Introduction
The nucleolus is an intra-nuclear organelle that
contains ribosomal DNA (rDNA) that encodes ribosomal RNA (rRNA),
which is an essential component of ribosomes. The rRNA and a
variety of ribosomal proteins form ribosomes, which serve as the
site of biological protein synthesis. Previous studies have
demonstrated that ribosome biogenesis is associated with multiple
cellular activities. For example, limiting ribosome biogenesis
extends the lifespan, whereas increased ribosome biogenesis has
been reported as a hallmark of premature aging (1). In the context of cancer, alterations in
ribosome biogenesis have been suggested to be involved in
tumorigenesis by downregulating tumor suppressors (2,3). In
addition, rDNA rearrangements have been observed in over half of
adult solid tumors (4). This
observation suggests that rDNA restructuring is one of the most
common chromosomal alterations in adult solid tumors and may be
involved in tumorigenesis or malignancy. Conversely, for cancer
treatment, an inhibitor targeting RNA polymerase I, which is
required for rDNA transcription, induces apoptosis in cancer cells
by activating the ATM/ATR pathway (5).
Ionizing radiation (IR) generates multiple types of
DNA damage, such as DNA double-strand breaks (DSBs), single-strand
breaks (SSBs), and base damage (6).
Among the various types of DNA damage, DSBs are considered to be a
critical factor causing severe genome instability, such as
deletion, duplication, inversion and translocation (7). Linear energy transfer (LET) is a
parameter used to describe the pattern of energy deposition. X-rays
or γ-rays are known as low LET IR, whereas heavy charged particle
IR is high LET radiation. High LET particle irradiation causes
complex DNA lesions, defined as DNA damage containing both DSBs and
SSBs, as well as base damage, within 1–2 helical turns (<3–4 nm)
(8).
Another feature of high LET particle
irradiation-dependent DNA damage was recently identified, observed
as clustered γH2AX foci along the particle track (9–11). A
combination of complex DNA lesions within clustered DSBs is not
efficiently repaired and is associated with a high cell-killing
effect (9,12). IR is a critical risk factor for human
health, particularly for astronauts during periods of space travel
or when radiation accidents occur, whereas it is significantly
beneficial for cancer treatment (13).
In human cells, ~150–200 copies of rDNA encoding
rRNA is located on chromosomes 13, 14, 15, 21 and 22, and each
chromosome contains repetitive rDNA sequences (14). These repetitive sequences are
genetically unstable and highly sensitive when cells are exposed to
exogenous DNA stress (15). Classical
studies have demonstrated that morphological changes in the
nucleolus have been observed following IR under a phase-contrast
microscope (16). However, the
stability of the nucleolus and the transcriptional activity of rDNA
following IR in the context of DSBs and its signaling have not been
thoroughly investigated. Moreover, although irradiation-induced
dose-dependent increase in polysomal aggregates has been reported
(17), the irradiation-dependent
morphological changes of individual ribosomes have not been
investigated.
The aim of the present study was to examine the
alterations in the amount of nucleolin, a marker of the nucleolus,
and its morphological changes following IR, in order to investigate
nucleolar stability in response to DSBs. We also investigated the
role of p53 in preventing nucleolar instability following IR and
protecting the transcriptional activity of rDNA in irradiated
cells.
Materials and methods
Cell culture and irradiation
HCT116 (human colorectal carcinoma, wild-type (WT)
p53+/+ and isogenic p53−/− null
derivative, provided by Dr Vogelstein of Johns Hopkins University)
and U2OS (human osteosarcoma) cells were cultured in Eagle's
minimum essential medium (MEM) with 10% fetal calf serum (FCS)
(Sigma-Aldrich; Merck KGaA). 1BR (normal human dermal fibroblast
strain) hTERT cells were cultured in the alpha modification of MEM
(Wako Pure Chemical Industries, Ltd.) with 10% FCS. X-ray
irradiation was performed using a Faxitron RX-650 at 100 kVp and 20
mA with a dose rate of 1.14 Gy/min (Faxitron Bioptics). Carbon ion
irradiation was performed at Gunma University Heavy Ion Medical
Center using a spread-out Bragg-peak (SOBP) beam (290 MeV/nucleon;
mean LET at the center of a 6-cm SOBP, 50 keV/µm).
siRNA knockdown and p53
overexpression
In U2OS cells, p53 siRNA transfection was performed
using HiPerFect (Qiagen GmbH), as previously described (18). The efficiency of the knockdown was
confirmed by immunoblotting for the detection of p53. The sequence
of the p53 siRNA oligonucleotide used was
5′-GUGCAGCUGUGGGUUGAUUdTdT-3′. To examine the effect of p53
overexpression on nucleolar stability after IR, Flag-p53 WT was
transfected in HCT116 cells using the Neon® Transfection
System (Thermo Fisher Scientific, Inc.). Transfection efficiency
was confirmed by immunoblotting using p53 antibodies (1:500
anti-p53 antibody, sc-6243, Santa Cruz Biotechnology, Inc.) and
flag (1:1,000 anti-flag antibody, F3165, Sigma-Aldrich; Merck
KGaA). Cells were incubated in fresh MEM for 24 h and then
irradiated and processed for immunofluorescence analysis.
Immunofluorescence staining
Cells were seeded on Fisherbrand microscope cover
glasses (12-545-80 12CIR-1, Thermo Fisher Scientific, Inc.) 48 h
prior to irradiation. At the indicated time points after
irradiation, cells were fixed with 3% paraformaldehyde-2% sucrose
for 10 min. After washing with phosphate-buffered saline (PBS),
permeabilization was carried out by treatment with 0.2%
TritonX-100-PBS (Sigma-Aldrich; Merck KGaA) for 2 min and 30 sec.
Cells were then washed twice with PBS and incubated with the
primary antibody in 2% bovine serum albumin (BSA)-PBS (1:1,000
anti-nucleolin antibody, ab22758, Abcam) at 37°C for 30 min. The
cells were washed twice with PBS before being incubated with the
appropriate secondary antibody conjugated to Alexa-Fluor-555 (1:500
in 2% BSA-PBS) at 37°C for 30 min. The cells were again washed
twice with PBS and mounted onto slides using ProLong Gold antifade
mountant (Thermo Fisher Scientific, Inc.).
Morphological evaluation of
nucleoli
Immunofluorescence slides were examined with a Nikon
microscope (ECLIPSE Ni-U with DS-Qi2 camera and NIS-Elements D)
using a 60× magnification lens. From a representative area of each
sample, at least 100 consecutive cells were evaluated by direct
visualization. First, each cell was examined simultaneously for
nuclear staining and nucleolar morphology, and then categorized
into one of the seven nucleolar types. The number of nucleoli per
cell was noted for all cells, excluding type 7 cells (cells
undergoing apoptosis, mitosis, or mitotic catastrophe). Nucleolin
signal within micronuclei (types 3 and 6) were also counted
individually, and the total number was a combination of the signal
within the main nuclei and micronuclei.
Quantification of 47S rRNA expression
levels using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) analysis
Cells were seeded and incubated for 24 h prior to
irradiation. At 72 h of irradiation, total RNA was extracted from
the cells using NucleoSpin RNA (Macherey-Nagel GmbH). PrimeScript
RT Reagent Kit (Perfect Real Time; Takara) was used to
reverse-transcribe cDNA from total RNA, according to the
manufacturer's instructions, and qPCR was performed using
StepOnePlus (Thermo Fisher Scientific, Inc.). Reactions (20
µl each) were prepared in duplicate in a MicroAmp Fast
Optical 96-Well Reaction Plate (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Each reaction contained 0.5 µM of each
primer, 0.2 µM probe, 10 µl of TaqMan Universal PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.),
and cDNA as a template. The expression levels were normalized to
GAPDH and were calculated using the 2−ΔΔCq method
(19). The qPCR settings were as
follows: Initial denaturation at 95°C for 10 min, followed by 45
cycles of denaturation at 95°C for 15 sec, and annealing and
extension at 60°C for 1 min. The primers and probes used for the
qPCR are listed below:
47S rRNA forward: 5′-GAACGGTGGTGTGTCGTTC-3′
47S rRNA reverse: 5′-GCGTCTCGTCTCGTCTCACT-3′
47S rRNA probe: 5′-CGGTGTGGGGTTCGAGGCGGTTT-3′
GAPDH forward: 5′-CTCCTCTGACTTCAACAGCGA-3′
GAPDH reverse: 5′-CCAAATTCGTTGTCATACCAGGA-3′
GAPDH probe: 5′-ATGCCAGCCCCAGCGTCAAAGGT-3′
Statistical analysis
Box plots were constructed using SigmaPlot 12.3
(Systat Software, Inc.) from the combined data of three independent
experiments. The statistical significance was analyzed using the
Student's two-tailed t-test or Mann-Whitney U test. For datasets
where multiple comparisons were performed using single group, to
avoid the high rate of familywise error, Bonferroni's correction
was applied. The level of significance was determined using the
α-value obtained by Bonferroni's correction and is specified in
each individual figure legend.
Results
X-rays cause nucleolar fragmentation
in human cancer cell lines
To investigate the nucleolar stability in response
to IR in cancer cells, the number of nucleoli per cell and its
morphological changes were examined following X-ray irradiation
(n.b., nucleolin is localized within the nucleolus; therefore, it
is used as a marker of nucleolus). In HCT116
p53+/+ and U2OS cell lines, there was an increase
in the number of nucleoli per cell over time after 2 Gy of X-ray
irradiation (Fig. 1A and B). Next,
the morphological patterns were categorized to investigate the
morphological changes after IR (Fig.
1C). Interestingly, a marked increase in type 3 HCT116
p53+/+ cells was observed following IR (Fig. 1D). Type 3 represents a cell containing
micronuclei with a nucleolin signal. The peak of type 3 among
HCT116 p53+/+ cells was observed at 24 h
following IR. By contrast, U2OS cells exhibited an increase in
types 2 and 3 following X-ray irradiation (Fig. 1E). The majority of the nucleolar
aberrations in U2OS cells were type 2 (cell containing micronuclei
without nucleolin signal). These data suggest that nucleolar
instability is caused in cancer cells following IR, and the type of
morphological changes is cell type-dependent (Fig. 1F-H, supplementary information Fig. S1A). Furthermore, the levels of
nucleolar instability were examined after different doses of X-ray
irradiation. Importantly, the incidence of nucleolar fragmentation
and type 3 morphology increased with the dose of X-ray irradiation
until it peaked at 2–4 Gy, followed by a modest decline at >8 Gy
at all the evaluated time points (Fig. 2A
and B).
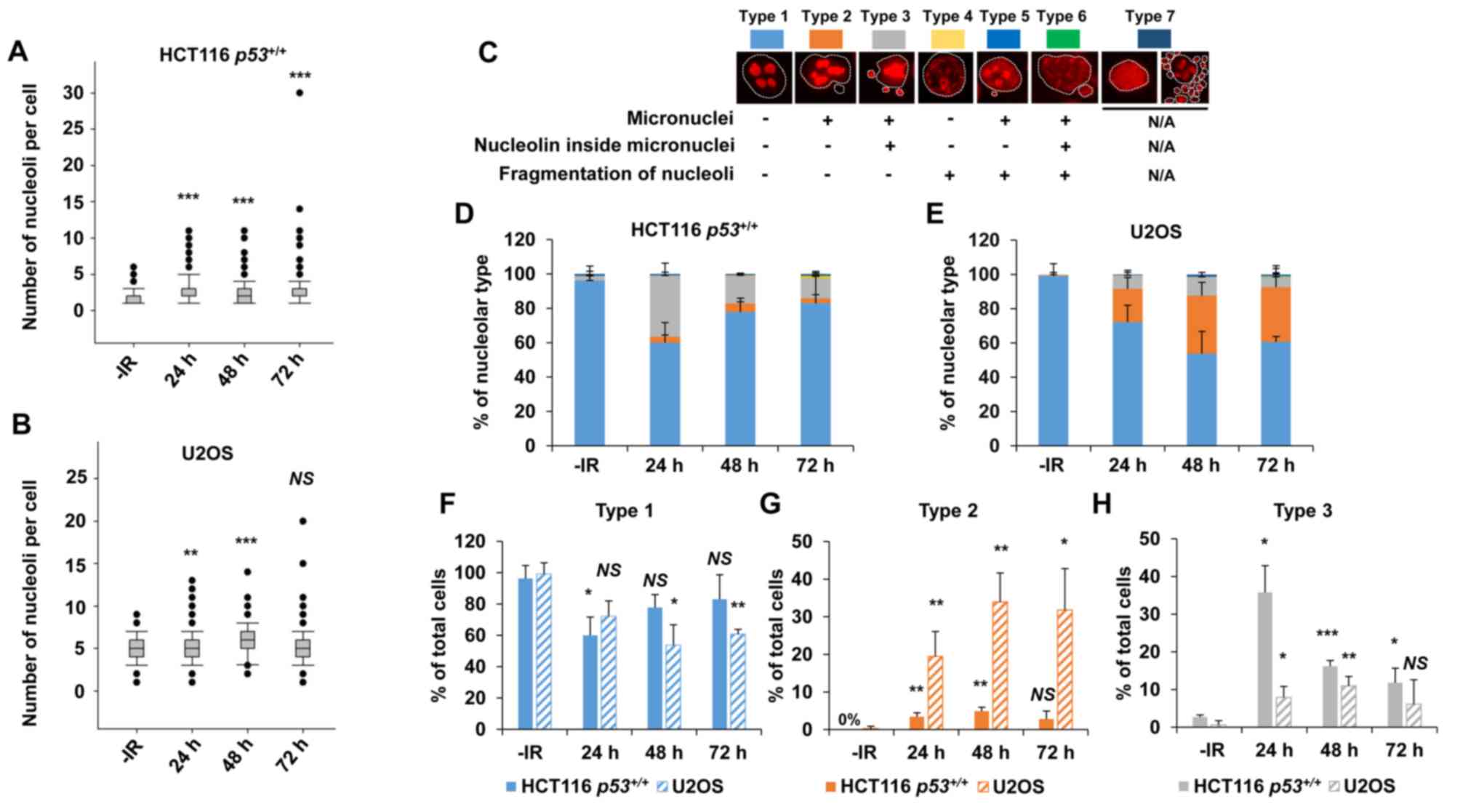 | Figure 1.X-rays cause nucleolar fragmentation
in human cancer cell lines. (A and B) Distribution of the number of
nucleoli per cell at 24, 48 and 72 h after irradiation with 2 Gy of
X-rays in (A) HCT116 p53+/+ and (B) U2OS cells.
Box plot: Upper horizontal line of the box, 75th percentile; lower
horizontal line of the box, 25th percentile; horizontal line within
the box, median; upper horizontal bar outside the box, highest
observation; lower horizontal bar outside the box, smallest
observation. Circles represent outliers. (C) Cells were categorized
into seven types based on the morphological characteristics of the
nuclei (presence of micronuclei, as seen by DAPI staining
corresponding to the white dashed line) and nucleoli (presence of
fragmentation and localization inside micronuclei, if the latter is
present). Type 7 includes apoptotic, mitotic, and mitotic
catastrophe cells, which were subsequently excluded from the
nucleolar quantification. (D and E) Percentage of nucleolar types
in the total set of evaluated cells as shown in (D) HCT116
p53+/+ and (E) U2OS cells. Cells were analyzed at
24, 48 and 72 h after irradiation with 2 Gy of X-rays. (F-H)
Percentage of predominant individual nucleolar types. Types (F) 1,
(G) 2 and (H) 3 in the total set of evaluated HCT116
p53+/+ and U2OS cells at 24, 48 and 72 h after
irradiation with 2 Gy of X-rays. The error bars represent the
standard deviation of three independent experiments. The
statistical significance in A, B and F-H was examined by comparison
with non-irradiated cells of the corresponding cell line, using
Bonferroni's correction. *P<0.0167, **P<0.0033,
***P<0.0003. NS, not significant; IR, ionizing radiation. |
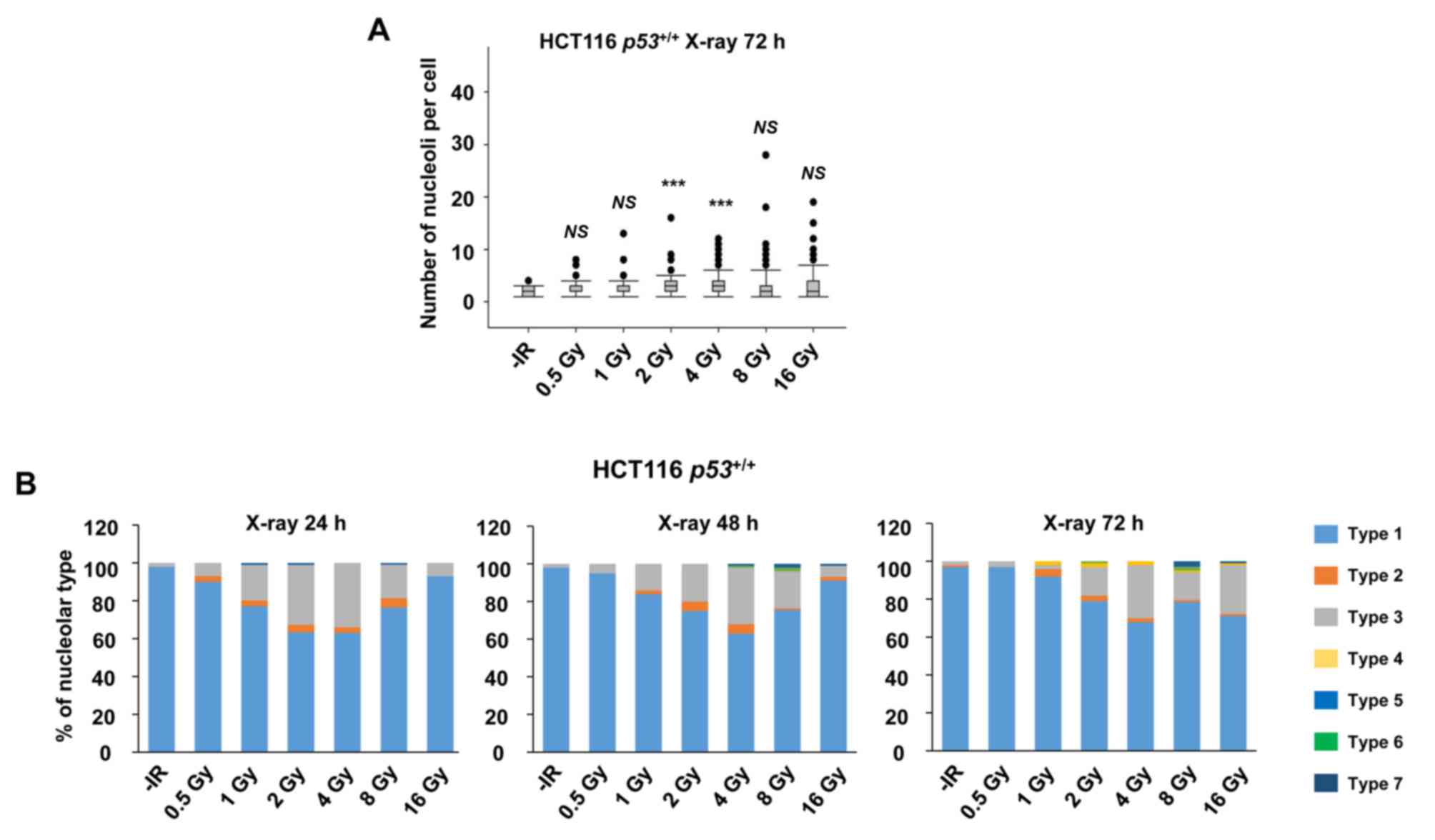 | Figure 2.X-rays cause dose-dependent nucleolar
fragmentation. (A) Distribution of the number of nucleoli per cell
at 72 h after irradiation with 0.5, 1, 2, 4, 8, or 16 Gy of X-rays
in HCT116 p53+/+ cells. (B) Percentage of
nucleolar types in the total set of evaluated cells after 0.5, 1,
2, 4, 8 and 16 Gy of X-rays at 24, 48 and 72 h after irradiation in
HCT116 p53+/+ cells. Similar results were
obtained in two independent experiments. Statistical significance
was examined by comparison of samples irradiated with different
doses individually with non-irradiated cells, using Bonferroni's
correction. ***P<0.00017. NS, not significant; IR, ionizing
radiation. |
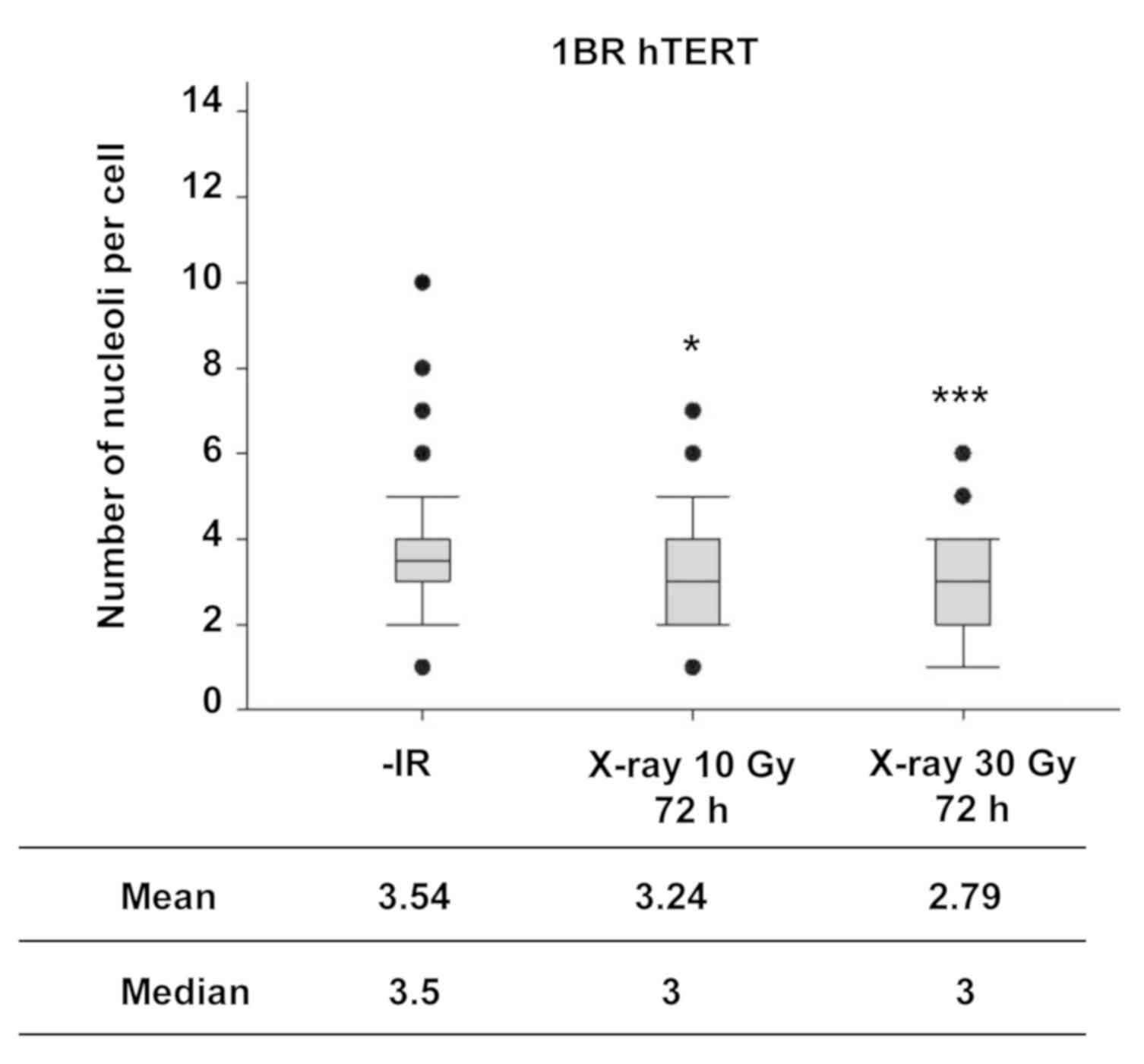
Next, the number of nucleoli in normal human
fibroblasts was examined following IR to investigate the nucleolar
stability in normal cells. In contrast to the observation of the
increase in the number of nucleoli in cancer cell lines, X-ray
irradiation did not increase the number of nucleoli in normal human
fibroblasts, even at markedly higher radiation doses (Fig. 3).
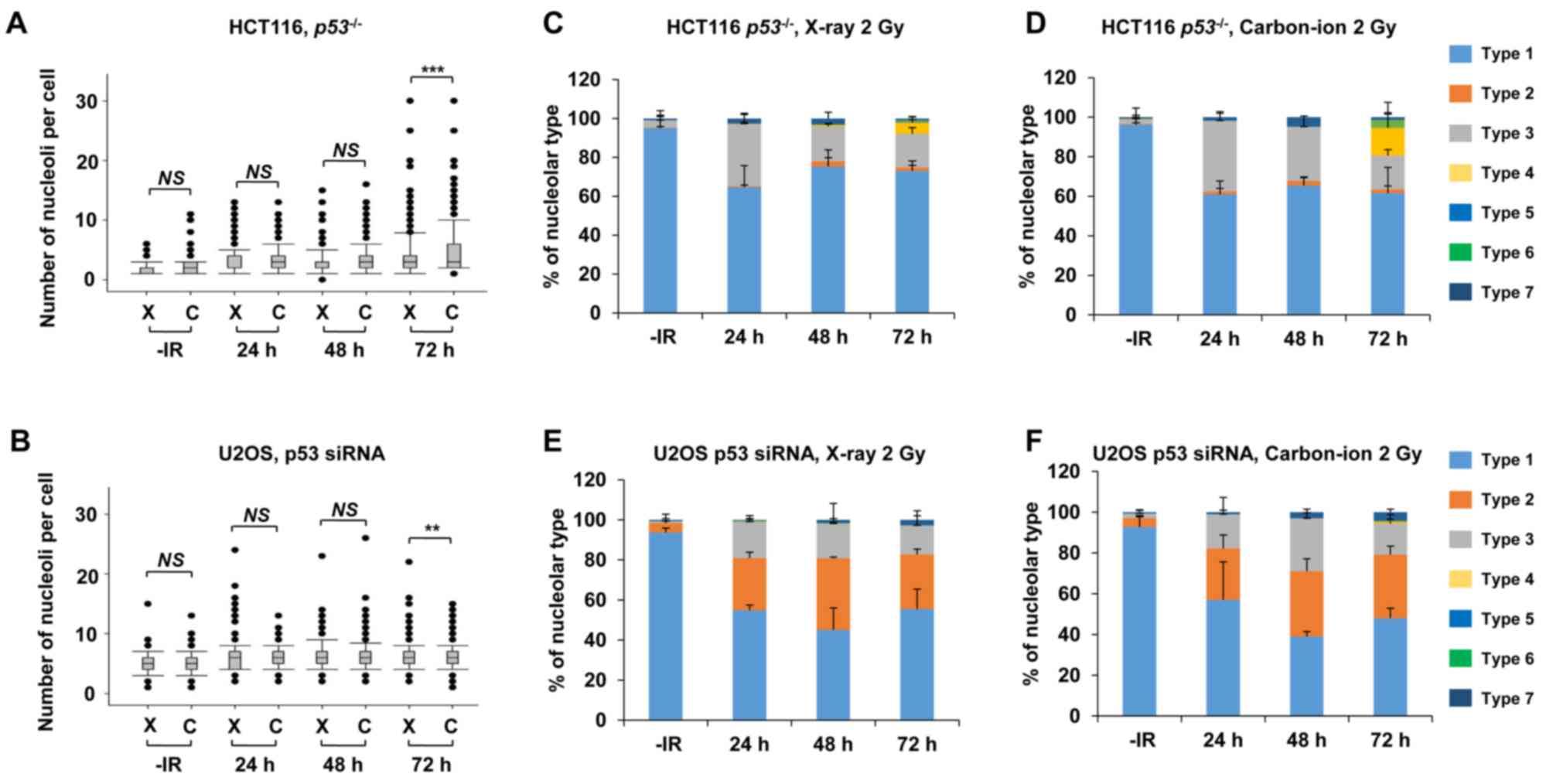
p53 deficiency augments nucleolar
fragmentation following exposure to X-rays
Furthermore, to address whether the genetic status
involving genome stability affects nucleolar fragmentation after
IR, the number of nucleoli per cell and any morphological changes
were examined in cancer cells with p53 deficiency. Then, we
compared the nucleolin signal in HCT116 p53+/+
and HCT116 p53−/− cells to investigate the impact
of the p53 gene status on nucleolar stability. Importantly,
a statistically significant (P=0.003) increase in nucleolar number
was observed in HCT116 p53−/− cells at 72 h
following exposure to X-rays (Fig.
4A). Furthermore, a dose-dependent increase in the nucleolar
number of HCT116 p53−/− cells up to 8 Gy was
found at 72 h (Fig. 4B). To
investigate the effect of p53 on U2OS cells, siRNA knockdown
targeting p53 was performed in U2OS cells. Similar to the results
in HCT116 cells, there was a significant increase in the number of
nucleoli in p53-depleted U2OS cells (Fig.
4C), with this increase being evident at later time points of
48–72 h after X-ray irradiation.
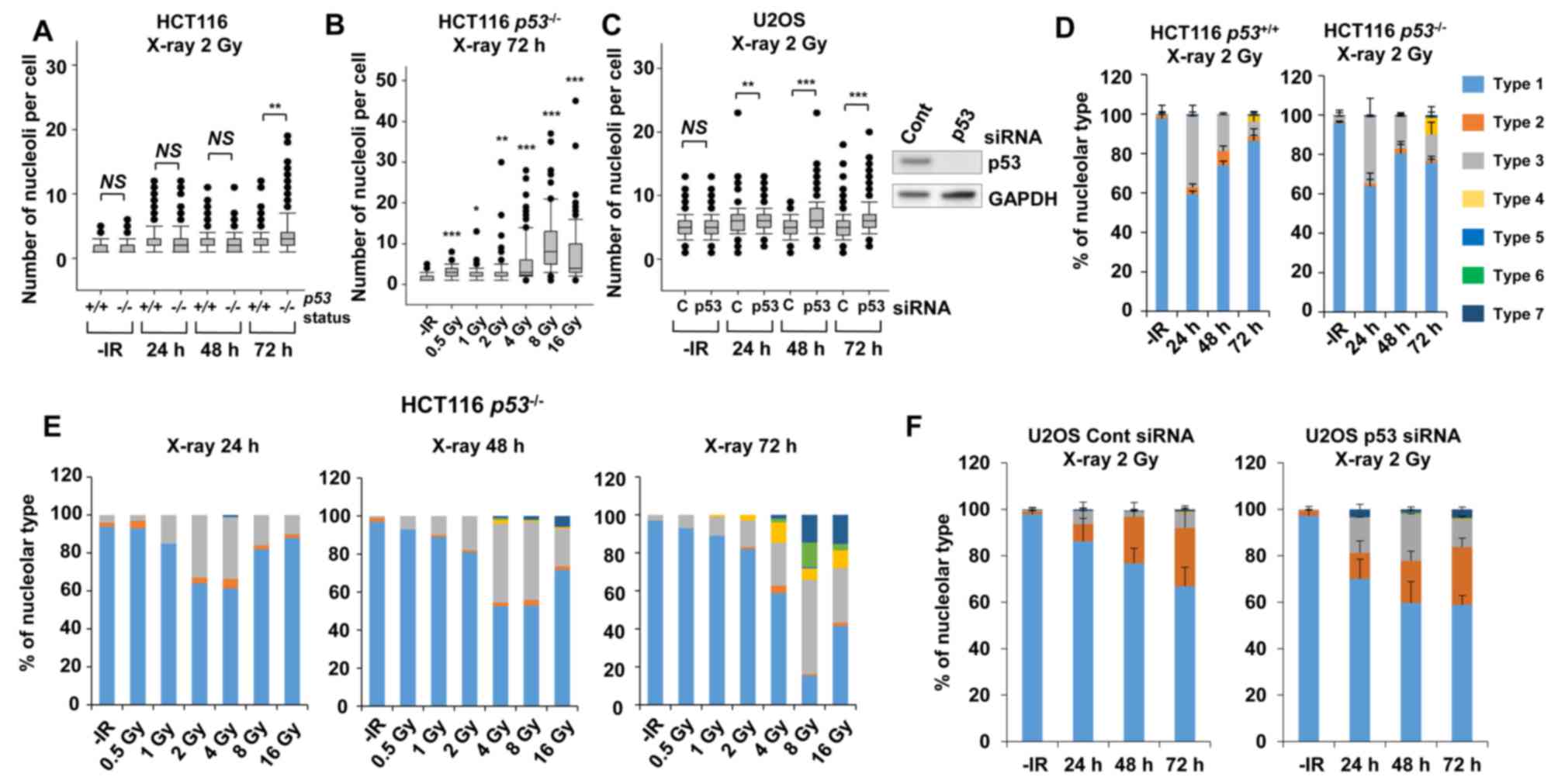 | Figure 4.p53 deficiency augments nucleolar
fragmentation following X-rays. (A) The effect of p53 deficiency
was compared between p53+/+ and
p53−/− variants of HCT116 cells. Cells were
irradiated with 2 Gy of X-rays and the distribution of the number
of nucleoli per cell at 24, 48 and 72 h after irradiation is shown.
(B) Distribution of the number of nucleoli per cell at 72 h after
irradiation with 0.5, 1, 2, 4, 8, or 16 Gy of X-rays in HCT116
p53−/− cells. Statistical significance was
examined by comparison with non-irradiated cells, using
Bonferroni's correction. *P<0.0083, **P<0.0017,
***P<0.00017. n.s., not significant. (C) Distribution of the
number of nucleoli per cell at 24, 48 and 72 h after irradiation in
p53-depleted U2OS cells. The knockdown efficiency is shown in the
right panel. C: siControl, p53: siRNA of p53. (D) Percentage of
nucleolar types in the total set of HCT116 cells. HCT116
p53+/+ (left panel) and p53−/−
(right panel) cells were examined at 24, 48 and 72 h after
irradiation with 2 Gy of X-rays. The comparison of individual
nucleolar types is shown in supplementary information (Fig. S2A-E). (E) Percentage of nucleolar
types at 24, 48 and 72 h after irradiation with 0.5, 1, 2, 4, 8, or
16 Gy of X-rays in HCT116 p53−/− cells. Similar
results were obtained by two independent experiments. (F)
Percentage of nucleolar types in the entire set of U2OS cells.
Cells after transfection with control siRNA (left panel) or p53
siRNA (right panel) were harvested at 24, 48 and 72 h after
irradiation with 2 Gy of X-rays. The comparison of individual
nucleolar types is shown in the supplementary information (Fig. S2F-J). The error bars represent the
standard deviation of three independent experiments. *P<0.05,
**P<0.01, ***P<0.001. NS, not significant, for panels A and
C. IR, ionizing radiation. |
Next, morphological changes following the
categorization shown in Fig. 1C in
p53-defective cells were analyzed following IR. A morphological
categorization analysis revealed the appearance of type 4
morphology (multiple nucleolar fragmentations without micronuclei)
in HCT116 p53−/− cells at 72 h (Fig. 4D, supplementary information Figs. S1B and S2A-E). In addition, dose-dependent analysis
confirmed more prominent type 4, type 6 and type 7 characteristics
in HCT116 p53−/− cells (Fig. 4E). Alternatively, the morphological
analysis in p53-depleted U2OS cells revealed a change in the
nucleolar structure from type 2 to types 3 and 7 (Fig. 4F, supplementary information Figs. S1B and S2F-J). These results suggest that p53 is an
important gene for suppressing nucleolar fragmentation following
X-rays.
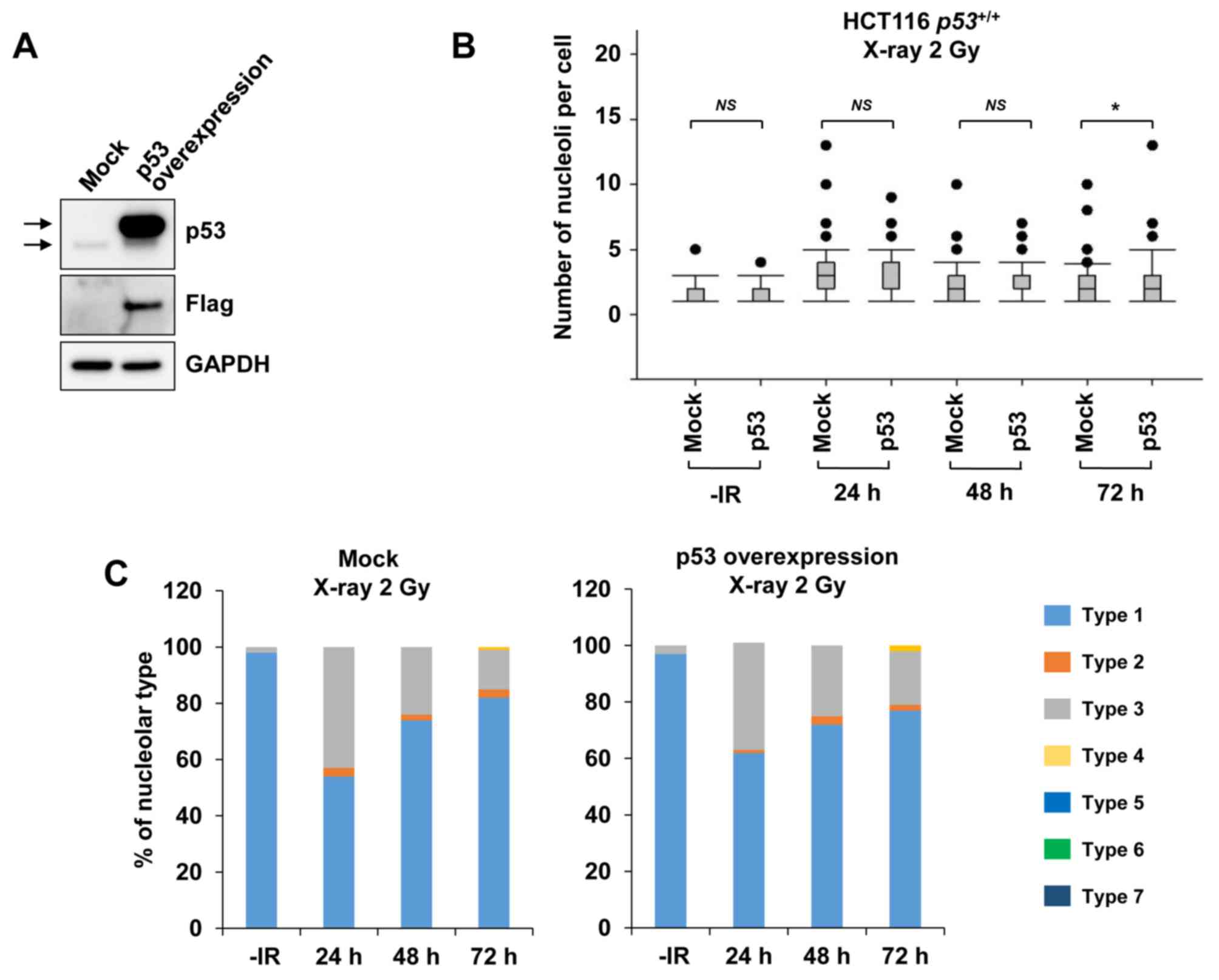
By contrast, when examining nucleolar fragmentation
levels in HCT116 p53+/+ cells exogenously
overexpressing p53 (Fig. 5A), it was
observed that, compared with mock cells, the HCT116
p53+/+ cells overexpressing p53 exhibited a
marginal increase in nucleolar fragmentation at 72 h after
irradiation with 2 Gy of X-rays (Fig. 5B
and C).
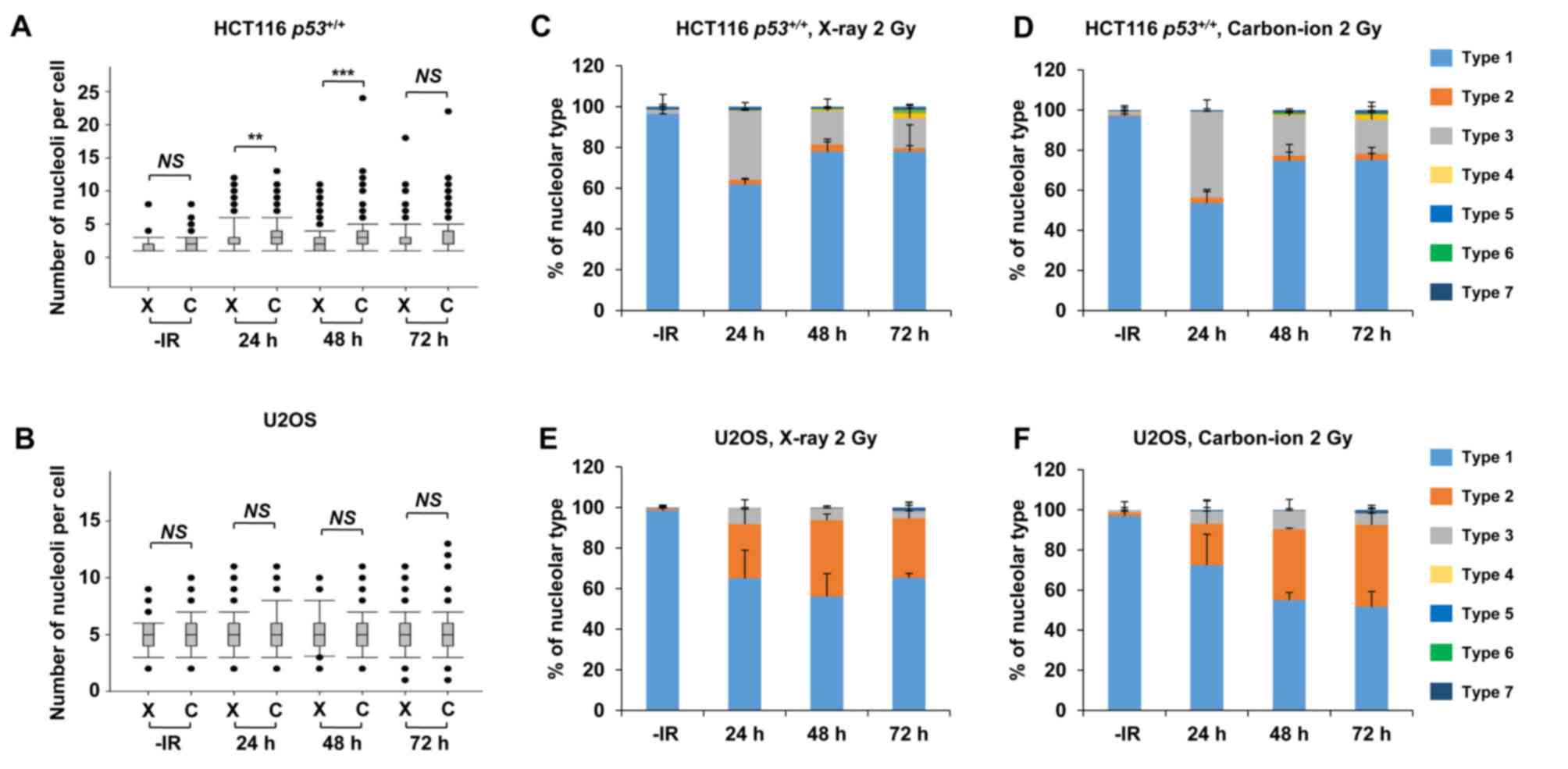
Carbon ions induce moderately greater
nucleolar fragmentation compared with X-rays
High LET carbon ion irradiation is one of the most
promising next-generation radiotherapies (20). Following high LET carbon ion
irradiation, critical genome instability causes greater
cell-killing effect due to the formation of complex DNA lesions and
clustered DSBs, leading to large deletions or chromosomal
translocations (21,22). To evaluate the biological effects of
high LET carbon ion irradiation on nucleolar stability, the number
of nucleoli per cell and the morphology were examined after 2 Gy of
X-ray or carbon ion irradiation. Similar to the results following
X-ray irradiation (Fig. 1), the
number of nucleoli per cell increased after carbon ion irradiation
in HCT116 p53+/+ cells and U2OS cells (Fig. 6A and B). The increase in the number of
nucleoli after carbon ion irradiation was modestly but
statistically significantly higher at 24 h (P=0.001) and at 48 h
(P<0.001) compared with that after X-ray irradiation in HCT116
p53+/+ cells. Compared with HCT116
p53+/+ cells, U2OS cells exhibited a modest
increase in the number of nucleoli following carbon ion irradiation
compared with that after X-ray irradiation; however, it was not
statistically significant. Although the type distribution was
similar between X-rays and carbon ion irradiation, the
morphological changes in HCT116 p53+/+ and U2OS
cells were also observed after carbon ion irradiation (Fig. 6C-F, supplementary information Figs. S1C and S3A-H). Next, we examined the number of
nucleoli per cell and the morphological changes under a p53
defective background following carbon ion irradiation. At 24–48 h
after IR, there was no significant alteration in the number of
nucleoli due to p53 deficiency in either HCT116 or U2OS cells, as
observed after X-ray exposure (Fig. 7A
and B). However, a higher percentage of type 4 morphology was
found in HCT116 p53−/− cells at 72 h after carbon
ion irradiation, compared with after X-rays or HCT116
p53+/+ cells after carbon ion irradiation
(Fig. 7C and D, supplementary
information Figs. S1D and S4D). Type 6, which was rarely observed in
other conditions, was detected in HCT116 p53−/−
cells after carbon ion irradiation (Fig.
7D, supplementary information Fig.
S4F). By contrast, a similar type of morphological change was
observed in U2OS cells irrespective of the type of IR (Fig. 7E and F, supplementary information
Fig. S4G-K). In the normal human
fibroblast, similar to X-ray irradiation, carbon ion irradiation
did not increase the number of nucleoli (supplementary information
Fig. S5).
Transcriptional activity of rDNA is
reduced under conditions of p53 deficiency following IR
To address the question whether nucleolar
fragmentation after IR affects transcriptional activity of rDNA,
qPCR was performed to measure the level of 47S rRNA, an immediate
transcript of rDNA, which ultimately yields mature rRNA after RNA
processing. Importantly, consistent with the results of nucleolar
fragmentation, a significant decrease in rRNA synthesis was
observed in HCT116 p53−/− cells 72 h after
irradiation with 4–16 Gy of X-rays; however, X-ray irradiation did
not change rRNA synthesis in HCT116 p53+/+ cells
after 2, 4 or 8 Gy (Fig. 8A and B).
Instead, an increase was observed in HCT116
p53+/+ cells after 16 Gy.
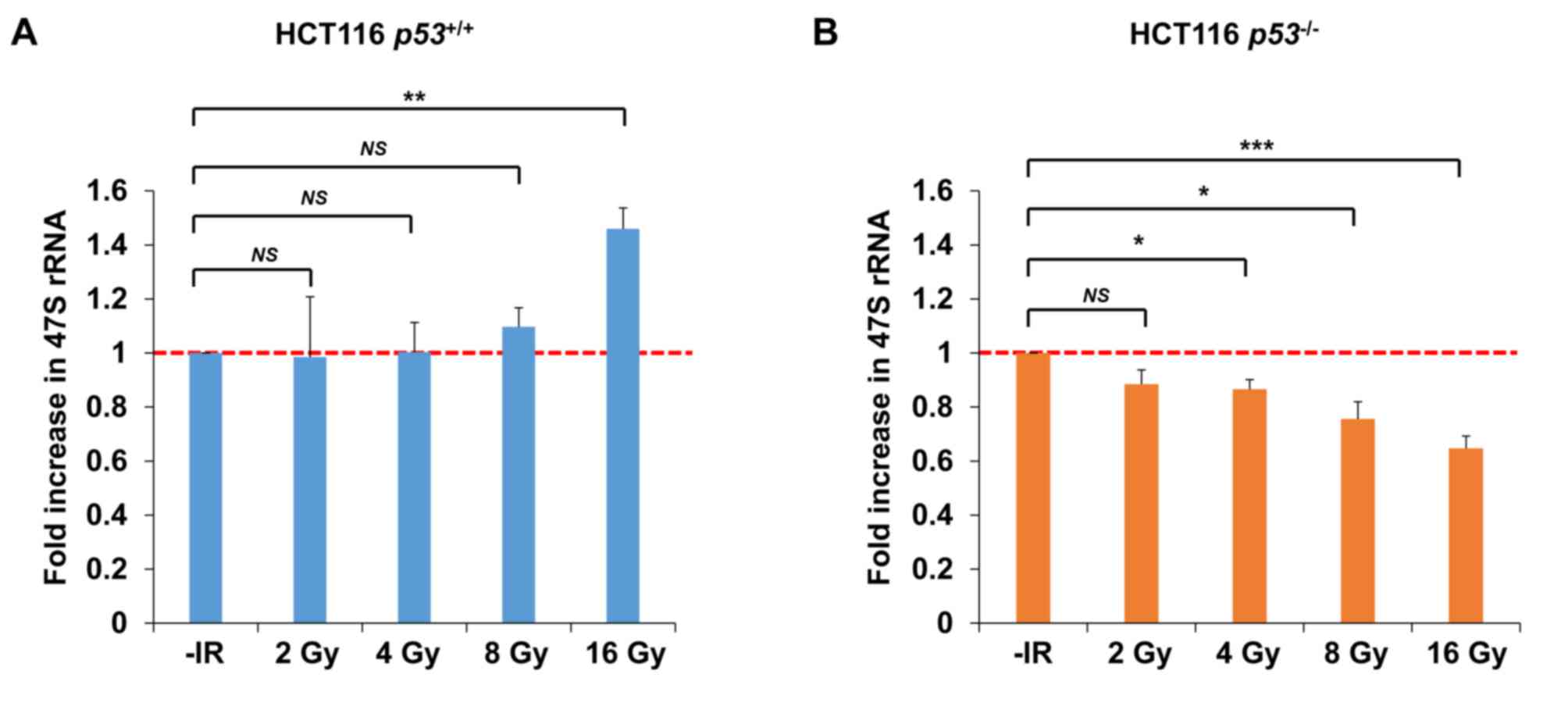 | Figure 8.Transcriptional activity of rDNA is
reduced under conditions of p53 deficiency following IR. (A and B)
Levels of 47S precursor rRNA, quantified by qPCR analysis 72 h
after irradiation of HCT116 (A) p53+/+ (B) or
p53−/− cells with 2, 4, 8, or 16 Gy of X-rays.
The error bars represent the standard deviation of three
independent experiments. The statistical significance was examined
between irradiated cells at the indicated dose and non-irradiated
cells, using Bonferroni's correction. *P<0.0125, **P<0.0025,
***P<0.00025. NS, not significant; IR, ionizing radiation; qPCR,
quantitative polymerase chain reaction. |
Discussion
In the present study, it was observed that IR causes
nucleolar fragmentation, suggesting that DNA damage disrupts the
nucleolar structure. Furthermore, it was observed that cancer cells
lacking p53 or subjected to p53 siRNA display increased nucleolar
fragmentation following IR. Compared with X-ray irradiation, carbon
ion irradiation, generating complex DNA lesions and clustered DSBs,
caused only slightly greater nucleolar fragmentation. In addition,
the analysis of 47S rRNA using qPCR revealed a significant
reduction of ribosomal RNA expression in p53-defective cells
following IR. These results suggest that IR causes nucleolar
instability, and p53 plays a key role in its prevention.
A chromosomal rearrangement, such as a large
deletion, duplication, inversion or translocation, is a
characteristic manifestation of dynamic genome instability
following IR. The nucleolar fragmentation detected in the present
study likely occurs during chromosomal rearrangements. In human
cells, ~150–200 copies of rDNA encoding rRNA are located on five
distinct chromosomes. Each chromosome contains a repetitive rDNA
sequence. As rRNA forms secondary structures, such as hairpins,
stem-loops, or cruciform structures (23), such loop structures are also expected
to be formed in rDNA when DNA becomes single-stranded (e.g., during
DNA replication or transcription). In the present study, however,
it may be inferred that DSBs at rDNA regions are unlikely to be
induced by IR, since the percentage of rDNA in the total genome is
<1%. For example, 2 Gy of X-rays induce ~60 DSBs per cell in G1
phase cells and ~120 DSBs in G2 phase cells (24). Therefore, only 1 or <1 DSB per cell
are induced at the rDNA region after 2–8 Gy of X-rays. Since such
isolated DSBs are unlikely to be the cause of nucleolar
fragmentation, it may be inferred that additional steps are
required to induce nucleolar fragmentation following IR. One
possibility is that DNA replication-associated DSBs in S phase
cells may be the cause of nucleolar fragmentation. IR causes
multiple types of DNA damage, such as DSBs, base damage, SSBs and
cross-link damage. The amount of base damage and SSBs is estimated
to be ~1,000-3,000 per cell per Gy (25). The number of direct DSBs after 1 Gy
X-ray is ~30 per G1 cell (24).
However, if a DNA replication fork encounters unrepaired base
damage or SSBs, the damage is converted into DSBs, namely
replication-associated DSBs (i.e., several thousand DSBs may be
formed at replication sites) (26).
The generation of DNA replication-associated DSBs may be augmented
under p53 deficiency. Since p53 plays an important role in the G1/S
checkpoint arrest following IR (27),
the lack of a p53-dependent G1 arrest leads to the enhancement of
the replication-associated DSBs. This model is supported by the
findings of the present study, showing that nucleolar fragmentation
following IR is augmented in p53-defective cells.
At the beginning of this study, it was expected that
carbon ion irradiation may cause substantially greater nucleolar
fragmentation, as it is associated with a higher frequency of
chromosomal rearrangements compared with that observed after
X-rays; however, the present analysis uncovered only a subtle
enhancement of nucleolar fragmentation following carbon ion
exposure. Replication-associated DSBs from base damage and SSBs may
be the primary source of nucleolar fragmentation, as carbon
ion-specific DSBs may not be implicated in the enhancement of
nucleolar fragmentation. Consistent with this model, it may be
inferred that p53 deficiency, which causes failure of G1/S
checkpoint arrest (i.e., increase in S phase entry), led to
nucleolar fragmentation. Additionally, G2/M checkpoint and
mis-segregation during mitosis may be a major factor inducing
nucleolar fragmentation. On morphological analysis, an increase in
the formation of micronuclei with nucleolin signals was observed
following IR, particularly in HCT116 cells and p53-depleted U2OS
cells. As micronuclei are formed following mis-segregation in
mitosis (28,29) the sensitivity of the G2/M checkpoint
arrest is associated with nucleolar fragmentation. Although the
role of p53 in G2/M checkpoint arrest following IR may be less
significant, the lack of p53 may promote the formation of
micronuclei with nucleolin signals through mis-segregation in
mitosis following inefficient G2/M checkpoint arrest, particularly
in U2OS cells. Additionally, it was observed that the primary type
of fragmented nucleoli following IR in HCT116 cells was type 3,
whereas in U2OS cells it was type 2. Although the precise mechanism
underlying such a distinct type of formation is unclear, different
responses between HCT116 and U2OS cells against the G1/S and G2/M
checkpoint arrest may be involved. In contrast to the results
observed in cancer cell lines, the normal human fibroblasts (1BR
hTERT) did not exhibit an increase in the number of nucleolar
fragments. Since normal fibroblasts display an intact G1/S and G2/M
checkpoint arrest, nucleolar fragmentation and formation of
micronuclei may not be efficiently induced via DNA replication and
mitosis, respectively. In addition, following persistent checkpoint
arrest, fibroblast cells tend to undergo cellular senescence rather
than apoptosis (30); these distinct
characteristics may explain the different levels of nucleolar
fragmentation following IR. By contrast, cancer cells, which
normally exhibit the predominant modes of cell death after
irradiation, preferentially undergo apoptosis and mitotic
catastrophe under p53 intact and deficient conditions, respectively
(12). Thus, it appears that the
checkpoint machinery involving G1/S and G2/M checkpoint arrest and
apoptosis under the regulation of p53 plays a critical role in
preventing nucleolar fragmentation following IR (Fig. 9).
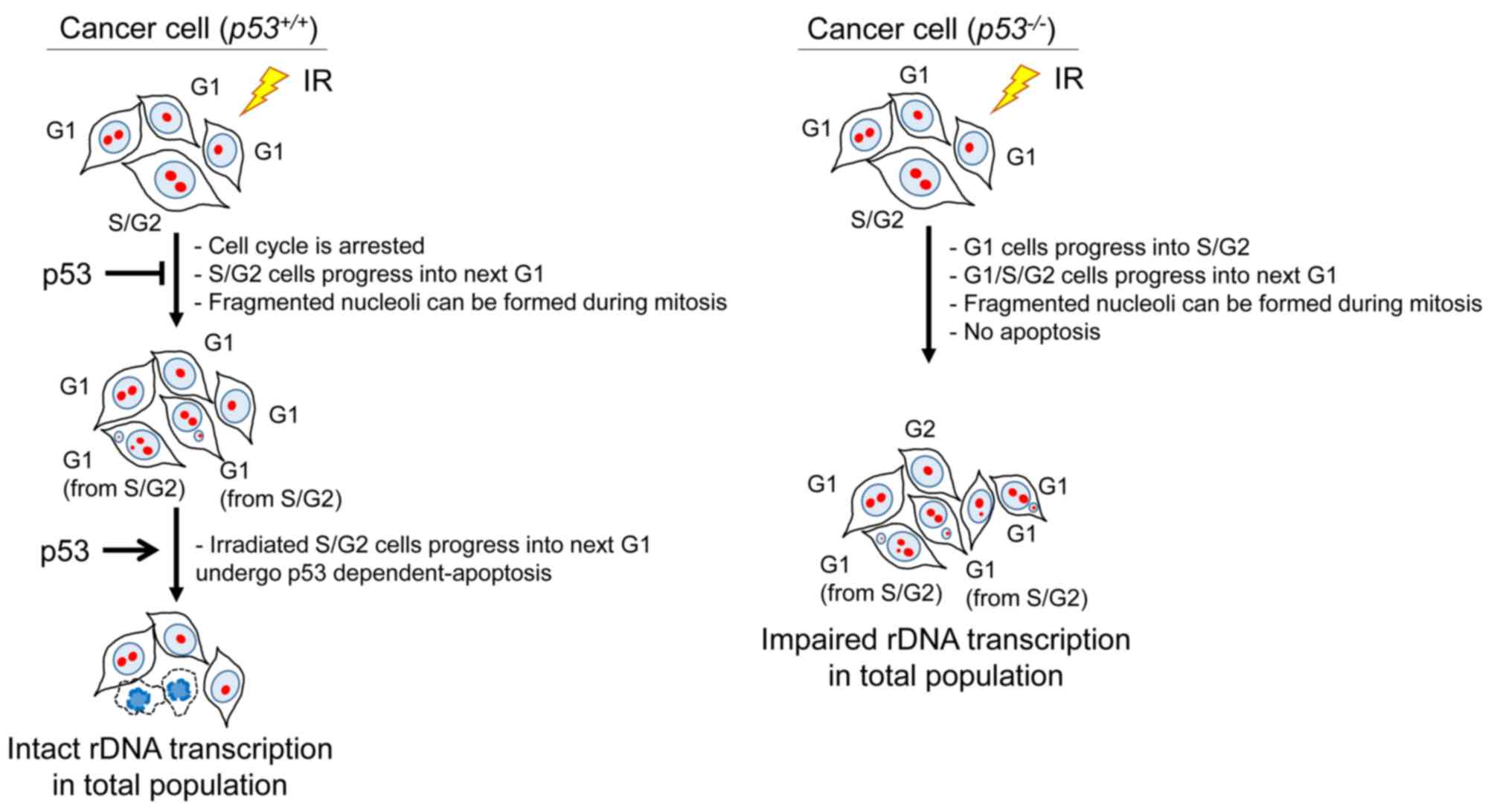 | Figure 9.Model for the pathway of nucleolar
instability following IR. Following activation of cell cycle
checkpoint arrest after IR, irradiated cells are arrested at each
cell cycle phase. Since the G1/S checkpoint arrest is sensitive, G1
cells are arrested until most DSBs are repaired, if p53 is present.
G2/M checkpoint arrest is released when the number of DSBs in G2
cells becomes 10–20, and cells progress into the M phase. During
mitosis, micronuclei with or without a nucleolin signal may form.
After mitosis, G1 cells that have progressed from irradiated S/G2
cells may undergo p53-dependent apoptosis. Thus, surviving cells
may exhibit normal transcriptional activity of rDNA. By contrast,
as irradiated cells may not be effectively arrested under
conditions of p53 deficiency, the formation of micronuclei is a
frequent occurrence following mitosis. In addition, as
p53-deficient G1 cells with micronuclei are not able to undergo
apoptosis, surviving cells displaying a damaged nucleolus may
exhibit low transcriptional activity of rDNA. IR, ionizing
radiation, DSBs, double-strand breaks. |
In addition to nucleolar fragmentation, rRNA
production was found to be reduced in p53-defective cells following
IR. Since rRNA is an essential component of the ribosome, which is
required for all protein synthesis, the reduction of ribosomal
biogenesis would impair cellular activity and viability. In
contrast to p53-defective cells, rRNA production in p53-proficient
cells was not reduced, but rather enhanced, following high-dose IR.
As described above, a proportion of cells undergo mitosis due to a
checkpoint release following a prolonged G2/M checkpoint arrest,
which results in micronuclei formation. Following mitosis, cells
with micronuclei may undergo apoptosis in the next G1 phase if the
p53-dependent apoptosis pathway is intact. Thus, in p53-proficient
cells, normal levels of rDNA transcription may be sustained in the
remaining cells with intact nucleoli, as cells with fragmented
nucleoli are eliminated by apoptosis. On the other hand, in
p53-deficient cancer cells, the remaining cells exhibit inefficient
rDNA transcriptional activity due to the nucleolar fragmentation,
as cells displaying fragmented nucleoli with micronuclei cannot
efficiently undergo apoptosis.
Consistently with the present findings, evidence
supporting the involvement of p53 in nucleolar biogenesis has also
been provided by previous studies (31,32). The
downregulation of ribosome biogenesis leads to the release of
ribosomal proteins, including Arf, from the nucleolus to the
nucleoplasm, which promotes the binding between Murine Double
Minute 2 (MDM2) and released ribosomal proteins (31). Thus, the prevention of MDM2-dependent
p53 degradation results in p53 activation. In addition, nucleolin
and ribosomal protein L26 (RPL26) bind to the 5′ untranslated
region of p53 mRNA after DNA damage, thereby controlling the
translation of p53 (32). In
conclusion, the present study demonstrated that p53 plays an
important role in maintaining the transcriptional activity of rDNA
in response to DNA damage. The downregulation of the rDNA-rRNA axis
(i.e., the pathway of ribosomal biogenesis) under conditions of p53
deficiency may be involved in cancer development via the impairment
of cellular homeostasis, particularly after IR or genotoxic stress.
These pathways play a significant role in the maintenance of
ribosomal biogenesis as one of the mechanisms of cancer prevention,
as the downregulation of the rDNA-rRNA axis may be caused by the
two key functions of p53, including G1/S and G2/M checkpoint arrest
and apoptosis.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Yoshimi Omi, Akiko
Shibata, Yoko Hayashi, Shiho Nakanishi, Yukihiko Yoshimatsu, Itaru
Sato and Yuka Hirota for assisting with the laboratory work.
Funding
The present study was supported by Grants-in-Aid
from the Japan Society for the Promotion of Science for KAKENHI
(JP17H04713 to AS), the Takeda Science Foundation, the Uehara
Memorial Foundation, and the Program of the network-type Joint
Usage/Research Center for Radiation Disaster Medical Science of
Hiroshima University, Nagasaki University and Fukushima Medical
University. The present study was also supported by Grants-in-Aid
from the Ministry of Education, Culture, Sports, Science and
Technology of Japan for programs for Leading Graduate Schools,
Cultivating Global Leaders in Heavy Ion Therapeutics and
Engineering.
Availability of materials and data
All the datasets generated and analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
AS designed the experiments and wrote the paper with
SK. The experiments including immunofluorescence, immunoblotting
and qPCR were performed by SK, YH and AS. Acquired data were
analyzed and interpreted by SK, WG, YH, TO, HS, MY, RK, TY and AS.
The manuscript was reviewed by TO, HS, KDH, MY and TY.
Administrative, technical, or material support was provided by TO,
SL and TN. The study was supervised by AS.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interest.
References
|
1
|
Buchwalter A and Hetzer MW: Nucleolar
expansion and elevated protein translation in premature aging. Nat
Commun. 8:3282017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ruggero D: Revisiting the nucleolus: From
marker to dynamic integrator of cancer signaling. Sci Signal.
5:pe382012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Montanaro L, Trere D and Derenzini M:
Changes in ribosome biogenesis may induce cancer by down-regulating
the cell tumor suppressor potential. Biochim Biophysica Acta.
1825:101–110. 2012.
|
|
4
|
Stults DM, Killen MW, Williamson EP,
Hourigan JS, Vargas HD, Arnold SM, Moscow JA and Pierce AJ: Human
rRNA gene clusters are recombinational hotspots in cancer. Cancer
Res. 69:9096–9104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Negi SS and Brown P: rRNA synthesis
inhibitor, CX-5461, activates ATM/ATR pathway in acute
lymphoblastic leukemia, arrests cells in G2 phase and induces
apoptosis. Oncotarget. 6:18094–18104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shibata A and Jeggo P: A historical
reflection on our understanding of radiation-induced DNA double
strand break repair in somatic mammalian cells; interfacing the
past with the present. Int J Radiat Biol. 95:945–956. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Gent DC, Hoeijmakers JH and Kanaar R:
Chromosomal stability and the DNA double-stranded break connection.
Nat Rev Genet. 2:196–206. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hada M and Georgakilas AG: Formation of
clustered DNA damage after high-LET irradiation: A review. J Radiat
Res. 49:203–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakajima NI, Brunton H, Watanabe R,
Shrikhande A, Hirayama R, Matsufuji N, Fujimori A, Murakami T,
Okayasu R, Jeggo P and Shibata A: Visualisation of gammaH2AX foci
caused by heavy ion particle traversal; distinction between core
track versus non-track damage. PLoS One. 8:e701072013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hagiwara Y, Niimi A, Isono M, Yamauchi M,
Yasuhara T, Limsirichaikul S, Oike T, Sato H, Held KD, Nakano T and
Shibata A: 3D-structured illumination microscopy reveals clustered
DNA double-strand break formation in widespread γH2AX foci after
high LET heavy-ion particle radiation. Oncotarget. 8:109370–109381.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hagiwara Y, Oike T, Niimi A, Yamauchi M,
Sato H, Limsirichaikul S, Held KD, Nakano T and Shibata A:
Clustered DNA double-strand break formation and the repair pathway
following heavy-ion irradiation. J Radiat Res. 60:69–79. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Amornwichet N, Oike T, Shibata A, Ogiwara
H, Tsuchiya N, Yamauchi M, Saitoh Y, Sekine R, Isono M, Yoshida Y,
et al: Carbon-ion beam irradiation kills X-ray-resistant p53-null
cancer cells by inducing mitotic catastrophe. PLoS One.
9:e1151212014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Durante M and Cucinotta FA: Heavy ion
carcinogenesis and human space exploration. Nat Rev Cancer.
8:465–472. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Henderson AS, Warburton D and Atwood KC:
Location of ribosomal DNA in the human chromosome complement. Proc
Natl Acad Sci USA. 69:3394–3398. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Durkin SG and Glover TW: Chromosome
fragile sites. Annu Rev Genet. 41:169–192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Montgomery PO, Karney D, Reynolds RC and
McClendon D: Cellular and subcellular effects of ionizing
radiations. Am J Pathol. 44:727–746. 1964.PubMed/NCBI
|
|
17
|
Hidvegi EJ, Holland J, Boloni E, Lonai P,
Antoni F and Varteresz V: The effect of whole-body x-irradiation of
guinea pigs on liver ribosomes. Biochem J. 109:495–505. 1968.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shibata A, Conrad S, Birraux J, Geuting V,
Barton O, Ismail A, Kakarougkas A, Meek K, Taucher-Scholz G,
Löbrich M and Jeggo PA: Factors determining DNA double-strand break
repair pathway choice in G2 phase. EMBO J. 30:1079–1092. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ohno T: Particle radiotherapy with carbon
ion beams. EPMA J. 4:92013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oike T, Niimi A, Okonogi N, Murata K,
Matsumura A, Noda SE, Kobayashi D, Iwanaga M, Tsuchida K, Kanai T,
et al: Visualization of complex DNA double-strand breaks in a tumor
treated with carbon ion radiotherapy. Sci Rep. 6:222752016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Niimi A, Yamauchi M, Limsirichaikul S,
Sekine R, Oike T, Sato H, Suzuki K, Held KD, Nakano T and Shibata
A: Identification of DNA double strand breaks at chromosome
boundaries along the track of particle irradiation. Genes
Chromosomes Cancer. 55:650–660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Petrov AS, Bernier CR, Gulen B, Waterbury
CC, Hershkovits E, Hsiao C, Harvey SC, Hud NV, Fox GE, Wartell RM
and Williams LD: Secondary structures of rRNAs from all three
domains of life. PLoS One. 9:e882222014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lobrich M, Shibata A, Beucher A, Fisher A,
Ensminger M, Goodarzi AA, Barton O and Jeggo PA: gammaH2AX foci
analysis for monitoring DNA double-strand break repair: Strengths,
limitations and optimization. Cell Cycle. 9:662–669. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Powell S and McMillan TJ: DNA damage and
repair following treatment with ionizing radiation. Radiother
Oncol. 19:95–108. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vilenchik MM and Knudson AG: Endogenous
DNA double-strand breaks: Production, fidelity of repair, and
induction of cancer. Proc Natl Acad Sci USA. 100:12871–12876. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amundson SA, Myers TG and Fornace AJ Jr:
Roles for p53 in growth arrest and apoptosis: Putting on the brakes
after genotoxic stress. Oncogene. 17:3287–3299. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fenech M, Kirsch-Volders M, Natarajan AT,
Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD
and Thomas P: Molecular mechanisms of micronucleus, nucleoplasmic
bridge and nuclear bud formation in mammalian and human cells.
Mutagenesis. 26:125–132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang H, Fletcher L, Beeharry N, Daniel R,
Kao G, Yen TJ and Muschel RJ: Abnormal cytokinesis after
x-irradiation in tumor cells that override the G2 DNA damage
checkpoint. Cancer Res. 68:3724–3732. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johmura Y, Shimada M, Misaki T, Naiki-Ito
A, Miyoshi H, Motoyama N, Ohtani N, Hara E, Nakamura M, Morita A,
et al: Necessary and sufficient role for a mitosis skip in
senescence induction. Mol Cell. 55:73–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
James A, Wang Y, Raje H, Rosby R and
DiMario P: Nucleolar stress with and without p53. Nucleus.
5:402–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takagi M, Absalon MJ, McLure KG and Kastan
MB: Regulation of p53 translation and induction after DNA damage by
ribosomal protein L26 and nucleolin. Cell. 123:49–63. 2005.
View Article : Google Scholar : PubMed/NCBI
|