Introduction
Prostate cancer is the most common malignancy of the
reproductive system in male patients, and is one of the leading
causes of cancer-related mortalities in men worldwide. Prostate
function is dependent on androgen (1). When the androgen receptor (AR) binds its
ligands, including dihydrotestosterone, AR translocates into the
nucleus, binds to androgen-responsive elements and regulates the
expression of its downstream genes by interacting with various
transcription factors. The AR signaling pathway is essential for
the development, function and homeostasis of the prostate (2). However, the constitutive transactivation
of the AR signaling pathway may result in prostate cancer
initiation and progression. Although localized prostate cancer is
treated with surgery or radiation (3), cancers at advanced and metastatic stages
are typically treated using androgen-deprivation therapy (ADT)
(4). However, the majority of
patients may develop castration-resistant prostate cancer (CRPC)
after months, or even years of ADT. At present, the progression of
CRPC is believed to be caused by constitutive transactivation of
the AR signaling pathway that may be dependent on low-levels of
androgen or androgen-independent. Various activation mechanisms
depend on AR activity and function, including AR amplification, AR
mutations, AR splice variants and the abnormal activation of the AR
(5). Therefore, numerous ADT agents
have been identified over the years. However, AR activation
mechanisms constantly evolve to antagonize these agents and may
vary markedly among individuals. Therefore, in order to investigate
novel therapeutic targets in addition to ADT, the mechanism
underlying the AR signaling pathway requires further
examination.
Protein fatty acid acylation is a newly identified
co-translational or post-translational protein modification
mechanism. This covalent attachment of lipids onto proteins can
control protein-protein and protein-membrane interactions, thus
serving important roles in human physiology and pathology. Protein
palmitoylation is one form of fatty acid acylation and involves the
covalent attachment of a 16-carbon fatty acid moiety, palmitate,
onto amino acid residues (6). There
are three types of palmitoylation, the occurrence of which depend
on the type of bond formed between palmitate and amino acid
residues: i) S-palmitoylation, presenting thioester bonds with
cysteines; ii) O-palmitoylation, exhibiting ester bonds with
serines or threonines; and iii) N-palmitoylation, forming amide
bonds with lysines (7). There is a
dynamic cycle between palmitoylated and non-palmitoylated protein
forms. The regulation of this cycle depends on acyltransferases. In
mammalian cells, S-palmitoylation, which is found in N-Ras and
H-Ras, is catalyzed by a family of palmitoyltransferases with DHHC,
or Asp-His-His-Cys, motifs, and is removed by fatty acyl protein
thioesterases (8). By contrast,
O-palmitoylation and N-palmitoylation are found on secretory
proteins, including Wnt and Hedgehog, respectively (9). These palmitoylations are catalyzed by
membrane-bound O-acyltransferases (10).
Palmitoylation promotes protein hydrophobicity,
increasing membrane affinity, and alters the proteins subcellular
localization, thus regulating protein trafficking, stability and
activity (11). Accumulating evidence
has indicated that many palmitoylated proteins are associated with
tumorigenesis and metastasis in prostate cancer. For example,
Src-family tyrosine kinases (SFKs) are involved in signaling
pathways that regulate cell proliferation, survival, motility and
adhesion (12). The efficient
activity of SFKs, including FYN proto-oncogene, Src family tyrosine
kinase, LYN proto-oncogene, Src family tyrosine kinase, YES
proto-oncogene 1, Src family tyrosine kinase and HCK
proto-oncogene, Src family tyrosine kinase, require association
with the plasma membrane, and this association is mediated by
palmitoylation (13). Neurotensin
receptor 1 is a G-protein-coupled receptor that mediates cancer
progression in prostate cancer. Palmitoylation is essential for the
localization of certain proteins in specific membrane microdomains
and for the activation of their downstream signaling pathways
(14). Considering the key roles of
palmitoylated proteins in tumorigenesis and the limitations of the
currently available treatments for CRPC, the present study aimed to
identify the androgen-induced palmitoylated proteins and examine
potential novel regulatory mechanisms of the AR signaling pathway
in order to develop novel therapies for prostate cancer.
Click-chemistry-based chemical probes for the
detection of protein palmitoylation may allow the rapid discovery
of novel palmitoylated proteins and the understanding of their
biological functions (15). The
combination of mass spectrometry (MS)-based quantitative proteomics
with click-chemistry-probes can be used to examine the dynamics of
protein palmitoylation under different physiological or
pathological conditions (16,17). In the present study, chemical tools
were used, and palmitoylated proteins between androgen-treated
LNCaP cells and untreated LNCaP cells were investigated using
palmitoylome profiles. The present study aimed to identify
candidate proteins exhibiting palmitoylation levels that could be
promoted by androgen treatment. The regulation of the
palmitoylation of candidate proteins may provide new directions for
the development of novel therapies to treat prostate cancer.
Moreover, the palmitoylation of specific proteins induced by
androgen treatment may represent a novel biomarker for prostate
cancer.
Materials and methods
Cell culture
LNCaP and PC3 cells were purchased from ATCC and
cultured in RPMI-1640 medium supplemented with 10% FBS. RWPE-1
cells were provided by The Stem Cell Bank, Chinese Academy of
Sciences, and were cultured in Keratinocyte-SFM media. All cells
were incubated in a humidified incubator at 37°C with 5%
CO2 for 48 h prior to further experimentation.
Antibodies and reagents
The antibodies used in the present study were as
follows: Rabbit anti-α-tubulin (dilution 1:2,000; cat. no.
11224-1-AP), rabbit anti-Rab7a (dilution 1:2,000; cat. no.
55469-1-AP), mouse anti-GAPDH (dilution 1:2,000; cat. no.
60004-1-Ig; all from ProteinTech Group, Inc.). The reagents used in
the present study were as follows: Protein (A/G) UltraLink Resin
(Thermo Fisher Scientific, Inc.), azide agarose beads (Nanocs),
Thiopropyl sepharose 6B (Sigma-Aldrich; Merck KGaA), hydroxylamine
(Sigma-Aldrich; Merck KGaA), N-Ethylmaleimide (NEM; Sigma-Aldrich;
Merck KGaA), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP;
Sigma-Aldrich; Merck KGaA),
Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA;
Sigma-Aldrich; Merck KGaA), R1881 (Shanghai Yuanye Bio-Technology
Co., Ltd.), and 2-Brp (Sigma-Aldrich; Merck KGaA).
Protein S-palmitoylation assay in
cells or supernatants
Before the S-palmitoylation assay, the cells were
treated with R1881 or 2-Brp. Prior to R1881 treatment, cells were
seeded with complete medium onto 10-cm-dishes (4×106
cells/dish) and incubated for 24 h. Then, the LNCaP or PC3 cells
were treated with R1881 (5 nM) or DMSO in RPMI-1640 medium
supplemented with 0.2% FBS. The RWPE-1 cells were treated with
R1881 (5 nM) or DMSO in Keratinocyte-SFM media. After 24 h, the
cells or supernatants were harvested for the S-palmitoylation
assay. Prior to 2-Brp treatment, LNCaP cells were seeded with
complete media onto 10-cm-dishes (4×106 cells/dish) and
incubated for 40 h. The cells were then treated with 2-Brp (25 µM)
or DMSO in RPMI-1640 medium supplemented with 0.2% FBS. After 4–6
h, the cells were harvested for S-palmitoylation assessment.
The S-palmitoylation assay was performed as
previously described (18) with some
minor modifications. In brief, the cells were homogenized in lysis
buffer [10 mM sodium phosphate, 2 mM Na2-EDTA, 0.32 M
sucrose, 1% Triton X-100, 50 mM N-ethylmaleimide and Pierce
protease and phosphatase inhibitor cocktail (Pierce; Thermo Fisher
Scientific, Inc.); pH 7.4] for 30 min. Then, the lysates were
immunoprecipitated overnight using protein A/G resin preloaded with
α-tubulin antibody. The supernatants were harvested and directly
incubated with 50 mM N-ethylmaleimide and Pierce protease and
phosphatase inhibitor cocktail. Then, the supernatants were
immunoprecipitated overnight using protein A/G resin preloaded with
α-tubulin antibody.
After overnight incubation, the protein A/G resin
was washed three times and incubated with elution buffer (1% SDS,
10 mM sodium phosphate, 2 mM Na2-EDTA, 0.32 M sucrose)
at 50°C for 5 min to release α-tubulin. Eluted samples were divided
into two equal portions: i) One treated with 1 M hydroxylamine and
thiopropyl sepharose 6B; and ii) the other, used as the control,
with 1 M Tris·HCl (pH 7.4) and thiopropyl sepharose 6B
(Sigma-Aldrich; Merck KGaA). After a 2-h incubation at room
temperature, sepharose beads were washed three times with washing
buffer (10 mM sodium phosphate, 2 mM Na2-EDTA, 0.32 M
sucrose, 1% Triton X-100, 500 mM NaCl and 0.2% SDS). Western blots
analysis was performed to determine the presence of α-tubulin.
Synthesis of palmitate probe,
metabolic labeling and click chemistry
In the present study, an alkyne analogue was
selected due to its physicochemical properties, similar to the
wild-type fatty acid carbon chain and presented a degree of
hydrophobicity that allowed a high affinity for the cell membrane
(19). The alkyne group was added at
the ω-position of the fatty acid. The ω-alkynyl palmitate analogue
(Alk-C16) was synthesized from alcohols with internal alkynes via a
zipper reaction. Subsequently, the internal alkyne was isomerized
to a terminal alkyne via Jones oxidation (19).
Before the experiments, the LNCaP cells were seeded
with complete media onto 6-cm-dishes (5×105 cells/dish)
and incubated for 48 h. The metabolic labeling and click chemistry
were performed as previously described (19) with minor modifications. The ω-alkynyl
fatty acid analogue, Alk-C16, was dissolved in DMSO to generate a
50-mM stock solution and stored at −80°C. Before cell treatment,
Alk-C16 was dissolved in RPMI-1640 serum-free media supplemented
with 5% BSA (fatty-acid free) at a final concentration of 100 µM.
Alk-C16 was added to RPMI-1640 medium and sonicated for 15 min at
room temperature. In addition, an equal volume of DMSO was added to
RPMI-1640 medium as a negative control. Then, the solution
containing RPMI-1640 medium and Alk-C16 or DMSO was divided into
two parts: i) One was supplemented with R1881 (5 nM); and ii) the
other with DMSO. The seeded LNCaP cells were washed once with PBS
and treated with the four media for 24 h at 37°C with 5%
CO2. After a 24-h incubation, the cells were washed
three times and homogenized in 500 µl lysis buffer (1% Nonidet
P-40, 150 mM NaCl, Pierce protease and phosphatase inhibitor
cocktail and 100 mM sodium phosphate; pH 7.5) for 30 min at 4°C.
Then, the protein extracts were subjected to a probe labeling
reaction for 1 h at room temperature using the following reagents:
1 mM azide agarose beads, 1 mM TCEP dissolved in water, 0.2 mM TBTA
dissolved in DMSO/tert-butanol (20/80% v/v) and 1 mM
CuSO4 in PBS. The order of addition of the reagents to
the protein extracts was critical for the reaction. Following the
click-chemistry reaction, the Alk-C16-conjugated proteins were
bound to the azide agarose beads. The agarose beads were washed
three times with lysis buffer at room temperature. Then, the
proteins bound to the azide agarose beads were digested for MS
analysis.
nanoLC-MS/MS analysis
Proteomic profiling was performed on an Easy nLC1000
(Thermo Fisher Scientific, Inc.) system combined with the
LTQ-Orbitrap-Elite (Thermo Fisher Scientific, Inc.) mass
spectrometer as previously described (20).
Database searching
The recorded MS spectra were analyzed using MaxQuant
Software (version 1.5.5.1; http://www.Maxquant.org/). The MS/MS peak list
analysis was performed by searching against a forward and reverse
version of the UniProtKB/Swiss-Prot human database (generated from
version 2017_05; human taxonomy; 20,316 entries; http://www.uniprot.org/). The cutoff of the false
discovery rate for peptide and protein identification was set to
0.01, and only peptides with ≥7 amino acidic residues were
analyzed. Label-free quantitation (LFQ) was performed using the
MaxQuant software on the identified razor and unique peptides in
order to quantify the identified proteins (20). Gene Ontology (GO) analysis was
performed using the database for annotation, visualization and
integrated discovery (DAVID) software (National Institutes of
Health) to assign the identified proteins with the corresponding GO
terms within the category ‘cellular function’.
Data processing and statistical
analysis
Protein abundances normalized by the LFQ algorithm
integrated in MaxQuant were log2-transformed for further
analyses. The filtering steps were performed using Microsoft Excel
2010. DanteR (version 1.0.1.1) and Perseus (version 1.5.5.3) were
used to perform various types of statistical analyses including
log2 transformation, correlation plot, statistical tests
and P-value adjustments (20).
Fractionation assays
Before the experiments, the cells were seeded with
complete media onto 10-cm-dishes (4×106 cells/dish) and
incubated for 24 h. Then, the cells were treated with R1881 (5 nM)
or DMSO in RPMI-1640 medium supplemented with 0.2% FBS. After 24 h,
the cells were harvested for fractionation. An Invent
Biotechnologies Minute™ Plasma Membrane Protein Isolation kit
(Invent Biotechnologies, Inc.) was used for total membrane
fractionation according to the manufacturer's protocol. Western
blot analysis was performed to analyze the protein level of
α-tubulin.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using a CCK-8 assay
according to the manufacturer's protocol [MedchemExpress, (MCE)].
LNCaP cells (5×103) were seeded onto 96-well plates in
RPMI-1640 medium supplemented with 0.2% FBS. After 24 h, which was
selected as day 0, the cells were treated with R881 (5 nM), 2-Brp
(5 µM) or DMSO. The changes in cell proliferation were analyzed for
3 consecutive days. At day 2, the cells were supplemented with 0.2%
FBS. The cells were incubated with CCK-8 solution (10 µl/well) at
37°C with 5% CO2 for 1.5 h. The optical density value of
each well was detected using a microplate reader, and the
absorbance was measured at 450 nm.
Statistical analysis
Statistical analyses were performed using SPSS
Statistics (version 21.0; IBM Corp.). The numerical data of each
group are presented as the mean ± standard deviation. For the
proteins whose palmitoylation levels was increased following
androgen treatment, the significant differences in their LFO values
between androgen-treated vs. untreated LNCaP cells were analyzed
using a paired Student's t-test. For the cell proliferation
examined by CCK-8 assay, the significant differences in cell
viability between untreated vs. R1881 treated or 2-Brp treated
LNCaP cells were analyzed using a paired Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of androgen-induced
palmitoylome using Alk-C16
In order to compare the palmitoylated protein
profiles between androgen-treated and untreated LNCaP cells,
Alk-C16, a palmitate fatty acid analogue, was metabolically
incorporated onto the cellular proteins. Then, the Alk-C16
incorporated onto proteins was chemoselectively ligated to azide
agarose beads via a Cu1-catalyzed alkyne-azide [3+2]
cycloaddition reaction, a type of click chemistry reaction. The
Alk-C16-conjugated proteins were then ligated to the azide agarose
beads. In theory, only palmitoylated proteins were conjugated to
Alk-C16 and bound to the azide agarose beads. Subsequently, the
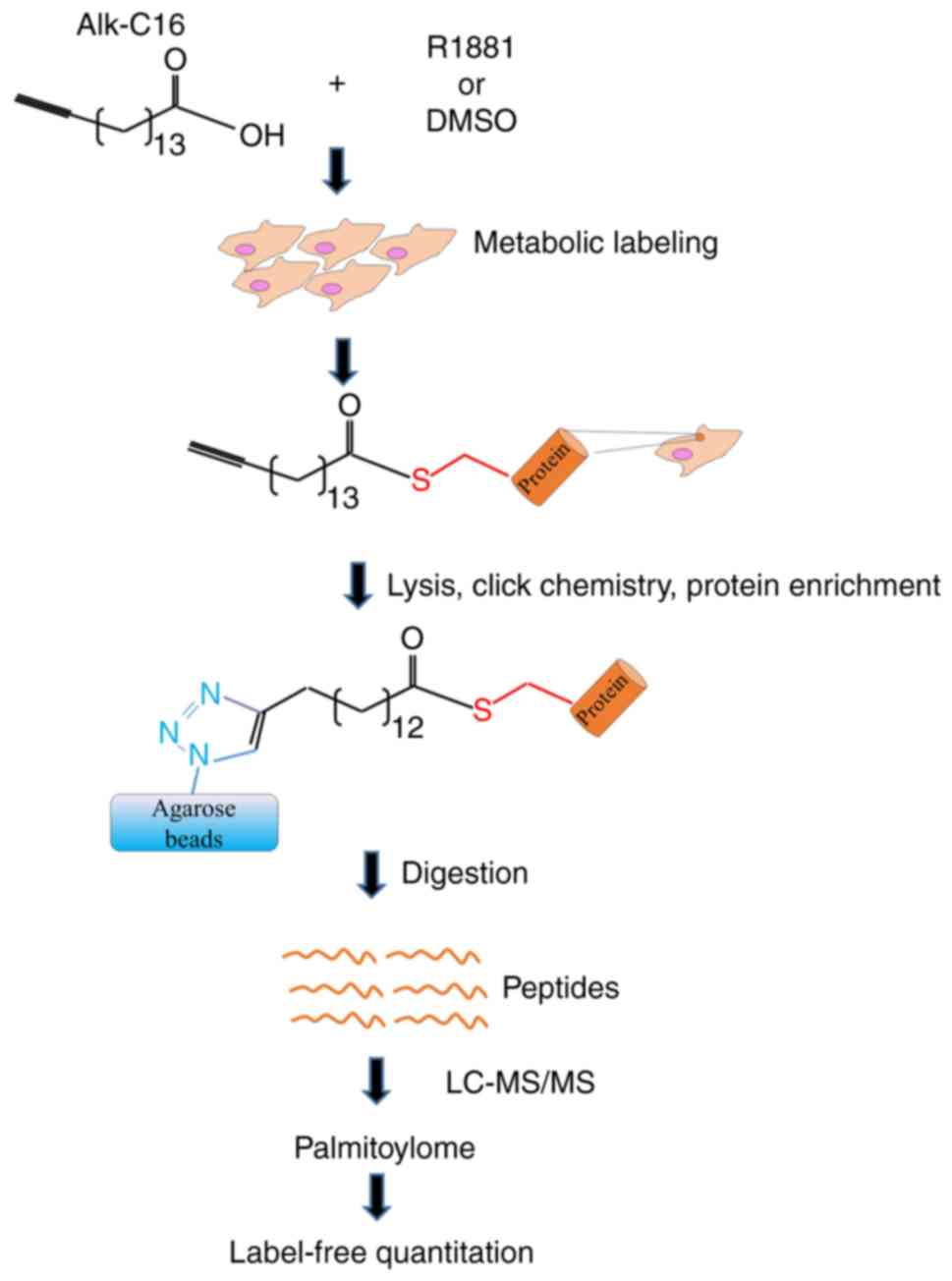
proteins were digested on the agarose beads and subjected to MS and
to an informatics-assisted label-free quantitation (Fig. 1).
To identify the proteins exhibiting increased levels
of palmitoylation following androgen treatment, three groups of
androgen-treated and untreated LNCaP cells were investigated. In
total, 927 proteins exhibiting a Mascot score >2 were identified
(P<0.05). The outlier proteins and contaminant proteins were
removed. The 927 proteins were subjected to LFQ using MaxQuant
software. In these 927 proteins, the LFQ values were positively
correlated with the palmitoylation levels. Among these 927
proteins, 504 proteins were identified to be palmitoylated
preliminarily, and their LFQ values were >0 in ≥2 replicates of
untreated LNCaP cells. Among these 504 proteins, there were a
number of well-known palmitoylated proteins, including α- and
β-tubulin, fatty acid synthase, catenin ∆1 and ubiquinol-cytochrome
c reductase core protein 1 (21,22). Among
the 504 proteins, 96 candidates exhibited palmitoylation levels
that were increased following androgen treatment, and their LFQ
values were significantly upregulated (fold change >1.5;
P<0.05) in ≥2 samples of androgen-treated vs. untreated LNCaP
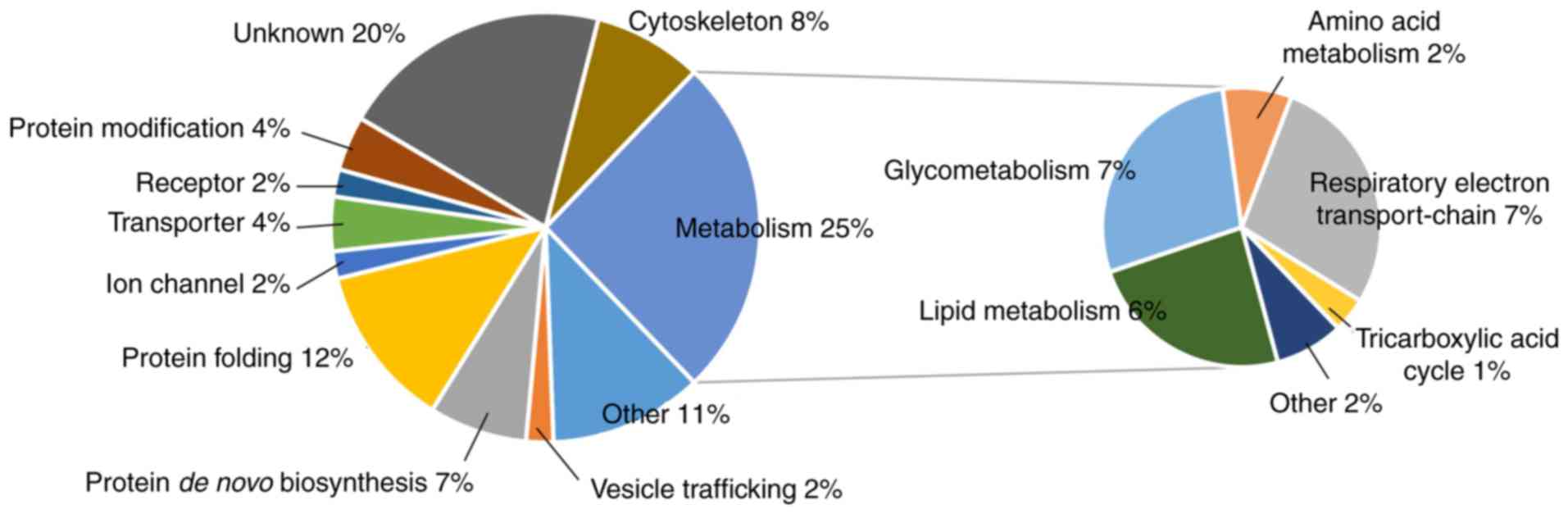
cells (Table SI). GO analysis
revealed that the functions of the 96 proteins were categorized as:
25% ‘metabolism’, 12% ‘protein folding’, 8% ‘cytoskeleton’, 7%
‘protein de novo biosynthesis’, 4% ‘protein modification’
and 2% ‘vesicle trafficking’, among others (Fig. 2). The proteins involved in
‘metabolism’ were included in the following categories: 7%
‘glycometabolism’, 7% ‘respiratory electron transport-chain’, 6%
‘lipid metabolism’, 2% amino acid metabolism’ and 1% ‘tricarboxylic
acid cycle’, among others.
Androgen treatment promotes the
palmitoylation level of α-tubulin
Among the 96 proteins exhibiting increased
palmitoylation following androgen treatment, 7% were
cytoskeleton-related proteins including tubulin α 4A (TUBA4A),
TUBA1B, tubulin β 2C, tubulin β chain, myosin heavy chain 9,
profilin-1, actinin α 4 and filamin-B (Fig. 3A-H). Notably, the palmitoylation
levels of both α-tubulin and β-tubulin were significantly
(>1.5-fold) higher in androgen-treated LNCaP cells compared with
untreated LNCaP cells (Fig. 3A-D).
The protein with the highest LFQ value was TUBA1B in both
androgen-treated and untreated LNCaP cells.
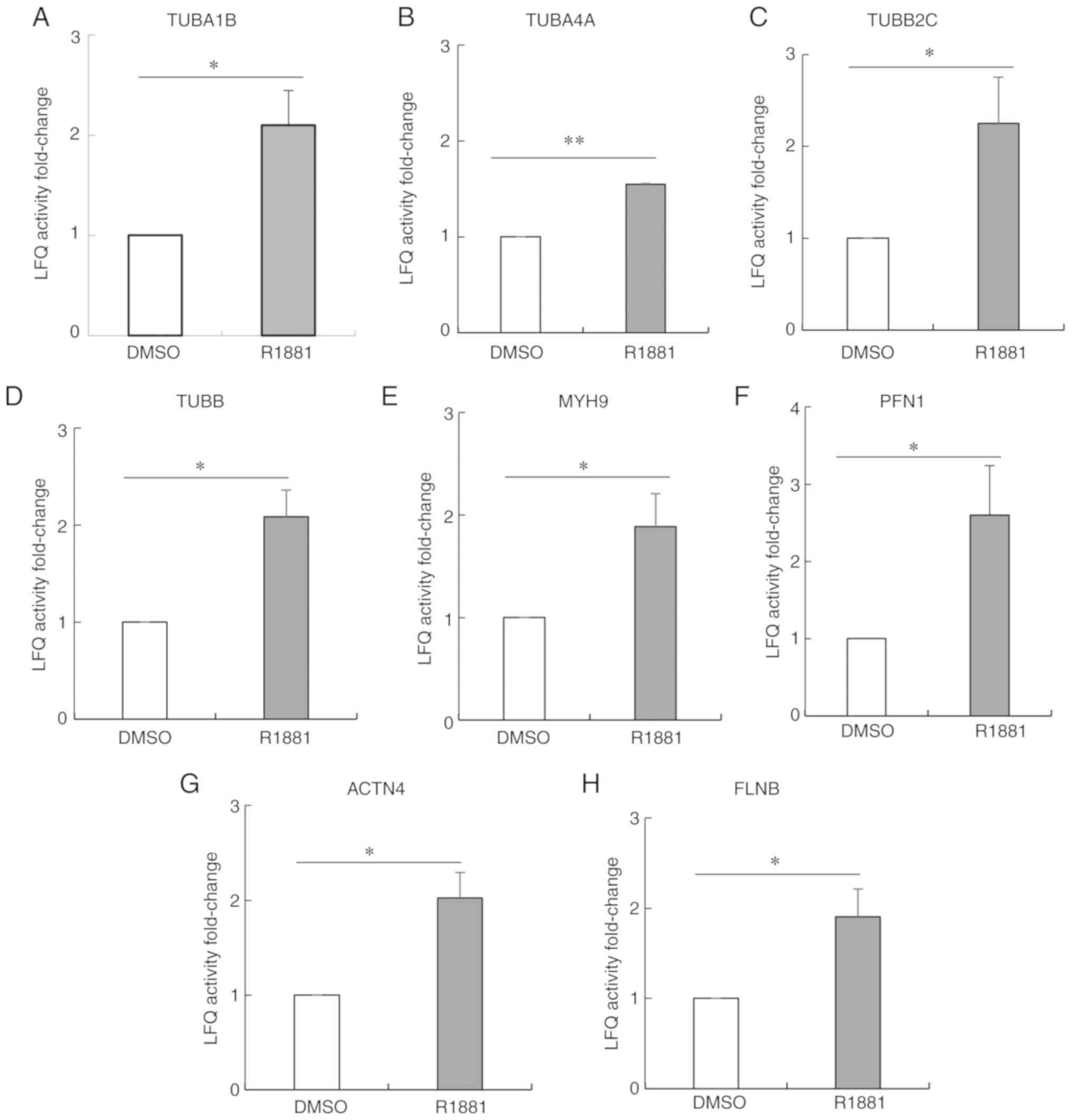 | Figure 3.Androgen-enhanced palmitoylation of
cytoskeleton-related proteins in LNCaP cells identified following
palmitoylome analysis. The palmitoylation levels of
cytoskeleton-related proteins (A-H) were significantly
(>1.5-fold) higher in androgen-treated LNCaP cells compared with
untreated cells. (A) TUBA1B, (B) TUBA4A, (C) TUBB2C, (D) TUBB, (E)
MYH9, (F) PFN1, (G) ACTN4, (H) FLNB. Data are presented as the mean
± SD from three independent experiments. Data were analyzed using a
paired Student's t-test. *P<0.05, **P<0.01. TUBA1B, tubulin α
1b; TUBA4A, tubulin α 4a; TUBB2C, tubulin β 4B class IVb; TUBB,
tubulin β class I; MYH9, myosin heavy chain 9; PFN1, profilin-1;
ACTN4, actinin α 4; FLNB, filamin B. |
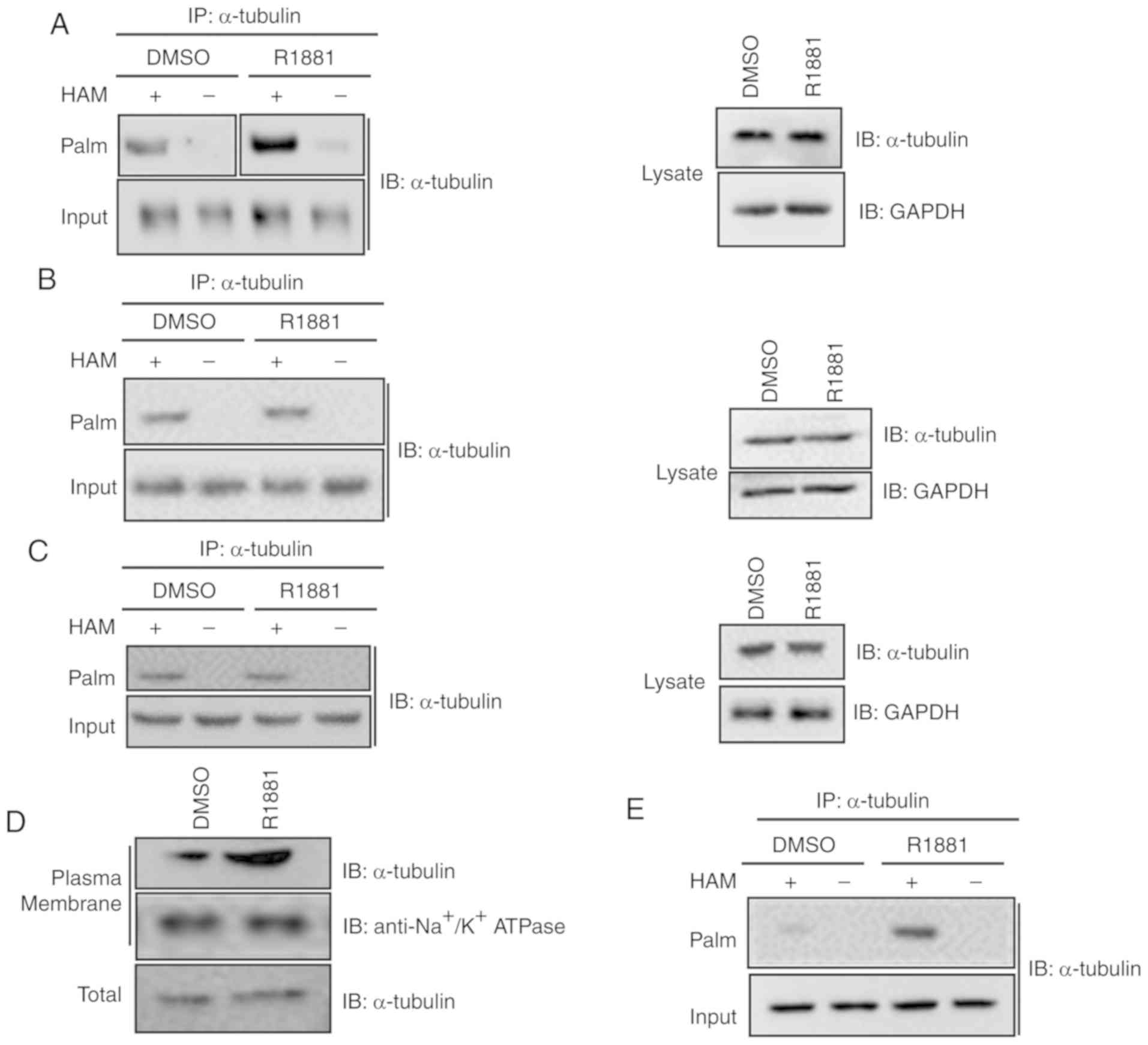
α-tubulin is a well-known S-palmitoylated protein
(22). To further investigate the
role of androgen treatment on α-tubulin palmitoylation in LNCaP
cells, a thiopropyl protein captivation assay was performed. As
revealed in Fig. 4A, α-tubulin
proteins were observed to be associated with the thiopropyl beads
following treatment with hydroxylamine, but not following treatment
with Tris·HCl, used as the control. Following treatment with R1881,
an androgen, the palmitoylation level of α-tubulin was
significantly upregulated. The present results were consistent with
the aforementioned MS results. Collectively, the present results
indicated that androgen promoted the palmitoylation level of
α-tubulin in LNCaP cells.
In addition to the results observed in LNCaP cells,
thiopropyl captivation assay of S-palmitoylated proteins was
performed in other cell types in order to investigate whether
androgen promoted the levels of tubulin palmitoylation in RWPE-1
cells, normal prostate epithelial cells and in PC3 cells, which
exhibit a decreased expression level of AR (23). As revealed in Fig. 4B and C, there were no significant
differences in the palmitoylation levels of α-tubulin between
RWPE-1 or PC3 cells treated with R1881 or DMSO.
Since androgen significantly increased the
palmitoylation level of α-tubulin, the effect of androgen on the
function of α-tubulin was investigated. Considering that
palmitoylation is essential for membrane association, plasma
membrane fractions were separated from androgen-treated and
untreated cells. The protein expression levels of plasma
membrane-associated α-tubulin were compared between the two
samples. As revealed in Fig. 4D, the
protein expression level of plasma membrane-associated α-tubulin in
androgen-treated cells was higher compared with untreated cells.
The present result indicated that androgen treatment enhanced the
plasma membrane association of α-tubulin, in line with the
aforementioned results indicating that androgen treatment increased
the palmitoylation levels of α-tubulin (Fig. 4A).
In addition, thiopropyl captivation of
S-palmitoylated proteins was performed to examine whether androgen
promoted the level of α-tubulin palmitoylation in the supernatants
of LNCaP cells. As revealed in Fig.
4E, the palmitoylation level of α-tubulin was significantly
higher in R1881-treated cells compared with untreated cells, in
line with the aforementioned results.
In summary, androgen treatment promoted the
palmitoylation level of α-tubulin in LNCaP cells, an AR-dependent
prostate cancer deprived-cell line.
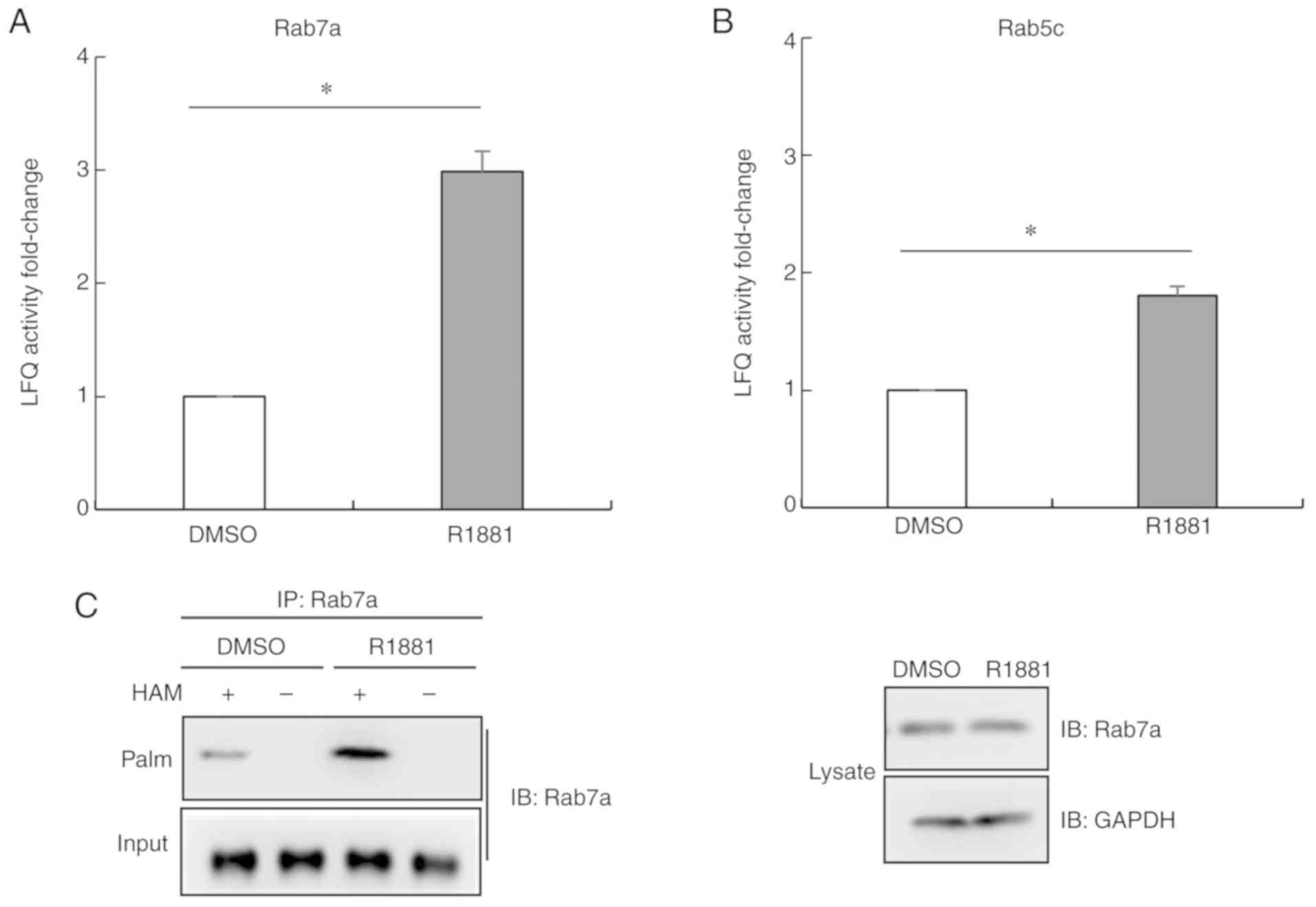
Androgen treatment promotes the
palmitoylation level of Ras-related protein Rab-7a (Rab7a)
Among the 96 proteins exhibiting increased
palmitoylation levels following androgen treatment, there were two
members of the Rab protein family, Rab7a and Rab5c (Fig. 5A and B). Their LFQ values were lower
compared with α-tubulin. In order to further examine whether
androgen promoted the palmitoylation level of the two members of
Rab family in LNCaP cells, thiopropyl captivation of
S-palmitoylated proteins was performed. As revealed in Fig. 5C, Rab7a proteins were associated with
thiopropyl beads following treatment with hydroxylamine, but not
following treatment with Tris·HCl, used as the control. Following
treatment with the androgen R1881, the palmitoylation level of
Rab7a was significantly upregulated. The present results were
consistent with the aforementioned MS results (Fig. 5A). Collectively, the present results
indicated that androgen treatment promoted the palmitoylation level
of Rab7a in LNCaP cells.
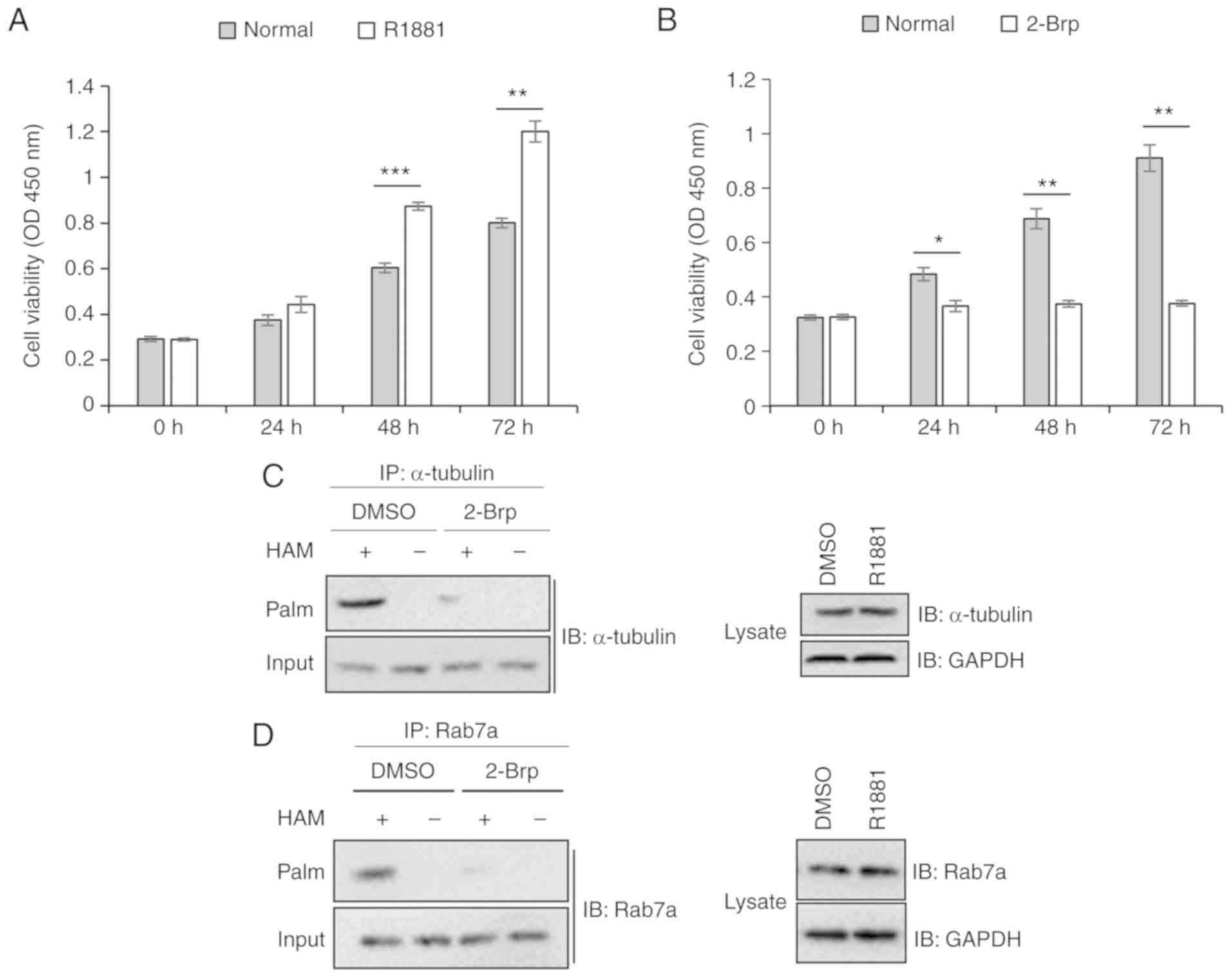
The proliferation of LNCaP cells is
promoted by androgen treatment and inhibited by palmitoylation
inhibitor
To investigate the effect of androgen-induced
palmitoylation of α-tubulin and Rab7a on the proliferation of LNCaP
cells, a CCK-8 assay was performed. As revealed in Fig. 6A, R1881 treatment induced the
proliferation of LNCaP cells. By contrast, following treatment with
2-Brp, a palmitoylation inhibitor, cell proliferation was inhibited
(Fig. 6B). S-palmitoylated assay
results indicated that 2-Brp treatment reduced the palmitoylation
levels of α-tubulin and Rab7a compared with untreated cells
(Fig. 6C and D). The present results
indicated that palmitoylation of α-tubulin and Rab7a were critical
for LNCaP cell proliferation.
Discussion
The clickable palmitate probe Alk-C16 can be used to
identify novel palmitoylated proteins, examine the subcellular
distribution of palmitoylated proteins and investigate fatty acid
substrates. The use of Alk-C16 presents certain advantages,
including the potential to be used in cultured cells to detect
proteins that have been palmitoylated during metabolic labeling in
different conditions, such as gene overexpression, gene knockdown
and drug treatment (16,17). Notably, Alk-C16 can be combined with
quantitative proteomics to quantify the levels of palmitoylated
proteins.
The present study aimed to identify the
androgen-induced palmitoylated proteins by comparing the
palmitoylome profile of androgen-treated LNCaP cells with untreated
cells. The present results indicated that androgen promoted the
palmitoylation of α-tubulin in LNCaP cells (Figs. 3A, B and 4A). In previous studies, α-tubulin was
revealed to be palmitoylated, and this modification is essential to
anchor microtubule filaments to the plasma membrane, influencing
cellular transport and cell division (24–26). In
addition, in the present study, androgen treatment was revealed to
increase the palmitoylation level of Rab7a, a member of the Rab
family (Fig. 5A and C). Rab7a
localizes in late endosomes and regulates the trafficking from
early endosomes to late endosomes and from late endosomes to
lysosomes (27,28). Rab7a is essential for lysosomal
biogenesis, localization and function, and lysosomes are involved
in vesicular trafficking and in the degradation of signaling
receptors (29). Rab7a palmitoylation
is required for the spatiotemporal recruitment of the retromer
complex and for an efficient endosome to trans-Golgi network
trafficking of the lysosomal sorting receptor; however, Rab7a
palmitoylation is not involved in the membrane anchoring (30). Rab7a controls vesicle trafficking by
cooperating with α-tubulin (31).
Therefore, the palmitoylation of α-tubulin and Rab7a is critical
for the vesicle trafficking from the plasma membrane to the
lysosomes, which are involved in the degradation of some signaling
receptors. Functional analysis indicated that androgen promoted the
palmitoylation of α-tubulin and Rab7a in LNCaP cells, inducing cell
proliferation (Fig. 6A). By contrast,
2-Brp, which reduced the palmitoylation of α-tubulin and Rab7a
(Fig. 6C and D), inhibited cell
proliferation (Fig. 6B).
Collectively, the present results indicated that androgen-induced
palmitoylation of α-tubulin and Rab7a promoted cell proliferation
by degrading inhibitors of cell proliferation. Therefore,
palmitoylation of α-tubulin and Rab7a may represent novel potential
therapeutic targets for treating prostate cancer. Notably, in the
supernatants of LNCaP cells, the palmitoylation level of α-tubulin
was significantly higher in the R1881-treated cells compared with
untreated cells (Fig. 4E). Therefore,
further studies are required to identify whether the palmitoylation
level of α-tubulin in the serum of patients with prostate cancer
may represent a potential novel biomarker for early-stage prostate
cancer.
The present results indicated that 25% of the
proteins exhibiting an increase in palmitoylation following
androgen treatment were involved in metabolism. However, it is
challenging to examine the regulatory mechanism underlying the
palmitoylation of all these proteins, which are involved in various
metabolic processes. In LNCaP cells, androgen treatment, which
promoted the palmitoylation of α-tubulin and Rab7a, induced cell
proliferation, whereas 2-Brp, which reduced the palmitoylation of
α-tubulin and Rab7a, inhibited cell proliferation. Palmitoylation
of α-tubulin and Rab7a were identified to be involved in cell
proliferation and may represent new targets for developing novel
treatments against prostate cancer. In addition, a high level of
α-tubulin palmitoylation may represent a novel biomarker for
early-stage prostate cancer.
Supplementary Material
Supporting Data
Acknowledgements
The proteomic experiments were performed using the
mass spectrometry facility of The State Key Laboratory of Membrane
Biology, Institute of Zoology, Chinese Academy of Sciences.
Funding
The proteomic experiments were supported by the
National Key Research and Development Program of China (grant no.
2016YFA0201500&2017YFC840102) and the National Natural Science
Foundation of China (grant no. 81571384). The palmitoylation assay,
fractionation assay and paper publication were supported by the Key
Research Program for Health Care in China (grant no. W2016ZD01) and
Capital Clinical Characteristics Applications Research Program
(grant no. Z171100001017201).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WL and GZ designed the study. WL, JZ, LZ, JC, FS and
JJ performed experiments. WL, FX, ML and GZ analyzed the data. WL,
FX, ML and GZ wrote the study. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lonergan PE and Tindall DJ: Androgen
receptor signaling in prostate cancer development and progression.
J Carcinog. 10:202011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Klein EA, Ciezki J, Kupelian PA and
Mahadevan A: Outcomes for intermediate risk prostate cancer: Are
there advantages for surgery, external radiation, or brachytherapy?
Urol Oncol. 27:67–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Loblaw DA, Virgo KS, Nam R, Somerfield MR,
Ben-Josef E, Mendelson DS, Middleton R, Sharp SA, Smith TJ, Talcott
J, et al: Initial hormonal management of androgen-sensitive
metastatic, recurrent, or progressive prostate cancer: 2006 update
of an American Society of Clinical Oncology Practice Guideline. J
clin Oncol. 25:1596–1605. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tilki D, Schaeffer EM and Evans CP:
Understanding mechanisms of resistance in metastatic
castration-resistant prostate cancer: The role of the androgen
receptor. Eur Urol Focus. 2:499–505. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aicart-Ramos C, Valero RA and
Rodriguez-Crespo I: Protein palmitoylation and subcellular
trafficking. Biochim Biophys Acta. 1808:2981–2994. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thinon E and Hang HC: Chemical reporters
for exploring protein acylation. Biochem Soc Trans. 43:253–261.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitchell DA, Vasudevan A, Linder ME and
Deschenes RJ: Protein palmitoylation by a family of DHHC protein
S-acyltransferases. J Lipid Res. 47:1118–1127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao X and Hannoush RN: Single-cell imaging
of Wnt palmitoylation by the acyltransferase porcupine. Nat Chem
Biol. 10:61–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pepinsky RB, Zeng C, Wen D, Rayhorn P,
Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski
K, et al: Identification of a palmitic acid-modified form of human
sonic hedgehog. J Biol Chem. 273:14037–14045. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hofmann K: A superfamily of membrane-bound
O-acyltransferases with implications for wnt signaling. Trends
Biochem Sci. 25:111–112. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smotrys JE and Linder ME: Palmitoylation
of intracellular signaling proteins: Regulation and function. Ann
Rev Biochem. 73:559–587. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Resh MD: Targeting protein lipidation in
disease. Trends Mol Med. 18:206–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Heakal Y, Woll MP, Fox T, Seaton K,
Levenson R and Kester M: Neurotensin receptor-1 inducible
palmitoylation is required for efficient receptor-mediated
mitogenic-signaling within structured membrane microdomains. Cancer
Biol Ther. 12:427–435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hannoush RN and Sun J: The chemical
toolbox for monitoring protein fatty acylation and prenylation. Nat
Chem Biol. 6:498–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hernandez JL, Davda D, Majmudar JD, Won
SJ, Prakash A, Choi AI and Martin BR: Correlated S-palmitoylation
profiling of snail-induced epithelial to mesenchymal transition.
Mol Biosyst. 12:1799–17808. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Greaves J, Munro KR, Davidson SC, Riviere
M, Wojno J, Smith TK, Tomkinson NC and Chamberlain LH: Molecular
basis of fatty acid selectivity in the zDHHC family of
S-acyltransferases revealed by click chemistry. Proc Natl Acad Sci
U S A. 114:E1365–E1374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li W, Li W, Zou L, Ji S, Li C, Liu K,
Zhang G, Sun Q, Xiao F and Chen D: Membrane targeting of inhibitory
Smads through palmitoylation controls TG-β/BMP signaling. Proc Natl
Acad Sci USA. 114:13206–132011. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hannoush RN and Arenas-Ramirez N: Imaging
the lipidome: Omega-alkynyl fatty acids for detection and cellular
visualization of lipid-modified proteins. ACS Chem Biol. 4:581–587.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu NQ, Braakman RB, Stingl C, Luider TM,
Martens JW, Foekens JA and Umar A: Proteomics pipeline for
biomarker discovery of laser capture microdissected breast cancer
tissue. J Mammary Gland Biol Neoplasia. 17:155–164. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen LF, Chen YJ, Liu KM, Haddad ANS, Song
IW, Roan HY, Chen LY, Yen JJY, Chen YJ, Wu JY and Chen YT: Role of
S-palmitoylation by ZDHHC13 in mitochondrial function and
metabolism in liver. Sci Rep. 7:21822017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Caron JM: Posttranslational modification
of tubulin by palmitoylation: I. In vivo and cell-free studies. Mol
Biol Cell. 8:621–636. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Niu Y, Altuwaijri S, Lai KP, Wu CT, Ricke
WA, Messing EM, Yao J, Yeh S and Chang C: Androgen receptor is a
tumor suppressor and proliferator in prostate cancer. Proc Natl
Acad Sci USA. 105:12182–12187. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caron JM, Vega LR, Fleming J, Bishop R and
Solomon F: Single site alpha-tubulin mutation affects astral
microtubules and nuclear positioning during anaphase in
Saccharomyces cerevisiae: Possible role for palmitoylation of
alpha-tubulin. Mol Biol Cell. 12:2672–2687. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Caron JM and Herwood M: Vinblastine, a
chemotherapeutic drug, inhibits palmitoylation of tubulin in human
leukemic lymphocytes. Chemotherapy. 53:51–58. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heuer TS, Ventura R, Mordec K, Lai J,
Fridlib M, Buckley D and Kemble G: FASN inhibition and taxane
treatment combine to enhance anti-tumor efficacy in diverse
xenograft tumor models through disruption of tubulin palmitoylation
and microtubule organization and FASN inhibition-mediated effects
on oncogenic signaling and gene expression. EBioMedicine. 16:51–62.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ng EL, Gan BQ, Ng F and Tang BL: Rab
GTPases regulating receptor trafficking at the late
endosome-lysosome membranes. Cell Biochem Function. 30:515–523.
2012. View
Article : Google Scholar
|
|
28
|
Rink J, Ghigo E, Kalaidzidis Y and Zerial
M: Rab conversion as a mechanism of progression from early to late
endosomes. Cell. 122:735–749. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bucci C, Thomsen P, Nicoziani P, McCarthy
J and van Deurs B: Rab7: A key to lysosome biogenesis. Mol Biol
Cell. 11:467–480. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Modica G, Skorobogata O, Sauvageau E,
Vissa A, Yip CM, Kim PK, Wurtele H and Lefrancois S: Rab7
palmitoylation is required for efficient endosome-to-TGN
trafficking. J Cell Sci. 130:2579–2590. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guerra F and Bucci C: Multiple roles of
the small GTPase Rab7. Cells. 5(pii): E342016. View Article : Google Scholar : PubMed/NCBI
|