels of phosphorylated Smad 1/5/8 (36). The adhesion of MSCs has previously
been shown to be primarily mediated through the ERK and BMP signal
pathways (65). This is consistent
with the results of the decreased BMP4 and Smad1/5 protein levels
found in this study (Figs. 6 and
7) and lends further support to the
role of CTGF in the BMP pathway in the development of drug
resistance.
In this study, we only tested the classical BMP4
protein of the BMP family. Further research is required to assess
the role of additional members of the BMP family as regards CTGF
expression and the BMP pathway. Future investigations may
additionally shed light onto the development and chemoresistance of
AML in response to drug targeting the BMP pathway.
Soluble factors involved in stem cell renewal are
the primary targets within the BMM, and MSCs may antagonize
chemotherapy within this microenvironment by acting on the cytokine
network (58,66). As a core player, IL-6 has shown to be
capable of driving the initiation, growth and metastasis of tumors
(67–69). Simultaneously, IL-6 is secreted by
MSCs, which activates signal transduction and transcriptional
activator 3 (STAT3), further leading to activation of TGF-β
signaling, thereby affecting the growth, proliferation and
cytoprotection of stem and progenitor cells (70). In combination with BMPs present in
serum, IL-6 may additionally function to enhance the self-renewal
and affect the pluripotency of embryonic stem cells (71). IL-6 and its downstream signaling
molecules are responsible for multidrug resistance (72). Similarly, changes in IL-32 levels can
alter the chemical protective effects of cells on
cytarabine-induced apoptosis (20).
In this study, our co-culture experiments further implicated the
role of IL-6 and IL-32 in the development of chemoresistance as
mediated by CTGF and the BMP pathway (Figs. 6, 8 and
9), which further indicates that the
BMP pathway and CTGF may alter the BMM and participate in tumor
resistance.
In conclusion, the findings of this study suggest
that the BMM, which is largely composed of MSCs undergo various
changes that confer drug resistance. These changes occur at the
genetic and transcriptional level involving changes in cytokines,
adhesion, and immunity (18,19,52,73,74).
Precipitating factors, include those from the environment,
radiation and chemotherapy (75), as
well as from leukemia cells themselves (8). The extent to which these changes are
observed depends on a combination of factors, which cumulatively
contributes to a heterogenous leukemic microenvironment and
plausibly confer chemoresistance.
The findings of this study suggest that changes in
AML-MSCs in the BMM result in the dysregulation of the BMP pathway.
This dysregulation further modifies the secretion and expression of
CTGF, which induces morphological changes in K562-ADM cells. This
shift may be the result of an increased CTGF expression or the
conversion of K562-ADM cells to leukemic stem cells. Moreover, IL-6
and IL-32, which are known to promote the proliferation of HSPCs,
were elevated in the K562-ADM co-cultured with AML-MSCs. Therefore,
this transformation may be key for the recurrence of leukemia, and
its inhibition may be essential for eradicating minimal residual
disease. Lastly, this study provides further support for the role
of cell adhesion in drug resistance, and demonstrated the
importance of the BMP pathway in the BMM, providing a novel
therapeutic target for the treatment of leukemia and prevention of
its recurrence.
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science
Foundation of Gansu province (grant no. 18JR3RA342).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
HL was responsible for the design and implementation
of the entire study, and also participated in the acquisition,
analysis and writing of all the experimental data. JL participated
in the experimental design, and put forward important verification
methods for the problems found in the experimental process. JC was
involved in the collection and provision of the experimental
samples, and participated in the application and implementation of
experimental ethics certification. XC was involved in cell
experiment technical guidance and assistance. LZ was involved in
flow cytometry technical guidance and assistance. ZL was
responsible for the design and implementation of the entire study,
and participated in the revision of the final version of the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional Ethics
Committee of the First Hospital of Lanzhou University and written
informed consent was obtained from patients and/or their legal
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schofield R: The relationship between the
spleen colony-forming cell and the haemopoietic stem cell. Blood
Cells. 4:7–25. 1978.PubMed/NCBI
|
|
2
|
Krause DS and Scadden DT: A hostel for the
hostile: The bone marrow niche in hematologic neoplasms.
Haematologica. 100:1376–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Korn C and Méndez-Ferrer S: Myeloid
malignancies and the microenvironment. Blood. 129:811–822. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geyh S, Rodríguez-Paredes M, Jäger P,
Khandanpour C, Cadeddu RP, Gutekunst J, Wilk CM, Fenk R, Zilkens C,
Hermsen D, et al: Functional inhibition of mesenchymal stromal
cells in acute myeloid leukemia. Leukemia. 30:683–691. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Medyouf H: The microenvironment in human
myeloid malignancies: Emerging concepts and therapeutic
implications. Blood. 129:1617–1626. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schepers K, Campbell TB and Passegué E:
Normal and leukemic stem cell niches: Insights and therapeutic
opportunities. Cell Stem Cell. 16:254–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar A, Anand T, Bhattacharyya J, Sharma
A and Jaganathan BG: K562 chronic myeloid leukemia cells modify
osteogenic differentiation and gene expression of bone marrow
stromal cells. J Cell Commun Signal. 12:441–450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Battula VL, Le PM, Sun JC, Nguyen K, Yuan
B, Zhou X, Sonnylal S, McQueen T, Ruvolo V, Michel KA, et al:
AML-induced osteogenic differentiation in mesenchymal stromal cells
supports leukemia growth. JCI Insight. 2:900362017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Méndez-Ferrer S, Michurina TV, Ferraro F,
Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A,
Enikolopov GN and Frenette PS: Mesenchymal and haematopoietic stem
cells form a unique bone marrow niche. Nature. 466:829–834. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frenette PS, Pinho S, Lucas D and
Scheiermann C: Mesenchymal stem cell: Keystone of the hematopoietic
stem cell niche and a stepping-stone for regenerative medicine.
Annu Rev Immunol. 31:285–316. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sacchetti B, Funari A, Michienzi S, Di
Cesare S, Piersanti S, Saggio I, Tagliafico E, Ferrari S, Robey PG,
Riminucci M and Bianco P: Self-renewing osteoprogenitors in bone
marrow sinusoids can organize a hematopoietic microenvironment.
Cell. 131:324–336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Song N, Gao L, Qiu H, Huang C, Cheng H,
Zhou H, Lv S, Chen L and Wang J: Mouse bone marrow-derived
mesenchymal stem cells inhibit leukemia/lymphoma cell proliferation
in vitro and in a mouse model of allogeneic bone marrow transplant.
Int J Mol Med. 36:139–149. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Tang X, You Y, Li W, Liu F and Zou
P: Assessment of bone marrow mesenchymal stem cell biological
characteristics and support hemotopoiesis function in patients with
chronic myeloid leukemia. Leuk Res. 30:993–1003. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JC, Basu SK, Zhao X, Chien S, Fang
M, Oehler VG, Appelbaum FR and Becker PS: Mesenchymal stromal cells
derived from acute myeloid leukemia bone marrow exhibit aberrant
cytogenetics and cytokine elaboration. Blood Cancer J. 5:e3022015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fracchiolla NS, Fattizzo B and Cortelezzi
A: Mesenchymal stem cells in myeloid malignancies: A focus on
immune escaping and therapeutic implications. Stem Cells Int 2017.
67205942017.
|
|
16
|
Schroeder T, Geyh S, Germing U and Haas R:
Mesenchymal stromal cells in myeloid malignancies. Blood Res.
51:225–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geyh S, Oz S, Cadeddu RP, Fröbel J,
Brückner B, Kündgen A, Fenk R, Bruns I, Zilkens C, Hermsen D,
Gattermann N, et al: Insufficient stromal support in MDS results
from molecular and functional deficits of mesenchymal stromal
cells. Leukemia. 27:1841–1851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
von der Heide EK, Neumann M, Vosberg S,
James AR, Schroeder MP, Ortiz-Tanchez J, Isaakidis K, Schlee C,
Luther M, Jöhrens K, et al: Molecular alterations in bone marrow
mesenchymal stromal cells derived from acute myeloid leukemia
patients. Leukemia. 31:1069–1078. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Blau O, Hofmann WK, Baldus CD, Thiel G,
Serbent V, Schümann E, Thiel E and Blau IW: Chromosomal aberrations
in bone marrow mesenchymal stroma cells from patients with
myelodysplastic syndrome and acute myeloblastic leukemia. Exp
Hematol. 35:221–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lopes MR, Pereira JK, de Melo Campos P,
Machado-Neto JA, Traina F, Saad ST and Favaro P: De novo AML
exhibits greater microenvironment dysregulation compared to AML
with myelodysplasia-related changes. Sci Rep. 7:407072017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang EA, Rosen V, D'Alessandro JS, Bauduy
M, Cordes P, Harada T, Israel DI, Hewick RM, Kerns KM, LaPan P, et
al: Recombinant human bone morphogenetic protein induces bone
formation. Proc Nati Acad Sci USA. 87:2220–2224. 1990. View Article : Google Scholar
|
|
22
|
Scarfi S: Use of bone morphogenetic
proteins in mesenchymal stem cell stimulation of cartilage and bone
repair. World J Stem Cells. 8:1–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He Y, Yu L, Liu J, Li Y, Wu Y, Huang Z, Wu
D, Wang H, Wu Z and Qiu G: Enhanced osteogenic differentiation of
human bone-derived mesenchymal stem cells in 3-dimensional printed
porous titanium scaffolds by static magnetic field through
up-regulating Smad4. FASEB J. 33:6069–6081. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Chen B, Li D, Zhou X and Chen Z:
LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic
differentiation in human bone marrow-derived mesenchymal stem
cells. Pathol Res Pract. 215:525–531. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vicente López MA, Vázquez García MN,
Entrena A, Olmedillas Lopez S, García-Arranz M, García-Olmo D and
Zapata A: Low doses of bone morphogenetic protein 4 increase the
survival of human adipose-derived stem cells maintaining their
stemness and multipotency. Stem Cells Dev. 20:1011–1019. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Toofan P and Wheadon H: Role of the bone
morphogenic protein pathway in developmental haemopoiesis and
leukaemogenesis. Biochem Soc Trans. 44:1455–1463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zylbersztejn F, Flores-Violante M,
Voeltzel T, Nicolini FE, Lefort S and Maguer-Satta V: The BMP
pathway: A unique tool to decode the origin and progression of
leukemia. Exp Hematol. 61:36–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao X, Liu J, Peng M, Liu J and Chen F:
BMP4 is involved in the chemoresistance of myeloid leukemia cells
through regulating autophagy-apoptosis balance. Cancer Invest.
31:555–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goldman DC, Bailey AS, Pfaffle DL, Al
Masri A, Christian JL and Fleming WH: BMP4 regulates the
hematopoietic stem cell niche. Blood. 114:4393–4401. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Voeltzel T, Flores-Violante M,
Zylbersztejn F, Lefort S, Billandon M, Jeanpierre S, Joly S,
Fossard G, Milenkov M, Mazurier F, et al: A new signaling cascade
linking BMP4, BMPR1A, DeltaNp73 and NANOG impacts on stem-like
human cell properties and patient outcome. Cell Death Dis.
9:10112018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bradham DM, Igarashi A, Potter RL and
Grotendorst GR: Connective tissue growth factor: A cysteine-rich
mitogen secreted by human vascular endothelial cells is related to
the SRC-induced immediate early gene product CEF10. J Cell Biol.
114:1285–1294. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aguiar DP, de Farias GC, de Sousa EB, de
Mattos Coelho- Aguiar J, Lobo JC, Casado PL, Duarte ME and Abreu JG
Jr: New strategy to control cell migration and metastasis regulated
by CCN2/CTGF. Cancer Cell Int. 14:612014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jun JI and Lau LF: Taking aim at the
extracellular matrix: CCN proteins as emerging therapeutic targets.
Nat Rev Drug Discov. 10:945–963. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen CC and Lau LF: Functions and
mechanisms of action of CCN matricellular proteins. Int J Biochem
Cell Biol. 41:771–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Istvánffy R, Vilne B, Schreck C, Ruf F,
Pagel C, Grziwok S, Henkel L, Prazeres da Costa O, Berndt J,
Stümpflen V, et al: Stroma-derived connective tissue growth factor
maintains cell cycle progression and repopulation activity of
hematopoietic stem cells in vitro. Stem Cell Rep. 5:702–715. 2015.
View Article : Google Scholar
|
|
36
|
Mundy C, Gannon M and Popoff SN:
Connective tissue growth factor (CTGF/CCN2) negatively regulates
BMP-2 induced osteoblast differentiation and signaling. J Cell
Physiol. 229:672–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johansen S, Brenner AK, Bartaula-Brevik S,
Reikvam H and Bruserud O: The possible importance of β3 integrins
for leukemogenesis and chemoresistance in acute myeloid leukemia.
Int J Mol Sci. 19:E2512018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Al-Asadi MG, Brindle G, Castellanos M, May
ST, Mills KI, Russell NH, Seedhouse CH and Pallis M: A molecular
signature of dormancy in CD34+CD38- acute myeloid leukaemia cells.
Oncotarget. 8:111405–111418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Liu X, Qiu Y, Shi Y, Cai J, Wang
B, Wei X, Ke Q, Sui X, Wang Y, et al: Cell adhesion-mediated
mitochondria transfer contributes to mesenchymal stem cell-induced
chemoresistance on T cell acute lymphoblastic leukemia cells. J
Hematol Oncol. 11:112018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meads MB, Gatenby RA and Dalton WS:
Environment-mediated drug resistance: A major contributor to
minimal residual disease. Nat Rev Cancer. 9:665–674. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Winkler IG, Barbier V, Nowlan B, Jacobsen
RN, Forristal CE, Patton JT, Magnani JL and Lévesque JP: Vascular
niche E-selectin regulates hematopoietic stem cell dormancy, self
renewal and chemoresistance. Nat Med. 18:1651–1657. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jacamo R, Chen Y, Wang Z, Ma W, Zhang M,
Spaeth EL, Wang Y, Battula VL, Mak PY, Schallmoser K, et al:
Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of
NF-κB mediates chemoresistance. Blood. 123:2691–2702. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Arber DA, Orazi A, Hasserjian R, Thiele J,
Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M and Vardiman JW:
The 2016 revision to the World Health Organization classification
of myeloid neoplasms and acute leukemia. Blood. 127:2391–2405.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
World Medical Association: World Medical
Association Declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nielsen EØ, Chen L, Hansen JO, Degn M,
Overgaard S and Ding M: Optimizing Osteogenic differentiation of
ovine adipose-derived stem cells by osteogenic induction medium and
FGFb, BMP2, or NELL1 in vitro. Stem Cells Int 2018.
97813932018.
|
|
46
|
Liu X and Harada S: RNA isolation from
mammalian samples. Curr Protoc Mol Biol Chapter. 4:Unit 4.16. 2013.
View Article : Google Scholar
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zor T and Selinger Z: Linearization of the
Bradford protein assay increases its sensitivity: Theoretical and
experimental studies. Anal Biochem. 236:302–308. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The International Society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gribble SM, Roberts I, Grace C, Andrews
KM, Green AR and Nacheva EP: Cytogenetics of the chronic myeloid
leukemia-derived cell line K562: Karyotype clarification by
multicolor fluorescence in situ hybridization, comparative genomic
hybridization, and locus-specific fluorescence in situ
hybridization. Cancer Genet Cytogenet. 118:1–8. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kouvidi E, Stratigi A, Batsali A, Mavroudi
I, Mastrodemou S, Ximeri M, Papadaki HA and Pontikoglou CG:
Cytogenetic evaluation of mesenchymal stem/stromal cells from
patients with myelodysplastic syndromes at different time-points
during ex vivo expansion. Leuk Res. 43:24–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Blau O, Baldus CD, Hofmann WK, Thiel G,
Nolte F, Burmeister T, Türkmen S, Benlasfer O, Schümann E, Sindram
A, et al: Mesenchymal stromal cells of myelodysplastic syndrome and
acute myeloid leukemia patients have distinct genetic abnormalities
compared with leukemic blasts. Blood. 118:5583–5592. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Soenen-Cornu V, Tourino C, Bonnet ML,
Guillier M, Flamant S, Kotb R, Bernheim A, Bourhis JH, Preudhomme
C, Fenaux P and Turhan AG: Mesenchymal cells generated from
patients with myelodysplastic syndromes are devoid of chromosomal
clonal markers and support short- and long-term hematopoiesis in
vitro. Oncogene. 24:2441–2448. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jootar S, Pornprasertsud N, Petvises S,
Rerkamnuaychoke B, Disthabanchong S, Pakakasama S, Ungkanont A and
Hongeng S: Bone marrow derived mesenchymal stem cells from chronic
myeloid leukemia t(9;22) patients are devoid of Philadelphia
chromosome and support cord blood stem cell expansion. Leuk Res.
30:1493–1498. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Flores-Figueroa E, Montesinos JJ,
Flores-Guzmán P, Gutiérrez-Espíndola G, Arana-Trejo RM,
Castillo-Medina S, Pérez-Cabrera A, Hernández-Estévez E, Arriaga L
and Mayani H: Functional analysis of myelodysplastic
syndromes-derived mesenchymal stem cells. Leuk Res. 32:1407–1416.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Aanei CM, Flandrin P, Eloae FZ, Carasevici
E, Guyotat D, Wattel E and Campos L: Intrinsic growth deficiencies
of mesenchymal stromal cells in myelodysplastic syndromes. Stem
Cells Dev. 21:1604–1615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chandran P, Le Y, Li Y, Sabloff M, Mehic
J, Rosu-Myles M and Allan DS: Mesenchymal stromal cells from
patients with acute myeloid leukemia have altered capacity to
expand differentiated hematopoietic progenitors. Leuk Res.
39:486–493. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Reikvam H, Brenner AK, Hagen KM, Liseth K,
Skrede S, Hatfield KJ and Bruserud Ø: The cytokine-mediated
crosstalk between primary human acute myeloid cells and mesenchymal
stem cells alters the local cytokine network and the global gene
expression profile of the mesenchymal cells. Stem Cell Res.
15:530–541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mohammadi S, Nikbakht M, Sajjadi SM, Rad
F, Chahardouli B, Sabour Takanlu J, Rostami Sh, Alimoghaddam K,
Ghavamzadeh A and Ghaffari SH: Reciprocal interactions of leukemic
cells with bone marrow stromal cells promote enrichment of leukemic
stem cell compartments in response to curcumin and daunorubicin.
Asian Pac J Cancer Prev. 18:831–840. 2017.PubMed/NCBI
|
|
60
|
Becker PS: Dependence of acute myeloid
leukemia on adhesion within the bone marrow microenvironment.
ScientificWorldJournal 2012. 8564672012.
|
|
61
|
Konopleva M, Konoplev S, Hu W, Zaritskey
AY, Afanasiev BV and Andreeff M: Stromal cells prevent apoptosis of
AML cells by up-regulation of anti-apoptotic proteins. Leukemia.
16:1713–1724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nwajei F and Konopleva M: The bone marrow
microenvironment as niche retreats for hematopoietic and leukemic
stem cells. Adv Hematol 2013. 9539822013.
|
|
63
|
Damiano JS, Hazlehurst LA and Dalton W:
Cell adhesion-mediated drug resistance (CAM-DR) protects the K562
chronic myelogenous leukemia cell line from apoptosis induced by
BCR/ABL inhibition, cytotoxic drugs, and gamma-irradiation.
Leukemia. 15:1232–1239. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Aguiar DP, Coelho-Aguiar JM and Abreu JG:
CCN2/CTGF silencing blocks cell aggregation in embryonal carcinoma
P19 cell. Braz J Med Biol Res. 44:200–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kumar A, Bhattacharyya J and Jaganathan
BG: Adhesion to stromal cells mediates imatinib resistance in
chronic myeloid leukemia through ERK and BMP signaling pathways.
Sci Rep. 7:95352017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kojima K, McQueen T, Chen Y, Jacamo R,
Konopleva M, Shinojima N, Shpall E, Huang X and Andreeff M: p53
activation of mesenchymal stromal cells partially abrogates
microenvironment-mediated resistance to FLT3 inhibition in AML
through HIF-1α-mediated down-regulation of CXCL12. Blood.
118:4431–4439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Naugler WE and Karin M: The wolf in
sheep's clothing: The role of interleukin-6 in immunity,
inflammation and cancer. Trends Mol Med. 14:109–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rose-John S: IL-6 trans-signaling via the
soluble IL-6 receptor: Importance for the pro-inflammatory
activities of IL-6. Int J Biol Sci. 8:1237–1247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fisher DT, Chen Q, Skitzki JJ, Muhitch JB,
Zhou L, Appenheimer MM, Vardam TD, Weis EL, Passanese J, Wang WC,
et al: IL-6 trans-signaling licenses mouse and human tumor
microvascular gateways for trafficking of cytotoxic T cells. J Clin
Invest. 121:3846–3859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Onishi K and Zandstra PW: LIF signaling in
stem cells and development. Development. 142:2230–2236. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Itoh F, Watabe T and Miyazono K: Roles of
TGF-β family signals in the fate determination of pluripotent stem
cells. Semin Cell Dev Biol. 32:98–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Conze D, Weiss L, Regen PS, Bhushan A,
Weaver D, Johnson P and Rincón M: Autocrine production of
interleukin 6 causes multidrug resistance in breast cancer cells.
Cancer Res. 61:8851–8858. 2001.PubMed/NCBI
|
|
73
|
Desbourdes L, Javary J, Charbonnier T,
Ishac N, Bourgeais J, Iltis A, Chomel JC, Turhan A, Guilloton F,
Tarte K, et al: Alteration analysis of bone marrow mesenchymal
stromal cells from de novo acute myeloid leukemia patients at
diagnosis. Stem Cells Dev. 26:709–722. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pontikoglou C, Kastrinaki MC, Klaus M,
Kalpadakis C, Katonis P, Alpantaki K, Pangalis GA and Papadaki HA:
Study of the quantitative, functional, cytogenetic, and
immunoregulatory properties of bone marrow mesenchymal stem cells
in patients with B-cell chronic lymphocytic leukemia. Stem Cells
Dev. 22:1329–1341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ogden A, Rida PC, Knudsen BS, Kucuk O and
Aneja R: Docetaxel-induced polyploidization may underlie
chemoresistance and disease relapse. Cancer Lett. 367:89–92. 2015.
View Article : Google Scholar : PubMed/NCBI
|